Dear Editor,
Most mitochondrial proteins are synthesized in the cytosol and transported into various mitochondrial subcompartments in a process that is mediated by intricate multimeric machineries. Tim23, the key component of the TIM23 complex forming a channel in the mitochondrial inner membrane (MIM), is believed to recognize and translocate precursor proteins into the mitochondrial matrix or to release them into the MIM.1 Previous study has demonstrated that purified Tim23 forms a protein-conducting channel that can be activated by an appropriate change in the membrane potential in the presence of a mimetic mitochondrial preprotein,2 a signaling peptide corresponding to the presequence of cytochrome c oxidase subunit IV (denoted as pCoxIV, primary sequence: MLSLRQSIRFFKPATRTLCSSRYLL). Here, we present the three-dimensional (3D) solution structure of the yeast Tim23 channel in complex with pCoxIV in micelles as determined by high-resolution nuclear magnetic resonance (NMR), providing structural insight into the molecular mechanism of presequence translocation.
Refolded Tim23 in lauryldimethylamine oxide (LDAO) detergent micelles forms dimer as determined via size exclusion chromatography and static light scattering (Supplementary information, Fig. S1a, b), with a predominantly α-helical secondary structure as shown by its far-ultraviolet circular dichroism spectra (Supplementary information, Fig. S1c). A well-dispersed 2D [15N]–transverse relaxation-optimized spectroscopy (TROSY)–heteronuclear single-quantum coherence (HSQC) spectrum of [2H, 15N]-labeled Tim23 can be obtained in LDAO micelles (Supplementary information, Fig. S2a–d). Stepwise addition of unlabeled pCoxIV into [2H, 15N]-labeled Tim23 yielded the best NMR spectrum with 1:1 stoichiometry between the Tim23 dimer and the peptide pCoxIV (Supplementary information, Fig. S3). Based on the 3D backbone experiments (Supplementary information, Fig. S4), we assigned 206 of the backbone NH cross peaks for Tim23 (Supplementary information, Fig. S5). The nuclear Overhauser effect (NOE) experiments and intermolecular paramagnetic relaxation enhancement experiments (Supplementary information, Fig. S6) were performed to acquire a large number of distance constraints (Supplementary information, Table S1). In addition, NOE experiments performed on [13C, 15N]-labeled pCoxIV premixed with unlabeled Tim23 allowed for the direct identification of 61 intermolecular NOE contacts between pCoxIV and Tim23 (Supplementary information, Fig. S7). Taken together, these constraints enabled the calculation of the Tim23–pCoxIV complex solution structure.
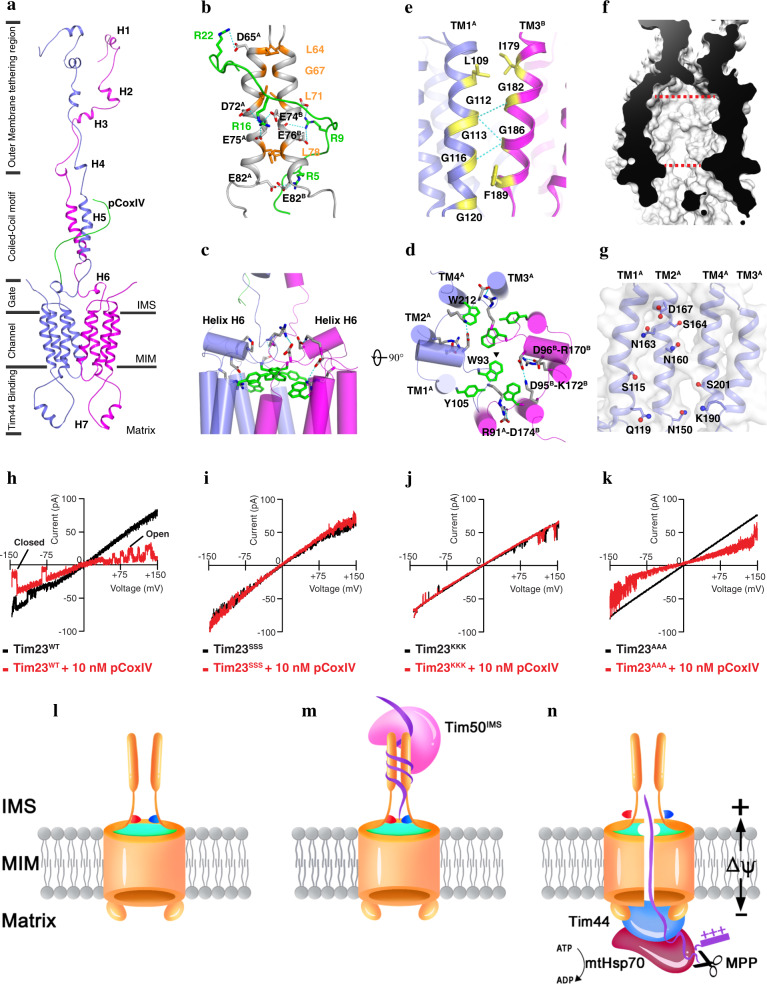
Our data indicated that the solution structure of the Tim23–pCoxIV complex is formed by two identical monomers and a completely disordered mitochondrial presequence peptide (Fig. 1a). In each Tim23 monomer, the N-terminal domain in the intermembrane space (IMS) is dynamic but has two distinct regions (Fig. 1a), and the mitochondrial outer membrane (MOM) tethering region3 contains helices H1 (A14–A17), H2 (L31–S34), H3 (N41–S45), H4 (T55–L58), and the coiled-coil helix H5 (L64–E82) interacting with pCoxIV. In the C-terminal portion of the Tim23 monomers, four transmembrane (TM) helices (TM1–TM4) pack together in the order of TM1–TM2–TM4–TM3 when viewed from the cytosol (Supplementary information, Fig. S8a). Among them, TM1 and TM3 interact with each other to form a dimer interface in the membrane, while TM2 and TM4 constitute the central channel (Supplementary information, Fig. S8b). Furthermore, a short helix H6 (D95–Y99) folding on the membrane interface in IMS, as well as the helix H7 (K131–N139) located in the matrix, have high sequence conservation across different organisms (Supplementary information, Fig. S9), indicating their important roles in channel function. Specifically, previous studies demonstrated that the helix H7 is responsible for anchoring the scaffold protein Tim44 to recruit heat-shock protein 70 (Hsp70)-based motor in matrix.4 Overall, the N-terminus of Tim23 channel is highly negatively charged in IMS, while the positive charges are present in matrix side (Supplementary information, Fig. S10).
Fig. 1. Solution structure of the Tim23-pCoxIV complex.
a Cartoon representation of the Tim23–pCoxIV structure. The seven soluble helices (H1–H7) of the monomers are indicated. The presequence peptide of CoxIV (green) is labeled as pCoxIV. The complex structure is divided into regions of the channel in the mitochondrial inner membrane (MIM); the gate, coiled-coil motif and outer membrane tethering region in the intermembrane space (IMS); and Tim44-binding helix H7 in the matrix. b Stick representation of the positively-charged arginine residues (R22, R16, R9, and R5) in pCoxIV (green) and the negatively-charged aspartate (D65, D72) and glutamate (E74, E75, E76, and E82) residues in the coiled-coil motif (gray) of the Tim23 channel. The superscripts A and B indicate monomer A and B, respectively. Potential interactions via an electrostatic effect are shown by dashed cyan lines. The residues forming the coiled-coil motif are shown and labeled in orange. c, d Side and top views of cylindrical representations of the gating zone of the Tim23 channel. Salt bridges are shown by dashed cyan lines. The large side chain residues W93, Y105, and W212 are shown in green. Half of the salt bridges and side chains are labeled at their positions. The triangle represents the center position of the Tim23 channel. e The residues in the Tim23 dimer interface are shown in yellow and labeled in TM1A (light blue) and TM3B (magenta) for each monomer. The H-bonds are indicated by dashed cyan lines. f Sliced-surface view of the Tim23 channel in membrane. The red dashed lines indicate the widest and narrowest diameters. g The hydrophilic and charged residues inside of the channel are shown on the TM helices of monomer A with side chains and labeled. h–k Representative traces of channel currents showing Tim23WT (h), and the Tim23 sequence variants Tim23SSS (i), Tim23KKK (j), and Tim23AAA (k) in the absence (black) or presence (red) of 10 nM pCoxIV. l A closed Tim23 channel in the idle state. The coiled-coil region interaction is relatively loose, and the channel is sealed by salt bridges (red and blue) and the aromatic rings of tryptophan and tyrosine (green). m Tim50IMS, the soluble domain of Tim50 located in the IMS, and the precursor protein bind to the coiled-coil region, changing it into a tight conformation. n In the presence of an appropriate change in the membrane potential (ΔΨ), the channel is opened when the salt bridges are broken and the aromatic rings are dispersed. The presequence is pulled through the channel by an electrophoresis effect and digested by protease MPP. The mature polypeptide is transported into the matrix by scaffold protein Tim44 and chaperone mtHsp70 using ATP.
The structure of the Tim23–pCoxIV complex includes several novel features. First, the presequence pCoxIV interacts with the coiled-coil motif in the IMS via electrostatic forces. The L64–E82 polypeptide from each monomer folds into a dimeric parallel coiled-coil motif (Fig. 1a, b). Each of the two helices can be depicted in a helical wheel diagram showing that the a and d residues constitute a predominantly hydrophobic interaction interface (Supplementary information, Fig. S11). The presequence pCoxIV binds to the coiled-coil motif in a C–N direction, ending at the surface of the Tim23 channel. A remarkable feature of the coiled-coil motif is the presence of a negatively-charged patch comprising six residues (D65, D72, E74, E75, E76, and E82) (Fig. 1b). The negatively-charged patches exposed to the IMS are responsible for interacting with the amphipathic positively-charged pCoxIV (Fig. 1b). Second, the described structure has two pairs of intramonomer salt bridges, D95–K172 and D96–R170, in the short helix H6 as well as an intermonomer salt bridge R91–D174 in the loop region, located on the border of the MIM (Fig. 1c, d). Given the results of a recent study that sequence variants of the two conserved negatively-charged residues, D95 and D96, reduce the mitochondrial membrane potential,5 we propose here that helix H6 may serve as a voltage sensor of the Tim23 channel associated with the polarization and depolarization of the MIM. Notably, residues W93, Y105, and W212 are positioned at the membrane interface of the Tim23 channel, where the large side chains are oriented toward the center of pore (Fig. 1c, d), suggesting that the tilting and rearrangement of indole and phenol may be associated with the transition from a closed to an open state of the channel. Third, the dimer interface of the Tim23 channel in the membrane is formed mainly via the hydrogen (H)-bonds between residues of the conserved GxxxG motif6 in TM1 and TM3 of each monomer (Fig. 1e). The Tim23 channel has a diameter ranging from about 12 Å at its narrowest, to 22 Å at its widest (Fig. 1f and Supplementary information, Fig. S12), consistent with the previous biochemical study result.2 The TM2-TM4 is organized to constitute the central pore module, with residues D167, S164, N163, N160, and N150 in TM2, residues S115 and Q119 in TM1, residue K190 in TM3, and residue S201 in TM4 facing the interior of the pore (Fig. 1g), consistent with previous studies.7,8 The charged residues D167 and K190 located at the interface of the membrane may have important roles in regulating the Tim23 channel.
Next, we designed three sequence variants, with the goal to destruct the coiled-coil motif in the IMS (substituting Y70, L71, and L78 with serine; denoted as Tim23SSS), the salt bridges of the voltage sensor (D95, D96, and D174 with lysine; denoted as Tim23KKK), and the large side chain residues blocking the channel (W93, Y105, and W212 with alanine; denoted as Tim23AAA). We used cell-attached patch-clamp electrophysiology techniques to record single-channel currents of the wild-type Tim23 (denoted as Tim23WT) and the three sequence variants that had been reconstituted in a giant liposome in the absence or presence of pCoxIV. Single-channel recording of Tim23WT at a membrane potential from −150 to 150 mV showed a large current indicative of a pore (Fig. 1h, black curve). As expected, the addition of pCoxIV at the same membrane potential opened the Tim23 channel, and pCoxIV was pulled into the channel by electrophoresis and blocked the current in channel, therefore, a lower current was recorded (Fig. 1h, red curve). Notably, the movement of pCoxIV into the channel induced a flickering between the open and closed states (Fig. 1h, red curve). By contrast, the inhibitory effect of pCoxIV on the current was nearly abolished in sequence variants Tim23SSS and Tim23KKK (Fig. 1i, j). Substituting Y70, L71, and L78 with serine destabilized the coiled-coil motif in the IMS, affecting the binding with pCoxIV. Replacing conserved D95, D96, and D174 with lysine completely disrupted the salt bridges in the voltage sensor, leading to the defective membrane depolarization. Therefore, pCoxIV did not pass into the channel of either the Tim23SSS or Tim23KKK sequence variants. For the third variant, Tim23AAA, we observed a current and inhibitory pattern similar to those observed with Tim23WT, but no closed state was detected in Tim23AAA in the presence of pCoxIV (Fig. 1k). A plausible explanation is that substituting tryptophan and tyrosine with alanine led to a failure to seal the gate when the channel was occupied by pCoxIV, indicating an important role for residues W93, Y105, and W212 in preventing the leakage of small ions. Overall, the results from our electrophysiological experiments are consistent with the Tim23–pCoxIV complex solution structure determined herein.
On the basis of the structure determined in the present study and extensive data in the literature, we propose the following model for the translocation of precursor proteins into the mitochondrial matrix. First, the coiled-coil motif exists in a relatively loose conformation in the idle Tim23 channel while the channel in the membrane is sealed by intramonomer and intermonomer salt bridges and the clustering of indole and phenol at the membrane interface (Fig. 1l). With the aid of Tim50,9 the precursor proteins bind to the coiled-coil region and cause a tight conformation of this motif (Fig. 1m). In the next step, depolarization of the membrane evoked by a change in the membrane potential results in breakage of the salt bridges and induction of an outward movement of the putative voltage sensor helices H6. These changes effectively lead to separation of the membrane interfacial clustering of the indole and phenol fragments of tryptophan and tyrosine, respectively. The precursor proteins move into the channel via an electric field, expanding the channel width by disrupting the weak H-bonds at the dimer interface (Fig. 1e). This disruption could lead to the dissociation of the dimer into monomers during the transport of a polypeptide chain through the channel as suggested previously (Fig. 1n).10 The presequence peptide is cleaved by the matrix processing protease (MPP) when moving from the channel to the matrix, and the mature region of the polypeptide chain is then transported through the channel by scaffold protein Tim44 and chaperone mitochondrial Hsp70 (mtHsp70) using ATP (Fig. 1n).4,11 Finally, when the precursor protein has passed through, the re-formation of salt bridges and re-clustering of the large side chains of tryptophan and tyrosine return the channel to its closed state (Fig. 1l).
In summary, the solution structure of the Tim23–pCoxIV complex may shed light on the understanding of how precursor proteins are imported into the mitochondrial matrix.
Supplementary information
Supplementary information, Table S1 and Methods
Acknowledgements
We acknowledge the High Magnetic Field Laboratory of Chinese Academy of Sciences and the Biomolecular Magnetic Resonance Center supported by Hesse State at University of Frankfurt for allowing us to access NMR spectrometers. A portion of this work was supported by the High Magnetic Field Laboratory of Anhui Province. This study was supported by grants from the Ministry of Science and Technology of China (Grant No., 2016YFA0400901), the National Natural Science Foundation of China (Grant No., U1632274), Hefei Science Center of Chinese Academy of Sciences (Grant No., 2020HSC-CIP011) and by iNEXT-Discovery, project number 871037, funded by the Horizon 2020 program of the European Commission.
Author contributions
M.R. purified the NMR samples. S.Z., M.R., Y.L., J.Y., and C.R. performed the NMR experiments and analyzed the NMR data, S.Z. and M.R. determined the NMR structure. B.S. and S.B. performed the electrophysiological experiments. S.Z., H.S., C.X., and J.W. designed the experiments, analyzed the data and wrote the manuscript.
Data availability
The structural restraints and coordinates have been deposited in the Protein Data Bank (PDB: 7CLV). The chemical shift values have been deposited in the Biological Magnetic Resonance Data Bank (36365).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shu Zhou, Maosen Ruan
Contributor Information
Shu Zhou, Email: shu.zhou@hmfl.ac.cn.
Bing Shen, Email: shenbing@ahmu.edu.cn.
Junfeng Wang, Email: junfeng@hmfl.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-00452-y.
References
- 1.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truscott KN, et al. Nat. Struct. Mol. Biol. 2001;8:1074–1082. doi: 10.1038/nsb726. [DOI] [Google Scholar]
- 3.Donzeau M, et al. Cell. 2000;101:401–412. doi: 10.1016/S0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- 4.Ting S, Yan NL, Schilke BA, Craig EA. Elife. 2017;6:e23609. doi: 10.7554/eLife.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Günsel U, et al. J. Mol. Biol. 2020;432:3326–3337. doi: 10.1016/j.jmb.2020.03.031. [DOI] [PubMed] [Google Scholar]
- 6.Demishtein-Zohary K, Marom M, Neupert W, Mokranjac D, Azem A. FEBS J. 2015;282:2178–2186. doi: 10.1111/febs.13266. [DOI] [PubMed] [Google Scholar]
- 7.Alder NN, Jensen RE, Johnson AE. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Malhotra K, Sathappa M, Landin JS, Johnson AE, Alder NN. Nat. Struct. Mol. Biol. 2013;20:965–972. doi: 10.1038/nsmb.2613. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto H, et al. Cell. 2002;111:519–528. doi: 10.1016/S0092-8674(02)01053-X. [DOI] [PubMed] [Google Scholar]
- 10.Bauer MF, Sirrenberg C, Neupert W, Brunner M. Cell. 1996;87:33–41. doi: 10.1016/S0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, D’Silva P, Walter W, Marszalek J, Craig EA. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Table S1 and Methods
Data Availability Statement
The structural restraints and coordinates have been deposited in the Protein Data Bank (PDB: 7CLV). The chemical shift values have been deposited in the Biological Magnetic Resonance Data Bank (36365).