Abstract
DNA damage-inducible transcript 4 (DDIT4) is induced in various cellular stress conditions. This study was conducted to investigate expression and prognostic significance of DDIT4 protein as a biomarker in the patients with colorectal cancer (CRC). PPI network and KEGG pathway analysis were applied to identify hub genes among obtained differentially expressed genes in CRC tissues from three GEO Series. In clinical, expression of DDIT4 as one of hub genes in three subcellular locations was evaluated in 198 CRC tissues using immunohistochemistry method on tissue microarrays. The association between DDIT4 expression and clinicopathological features as well as survival outcomes were analyzed. Results of bioinformatics analysis indicated 14 hub genes enriched in significant pathways according to KEGG pathways analysis among which DDIT4 was selected to evaluate CRC tissues. Overexpression of nuclear DDIT4 protein was found in CRC tissues compared to adjacent normal tissues (P = 0.003). Furthermore, higher nuclear expression of DDIT4 was found to be significantly associated with the reduced tumor differentiation and advanced TNM stages (all, P = 0.009). No significant association was observed between survival outcomes and nuclear expression of DDIT4 in CRC cases. Our findings indicated higher nuclear expression of DDIT4 was significantly associated with more aggressive tumor behavior and more advanced stage of disease in the patients with CRC.
Subject terms: Tumour biomarkers, Colorectal cancer, Bioinformatics, Immunological techniques, Medical research
Introduction
Colorectal cancer (CRC) as a heterogeneous disease is considered as the second leading cause of cancer-related deaths worldwide1. Molecular heterogeneity of CRC is due to genetic modifications commonly driving advancement in cancer and development of the disease2,3. Overall survival varies significantly in the patients with CRC depending on stage of the disease at the time of diagnosis and treatment decisions4–6. Despite the recent advances in screening and treatment of CRC, there is still a need to identify new markers for early diagnosis and prognosis in order to improve treatment decisions6. Therefore, diagnostic and prognostic biomarkers are required that could be more clinically applicable rather than conventional biomarkers7 and stratify the patients with CRC for cancer therapy6,8,9. In this regard, studying character and frequency of genetic and epigenetic alterations in cancer has been used as a great tool to investigate and identify those biomarkers which are linked to tumor development and medicine arrangements3,9.
Omics technologies have been used in order to characterize molecular features of tumor cells and their functional irregularities in cancer findings of which have been utilized in clinical settings to help cancer treatment10. Bioinformatics and computational biology are also necessary for analysis of Omics data and discovering biomarkers that will become an important part of medical research as well as clinical routine11. Therefore, in the present study, bioinformatics tools were applied to identify hub genes among the differentially expressed genes (DEGs) influencing CRC from transcriptome data, which led to identification of DNA damage-inducible transcript 4 (DDIT4) to investigate in CRC tissues as prognostic biomarker.
DDIT4 also known as a regulated in development and DNA damage response 1 (REDD1) protein and hypoxia-inducible factor 1 (HIF1)-responsive protein RTP801 (RTP801) was discovered and cloned in 200212,13. Generally, DDIT4 is rapidly induced in various cellular stresses12,14, such as hypoxia15,16, heat shock17, endoplasmic reticulum stress, and chemical molecules18. Most reports on DDIT4 function have indicated that DDIT4 protein suppresses mammalian target of rapamycin complex 1 (mTORC1) and regulates cell growth, tumorigenesis, cell aging, and autophagy12,14,19–21.
Upregulation of DDIT4 has been shown to promote cell proliferation, reduce apoptotic rate, and S phase arrest in gastric epithelial cells22. Moreover, upregulation of DDIT4 protein has been found to be associated with the decreased expression of pro-apoptotic proteins and at the same time the increased levels of anti-apoptotic proteins after inducing activity of RAS oncogene in ovarian epithelial cells23,24.
Prominently, the previous studies have revealed that dysregulation of DDIT4 occurs in various human cancers with paradoxical roles. A number of studies have attributed DDIT4 to tumor suppressor process, through suppression of mTORC1 in CRC25, breast cancer26, sporadic clear cell renal cell carcinoma (ccRCC)27, and non-small cell lung cancer28. While as an oncogene, upregulation of DDIT4 contributes to reduction of apoptosis processes, promotion of proliferation, migration, and invasion of cancer cells in in-vitro and in-vivo cancer studies22,29–31. High expression levels of DDIT4 protein have been observed in ovarian cancer (OC)31, bladder urothelial carcinoma (BUC)30, ccRCC (patients with von Hippel Lindau-deficient)27, and gastric cancer (GC)22 tissues compared to adjacent normal tissues. Increased expression of DDIT4 has been remarked as a prognosis factor in the patients with OC and BUC30–32. Additionally, DDIT4 gene is introduced as a cell intrinsic regulator for cancer therapy resistance in some cancers like brain, lung and gastric because it confers protection of tumor cells from therapy19,22,33. In contrast, in CRC, therapy with baicalein and polyisoprenylated benzophenones as anticancer agents in in-vitro indicated upregulation of DDIT4 associated with growth inhibition25,34.
In the present study, publicly available transcriptomic series (GSE74602, GSE110223, and GSE110224) were applied to detect genes that were differentially expressed in tumor tissues compared to adjacent normal tissues from the CRC patients. For identifying hub genes based on protein information, a protein–protein interaction (PPI) network was created for common upregulated DEGs among these three series. Then, Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis was done in order to better understand the pathways in which hub genes are involved. The bioinformatics analysis and literature review of these hub genes led to selection of DDIT4 as a biomarker in CRC tissues. Besides, in our other study, we have evaluated and detected mRNA expression levels of DDIT4 as a predictor biomarker for advanced disease in fresh CRC tissue samples35, herein, expression of DDIT4 protein was further confirmed as a biomarker in clinical CRC samples. Expression levels and localization of DDIT4 protein, for the first time were investigated in nucleus, cytoplasm, and plasma membrane for a series of formalin-fixed paraffin-embedded (FFPE) tissues from CRC patients using the immunohistochemistry (IHC) method on tissue microarrays (TMAs). Then, the associations between expression levels of DDIT4 protein at different subcellular locations with clinicopathological features as well as survival outcomes were analyzed.
Results
Bioinformatics approach
Identification of the upregulated DEGs and PPI network
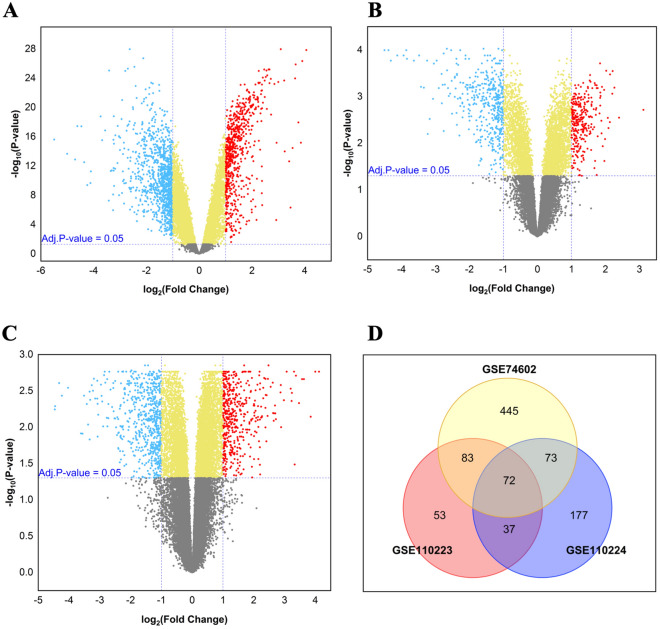
The workflow of our research is presented in Fig. 1. Based on the preset criteria of log FC ≥ ± 1.0 and adjusted P value < 0.05, 1672 DEGs (673 upregulated and 999 downregulated), 589 DEGs (245 upregulated and 344 downregulated), and 814 DEGs (359 upregulated and 455 downregulated) were extracted from GSE74602, GSE110223, and GSE110224 series, respectively, using volcano plots as shown in Fig. 2A–C. Further investigation of the obtained upregulated DEGs from three series using Venn diagram analysis displayed that there were 72 common upregulated DEGs between these series (Fig. 2D and Supplementary Table 1). Despite the most of DEGs in the current study were in line with our previous results in other CRC series of GEO36, we rechecked 72 common upregulated DEGs from this analysis with obtained common DEGs of merged three series (GSE74602, GSE110223, and GSE110224) by MINT tool method (Supplementary Fig. S1 and Table S1). Comparison between these DEGs by MINT and 72 common upregulated genes showed that total 72 common upregulated genes exist in common upregulated DEGs by MINT method.
Figure 1.
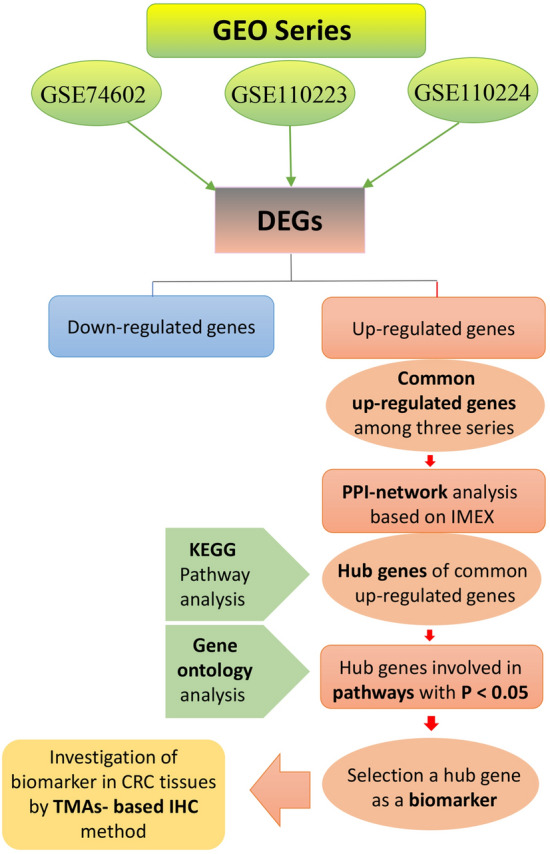
The workflow of the present study. This flowchart is a schematic overview of the bioinformatics analysis for selection of biomarker to investigate in CRC samples using the tissue microarrays-based immunohistochemistry (TMAs-based IHC).
Figure 2.
Identification of the differentially expressed genes (DEGs) in tumor tissues compared to adjacent normal tissues. Volcano plots of the gene expression profile from the (A) GSE74602, (B) GSE110223, and (C) GSE110224 series. The red dots at the top right represent the upregulated DEGs and the blue dots at the top left represent the downregulated DEGs (log FC ≥ ± 1, adjusted P value < 0.05). (D) Venn diagram displayed common upregulated DEGs among three series for subsequent analysis.
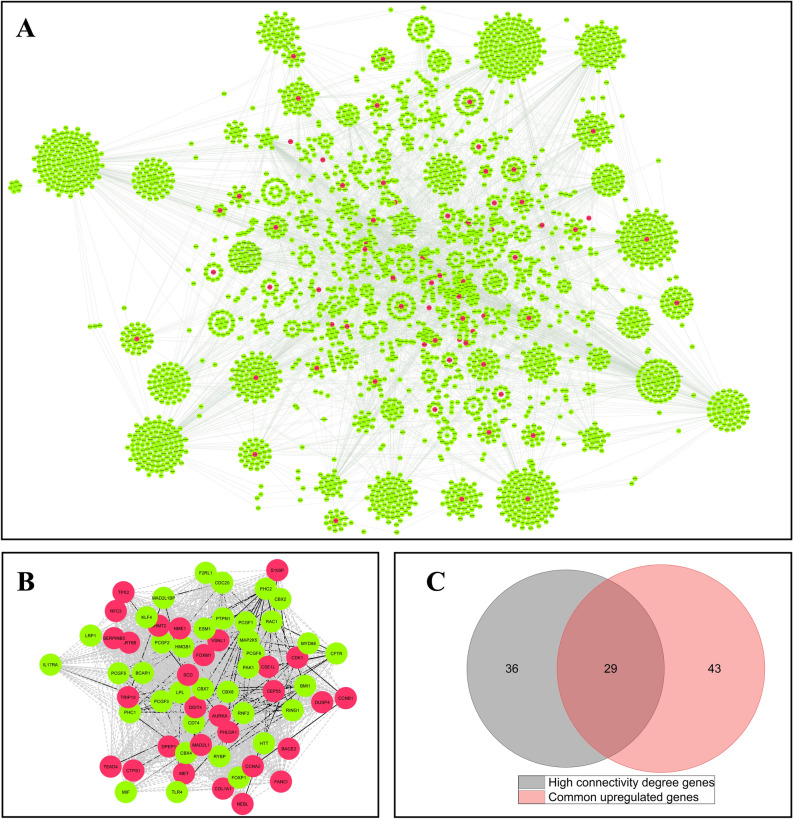
For investigating all the protein interactions available for 72 common upregulated DEGs, a PPI network was constructed for these genes according to IMEX database, which included 3831 nodes and 9911 edges (Fig. 3A). Sixty-five genes with high connectivity degree were extracted from this network by cyto-Hubba in which 29 genes overlapped with 72 common upregulated DEGs (Fig. 3B,C). Therefore, these 29 genes were considered as hub genes of common upregulated DEGs for subsequent analysis (Supplementary Table 1).
Figure 3.
Protein–protein interaction (PPI) analysis based on IMEX database for common upregulated DEGs and detection of hub genes. (A) The nodes represent genes and edges represent the interactions between genes. The red nodes signify 72 common upregulated DEGs that are in the interaction with each other and other genes (green nodes). (B) PPI network analysis revealed the genes with high connectivity degree (> 75 percentile for the genes with degree > 10) by cyto-Hubba plug-in using Cytoscape software. The dashed lines indicate the connection of genes that are not direct. (C) The genes with high connectivity degree that overlapped with common upregulated DEGs were extracted as hub genes by Venn diagram.
KEGG pathway and gene ontology (GO) analysis for hub genes
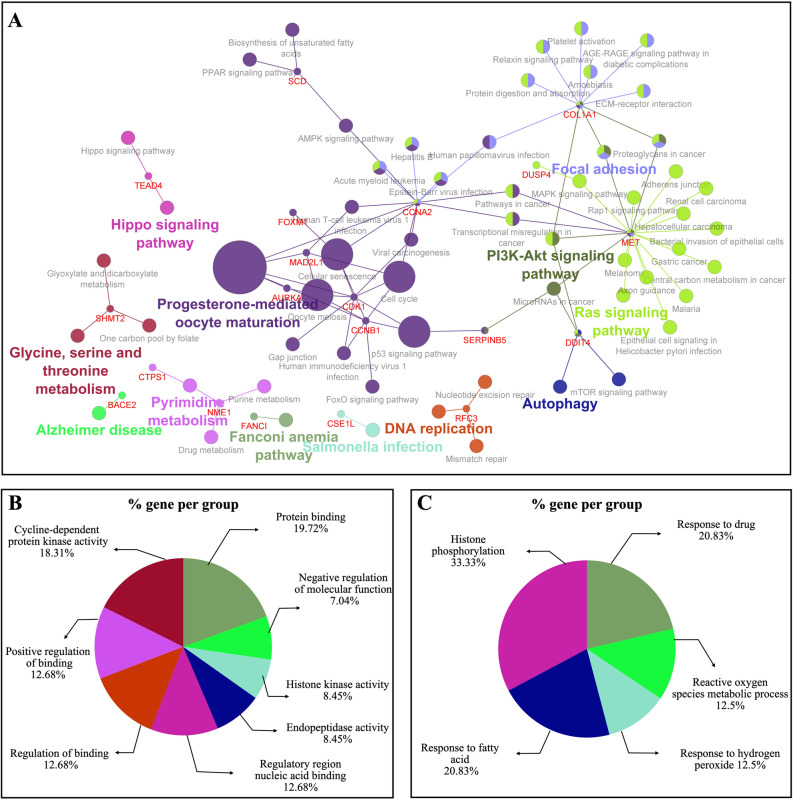
The KEGG pathway analysis for 29 hub genes indicated that these genes were involved and clustered in different pathways, such as ‘progesterone-mediated oocyte maturation’, ‘cell cycle’ and ‘PI3K-Akt signaling pathway’ as summarized in Fig. 4A. (Supplementary Table 2). Moreover, 14 hub genes were found in 10 pathway terms with P < 0.05 (Table 1) that GO enrichment analysis was performed in order to obtain more comprehensive and deep understanding of the biological process and function of these hub genes. GO enrichment analysis in MF domain indicated that most 14 hub genes were clustered in functional groups of ‘protein binding’ and ‘cyclin-dependent protein kinase activity’ (Fig. 4B). These hub genes were mainly enriched in ‘histone phosphorylation’ and ‘response to drug’ groups of BP domain (Fig. 4C). Also, KEGG pathways involved in CRC diseases were obtained from the KEGG DISEASE Database, which included ‘mucin type O-glycan biosynthesis’, ‘colorectal cancer pathway’, and ‘microRNAs in cancer’ pathways as summarized in Supplementary Fig. S2. Then, accurate scrutiny of hub genes҆ pathways displayed those 3 hub genes (MET, DDIT4, and SERPINB5) were involved in ‘microRNAs in cancer’ pathway, a part of pathways in CRC diseases based on KEGG database. Finally, literature review and enrichment analysis led to selection of DDIT4 to evaluate protein expression level of this marker in CRC tissues. According to the KEGG pathway analysis, DDIT4 was found to be related to PI3k-Akt/mTOR signaling and microRNAs in cancer pathway, as a part of CRC disease using the KEGG DISEASE Database. PI3k-Akt/mTOR signaling is frequently deregulated in human cancer that is one of the primary mechanisms for sustaining tumor outgrowth and metastasis. Targeting these pathways have been considered as a strategy for cancer therapy37,38. DDIT4 was participated in important BP and MF, such as cell death, response to drug, protein binding, and cellular response to stress in the cell. Moreover, our review of literature indicated that DDIT4 protein expression was not evaluated in CRC patients with various stages by IHC.
Figure 4.
KEGG pathways and gene ontology (GO) analysis for hub genes in Cytoscape software by ClueGO plug-in. (A) KEGG pathway analysis for 29 obtained hub genes of protein–protein interaction network. (B) Gene ontology analysis for domains of molecular function (MF) and (C) biological process (BP) for 14 hub genes involved in KEGG pathways with P value < 0.05.
Table 1.
Hub genes involved in pathways with P value < 0.05 based on KEGG pathway analysis.
| KEGG number | KEGG pathways | Gene names | P value |
|---|---|---|---|
| KEGG:04914 | Progesterone-mediated oocyte maturation | AURKA, CCNA2, CCNB1, CDK1, MAD2L1 | < 0.00001 |
| KEGG:04110 | Cell cycle | CCNA2, CCNB1, CDK1, MAD2L1 | < 0.001 |
| KEGG:04114 | Oocyte meiosis | AURKA, CCNB1, CDK1, MAD2L1 | < 0.001 |
| KEGG:04218 | Cellular senescence | CCNA2, CCNB1, CDK1, FOXM1 | < 0.001 |
| KEGG:04115 | P53 signaling pathway | CCNB1, CDK1, SERPINB5 | < 0.001 |
| KEGG:00240 | Pyrimidine metabolism | CTPS1, NME1 | < 0.01 |
| KEGG:04152 | AMPK signaling pathway | CCNA2, SCD | 0.035 |
| KEGG:05206 | MicroRNAs in cancer | DDIT4, MET, SERPINB5 | 0.040 |
| KEGG:04151 | PI3K-Akt signaling pathway | COL1A1, DDIT4, MET | 0.045 |
| KEGG:00670 | One carbon pool by folate | SHMT2 | 0.048 |
Subcellular localization study for DDIT4 showed that this marker is mostly enriched in the cytosol and cell nucleus and also to some extent; it is found in the plasma membrane and extracellular environment based on COMPARTMENTS database.
Findings related to the study population
Baseline characteristics of the study population
A total of 198 samples from CRC population were included in the current study, in which 104 (52.5%) samples belonged to males and 94 (47.5%) of them were from females with a male/female ratio of 1.1. Mean age of the patients was equal to 59 (SD = 13) years old, (ranging from 25 to 88); 98 (49.5%) patients were younger than 59 and 100 (50.5%) of them were over 59 years old. Tumor size, ranging from 1 to 24 cm was categorized based on mean size into two groups: Group 1: ≤ 5 cm [134 cases (67.7%)] and Group 2: > 5 cm [64 cases (32.3%)]. In this study, 90 (45.5%) patients had well differentiated, 95 (48.0%) patients had moderately differentiated, and 13 (6.6%) of them had poorly differentiated. Moreover, 31 (15.7%) cases were at stage I of the disease, 81 (40.9%) cases were at stage II, 79 (39.9%) cases were at stage III, and 7 (3.5%) cases were at stage IV according to TNM stage system. Vascular and perineural invasion were observed in 33 (16.7%) and 48 (24.2%) of the patients with CRC, respectively while lymph node metastasis was found in 84 (42.4%) cases.
Expression and localization of DDIT4 in CRC tissues
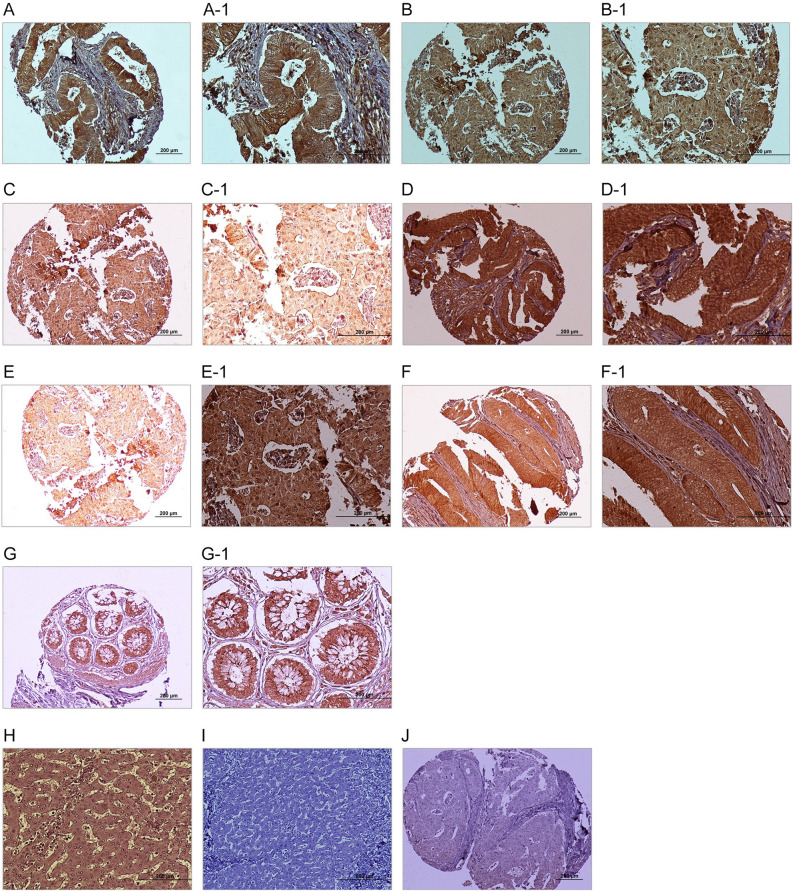
For estimating expression levels of DDIT4, three scoring systems were employed incorporating intensity of staining, percentage of positive tumor cells, and the H-score. Furthermore, expression of DDIT4 was detected at different subcellular locations including nucleus, cytoplasm, and plasma membrane in both CRC tumor cells and adjacent normal tissues by IHC analysis. However, a significant overexpression of DDIT4 at nucleus was observed in tumor cells compared to adjacent normal tissue samples (P = 0.003). This comparison was not statistically significant in terms of cytoplasmic and membranous expression of DDIT4. Measurement of staining based on the median H-score as the cut-off was done for lower DDIT4 expression versus higher DDIT4 expression in the nucleus, cytoplasm, and plasma membrane of cells. Expression of DDIT4 protein in CRC and adjacent normal tissue samples are exhibited in Fig. 5A–G1. Also, DDIT4 expression was observed with various intensities amongst the CRC tissues in various locations (Table 2).
Figure 5.
Immunohistochemical analysis of DDIT4 expression in colorectal cancer (CRC) samples. Nuclear expression of DDIT4 in CRC: (A, A-1) low expression and (B, B-1) high expression. Cytoplasmic expression of DDIT4 in CRC: (C, C-1) low expression and (D, D-1) high expression. Membranous expression of DDIT4 in CRC: (E, E1) low expression and (F, F1) high expression. (G, G1) DDIT4 expression in adjacent normal tissue. (H) DDIT4 expression in normal human liver as a positive control, (I) DDIT4 expression in normal human liver as a negative control, and (J) Isotype control. (Figures shown with magnification of 100 × and 200 ×).
Table 2.
Nuclear, cytoplasmic, and membranous DDIT4 expression in colorectal cancer (CRC) tissues and their adjacent normal tissue samples (Intensity of staining, percentage of positive tumor cells, and H-score).
| Scoring system | Nuclear DDIT4 N (%) | Adjacent normal N (%) | P value | Cytoplasmic DDIT4 N (%) | Adjacent normal N (%) | P value | Membranous DDIT4 N (%) | Adjacent normal N (%) | P value |
|---|---|---|---|---|---|---|---|---|---|
| Intensity of staining | 0.004 | 0.023 | 0.013 | ||||||
| No staining (0) | 0 (0.0) | 1 (2.6) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 0 (0.0) | |||
| Weak (+ 1) | 6 (3.0) | 1 (2.6) | 12 (6.1) | 7 (17.9) | 6 (3.0) | 2 (5.1) | |||
| Moderate (+ 2) | 86 (43.4) | 26 (66.7) | 134 (67.7) | 26 (66.7) | 101 (51.0) | 28 (71.8) | |||
| Strong (+ 3) | 106 (53.5) | 11 (28.1) | 52 (26.2) | 6 (15.4) | 89 (45.0) | 9 (23.1) | |||
| Percentage of positive tumor cells | 0.781 | 0.533 | 0.086 | ||||||
| < 25% | 1 (0.5) | 1 (2.6) | 0 (0.0) | 0 (0.0) | 4 (2.0) | 0 (0.0) | |||
| 25–50% | 4 (2.0) | 1 (2.6) | 6 (3.0) | 3 (7.7) | 7 (3.5) | 1 (2.6) | |||
| 51–75% | 53 (26.8) | 10 (25.6) | 73 (36.9) | 14 (35.9) | 69 (34.8) | 9 (23.1) | |||
| > 75% | 140 (70.7) | 27 (69.2) | 119 (60.1) | 22 (56.4) | 118 (59.6) | 29 (74.4) | |||
| H-score cut off | 0.003 | 0.142 | 0.211 | ||||||
| Low | 93 (47.0) | 22 (56.4) | 120 (60.6) | 25 (64.1) | 115 (58.1) | 31 (79.5) | |||
| High | 105 (53.0) | 17 (43.6) | 78 (39.4) | 14 (35.9) | 83 (41.9) | 8 (20.5) | |||
| Total | 198 | 39 | 198 | 39 | 198 | 39 |
P value is based on Mann–Whitney U test.
H-score histological score.
Besides, human normal liver tissue as a positive control exhibited moderate staining in cytoplasm of all the hepatocyte and bile duct cells as well as nucleus of a few cells lining the sinusoids and results related to positive, negative, and isotype controls are shown in Fig. 5H–J.
Association between expressions of DDIT4 and clinicopathological features
Nuclear and cytoplasmic expressions of DDIT4 were observed in all CRC samples while membranous expression of DDIT4 was not found in 2 (1%) cases of total patients with CRC. In this study, Pearson's chi-squared test was used to investigate the association between expression levels of DDIT4 and various clinicopathological features in the patients with CRC. The results of TMA-based IHC analysis were prepared according to the patients҆ clinicopathological features and are presented in Tables 3, 4, and 5 based on subcellular locations of DDIT4 marker staining. The results of Pearson’s chi-square test revealed that nuclear expression of DDIT4 in terms of intensity of staining (P = 0.031) and H-score (P = 0.009) had a significant association with tumor differentiation. In addition, a significant association was found between nuclear expression of DDIT4 and TNM stages (intensity of staining; (P = 0.013) and H-score [(P = 0.009), respectively]. The Spearman’s correlation test indicated a significant direct correlation between nuclear expression of DDIT4 with tumor differentiation and TNM stages (P < 0.05).
Table 3.
The association between nuclear DDIT4 expression and clinicopathological features of colorectal cancer (CRC) samples (Intensity of staining and H-score).
| Patients and tumor characteristics | Total samples N (%) | Intensity of staining N (%) | P value | H-score (cut off = 210) N (%) | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0 (Negative) | 1+ (Weak) | 2+ (Moderate) | 3+ (Strong) | Low (≤ 210) | High (> 210) | ||||
| Mean age, years (Range) | 59 (25–88) | 0.991 | 0.769 | ||||||
| ≤ Mean age | 98 (49.5) | 0 (0.0) | 3 (50.0) | 43 (50.0) | 52 (49.1) | 45 (48.4) | 53 (50.5) | ||
| > Mean age | 100 (50.5) | 0 (0.0) | 3 (50.0) | 43 (50.0) | 54 (50.9) | 48 (51.6) | 52 (49.5) | ||
| Gender | 0.760 | 0.966 | |||||||
| Male | 104 (52.5) | 0 (0.0) | 4 (66.7) | 44 (51.2) | 56 (52.8) | 49 (52.7) | 55 (52.4) | ||
| Female | 94 (47.5) | 0 (0.0) | 2 (33.3) | 42 (48.8) | 50 (47.2) | 44 (47.3) | 50 (47.6) | ||
| (Male/Female) | 1.1 | - | 2 | 1.0 | 1.1 | 1.1 | 1.1 | ||
| Mean tumor size (cm) | 5 | 0.159 | 0.985 | ||||||
| ≤ Mean | 134 (67.7) | 0 (0.0) | 2 (33.3) | 61 (70.9) | 71 (67.0) | 63 (67.7) | 71 (67.6) | ||
| > Mean | 64 (32.3) | 0 (0.0) | 4 (66.7) | 25 (29.1) | 35 (33.0) | 30 (32.3) | 34 (32.4) | ||
| Tumor differentiation | 0.031 | 0.009 | |||||||
| Well | 90 (45.5) | 0 (0.0) | 2 (33.3) | 50 (58.1) | 38 (35.8) | 53 (57.0) | 37 (35.2) | ||
| Moderately | 95 (48.0) | 0 (0.0) | 4 (66.7) | 31 (36.0) | 60 (56.6) | 35 (37.6) | 60 (57.1) | ||
| Poorly | 13 (6.6) | 0 (0.0) | 0 (0.0) | 5 (5.8) | 8 (7.5) | 5 (5.4) | 8 (7.6) | ||
| TNM stages | 0.013 | 0.009 | |||||||
| I | 31 (15.7) | 0 (0.0) | 0 (0.0) | 10 (11.6) | 21 (19.8) | 11 (11.8) | 20 (19.0) | ||
| II | 81 (40.9) | 0 (0.0) | 3 (50.0) | 46 (53.5) | 32 (30.2) | 48 (51.6) | 33 (31.4) | ||
| III | 79 (39.9) | 0 (0.0) | 3 (50.0) | 25 (29.1) | 51 (48.1) | 29 (31.2) | 50 (47.6) | ||
| IV | 7 (3.5) | 0 (0.0) | 0 (0.0) | 5 (5.8) | 2 (1.9) | 5 (5.4) | 2 (1.9) | ||
| Vascular invasion (VI) | 0.874 | 0.848 | |||||||
| Present | 33 (16.7) | 0 (0.0) | 1(16.7) | 13 (15.1) | 19 (17.9) | 15 (16.1) | 18 (17.1) | ||
| Absent | 165 (83.3) | 0 (0.0) | 5 (83.3) | 73 (84.9) | 87 (82.1) | 78 (83.9) | 87 (82.9) | ||
| Lymph node invasion (LNI) | 0.343 | 0.199 | |||||||
| Involved | 84 (42.4) | 0 (0.0) | 2 (33.3) | 32 (37.2) | 50 (47.2) | 35 (37.6) | 49 (46.7) | ||
| None | 114 (57.6) | 0 (0.0) | 4 (66.7) | 54 (62.8) | 56 (52.8) | 58 (62.4) | 56 (53.3) | ||
| Perineural invasion (PI) | 0.314 | 0.629 | |||||||
| Involved | 48 (24.2) | 0 (0.0) | 3 (50.0) | 21 (24.4) | 24 (22.6) | 24 (25.8) | 24 (22.9) | ||
| None | 150 (75.8) | 0 (0.0) | 3 (50.0) | 65 (75.6) | 82 (77.4) | 69 (74.2) | 81 (77.1) | ||
| Distant metastasis | 0.500 | 0.989 | |||||||
| Present | 28 (14.1) | 0 (0.0) | 0 (0.0) | 11 (25.6) | 17 (26.2) | 12 (25.0) | 16 (25.0) | ||
| Absent | 84 (42.4) | 0 (0.0) | 4 (100.0) | 32 (74.4) | 48 (73.8) | 36 (75.0) | 48 (75.0) | ||
| Tumor recurrence | 0.381 | 0.533 | |||||||
| Yes | 29 (14.6) | 0 (0.0) | 0 (0.0) | 10 (23.3) | 19 (29.2) | 11 (22.9) | 18 (28.1) | ||
| No | 83 (41.9) | 0 (0.0) | 4 (100.0) | 33 (76.7) | 46 (70.8) | 37 (77.1) | 46 (71.9) | ||
P value; Pearson’s χ2 test, and Values in bold are statistically significant.
H-score histological score.
Table 4.
The association between cytoplasmic DDIT4 expression and clinicopathological features of colorectal cancer (CRC) samples (Intensity of staining and H-score).
| Patients and tumor characteristics | Total samples N (%) | Intensity of staining N (%) | P value | H-score (cut off = 180) N(%) | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0 (Negative) | 1+ (Weak) | 2+ (Moderate) | 3+ (Strong) | Low (≤ 180) | High (> 180) | ||||
| Mean age, years (Range) | 59 (25–88) | 0.187 | 0.640 | ||||||
| ≤ Mean age | 98 (49.5) | 0 (0.0) | 8 (66.7) | 69 (51.5) | 21 (40.4) | 61 (50.8) | 37 (47.4) | ||
| > Mean age | 100 (50.5) | 0 (0.0) | 4 (33.3) | 65 (48.5) | 31 (59.6) | 59 (49.2) | 41 (52.6) | ||
| Gender | 0.557 | 0.377 | |||||||
| Male | 104 (52.5) | 0 (0.0) | 8 (66.7) | 68 (50.7) | 28 (53.8) | 60 (50.0) | 44 (56.4) | ||
| Female | 94 (47.5) | 0 (0.0) | 4 (33.3) | 66 (49.3) | 24 (46.2) | 60 (50.0) | 34 (43.6) | ||
| (Male/Female) | 1.1 | - | 2.0 | 1.0 | 1.1 | 1.0 | 1.2 | ||
| Mean tumor size (cm) | 5 | 0.679 | 0.578 | ||||||
| ≤ Mean | 134 (67.7) | 0 (0.0) | 9 (75.0) | 92 (68.7) | 33 (63.5) | 83 (69.2) | 51 (65.4) | ||
| > Mean | 64 (32.3) | 0 (0.0) | 3 (25.0) | 42 (31.3) | 19 (36.5) | 37 (30.8) | 27 (34.6) | ||
| Tumor differentiation | 0.554 | 0.987 | |||||||
| Well | 90 (45.5) | 0 (0.0) | 8 (66.7) | 60 (44.8) | 22 (42.3) | 54 (45.0) | 36 (46.2) | ||
| Moderately | 95 (48.0) | 0 (0.0) | 4 (33.3) | 64 (47.8) | 27 (51.9) | 58 (48.3) | 37 (47.4) | ||
| Poorly | 13 (6.6) | 0 (0.0) | 0 (0.0) | 10 (7.5) | 3 (5.8) | 8 (6.7) | 5 (6.4) | ||
| TNM stages | 0.0466 | 0.375 | |||||||
| I | 31 (15.7) | 0 (0.0) | 2 (16.7) | 21 (15.7) | 8 (15.4) | 15 (12.5) | 16 (20.5) | ||
| II | 81 (40.9) | 0 (0.0) | 5 (41.7) | 60 (44.8) | 16 (30.8) | 53 (44.2) | 28 (35.9) | ||
| III | 79 (39.9) | 0 (0.0) | 5 (41.7) | 47 (35.1) | 27 (51.9) | 47 (39.2) | 32 (41.0) | ||
| IV | 7 (3.5) | 0 (0.0) | 0 (0.0) | 6 (4.5) | 1 (1.9) | 5 (4.2) | 2 (2.6) | ||
| Vascular invasion (VI) | 0.207 | 0.696 | |||||||
| Present | 33 (16.7) | 0 (0.0) | 3 (25.0) | 18 (13.4) | 12 (23.1) | 21 (17.5) | 12 (15.4) | ||
| Absent | 165 (83.3) | 0 (0.0) | 9 (75.0) | 116 (86.6) | 40 (76.9) | 99 (82.5) | 66 (84.6) | ||
| Lymph node invasion (LNI) | 0.106 | 0.979 | |||||||
| Involved | 84 (42.4) | 0 (0.0) | 6 (50.0) | 50 (37.3) | 28 (53.8) | 51 (42.5) | 33 (42.3) | ||
| None | 114 (57.6) | 0 (0.0) | 6 (50.0) | 84 (62.7) | 24 (46.2) | 69 (57.5) | 45 (57.7) | ||
| Perineural invasion (PI) | 0.818 | 0.517 | |||||||
| Involved | 48 (24.2) | 0 (0.0) | 2 (16.7) | 33 (24.6) | 13 (25.0) | 31 (25.8) | 17 (21.8) | ||
| None | 150 (75.8) | 0 (0.0) | 10 (83.3) | 101 (75.4) | 39 (75.0) | 89 (74.2) | 61 (78.2) | ||
| Distant metastasis | 0.976 | 0.912 | |||||||
| Present | 28 (14.1) | 0 (0.0) | 2 (22.2) | 18 (25.0) | 8 (25.8) | 16 (24.6) | 12 (25.5) | ||
| Absent | 84 (42.4) | 0 (0.0) | 7 (77.8) | 54 (75.0) | 23 (74.2) | 49 (75.4) | 35 (74.5) | ||
| Tumor recurrence | 0.736 | 0.717 | |||||||
| Yes | 29 (14.6) | 0 (0.0) | 3 (33.3) | 17 (23.6) | 9 (29.0) | 16 (24.6) | 13 (27.7) | ||
| No | 83 (41.9) | 0 (0.0) | 6 (66.7) | 55 (76.4) | 22 (71.0) | 49 (75.4) | 34 (72.3) | ||
P value; Pearson’s χ2 test, and Values in bold are statistically significant.
H-score histological score.
Table 5.
The association between membranous DDIT4 expression and clinicopathological features of colorectal cancer (CRC) samples (Intensity of staining and H-score).
| Patients and tumor characteristics | Total samples N (%) | Intensity of staining N (%) | P value | H-score (cut off = 200) N(%) | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0 (Negative) | 1+ (Weak) | 2+ (Moderate) | 3+ (Strong) | Low (≤ 200) | High (> 200) | ||||
| Mean age, years (Range) | 59 (25–88) | 0.328 | 0.143 | ||||||
| ≤ Mean age | 98 (49.5) | 0 (0.0) | 4 (66.7) | 53 (52.5) | 41 (46.1) | 62 (53.9) | 36 (43.4) | ||
| > Mean age | 100 (50.5) | 2 (100.0) | 2 (33.3) | 48 (47.5) | 48 (53.9) | 53 (46.1) | 47 (56.6) | ||
| Gender | 0.781 | 0.686 | |||||||
| Male | 104 (52.5) | 1 (50.0) | 4 (66.7) | 50 (49.5) | 49 (55.1) | 59 (51.3) | 45 (54.2) | ||
| Female | 94 (47.5) | 1 (50.0) | 2 (33.3) | 51 (50.5) | 40 (44.9) | 56 (48.7) | 38 (45.8) | ||
| (Male/Female) | 1.1 | 1.0 | 2.0 | 0.9 | 1.2 | 1.0 | 1.1 | ||
| Mean tumor size (cm) | 5 | 0.674 | 0.958 | ||||||
| ≤ Mean | 134 (67.7) | 1 (50.0) | 3 (50.0) | 71 (70.3) | 59 (66.3) | 78 (67.8) | 56 (67.5) | ||
| > Mean | 64 (32.3) | 1 (50.0) | 3 (50.0) | 30 (29.7) | 30 (33.7) | 37 (32.2) | 27 (32.5) | ||
| Tumor differentiation | 0.198 | 0.817 | |||||||
| Well | 90 (45.5) | 1 (50.0) | 4 (66.7) | 47 (46.5) | 38 (42.7) | 54 (47.0) | 36 (43.4) | ||
| Moderately | 95 (48.0) | 0 (0.0) | 2 (33.3) | 47 (46.5) | 46 (51.7) | 53 (46.1) | 42 (50.6) | ||
| Poorly | 13 (6.6) | 1 (50.0) | 0 (0.0) | 7 (6.9) | 5 (5.6) | 8 (7.0) | 5 (6.0) | ||
| TNM stages | 0.927 | 0.989 | |||||||
| I | 31 (15.7) | 0 (0.0) | 1 (16.7) | 17 (16.8) | 13(14.6) | 18 (15.7) | 13 (15.7) | ||
| II | 81 (40.9) | 0 (0.0) | 3 (50.0) | 40 (39.6) | 38(42.7) | 46 (40.0) | 35 (42.2) | ||
| III | 79 (39.9) | 2 (100.0) | 2 (33.3) | 40 (39.6) | 35(39.3) | 47 (40.9) | 32 (38.6) | ||
| IV | 7 (3.5) | 0 (0.0) | 0 (0.0) | 4 (4.0) | 3(3.4) | 4 (3.5) | 3 (3.6) | ||
| Vascular invasion (VI) | 0.634 | 0.941 | |||||||
| Present | 33 (16.7) | 0 (0.0) | 0 (0.0) | 17 (16.8) | 16 (18.0) | 19 (16.5) | 14 (16.9) | ||
| Absent | 165 (83.3) | 2 (100.0) | 6 (100.0) | 84 (83.2) | 73 (82.0) | 96 (83.5) | 69 (83.1) | ||
| Lymph node invasion (LNI) | 0.400 | 0.724 | |||||||
| Involved | 84 (42.4) | 2 (0.0) | 2 (33.3) | 43 (42.6) | 37 (41.6) | 50 (43.5) | 34 (41.0) | ||
| None | 114 (57.6) | 0 (0.0) | 4 (66.7) | 58 (57.4) | 52 (58.4) | 65 (56.5) | 49 (59.0) | ||
| Perineural invasion (PI) | 0.647 | 0.294 | |||||||
| Involved | 48 (24.2) | 1 (50.0) | 1 (16.7) | 27 (26.7) | 19 (21.3) | 31 (27.0) | 17 (20.5) | ||
| None | 150 (75.8) | 1 (50.0) | 5 (83.3) | 74 (73.3) | 70 (78.7) | 84 (73.0) | 66 (79.5) | ||
| Distant metastasis | 0.878 | 0.912 | |||||||
| Present | 28 (14.1) | 0 (0.0) | 1 (25.0) | 14 (25.5) | 13 (25.5) | 16 (25.4) | 12 (24.5) | ||
| Absent | 84 (42.4) | 2 (100.0) | 3 (75.0) | 41 (74.5) | 38 (74.5) | 47 (74.6) | 37 (75.5) | ||
| Tumor recurrence | 0.759 | 0.568 | |||||||
| Yes | 29 (14.6) | 0 (0.0) | 1 (25.0) | 13 (23.6) | 15 (29.4) | 15 (23.8) | 14 (28.6) | ||
| No | 83 (41.9) | 2 (100.0) | 3 (75.0) | 42 (76.4) | 36 (70.6) | 48 (76.2) | 35 (71.4) | ||
P value; Pearson’s χ2 test, and Values in bold are statistically significant.
H-score histological score.
The data of the Kruskal–Wallis test exhibited statistically significant differences between the median nuclear expression level of DDIT4 and tumor differentiation groups as well as various TNM stages (I–IV) (P < 0.05). Mann–Whitney U test also showed a significant difference in median nuclear expression level of DDIT4 between well and moderately differentiated groups of tumor differentiation (P = 0.037) (Fig. 6A). Median nuclear expression level of DDIT4 was 225 in moderately differentiated and it was 200 in well differentiated. Moreover, the results of Mann–Whitney U test revealed a significant difference in median nuclear expression level of DDIT4 between stage II and stage III (P = 0.026). Similarly, median nuclear expression level of DDIT4 was 225 for stage III and 200 in stage II (Fig. 6B). The analysis did not reveal any significant differences in the association between cytoplasmic and membranous expression of DDIT4 with clinicopathological features.
Figure 6.
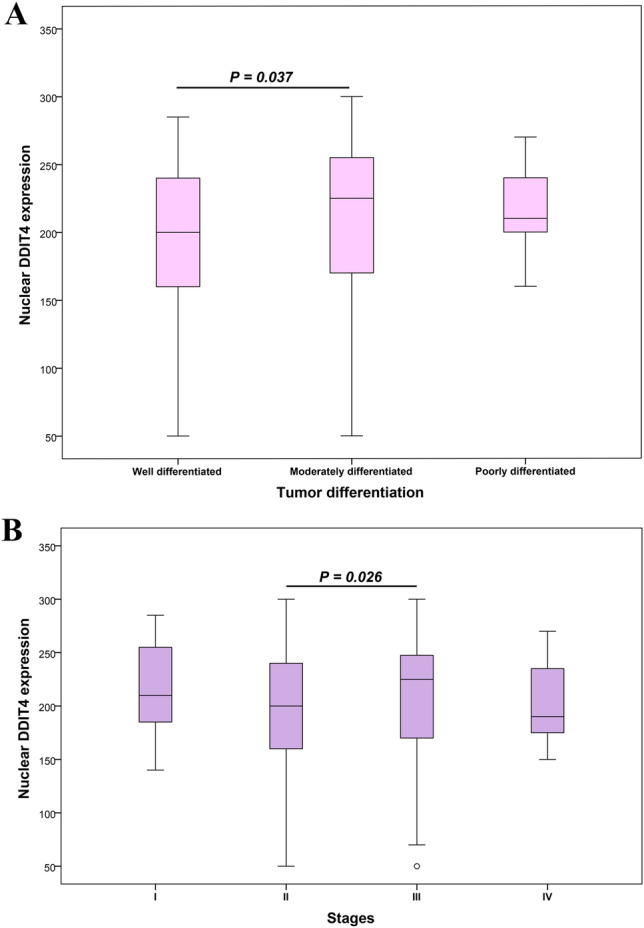
Box plot analysis of DDIT4 expression levels in tumor differentiation groups and TNM stages. The bold line precisely represents median expression levels of DDIT4. (A) The result of data analysis showed a statistically significant association in the median nuclear expression of DDIT4 between well and moderately tumor differentiation (P = 0.037). (B) A statistically significant association was observed in the median nuclear expression of DDIT4 between stages II and III (P = 0.026).
Survival outcomes based on expression of DDIT4 in the patients with CRC
The follow-up data were available for 112 out of 198 patients with CRC in this study. During the follow-up time, disease-related death was reported in 28 patients (14.1%), recurrence and metastasis happened in 29 (14.6%) and 28 (14.1%) patients, respectively, while 79 (39.9%) cases were absent for these two parameters. Median and mean follow-up time for these CRC patients were 34 months (Q1 = 21 and Q3 = 47) (ranging from 1 to 105 months) and 36 (SD = 24), respectively. Survival calculation was applied for disease-specific survival (DSS) and progression-free survival (PFS) terms. Kaplan–Meier survival analysis was implemented to compare survival (DSS and PFS) in two groups of the patients with CRC (high versus low nuclear expression of DDIT4 protein). Results of survival analysis showed no significant association between survival (DSS and PFS) and nuclear expression levels of DDIT4 (Log-rank test; DSS and PFS, P = 0.958, P = 0.911) (Fig. 7A,B).
Figure 7.
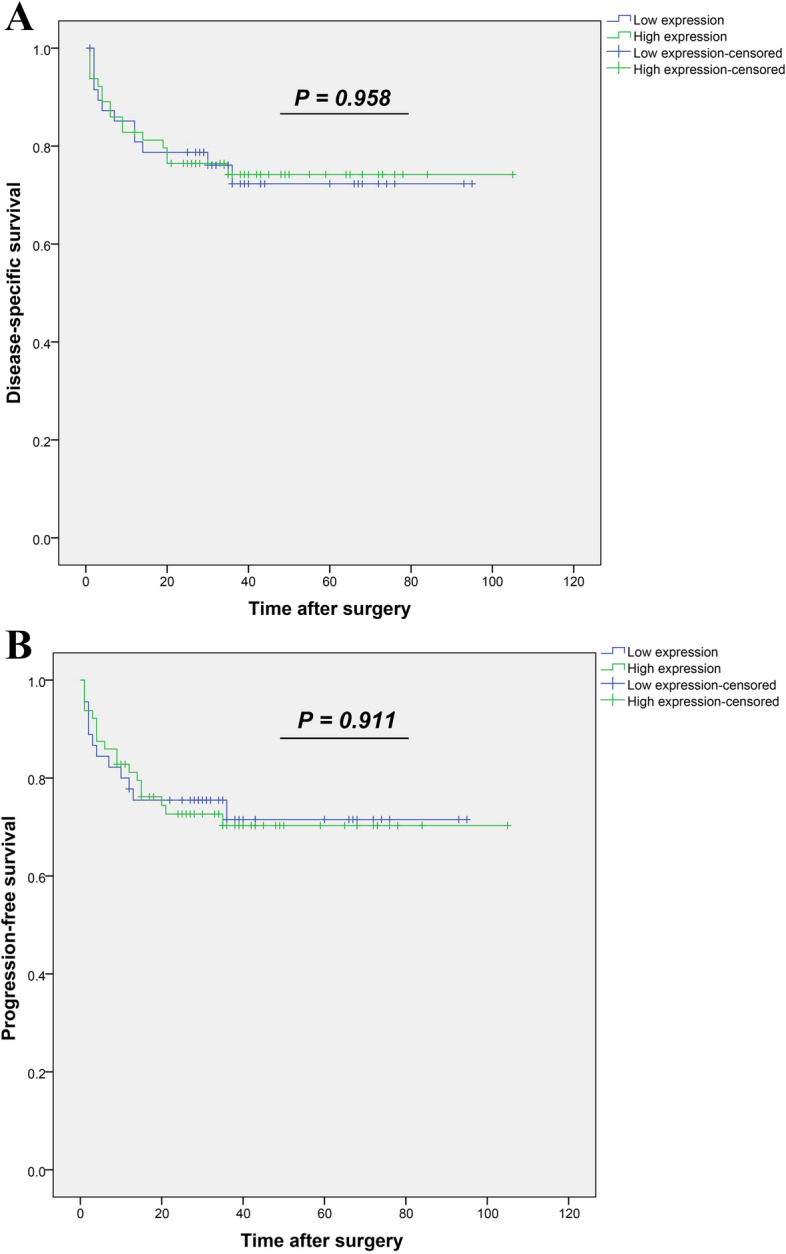
Kaplan–Meier survival analysis according to nuclear expression levels of DDIT4 protein in colorectal cancer (CRC). Log-rank test did not show any significant difference in (A) DSS and (B) PFS between two groups of CRC patients (high versus low nuclear expression of DDIT4 protein). DSS: Disease-specific survival and PFS: progression-free survival.
Discussion
Regarding study of cancer heterogeneity, biomarkers are valuable that can improve clinical management and the potential for recognition of therapeutic targets in drug development for CRC39. In this regard, development of markers and multiple therapeutics has led to an increase in survival up to 3 years for the patients with CRC at advanced stages of disease that is still not satisfactory40. For recognizing biomarkers in CRC, three series of the GEO were analyzed and common upregulated DEGs of these series were screened. Results of the PPI network and enrichment analyses determined hub genes that a few of them including DDIT4 were involved in ‘microRNAs in cancer’, a pathway in CRC disease based on the KEGG DISEASE Database. Moreover, KEGG pathway enrichment analysis revealed that DDIT4 participates in PI3K-Akt/mTOR signaling pathways as important regulators for properties of pluripotent stem cells41. These signaling pathways are responsible for various functions and biological processes in cell, such as proliferation, differentiation, and migration42. It is noteworthy that the defect of PI3K-Akt/mTOR signaling pathways has been reported in development and advances of CRC43. Our findings reviewing literature indicated protein expression of DDIT4 in CRC clinical samples has received much less attention. Also, a higher expression of DDIT4 in mRNA level was observed in fresh CRC tumors compared to adjacent normal tissue samples in our previous study35. To the best of our knowledge, this study is the first to investigate of protein levels and clinical significance of DDIT4 in the patients with CRC by TMA-based IHC method.
DDIT4 is an attractive potential target for therapeutic approach in cancer12 silencing of which leads to sensitization of tumor cells to cancer treatment and drugs in in-vitro and in-vivo studies19,22,30,33. Inconsistent roles have been reported for DDIT4 in cell death and carcinogenesis so that, upregulated expression of DDIT4 occurs due to DNA-damaging agents via nuclear p53 manner that inhibits mTORC144,45 and as a result can increase cell death21,44. On the other hand, high expression of DDIT4 can protect human cancer cells from hypoxia-induced cell death19,46,47 through stabilizing HIF1α in downstream of the suppressed mTOR pathway which followed by occurs increased cell survival and tumor growth47. Moreover, studies have indicated a relationship between expression of DDIT4 and an increase in expression of BCL2 as anti-apoptotic protein and also changes in p53 phosphorylation reducing apoptosis12,22.
In-silico findings have shown high expression of DDIT4 in several cancers that is significantly associated with a worse prognosis48. Our results using three GEO series illustrated the upregulated expression of DDIT4 in CRC tissues compared to adjacent normal tissues, corroborating with the previous study that indicated overexpression of DDIT4 as prognosis biomarker in the patients with CRC using bioinformatics tools49. For further considering this marker, expression level of DDIT4 protein was investigated in 198 tissue samples from patients after surgery. Expression levels of DDIT4 were noticed in nucleus, cytoplasm, and plasma membrane substantiating the evidence in the literature31,50,51, and COMPARTMENTS database. Previous results have demonstrated localization of DDIT4 in nucleus and cytoplasm of the cells before activation while its translocation to plasma membrane was observed during activation and probably, it becomes disabled by this mechanism51.
The evaluation of DDIT4 staining was obtained for each subcellular location in cells in a series of CRC tissues with a range of intensities from weak to strong. Our analysis showed a statistically higher nuclear expression of DDIT4 compared to adjacent normal tissues in CRC, which is in line with the study by Chang et al. that observed high nuclear expression of DDIT4 in OC tissues compared to normal tissues31. This result also validates the finding of a previous experiment that identified higher levels of DDIT4 protein in 10 CRC tissues compared to adjacent normal tissues by the western blot method47. In addition, results of the population study confirmed and supported upregulation of DDIT4 expression in our bioinformatics analysis. We observed positive correlates of nuclear DDIT4 expression with the TNM stages and tumor differentiation. Notably, TNM staging and tumor differentiation are generally considered as traditional and important prognostic factors advance in which is associated with worse outcome for the patients with CRC4,52–54. Generally, in solid tumors, an immature tumor is more aggressive than a tumor with a differentiated cell phenotype55. Interestingly, median nuclear expression of DDIT4 was significantly higher in more advanced stage (stage III) compared to stage II, showing the association of nuclear expression of DDIT4 protein with aggressiveness of CRC. In addition, a significant association was found between the increased expression of DDIT4 and the decreased tumor differentiation so that, the patients with CRC who had moderately differentiated tumor cells showed higher nuclear expression of DDIT4 rather than cases with well differentiated tumor cells. This result is in line with the study by Chen et al. that showed overexpression of DDIT4 may maintain seminomas in an undifferentiated state56. Furthermore, there was no significant association between cytoplasmic and membranous expression of DDIT4 with clinicopathological features. Therefore, our results suggested that nuclear expression of DDIT4, rather than its cytoplasmic or membranous expression is related to advances of malignancy and progression in CRC. There was no significant association between expression of DDIT4 and survival in the patients with CRC, which is consistent with earlier findings on GC22 but is contrary to the previous published studies on OC31,32. Given that our findings are based on follow-up time, it can be mentioned that a long-term follow-up may plausibly alter prognostic value of DDIT4 expression.
In CRC samples, more strong intensity and higher median expression of DDIT4 in nuclear staining for tumor tissues might be due to the role of DDIT4 in the cancer cells҆ nucleus. It has been proposed that DDIT4 may have different functions in subcellular localization31,50; nevertheless, little information exists about nuclear expression of DDIT4. Despite several questions regarding DDIT4 functions in different cell locations, DDIT4 is a key player within the mTOR pathway that is critical for the cells. It is important to note that inhibition of mTOR pathway leads to the increased expression of cancer stem cell (CSC) markers and CSC populations57, CSCs are a rare subpopulation of cancer cells within tumors with stemness, self-renewal, quiescence, and tumorigenicity properties that are resistant to cancer therapy58,59. Studies have indicated that expression of DDIT4 promotes stemness markers and is related to features of CSC60–62. Drug resistance and therapeutic failure of tumor cells seems to be closely related to various properties and indispensable functions of CSCs, such as quiescence, overexpression of anti-apoptotic proteins, and the ability of DNA repair63,64.
In conclusion, findings of this study highlighted overexpression of DDIT4 as a biomarker in CRC tissues especially in nucleus of tumor cells. Overexpression of DDIT4 protein may indicate more aggressive tumor behavior and more advanced disease in the patients with CRC. Although, nuclear expression of this marker has been observed in other tumors, function of the nuclear expression of DDIT4 protein has not been reported in the literature. Accordingly, evidence indicates that DDIT4 is a potential therapeutic marker. Therefore, there is a need to increase the knowledge regarding the role of DDIT4 in cell proliferation pathways and progression of cancer that may lead to improvement of prognosis and developing agents for targeted treatment.
Methods
Bioinformatics analysis
Microarray data source and screening for differentially expressed genes (DEGs)
Data source
Primary search was performed on the Gene Expression Omnibus (GEO) database65 of the National Center for Biotechnology Information (NCBI) using keywords of "Colorectal Neoplasms" OR "Colorectal Cancer" OR "Colorectal Tumor" as search terms. Obtained GEO series of primary search were limited to expression profiling by array, homo sapiens, tissues, and publication dates [1 year (OCT 2018–2019)]. The samples and experiments҆ condition of series were closely checked in order to minimize heterogeneity for selection of expression profiles. Also, the last series of colon cancer without time limitation was added because colon cancer is more common than rectal cancer1. Finally, three gene expression series including –GSE74602, GSE110223, and GSE110224—were selected from the GEO. Among series, GSE74602 had used 30 tumors and 30 adjacent normal colon samples based on the Agilent GPL6104 platform (Illumina humanRef-8 v2.0 expression beadchip). GSE110223 was based on GPL96 platform ([HG-U133A] Affymetrix Human Genome U133A Array) that included 13 CRC tumors and 13 adjacent normal samples. While GSE110224 consisted of 17 CRC tumors and 17 adjacent normal samples with GPL570 platform ([HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array).
Identification of DEGs
The classification of CRC samples for our analysis was only based on their tumor and adjacent normal tissues, because our CRC patients in the population study section was lacking molecular subtypes of CRC. Expression of the genes was compared between tumors and adjacent normal tissues using the GEO2R online analysis tool (http://www.ncbi.nlm.nih.gov/geo/geo2r) for each series. The DEGs in CRC tissues base cut-off criteria (logFC ≥ ± 1.0 and adjusted P value < 0.05) of each series were obtained and visualized by volcano plot using OriginPro 2019 (OriginLab Corporation, Northampton, MA, USA). Common upregulated DEGs in CRC tissues were extracted from three series for subsequent analysis by Venn diagram analysis in OriginPro 2019 software. In order to recheck the common upregulated genes obtained in this method, the results were compared with common DEGs obtained by MINT method. MINT is a powerful approach to solve the integrative classification framework by combining multiple independent studies and is a part of the mixOmics R CRAN package66. Therefore, 3 series of GEO were merged and detected common DEGs for them by MINT method in R platform.
Generation of PPI network and extraction of hub genes
PPI network
For finding more data about common upregulated DEGs and their interactions with other proteins in cellular activities, PPI network construction was performed using the International Molecular Exchange Consortium (IMEx) in Cytoscape software67. IMEx includes some databases, such as IntAct, DIP, HPIDB, MINT, and UniProtKB/Swiss-Prot without redundant set of interactions checking data quality and ensuring consistency between all the series68.
Hub genes
The genes with high connectivity degree (> 75 percentile for genes with degree > 10) were selected from PPI network in Cytoscape software by cyto-Hubba plug-in69. Among the obtained genes with high connectivity degree for this network, only the genes belonging to our common upregulated DEGs were selected as hub genes by Venn diagram.
Pathway and functional enrichment analysis for hub genes
The KEGG pathway analysis for all the hub genes was done in order to scrutinize the biological pathways in which these genes are involved. Then, gene ontology (GO) term enrichment analysis was performed in domains of molecular function (MF) and biological process (BP) only for the hub genes involved in pathways with P value < 0.05. Moreover, pathways similar to those mentioned in CRC disease according to the KEGG DISEASE Database were considered in order to select a biomarker70. These analyses were performed in Cytoscape software using ClueGO plug-in assisting the GO/pathway analysis and visualizing functionally grouped terms in networks and graphs71. Finally, one hub gene was selected for evaluation of protein expression levels using the IHC method in CRC tissue samples. Subcellular localization of this maker was investigated in COMPARTMENTS database72.
Population study
Collection of tissue samples and TMA construction
Patients
FFPE blocks as well as hematoxylin and eosin (H&E)-stained slides of 198 CRC and 39 adjacent normal tissues from patients that had undergone tumor resection were included in this study. These samples were collected from three university hospitals-Hashemi Nejad, Firoozgar, and Hazrate Rasoole Akram in Tehran, Iran during 2012–2018. Patients had not received any relevant radiotherapy, chemotherapy, or immunotherapy before surgery. Clinicopathological features including age, gender, tumor size, tumor differentiation, TNM stage, vascular invasion (VI), lymph node invasion (LNI), perineural invasion (PI), distant metastasis, and tumor recurrence were obtained by reviewing clinicopathological records for each sample defined according to the American Joint Committee on Cancer (AJCC) in 201852.
Outcome information of the patients after surgery was also recorded. Disease-specific survival (DSS) was described as the time from initial surgery to the date of CRC-related death. Progression-free survival (PFS) was determined as the time between the primary surgery and the last follow-up interview if the patient with CRC displayed no evidence of disease, recurrence, or metastasis.
TMA construction
TMA blocks were provided as described previously73. In brief, the most representative areas of the tumor and normal cells in different parts of tissue samples were selected by checking H&E slides. Then, a core of 0.6 mm diameter was separated from the selected areas in each donor block and was transferred to a new recipient paraffin block using a precision arraying instrument (Tissue Arrayer Minicore; ALPHELYS, Plaisir, France). In the current study, 3 cores of the selected areas randomly from each tissue sample were transferred and scored individually. These TMA blocks were built of different cores for each sample. TMA slides were obtained by cutting sections of TMA blocks, to a thickness of about 4 μm, which were transferred to an adhesive‑coated slide system (SuperFrost Plus, Thermo Scientific™, Germany). Mean expression of 3 cores was calculated from each tissue sample in order to increase accuracy and validity of the data analysis. The TMA-based IHC method is robust with respect to sampling that could increase this accuracy similar to conventional tissue sections, even analysis of 2 cores increased accuracy by more than 95%74,75.
Applying IHC method on TMA slides and immunostaining system scoring
IHC procedure
Immunostaining of TMA sections for DDIT4 was performed as described previously76. The slides were deparaffinized after 30 min at 60 °C and were rehydrated in xylene and serial ethanol dilutions, respectively. H2O2 (3%) was used for blockage of endogenous peroxidase on tissue sections for 20 min at room temperature. Following three times washing the tissue sections, antigen retrieval was carried out by covering the tissues in citrate buffer (pH 6.0) for 10 min in an autoclave. Subsequently, tissue sections with primary antibody specific for DDIT4 (1:80 dilution, Biorbyt, Cambridge, UK) were incubated overnight at 4 °C. Furthermore, rabbit immunoglobulin IgG (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was applied at a dilution of 1:500 for isotype control test. For visualizing bound antibody-antigen, TMA slides were incubated with the secondary antibody (EnVision Kit, Dako, Glostrup, Denmark) for 30 min and then 3, 30-diaminobenzidine as chromogen (DAB, Dako, Glostrup, Denmark). The hematoxylin (Dako, Glostrup, Denmark) was used a counterstain for all the slides. Finally, the tissues slides were dehydrated in alcohol, and were cleared in xylenes until scoring by pathologists. The normal liver tissue was applied as positive control while Tris-buffered saline (TBS) was replaced by the primary antibody as the negative control per each run of the test.
Evaluation of immunostaining
Positive cells for staining of DDIT4 were assessed using a semi-quantitative scoring at 40X magnifications by two pathologists (Z.HS. and M.M.) who were blinded to pathological parameters and the patients҆ outcomes. DDIT4 staining intensity was scored in a 4-point scale (staining intensity scores: absent: 0, weak: 1, moderate: 2, or strong: 3) and percentage was estimated according to < 25, 25–50%, 51–75%, and > 75% of positive tumor cells. In calculation of the scores, H-score was obtained for each core through multiplying the intensity score by percentage of the stained cells ranging from 0 to 30077. The final H-scores were calculated from average of three score spots for each sample and were classified into two groups (low or high expression) according to median expression of DDIT4.
Statistical analysis
The Chi-square and Spearman’s correlation tests were used to analyze the association and correlation between expression of DDIT4 and clinicopathological features. The Kruskal–Wallis and Mann–Whitney U tests were applied for comparison of DDIT4 expression between groups. DSS and PFS were calculated using the Kaplan–Meier method by the log-rank test to compare survival outcomes between patient groups (low and high expression of marker). The data were expressed as mean with standard deviation (SD) and median with quartile (Q1, Q3). All the statistical analyses for the obtained clinical data were performed using SPSS v.22.0 software (IBM Corp., Armonk, NY, USA) and P-value less than 0.05 was considered as statistically significant.
Ethical approval
This study was approved by the Iran University of Medical Sciences Research Ethics Committee (IR.IUMS.REC.1398.1043). All procedures performed in this study were in accordance with the 1964 Helsinki Declaration and its later amendments. Also, informed consent was obtained from all individual participants included in the study at the time of sample collection in accordance with the above-mentioned ethical standards.
Supplementary Information
MINT sPLS-DA plots for differentially expressed genes (DEGs) in CRC by MINT method as well aspathways related to colorectal cancer (CRC) recorded on the KEGG DISEASE Database.
Lists of up and down-regulated genes from GSE74602, GSE110223 and GSE110224, common upregulatedDEGs, high connectivity degree genes and hub genes as well as the obtained DEGs from merged 3 series by MINTmethod.
Results of enrichment analysis by ClueGO
Acknowledgements
This research was supported by a grant of the Iran University of Medical Sciences (Grant: 98-2-28-15643). Also, the authors are grateful to Dr. Saeideh Khodaee for good advice on bioinformatics analysis and Elham Kalantari for her excellent technical assistance.
Author contributions
Z.M. and M.N. were designed and supervised the study. F.F. wrote the manuscript, participated in tissue sample collection, performed the immunohistochemistry examinations, and data analysis. L.S.Z. analyzed and interpreted the data, edited the manuscript as well as helped to prepare the Figures and Tables. Z.H.S. marked the most representative areas in different parts of the tumor for preparing the TMA blocks. Z.H.S. and M.M. scored TMA slides after immunohistochemical staining. J.K. and M.N. interpreted the results of bioinformatics analysis and biomarker selection. All authors read and approved the final manuscript.
Data availability
The analyzed data during the current study are available from the corresponding authors on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Najafi, Email: nbsmmsbn@iums.ac.ir.
Zahra Madjd, Email: Zahra.madjd@yahoo.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-92720-z.
References
- 1.Siegel RL, et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Molinari C, et al. Heterogeneity in colorectal cancer: A challenge for personalized medicine? Int. J. Mol. Sci. 2018;19:3733. doi: 10.3390/ijms19123733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blank A, Roberts DE, 2nd, Dawson H, Zlobec I, Lugli A. Tumor heterogeneity in primary colorectal cancer and corresponding metastases. Does the apple fall far from the tree? Front. Med. (Lausanne) 2018;5:234–234. doi: 10.3389/fmed.2018.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joachim C, et al. Overall survival of colorectal cancer by stage at diagnosis: Data from the martinique cancer registry. Medicine (Baltimore) 2019;98:e16941–e16941. doi: 10.1097/MD.0000000000016941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carethers JM. Systemic treatment of advanced colorectal cancer: Tailoring therapy to the tumor. Ther. Adv. Gastroenterol. 2008;1:33–42. doi: 10.1177/1756283X08093607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koncina E, Haan S, Rauh S, Letellier E. Prognostic and predictive molecular biomarkers for colorectal cancer: Updates and challenges. Cancers (Basel) 2020;12:319. doi: 10.3390/cancers12020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh H-H, Joo Y-E. Novel biomarkers for the diagnosis and prognosis of colorectal cancer. Intest. Res. 2020;18:168–183. doi: 10.5217/ir.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vacante M, Borzì AM, Basile F, Biondi A. Biomarkers in colorectal cancer: Current clinical utility and future perspectives. World J. Clin. Cases. 2018;6:869–881. doi: 10.12998/wjcc.v6.i15.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Punt CJA, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2017;14:235–246. doi: 10.1038/nrclinonc.2016.171. [DOI] [PubMed] [Google Scholar]
- 10.Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int. J. Mol. Sci. 2019;20:4781. doi: 10.3390/ijms20194781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perera-Bel J, Leha A, Beibarth T. Predictive Biomarkers in Oncology. Springer; 2019. pp. 149–164. [Google Scholar]
- 12.Tirado-Hurtado I, Fajardo W, Pinto JA. DNA damage inducible transcript 4 gene: The switch of the metabolism as potential target in cancer. Front. Oncol. 2018;8:106. doi: 10.3389/fonc.2018.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamosh A, et al. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canal M, Romaní-Aumedes J, Martín-Flores N, Pérez-Fernández V, Malagelada C. RTP801/REDD1: a stress coping regulator that turns into a troublemaker in neurodegenerative disorders. Front. Cell. Neurosci. 2014 doi: 10.3389/fncel.2014.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoshani T, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801 involved in apoptosis. Mol. Cell Biol. 2002;22:2283. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J. Biol. Chem. 2003;278:27053–27058. doi: 10.1074/jbc.M303723200. [DOI] [PubMed] [Google Scholar]
- 18.Malagelada C, Ryu EJ, Biswas SC, Jackson-Lewis V, Greene LA. RTP801 is elevated in Parkinson brain substantia nigral neurons and mediates death in cellular models of Parkinson's disease by a mechanism involving mammalian target of rapamycin inactivation. J. Neurosci. 2006;26:9996–10005. doi: 10.1523/JNEUROSCI.3292-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foltyn M, et al. The physiological mTOR complex 1 inhibitor DDIT4 mediates therapy resistance in glioblastoma. Br. J. Cancer. 2019;120:481–487. doi: 10.1038/s41416-018-0368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff NC, McKay RM, Brugarolas J. REDD1/DDIT4-independent mTORC1 inhibition and apoptosis by glucocorticoids in thymocytes. Mol. Cancer Res. 2014;12:867–877. doi: 10.1158/1541-7786.MCR-13-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14–3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du F, et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun. (Lond.) 2018;38:45–45. doi: 10.1186/s40880-018-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith ER, Xu XX. REDD1, a new Ras oncogenic effector. Cell Cycle Georgetown Tex. 2009;8:675–676. doi: 10.4161/cc.8.5.8184. [DOI] [PubMed] [Google Scholar]
- 24.Chang B, et al. REDD1 is required for RAS-mediated transformation of human ovarian epithelial cells. Cell Cycle (Georgetown, Tex.) 2009;8:780–786. doi: 10.4161/cc.8.5.7887. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Baicalein upregulates DDIT4 expression which mediates mTOR inhibition and growth inhibition in cancer cells. Cancer Lett. 2015;358:170–179. doi: 10.1016/j.canlet.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horak P, et al. Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4675–4680. doi: 10.1073/pnas.0907705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kucejova B, et al. Interplay between pVHL and mTORC1 pathways in clear-cell renal cell carcinoma. Mol. Cancer Res. 2011;9:1255–1265. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin H-O, et al. Redd1 inhibits the invasiveness of non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 2011;407:507–511. doi: 10.1016/j.bbrc.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 29.Schwarzer R, et al. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene. 2005;24:1138–1149. doi: 10.1038/sj.onc.1208236. [DOI] [PubMed] [Google Scholar]
- 30.Zeng Q, et al. Inhibition of REDD1 sensitizes bladder urothelial carcinoma to paclitaxel by inhibiting autophagy. Clin. Cancer Res. 2018;24:445–459. doi: 10.1158/1078-0432.CCR-17-0419. [DOI] [PubMed] [Google Scholar]
- 31.Chang B, et al. Overexpression of the recently identified oncogene REDD1 correlates with tumor progression and is an independent unfavorable prognostic factor for ovarian carcinoma. Diagn. Pathol. 2018;13:87–87. doi: 10.1186/s13000-018-0754-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia W, et al. REDD1 and p-AKT over-expression may predict poor prognosis in ovarian cancer. Int. J. Clin. Exp. Pathol. 2014;7:5940–5949. [PMC free article] [PubMed] [Google Scholar]
- 33.Jin HO, et al. Induction of HSP27 and HSP70 by constitutive overexpression of Redd1 confers resistance of lung cancer cells to ionizing radiation. Oncol. Rep. 2019;41:3119–3126. doi: 10.3892/or.2019.7036. [DOI] [PubMed] [Google Scholar]
- 34.Protiva P, et al. Growth inhibition of colon cancer cells by polyisoprenylated benzophenones is associated with induction of the endoplasmic reticulum response. Int. J. Cancer. 2008;123:687–694. doi: 10.1002/ijc.23515. [DOI] [PubMed] [Google Scholar]
- 35.Fattahi, F. et al. Overexpression of DDIT4 and TPTEP1 are associated with metastasis and advanced stages in colorectal cancer patients: a study utilizing bioinformatics prediction and experimental validation. Cancer Cell Int21, 303. 10.1186/s12935-021-02002-x (2021). [DOI] [PMC free article] [PubMed]
- 36.Fattahi F, et al. Enrichment of Up-regulated and down-regulated gene clusters using gene ontology, miRNAs and lncRNAs in colorectal cancer. Comb. Chem. High Throughput Screen. 2019;22:534–545. doi: 10.2174/1386207321666191010114149. [DOI] [PubMed] [Google Scholar]
- 37.Danielsen SA, et al. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim. Biophys. Acta (BBA) Rev. Cancer. 2015;1855:104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Francipane MG, Lagasse E. mTOR pathway in colorectal cancer: an update. Oncotarget. 2014;5:49–66. doi: 10.18632/oncotarget.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley RK, Wang G, Venook AP. Biomarker use in colorectal cancer therapy. J. Natl. Comp. Cancer Netw. 2011;9:1293–1302. doi: 10.6004/jnccn.2011.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/s0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 41.Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development (Cambridge, England) 2016;143:3050–3060. doi: 10.1242/dev.137075. [DOI] [PubMed] [Google Scholar]
- 42.Xu F, Na L, Li Y, Chen L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 2020;10:54. doi: 10.1186/s13578-020-00416-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Koveitypour Z, et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019;9:97. doi: 10.1186/s13578-019-0361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Y, Vassetzky Y, Dokudovskaya S. mTORC1 pathway in DNA damage response. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2018;1865:1293–1311. doi: 10.1016/j.bbamcr.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 45.Ellisen LW, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell. 2002;10:995–1005. doi: 10.1016/S1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 46.Ronellenfitsch MW, et al. Antagonism of the mammalian target of rapamycin selectively mediates metabolic effects of epidermal growth factor receptor inhibition and protects human malignant glioma cells from hypoxia-induced cell death. Brain J. Neurol. 2009;132:1509–1522. doi: 10.1093/brain/awp093. [DOI] [PubMed] [Google Scholar]
- 47.Di Conza G, et al. The mTOR and PP2A pathways regulate PHD2 phosphorylation to fine-tune HIF1α levels and colorectal cancer cell survival under hypoxia. Cell Rep. 2017;18:1699–1712. doi: 10.1016/j.celrep.2017.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinto JA, et al. In silico evaluation of DNA damage inducible transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci. Rep. 2017;7:1526. doi: 10.1038/s41598-017-01207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, et al. Two precision medicine predictive tools for six malignant solid tumors: From gene-based research to clinical application. J. Transl. Med. 2019;17:405–405. doi: 10.1186/s12967-019-02151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin L, Stringfield TM, Shi X, Chen Y. Arsenite induces a cell stress-response gene, RTP801, through reactive oxygen species and transcription factors Elk-1 and CCAAT/enhancer-binding protein. Biochem. J. 2005;392:93–102. doi: 10.1042/BJ20050553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michel G, et al. Plasma membrane translocation of REDD1 governed by GPCRs contributes to mTORC1 activation. J. Cell Sci. 2014;127:773–787. doi: 10.1242/jcs.136432. [DOI] [PubMed] [Google Scholar]
- 52.Weiser MR. AJCC 8th edition: Colorectal cancer. Ann. Surg. Oncol. 2018;25:1454–1455. doi: 10.1245/s10434-018-6462-1. [DOI] [PubMed] [Google Scholar]
- 53.Fleming M, Ravula S, Tatishchev SF, Wang HL. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012;3:153–173. doi: 10.3978/j.issn.2078-6891.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derwinger K, Kodeda K, Bexe-Lindskog E, Taflin H. Tumour differentiation grade is associated with TNM staging and the risk of node metastasis in colorectal cancer. Acta Oncol. (Stockholm, Sweden) 2010;49:57–62. doi: 10.3109/02841860903334411. [DOI] [PubMed] [Google Scholar]
- 55.Jögi A, Vaapil M, Johansson M, Påhlman S. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Upsala J. Med. Sci. 2012;117:217–224. doi: 10.3109/03009734.2012.659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen KS, et al. EGF receptor and mTORC1 are novel therapeutic targets in nonseminomatous germ cell tumors. Mol. Cancer Ther. 2018;17:1079–1089. doi: 10.1158/1535-7163.mct-17-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhola NE, et al. Treatment of triple-negative breast cancer with TORC1/2 inhibitors sustains a drug-resistant and notch-dependent cancer stem cell population. Cancer Res. 2016;76:440–452. doi: 10.1158/0008-5472.CAN-15-1640-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012;44:2144–2151. doi: 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aponte PM, Caicedo A. Stemness in cancer: Stem cells, cancer stem cells, and their microenvironment. Stem Cells Int. 2017;2017:5619472. doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ho K-H, et al. A key role of DNA damage-inducible transcript 4 (DDIT4) connects autophagy and GLUT3-mediated stemness to desensitize temozolomide efficacy in glioblastomas. Neurotherapeutics. 2020 doi: 10.1007/s13311-019-00826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinto JA, Bravo L, Chirinos LA, Vigil CE. Expression of DDIT4 Is correlated with NOTCH1 and high molecular risk in acute myeloid leukemias. Blood. 2016;128:5254–5254. doi: 10.1182/blood.V128.22.5254.5254. [DOI] [Google Scholar]
- 62.Gharibi B, Ghuman M, Hughes FJ. DDIT4 regulates mesenchymal stem cell fate by mediating between HIF1α and mTOR signalling. Sci. Rep. 2016;6:36889. doi: 10.1038/srep36889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Das PK, Islam F, Lam AK. The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells. 2020;9:1392. doi: 10.3390/cells9061392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinogradov S, Wei X. Cancer stem cells and drug resistance: The potential of nanomedicine. Nanomedicine (Lond.) 2012;7:597–615. doi: 10.2217/nnm.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrett T, et al. NCBI GEO: archive for functional genomics data sets—Update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohart F, Eslami A, Matigian N, Bougeard S, Lê Cao K-AMINT. a multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. BMC Bioinform. 2017;18:128. doi: 10.1186/s12859-017-1553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Orchard S, et al. Protein interaction data curation: The International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chin C-H, et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11–S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bindea G, et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Binder JX, et al. COMPARTMENTS: Unification and visualization of protein subcellular localization evidence. Database. 2014 doi: 10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Erfani E, et al. Comparative expression analysis of putative cancer stem cell markers CD44 and ALDH1A1 in various skin cancer subtypes. Int. J. Biol. Mark. 2016;31:e53–61. doi: 10.5301/jbm.5000165. [DOI] [PubMed] [Google Scholar]
- 74.Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab. Investig. J. Tech. Methods Pathol. 2000;80:1943–1949. doi: 10.1038/labinvest.3780204. [DOI] [PubMed] [Google Scholar]
- 75.Rosen DG, et al. Validation of tissue microarray technology in ovarian carcinoma. Mod. Pathol. 2004;17:790–797. doi: 10.1038/modpathol.3800120. [DOI] [PubMed] [Google Scholar]
- 76.Shahin M, et al. Low level expression of human telomerase reverse transcriptase predicts cancer-related death and progression in embryonal carcinoma. J. Cancer Res. Clin. Oncol. 2020;146:2753–2775. doi: 10.1007/s00432-020-03319-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCarty KS, Jr, et al. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986;46:4244s–4248s. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MINT sPLS-DA plots for differentially expressed genes (DEGs) in CRC by MINT method as well aspathways related to colorectal cancer (CRC) recorded on the KEGG DISEASE Database.
Lists of up and down-regulated genes from GSE74602, GSE110223 and GSE110224, common upregulatedDEGs, high connectivity degree genes and hub genes as well as the obtained DEGs from merged 3 series by MINTmethod.
Results of enrichment analysis by ClueGO
Data Availability Statement
The analyzed data during the current study are available from the corresponding authors on reasonable request.