Abstract
Bacterial energy metabolism is now recognized as a critical factor for the efficacy of antibiotics. The F-type ATPase/ATP synthase (FOF1) is a central player in cellular bioenergetics of bacteria and eukaryotes, and its potential as a selective antibiotic target has been confirmed by the success of bedaquiline in combatting multidrug-resistant tuberculosis. Venturicidin macrolides were initially identified for their antifungal properties and were found to specifically inhibit FOF1 of eukaryotes and bacteria. Venturicidins alone are not effective antibacterials but recently were found to have adjuvant activity, potentiating the efficacy of aminoglycoside antibiotics against several species of resistant bacteria. Here we discovered more complex effects of venturicidins on the ATPase activity of FOF1 in bacterial membranes from Escherichia coli and Pseudomonas aeruginosa. Our major finding is that higher concentrations of venturicidin induce time– and ATP–dependent decoupling of F1-ATPase activity from the venturicidin-inhibited, proton-transporting FO complex. This dysregulated ATPase activity is likely to be a key factor in the depletion of cellular ATP induced by venturicidins in prior studies with P. aeruginosa and Staphylococcus aureus. Further studies of how this functional decoupling occurs could guide development of new antibiotics and/or adjuvants that target the F-type ATPase/ATP synthase.
Subject terms: Biochemistry, Microbiology
Introduction
The F-type ATPase/ATP synthase is a ubiquitous rotary motor enzyme involved in cellular bioenergetics in eukaryotes and bacteria. It couples proton transport through a transmembrane complex (FO) with hydrolysis/synthesis of ATP on a peripheral catalytic complex (F1)1–3. In eukaryotes and photosynthetic or respiratory bacteria, FOF1 functions primarily to synthesize ATP. In contrast, many anaerobic bacteria require FOF1 to work as an ATP-driven proton pump to generate the cell’s membrane potential (Δψ) and help maintain pH homeostasis4; this reverse function is critical even for some strongly aerobic bacteria like Pseudomonas aeruginosa under fermentative conditions5. Despite general conservation of structure and function with mitochondrial FOF1 (mitoFOF1), bacterial FOF1 is now recognized as a promising target in the fight against multidrug-resistant (MDR) pathogens6, as bedaquiline (BDQ) has become a key part of front-line therapy for MDR tuberculosis7. The antimycobacterial activity of BDQ is mainly due to its interaction with c-subunits of FO8. Each c-subunit has a conserved acidic residue involved in proton transport, and a ring of c-subunits spans the membrane; BDQ binds at multiple sites on the c-ring, close to the essential carboxylates9. BDQ’s bactericidal action correlates with dramatic depletion of cellular ATP10, but may also involve its ability to collapse transmembrane ΔpH through its interactions with FO11.
It is increasingly apparent that bioenergetic factors are promising targets for antibiotic development6 and that a bacterium’s metabolic state can greatly impact the efficacy of existing antibiotics12. Thus, compounds targeting bacterial FOF1 may lead to new antibiotics13 and/or adjuvants that enhance the efficacy of other antibiotics14. Genetic knockout of FOF1 in Escherichia coli15 and Staphylococcus aureus16 enhances their sensitivity to several antibiotics. Several antifungal macrolides, including oligomycins and venturicidins, target FO in membrane preparations isolated from mitochondria, chloroplasts, and bacteria. Mutations that make the enzyme resistant to these macrolides indicate that they bind at overlapping sites near the essential carboxylate on c-subunits of FO to block proton transport and thus inhibit ATP synthesis and hydrolysis on coupled F117. The structural binding site for oligomycin on FO has been determined recently and overlaps with the BDQ binding site noted above18. Oligomycin A is too toxic for clinical use but can act as a potent adjuvant for polymyxin B action against S. aureus16. Venturicidins have minimal toxicity in mice and dogs19,20; toxicity is minimal for some human cell lines but significant for others21,22. Alone, venturicidins do not exhibit antibacterial activity21–23. Recently, however, venturicidin A (ventA) was found to potentiate the action of aminoglycoside antibiotics against various MDR bacterial pathogens22; this adjuvant activity was suggested to be due to ventA’s direct inhibition of ATP synthesis by FOF1 and the subsequent increase in PMF, which should potentiate uptake of aminoglycosides. In particular, high concentrations of ventA dramatically enhanced bactericidal effects of gentamicin on S. aureus (MRSA) strains; reduction of cellular ATP content was considered a contributing factor, and was attributed to inhibition of FOF1. However, in the complex growth medium used, S. aureus can produce substantial ATP through substrate-level phosphorylation24,25 and FOF1 is not essential for growth16,26.
In this study, we report novel aspects of the interactions of venturicidins A and B (ventB) with FOF1-ATPase in inverted membrane vesicles from E. coli and P. aeruginosa. Adding ventA or ventB to membranes induces immediate inhibition of ATP hydrolysis that, at higher inhibitor concentrations, is followed by a time-dependent increase in ATPase activity. We show that the latter phase of ATPase recovery results from venturicidin-induced functional decoupling of F1-ATPase activity from the proton–transporting FO. Further, with E. coli membranes, we show that minimizing the fraction of MgADP-inhibited enzyme significantly increases the enzyme’s affinity for ventA and ventB. We discuss how these findings provide new insights into the likely mechanisms of venturicidins’ adjuvant activity for some antibiotics.
Results
Venturicidins exhibit complex, time-dependent effects on the rate of ATP hydrolysis by E. coli membranes
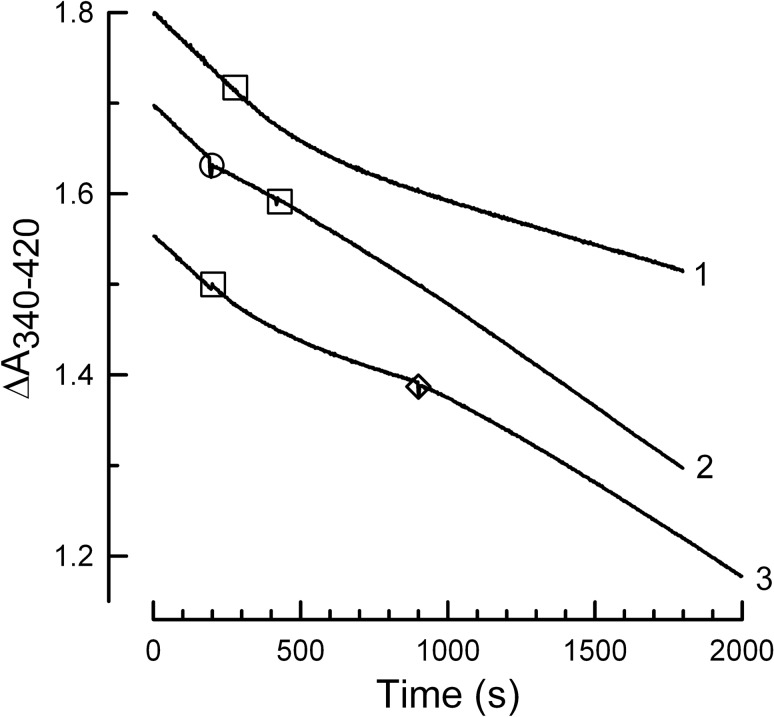
Figure 1 shows examples for the spectrophotometric assay used to monitor continuous hydrolysis of ATP by FOF1 on isolated membranes. ATP hydrolysis is enzyme-coupled to conversion of NADH to NAD+, which results in decreased light absorbance; thus, a larger rate of ATP hydrolysis is indicated by a steeper downward slope. The system also rapidly regenerates ATP, minimizing the concentration of product ADP during the assay. Unless noted, assays were preincubated to establish a steady state hydrolysis rate (see “Methods”) before the ‘zero’ point of measurements. Thus, control assays for each condition in Fig. 1 (traces 1, 5, 9) are essentially linear over the entire measurement period. With this method, we observed that ventA has complex effects on the kinetics of FOF1-ATPase activity of wild type (WT) E. coli membranes. Adding ventA at lower concentrations causes an immediate, small increase in the ATPase rate (Fig. 1, trace 2, + 11%, first minute post-addition), whereas an intermediate concentration immediately inhibits the rate (trace 3, –28%). At higher ventA concentrations, immediate inhibition is followed by time-dependent partial recovery of activity (trace 4) and, after completion of the recovery phase, a later ventA addition does not induce transient inhibition or further increase of the elevated rate (not shown). Immediate inhibition of ATPase activity by venturicidin has been reported for membranes from another bacterium, Paracoccus denitrificans27 but, for E. coli, assays typically included preincubating membranes with venturicidin (10–45 min) before adding ATP to start hydrolysis23,28. In the present study, effects of ventA on ATPase kinetics were similar when assays contained an optimal concentration of the activating anion selenite (traces 6–8), except that no increase in ATPase rate occurred at low concentrations of ventA. Similar effects of ventA were seen when assays contained the F1 inhibitor azide at a concentration that reduced the initial ATPase rate by ~ 50% (traces 9–12).
Figure 1.
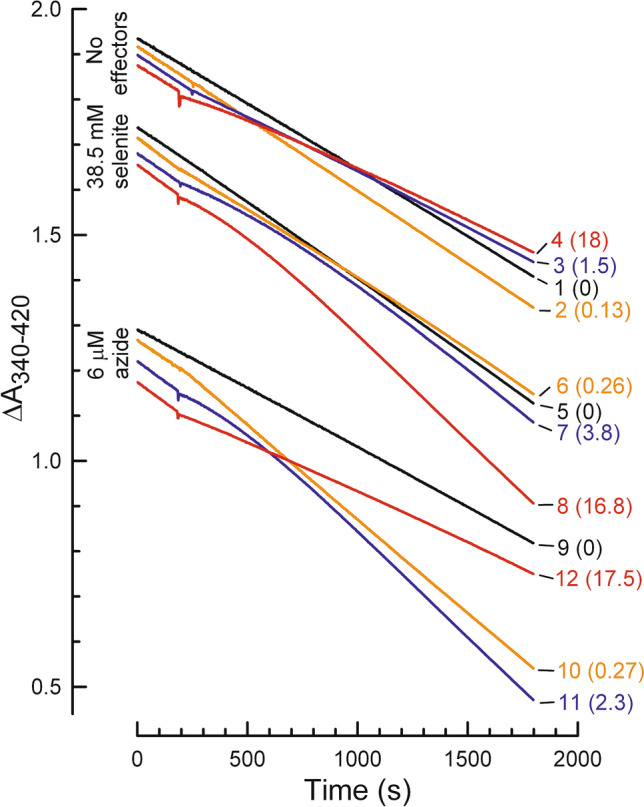
Venturicidin A has complex effects on ATPase kinetics of WT E. coli membranes. ATP hydrolysis was assayed as described in “Methods”. As noted beside the vertical axis, an additional effector was present initially for traces 5–8 (38.5 mM selenite) and 9–12 (6 µM azide). At 200 or 300 s, ventA was added to some assays; for each numbered trace, the final ventA concentration (μM) is in parentheses on the right, in the same color as the trace. Membrane protein per 1 ml assay: 3.36 µg (traces 1–4), 1.47 µg (traces 5–8) or 7.35 µg (traces 9–12). For each set of assay conditions (± selenite or azide), the specific ATPase activity (U/mg membrane protein) before adding ventA is listed in Table 1 under “100% ATPase”.
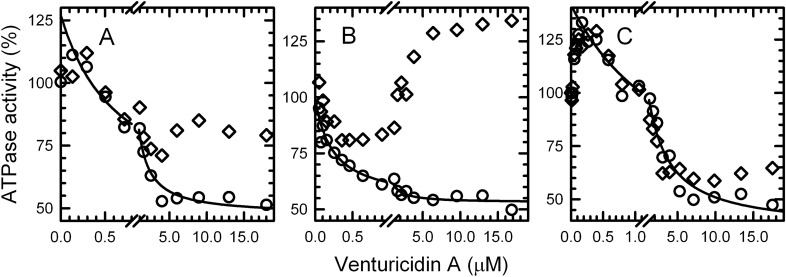
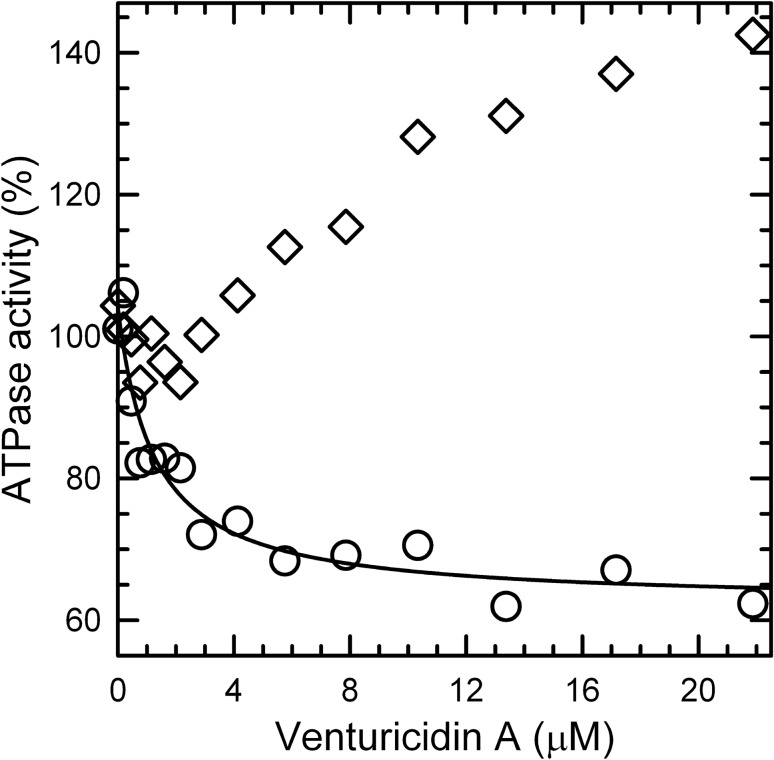
The complex dependence of ATPase rates on ventA concentration is illustrated in Fig. 2 by comparing rates measured within 1 min after adding ventA (‘early’, ○) and during the last 5 min of each 30-min assay (‘late’, ◊). Very low concentrations of ventA (< 1 μM, Fig. 2A) immediately increase the early hydrolysis rate, which then remains nearly constant for the remaining assay period (i.e., late rate is similar). Interestingly, this effect is absent in the presence of selenite (Fig. 2B) but is more pronounced in the presence of azide (Fig. 2C). At higher ventA concentrations, late rates deviate and become increasingly larger than early rates. Notably, the impacts of selenite and azide on the increasing late hydrolysis rates are opposite to their effects on early rates with low ventA: the time-dependent increase in late hydrolysis rates is enhanced by selenite and reduced by azide; the possible significance of these distinct impacts is addressed in the Discussion. The main point is that such a recovery phase in the action of venturicidins on FOF1-ATPases has not been reported before and would not have been detected in most earlier studies of venturicidin inhibition with E. coli membranes, which used end-point assays of ATPase over 3–6 min following addition of substrate ATP23,28.
Figure 2.
Concentration-dependence of venturicidin A’s immediate and time-dependent effects on the ATPase rate of WT E. coli membranes. WT membranes were assayed as in Fig. 1 and ATPase rates were measured ‘early’ (within 1 min after adding ventA, ○) and ‘late’ (during last 5 min of 30-min assay, ◊) in the absence (A) or presence of 38.5 mM selenite (B) or 6 μM azide (C). In each panel, the line shows the best fit of a hyperbolic equation, y = y0 + y1/(1 + x/Ki), to the range of early rates (○, y) that decrease with increasing ventA concentration (x); this includes all points with [ventA] > 0 for panel (A), all points for panel (B), and points with > 0.15 μM ventA for panel (C). The activity (U/mg) corresponding to 100% for each condition and the best fit parameters for each curve are in Table 1.
Focusing on the early ATPase rates of Fig. 2A and starting from the lowest ventA concentration that yielded the largest early rate, inhibition by venturicidin fits well to a hyperbolic equation. Although the forms of venturicidin used in some early studies were not specified (A, B, or a mixture), the half-maximal inhibitory concentration of venturicidin that can be obtained from the data of 28 is comparable to the Ki of 0.7 μM obtained here (Table 1) under similar assay conditions (excess of Mg2+ over ATP); the results of 23 yield a larger value, near 7 μM, which could be due to more divergent assay conditions and/or a predominance of ventB in that venturicidin sample (see Supplementary Fig. S1).
Table 1.
Best-fit parameters for early inhibition of E. coli membrane ATPase by venturicidin A.
| Membranes, condition | 100% ATPase (U/mg) | VentA-sensitive ATPase (y1) | ATPase at saturating ventA (y0) | VentA at half-max. inhibition (Ki, μM) |
|---|---|---|---|---|
| WT, alone (Fig. 2A) | 0.83 (± 0.07) | 80% (± 5) | 47% (± 2) | 0.7 (± 0.1) |
| WT + selenite (Fig. 2B) | 2.3 (± 0.2) | 46% (± 2) | 53% (± 1) | 0.23 (± 0.04) |
| WT + azide (Fig. 2C) | 0.42 (± 0.02) | 106% (± 5) | 36% (± 4) | 1.7 (± 0.3) |
| ε88stop (Fig. 3A) | 0.82 (± 0.05) | 67% (± 4) | 37% (± 3) | 0.5 (± 0.1) |
| ε88stop + selenite (Fig. 3B) | 3.4 (± 0.5) | 66% (± 4) | 37% (± 2) | 0.09 (± 0.02) |
Values for the best-fit parameters are listed for curves of Figs. 2 and 3 for nonlinear regression of the hyperbolic dependence [y = y0 + y1/(1 + x/Ki)] of relative ATPase early rates (y) on the concentration of ventA added (x). For each experiment, the 100% ATPase value is the average specific activity of all assays measured just before the addition of ventA.
Venturicidins A and B exhibit selective affinity for active states of E. coli FOF1-ATPase
As shown recently for assay conditions as in Fig. 2A without ventA, the measured ATPase rate for E. coli membranes actually reflects only ~ 20% of E. coli FOF1 complexes (EcFOF1) that are active on average, with most in transiently inactive states due to distinct actions of the ε subunit (≥ 50%) or inhibitory MgADP bound at one of the 3 catalytic sites on F1 (~ 30%)29. Certain anions stimulate ATPase activity of F1-ATPases30,31 including EcF132,33 by decreasing the MgADP-inhibited fraction of the enzyme population34, most likely due to accelerated dissociation of inhibitory MgADP35,36. Selenite is one of the most potent anion-activators31, and the presence of optimal selenite stimulates ATPase activity of WT membranes 2.8-fold (Table 1, ‘100%’ value). At the same time, selenite reduces the Ki value for early inhibition by ventA by threefold (Fig. 2B, Table 1). This suggests that ventA has a higher affinity for the enzyme actively hydrolyzing ATP than for MgADP-inhibited EcFOF1. If so, then increasing the fraction of MgADP-inhibited EcFOF1 should increase the Ki value for ventA. To test this, we measured inhibition by ventA in the presence of azide, a well-known inhibitor of F-ATPases from all sources. Azide inhibits F-ATPase by increasing the fraction of MgADP-inhibited enzyme34,37,38. Including 6 µM azide in the assay (Fig. 2C) inhibits ATPase activity ~ 50% and increases the early Ki value for ventA by 2.4-fold (Table 1), further supporting the suggestion that the active form of EcFOF1 has higher affinity for ventA than does the MgADP-inhibited form. In a prior study with membranes from P. denitrificans, ATPase activity was ~ threefold more sensitive to ventA inhibition when FOF1 was activated by the oxyanion sulfite than when it was activated by PMF, and that was interpreted as indicating two distinct active states27. However, consistent with our results, the ATPase activity was also increased 2- to fourfold more by sulfite than by PMF, which could indicate sulfite was simply more effective, driving a larger fraction of FOF1 out of the MgADP-inhibited state.
We next investigated whether the ε-inhibited state of EcFOF1 also impacts interactions of ventA with the enzyme. The N-terminal domain (NTD) of ε is required for FOF1 assembly and functional coupling of F1 to FO in eukaryotic and bacterial species; the C-terminal domain (CTD) of ε can auto-inhibit FOF1 and isolated F1 from some bacteria and from chloroplasts, but the mitochondrial homolog (δ) is not inhibitory39,40. Thus we measured the effects of ventA on ATPase activity of E. coli membranes expressing ε88-stop, which lacks the autoinhibitory εCTD (Fig. 3). From Table 1 (100% ATPase values), it is clear that selenite activates membrane ATPase more (fourfold) for ε88-stop than for WT (2.8-fold). This is because, without the inhibitory εCTD, more enzyme complexes shift to the MgADP-inhibited state, which can be activated by selenite29. Consistently, the results of Fig. 3 show that the Ki for early inhibition of ε88-stop membranes by ventA is fivefold lower in the presence of selenite than in its absence (Table 1). This further supports the suggestion that ventA binding differentiates between MgADP-inhibited and active forms of EcFOF1. In contrast, early inhibition of ATPase by ventA has similar Ki values for WT and ε88-stop membranes in the absence (0.7 vs 0.5 μM) or in the presence of selenite (0.23 vs 0.09 μM). This suggests that the εCTD-inhibited state has less impact on the enzyme’s affinity for ventA than does the MgADP-inhibited state. Also, the slow ATPase recovery period induced by higher concentrations of ventA is similar for membranes with WT ε (Fig. 2A,B) or ε88stop (Fig. 3A,B), indicating that effect is not significantly altered by the presence of the εCTD.
Figure 3.
Concentration-dependence of venturicidin A’s effects on the ATPase rate of E. coli membranes with FOF1 lacking the ε subunit’s CTD. The ε88-stop membranes were assayed for ATPase as in Fig. 1, but with 3.31 μg (A) or 0.93 μg (B) of membrane protein. As in Fig. 2, rates were measured ‘early’ (○) and ‘late’ (◊) after adding ventA in the absence (A) or presence of 38.5 mM selenite (B). Each line shows the best fit of all early rates (○) to the equation shown for Fig. 2. Activities corresponding to 100% and the best fit parameters for the lines are listed in Table 1.
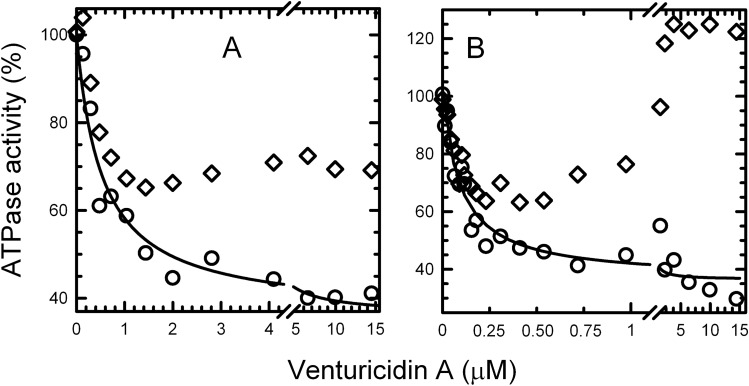
As noted earlier, we also tested the effects of venturicidin B (ventB) on ATP hydrolysis by WT E. coli membranes (see Supplementary Fig. S1). Without selenite, immediate activation induced by low concentrations of ventB is moderate but more significant than that observed for ventA (Fig. 2A); also similar to ventA, higher ventB concentrations induce immediate inhibition and time-dependent recovery of ATPase activity. With selenite present, as for ventA (Fig. 2B), low concentrations of ventB do not induce immediate activation. With or without selenite present, the Ki for inhibition of the early rate by ventB (see Supplementary Fig. S1) is ≥ tenfold higher than the respective Ki values for ventA, indicating higher affinity of EcFOF1 for ventA than for ventB. However, similar to ventA, the Ki of ventB is reduced (2.3-fold) when MgADP-induced inhibition is relieved by selenite. Thus, both venturicidins A and B exhibit lower affinity for EcFOF1 that is in the MgADP-inhibited state.
Venturicidin-induced recovery of ATPase activity is ATP–dependent
Time-dependent recovery of ATPase activity that follows immediate venturicidin-induced inhibition, reported here for the first time, occurs at higher venturicidin concentrations than required for initial inhibition. Thus it is likely that the venturicidin-binding site(s) that induce recovery of ATPase activity differ from the site(s) that cause initial inhibition. At high ventA concentrations, ATPase recovery is completed within ~ 20 min after adding ventA (traces 4 and 8 in Fig. 1, and Fig. 2). In the original study of venturicidin’s inhibition of E. coli membrane ATPase23, membranes were incubated with venturicidin for 45 min before adding ATP and assaying ATPase for 3 min. The fact that they did not observe decreased inhibition at high venturicidin concentrations suggests that MgATP is required for the recovery phase to occur after venturicidin-induced inhibition. To test whether this is the case, we incubated WT E. coli membranes at 30 °C in the assay medium containing optimal selenite but lacking ATP, PEP, and NADH in the presence and absence of 10 μM ventA for 30 min and then started the ATPase assay by adding a mixture of ATP, PEP, and NADH. In the absence of ventA, preincubating membranes without ATP does not affect kinetics of ATP hydrolysis significantly (see Supplementary Fig. S2, trace 1). If ATP were not required for the relatively fast recovery of ATPase activity after ventA-induced inhibition, then ventA-induced inhibition should not be observed after a 30-min incubation with ventA but without ATP. Supplementary Fig. S2 shows that this is not the case: preincubation with ventA decreases the initial ATPase rate by about 40%, which is similar to the extent of immediate inhibition by ventA under the same assay conditions (Fig. 2B). The recovery of activity to 130% after preincubation with ventA and subsequent addition of ATP (see Supplementary Fig. S2, trace 2) is also similar to that observed when ventA was added to membranes already hydrolyzing ATP (Fig. 2B). Thus, the time-dependent recovery of activity observed after ventA-induced inhibition requires ATP and likely catalytic turnover.
Venturicidin-induced recovery of ATPase activity involves functional decoupling of F1-ATPase from membrane-embedded FO
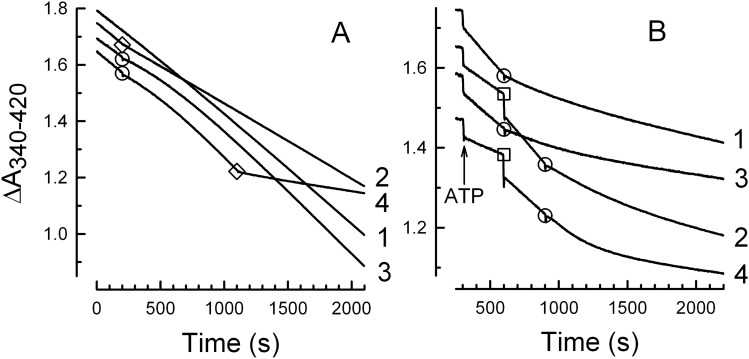
Since ventA targets FO within the membrane23, the ventA-induced, time-dependent recovery of ATPase activity could indicate that the EcF1-ATPase is functionally decoupling from EcFo. We tested this using DCCD (N,N′-dicyclohexylcarbodiimide), a well-known inhibitor of FOF1-ATPases that acts by covalently modifying c subunits of FO on a conserved acidic residue that is essential for proton transport41. Figure 4 shows that, within 10 min of adding DCCD, ATPase activity of WT membranes is inhibited by 73% (trace 1). Adding DCCD after 15 μM ventA (trace 2) does not induce inhibition and fails to prevent time-dependent recovery of ATPase activity, suggesting the recovery phase of ventA action is due to EcF1 that is no longer functionally coupled to EcFO. However, since an early study reported that venturicidin slows labeling of c subunits on EcFO by [14C]DCCD, at least in membranes stripped of EcF123, we investigated how ventA would affect ATPase activity of membranes that had been preincubated with DCCD. As shown by trace 3, adding 10 μM ventA to DCCD-inhibited membranes induces a time-dependent increase of ATPase activity from 37 to 70% of the activity observed before adding DCCD, analogous to the recovery phase observed after initial inhibition induced by high concentrations of ventA (Fig. 1, traces 4 and 8). This does not conflict with the ATP-dependence of ATPase recovery, as it has been shown that ATP can drive partial rotation of the c-ring in EcFOF1 that has been inhibited by [14C]-DCCD42. Results similar to those in Fig. 4 were also obtained in the presence of selenite (see Supplementary Fig. S3). Since DCCD inhibition involves irreversible covalent modification of c-subunit(s), the recovery phase induced by ventA must be due to decoupling of F1-ATPase from FO.
Figure 4.
VentA-induced increase in ATPase activity is not blocked by DCCD modification of FO. ATP hydrolysis was assayed as in “Methods”, using 3.36 µg of WT E. coli membrane protein. The mean of ATPase activity during the first 3 min of all assays is 0.83 (± 0.02) U/mg. DCCD was added to each assay at 0.1 mM (□) and ventA was added either before DCCD (○, trace 2, 10 μM ventA) or after DCCD (◊, trace 3, 15 μM ventA). During the last 3 min of assays, ATPase activity (± SD, n = 3) is 0.20 (± 0.02), 0.53 (± 0.05), or 0.49 (± 0.06) U/mg for conditions of representative traces 1–3, respectively.
In vitro, exposing inverted E. coli membranes to low ionic strength and chelators of divalent metals induces dissociation of EcF1 from the membrane as a soluble ATPase. Re-binding EcF1 to EcFO in EcF1-depleted membranes and coupling of EcF1-ATPase activity to proton translocation through FO require the presence of both δ and ε subunits43. Therefore, decoupling induced by high concentrations of ventA may be due to disrupting F1-FO interactions involving ε and/or δ, which leads to dissociation of EcF1 from EcFO. Since δ does not affect EcF1-ATPase activity but bound ε inhibits it44,45, we examined whether ATPase activity recovered after ventA action is a result of relieving inhibition by ε, which can dissociate from soluble EcF146. Thus we tested the effect of exogenous ε on the ATPase recovery induced by higher ventA concentrations (Fig. 5). Without ventA, adding excess ε to WT membranes hydrolyzing ATP in the presence of selenite leads to an immediate ~ 30% inhibition of ATPase activity (Fig. 5A, trace 2 vs 1). This suggests the slow increase in activity observed during initial ATP hydrolysis (~ 25%, as noted in Materials and Methods) occurs at least in part because a portion of the enzyme has lost inhibition by endogenous ε. Adding 9.5 μM ventA first (Fig. 5A, traces 3, 4) induces immediate inhibition followed by time-dependent recovery, and subsequent addition of excess ε immediately inhibits that ATPase activity by ~ 90% (trace 4), as occurs with soluble EcF146. Consequently, adding excess ε before ventA should eliminate the recovery phase. This was confirmed by assays done without the usual hydrolysis preincubation (Fig. 5B). With excess ε already present with membranes, hydrolysis was initiated by adding ATP (Fig. 5 B, arrow). Adding 10 μM ventA next immediately inhibited WT ATPase activity 55–60% in the absence (trace 1) or presence of selenite (trace 2) but without subsequent activation; instead, additional time-dependent inhibition of ~ 25% occurred. Similar results were obtained with ε88-stop membranes (Fig. 5B, traces 3, 4). Note that, in Fig. 5A, the slow ventA-induced recovery phase is complete before addition of excess WT ε (trace 4 rate is similar to final rate for trace 3), and maximum inhibition by ε occurs immediately. Thus, the same ventA-dependent limiting step is likely responsible for the slow inhibition observed with excess ε already present in the assays of Fig. 5B. This is supported by further assays without preincubation with ATP: once ventA-dependent recovery of activity is nearly complete, adding excess WT ε induces strong immediate inhibition without a significant time-dependent component, for both WT and ε88-stop membranes (see Supplementary Fig. S4).
Figure 5.
The ventA-induced increase in ATPase activity is inhibited by adding excess ε subunit. Panel (A): ATPase was assayed as described in “Methods” using 1.47 µg of WT membrane protein in the presence of 38.5 mM selenite. Symbols indicate addition of 67 nM ε (◊) or 9.5 μM ventA (○). Early rate (U/mg, first 3 min of assay): 2.30 (SD, ± 0.13 for traces 1–4). Late rates (U/mg, last 3 min of assay): #1, 2.2 (± 0.3, n = 3); #2, 1.4 (± 0.2, n = 3); #3, 2.7 (± 0.3, n = 3), #4, 0.28 (± 0.05, n = 3). For trace #4, during 100 s immediately before adding ε, the rate is 2.5 (± 0.4, n = 3) U/mg. Panel (B): traces representative of 2 experiments in which membranes were first added to the assay medium (see “Methods”) but lacking ATP and containing 88 nM ε. Membranes were either WT (traces: #1, 4.2 μg; #2, 2.1 μg), or ε88-stop (traces: #3, 3.1 μg; #4, 1.24 μg). Arrow indicates addition of 1 mM ATP to initiate each assay. Symbols indicate addition of 38.5 mM selenite (□) or 10 μM ventA (○).
Venturicidin-induced functional decoupling also occurs in FOF1 of P. aeruginosa
To investigate whether the complex effects of venturicidins are relevant to other species of pathogenic bacteria, we tested membranes from P. aeruginosa since, for three clinical isolates of aminoglycoside-resistant P. aeruginosa, ventA enhanced sensitivity to Gentamycin by 2-, 4- and 8-fold22. Compared to WT E. coli membranes (0.83 U/mg, Table 1), membranes from P. aeruginosa strain PAO1 exhibit low ATPase activity (~ 0.1 U/mg), but it is still attributable to PaFOF1 since it can be inhibited 80–90% by excess azide. The PAO1 membrane ATPase could also be activated ~ fourfold by selenite, indicating PaFOF1 is latent at least in part due to MgADP inhibition. Because of the low intrinsic ATPase activity of PAO1 membranes, we tested the effects of ventA with selenite present, and the results are very similar to those for E. coli membranes with selenite present (Figs. 2B, 3B, Table 1). VentA inhibits the early ATPase rate of PAO1 membranes (Fig. 6, ○) up to 42% and the late rate shows concentration-dependent recovery of activity up to 140% of the uninhibited rate. This indicates that PaFOF1 is also sensitive to the capacity of higher venturicidin concentrations to functionally decouple F1-ATPase activity from membrane-embedded FO.
Figure 6.
Complex effects of venturicidin A on ATPase rate are also observed with membranes from P. aeruginosa. PAO1 membranes (6.72 μg) were assayed in the presence of 38.5 mM selenite as for Fig. 1 and ATPase rates were measured at early (○) and late (◊) periods after adding ventA, as described for Fig. 2. The rate before ventA addition (100%) is 0.45 U/mg, and inhibition of the early rate fits a hyperbolic dependence on ventA concentration (see equation, Fig. 2 legend) with a Ki = 1.2 μM (± 0.4) for 42% (± 3) ventA-sensitive ATPase and 62% (± 2) ATPase remaining at saturating ventA.
Discussion
We report multiple effects of venturicidins on FOF1 in bacterial membranes, and these provide insights that could improve the efficacy of targeting bacterial FOF1 for developing new antibiotics and/or adjuvants for existing antibiotics. Figure 7 provides a schematic overview. First, venturicidins have higher affinity for FO when the coupled F1 is in an active state (Fig. 7, B) rather than shifted (step 1) to transiently inactive but significantly populated forms, the MgADP-inhibited or εCTD-inhibited states (Fig. 7, A). As evident from results summarized in Table 1, removing the εCTD has a small effect on the sensitivity of EcFOF1 to immediate inhibition by ventA, but the MgADP-inhibited state has greater impact. For example, when the proportion of WT EcFOF1 in the MgADP-inhibited state is minimized by selenite, ATPase activity is > fivefold higher and ~ sevenfold more sensitive to immediate inhibition by ventA than when the fraction of MgADP-inhibited complexes is increased by azide. Selectivity is also indicated in the modest activation of ATPase with sub-inhibitory concentrations of ventA (Fig. 2A) or ventB (see Supplementary Fig. S1): activation is eliminated by selenite (Fig. 2B, Supplementary Fig. S1) but enhanced by azide (Fig. 2C). These different impacts of the MgADP– and εCTD–inhibited states suggest that the affinity of FO for venturicidin is impacted by the rotational sub-states of F1: the orientation of γ’s central, asymmetric rotor shaft within the α3β3 assembly differs by ~ 30° for the εCTD-inhibited state of EcF1 vs the MgADP-inhibited state identified with mitoF146. The enzyme’s rotary coupling mechanism2,3, with 120° rotation per ATP hydrolyzed or synthesized, involves smaller angular sub-steps, and a range of rotational orientations have been identified in F1 structures (e.g., Fig. 9 of ref. 46). Thus, further analyses of distinct rotational states of FOF1 from different species may help identify specific states with the highest affinity binding of inhibitors to FO or F1, and/or sites that are most selective for binding to bacterial species of FOF1.
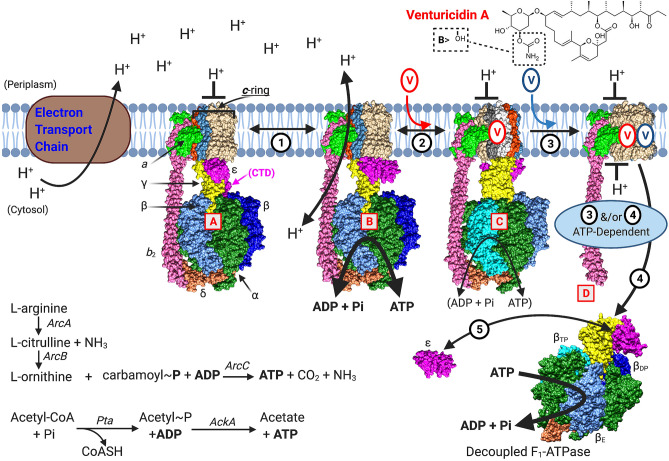
Figure 7.
Multiple actions of venturicidins on bacterial FOF1, can enhance depletion of cellular ATP. In respiring bacteria, the electron transport chain pumps protons (H+) out of the cell to generate PMF; return flux of H+ through FO of active FOF1 (B) drives synthesis of ATP at the 3 F1 catalytic sites. Active forms of FOF1 can convert (1) to transiently inactive states such as ε-inhibited (A) or MgADP-inhibited (not shown), and increased PMF favors return to the active state. Without respiration, active FOF1 can work in reverse as an ATPase-driven H+-pump to generate PMF, as long as sufficient cellular ATP is maintained by glycolysis or other pathways for substrate-level phosphorylation (e.g., lower left5,24,25). Active FOF1 (B) has the highest affinity site(s) for venturicidin binding to FO (2), which inhibits H+ transport and catalysis by coupled F1 (C). At higher concentrations, venturicidin binds (3) at additional sites and (4) induces functional decoupling of F1 (D), while venturicidin still blocks H+ transport through FO23. Decoupled F1 can only catalyze ATP hydrolysis; many F1 complexes may shift to the εCTD-inhibited state (not shown) but the portion of active F1-ATPases can increase if ε dissociates (KD < 1 nM)46 (5). Thus, decoupled F1-ATPase may significantly deplete levels of cellular ATP. In FO, 4 subunits of the c-ring are colored distinctly (steel blue, orange-red, light and dark grays) to indicate the major rotational states identified by Cryo-EM for E. coli FOF159. In F1, the catalytic β subunits are different shades of blue to reflect their distinct conformations (all 3 βs are partially visible, labeled in D). One state (PDB: 3OQR) is shown for (A,B) and decoupled F1 (D). State (C) (PDB: 3WNQ) follows one 120° rotary step (1 ATP hydrolysis) and state (D) shows FO after another 120° step (PDB: 6OQW). The compact, non-inhibitory conformation of ε (PDB: 1BSN) is shown in all states except (A). Molecular models were rendered with Chimera60; composite figure was prepared with BioRender.com.
Our more surprising and novel finding is that, although initially inhibitory (Fig. 7, step 2), venturicidin at higher concentrations binds at additional sites (step 3) and induces time– and ATP–dependent recovery of ATPase activity (steps 3, 4). For E. coli, we showed that recovered activity is due to decoupling of F1-ATPase from venturicidin–inhibited FO (Fig. 4). We then showed (Fig. 5) that decoupling is likely due to F1 dissociation from the membrane (Fig. 7, step 4). In our in vitro assays with dilute membranes, ε would dissociate from a significant portion of decoupled F1 (Fig. 7, step 5); this can explain why, with selenite present, recovered ATPase activity exceeded the activity present before adding venturicidin (Figs. 2B, 3B, and Supplementary Fig. S1). In assays without selenite (same Figs, panel A), ε dissociation would allow more decoupled F1 to become MgADP-inhibited46, resulting in lower levels of recovered activity. For growth of E. coli on glucose, decoupled cytosolic F1-ATPase lacking ε causes a greater growth defect than does a mutant completely lacking FOF1; however, the defect was reversed if ε was expressed with decoupled F147. Thus, the impact of venturicidin-decoupled F1 on bacterial ATP content should depend on the concentration of dissociated F1 and the fraction of F1 complexes that are active due to dissociation of ε (Fig. 7, step 5) and/or the presence of endogenous activating oxyanions. The prominence of ε– and MgADP–inhibited states can vary between bacterial species39 but has not been studied for many pathogens; in some species, inhibition by the εCTD may be supplemented or superseded by a unique subunit48 or by a unique segment of another F1 subunit49.
Venturicidin’s overall impact on cellular ATP should depend on the bacterium’s environmental and metabolic limitations. This could explain the variable efficacy observed in a recent study that identified ventA as an adjuvant that potentiates aminoglycoside antibiotics against several MDR pathogens22. VentA showed minimal adjuvant action against E. coli, which is capable of rapid substrate-level phosphorylation in complex growth medium. In contrast, ventA enhanced gentamycin action up to eightfold against clinical isolates of P. aeruginosa, which is highly dependent on oxidative phosphorylation since it lacks the Embden–Meyerhof–Parnas glycolytic pathway. We report ventA-induced recovery of ATPase activity with P. aeruginosa membranes (Fig. 6), and its likely importance for venturicidin’s adjuvant effect is supported by an early study that showed distinct effects of venturicidin on P. aeruginosa in different growth conditions50. During respiration, venturicidin reduced cellular ATP ~ fivefold, increased the membrane potential (Δψ), and cells retained PMF-driven flagellar motility. Inhibition of FOF1 alone could explain those effects but not venturicidin’s effects during anaerobic fermentation. In that case, most cellular ATP would be produced by fermenting added L-arginine (Fig. 7, lower left) and PMF would be maintained by FOF1 acting as an ATPase–driven proton pump5. Compared to respiratory conditions, anaerobic Δψ and cellular ATP were lower but supported flagellar motility for 45 min; venturicidin decreased cellular ATP ~ tenfold and eliminated Δψ and flagellar motility within 3.5 minutes50. Such rapid depletion of cellular ATP would not be expected from inhibition of FOF1–ATPase. We propose that the high venturicidin concentration decoupled F1, which rapidly hydrolyzed ATP generated by limited substrate-level phosphorylation. Like E. coli, S. aureus is capable of substantial substrate-level phosphorylation in complex growth media without glucose24,25 (e.g., Fig. 7, lower left). However, for several MRSA strains, ventA enhanced sensitivity to gentamycin 8- to 16-fold22, and some results suggest to us that ventA-induced decoupling of F1-ATPase is involved. For the MRSA strain tested further, the maximal effect on Δψ (by inhibiting FO) was achieved at 16 μg ventA/ml but cellular ATP was reduced by only ~ 40%; fourfold greater ventA was needed to reduce cellular ATP by ~ 90%22. Thus, the dual actions of ventA on FOF1 reported here may be important for optimal adjuvant efficacy of ventA against diverse bacterial pathogens.
Venturicidins inhibit FOF1-ATPase to varying extents in membranes from mitochondria, chloroplasts, and bacteria17 and, to our knowledge, the current study is novel in finding that higher venturicidin concentrations induce initial inhibition followed by time-dependent recovery of ATPase activity for membranes from two Gram-negative bacteria. One early study compared inhibitor sensitivities for mitoFOF1 in membranes vs detergent-solubilized mitoFOF1: oligomycin inhibited ATPase activity of both forms, whereas ventA inhibited membrane ATPase 95% but stimulated the solubilized ATPase up to 3-fold51. Thus, in membranes, mitoFOF1 likely resists decoupling due its more robust stator stalk and/or its usual dimeric state that is often disrupted by solubilization2,3. Nevertheless, this suggests a common mechanism for potential decoupling of F1-ATPase from FO. Results of a recent study suggest that decoupling FOF1 by an FO-targeted inhibitor is not unique to venturicidins: detergent-solubilized FOF1 from M. smegmatis (MsFOF1) was inhibited ~ 80% by nanomolar BDQ but most activity was restored by micromolar BDQ, although time dependence was not noted49. Recovery of ATPase activity by micromolar BDQ has not yet been observed with mycobacterial membranes [e.g., with 5 min assays52) but this could indicate BDQ-decoupled F1-ATPase activity contributes significantly to BDQ’s antibiotic efficacy: low (nM) BDQ is rapidly bacteriostatic for M. tuberculosis cultures but slow bactericidal action is greatly enhanced by higher (μM) BDQ, which also dramatically depletes cellular ATP10. The recent cryo-EM study49 determined high-resolution structures of MsFOF1 ± BDQ, with distinct high affinity sites at the “leading” and “lagging” interfaces of the c-ring with subunit a, and 5 lower affinity sites on c-subunits not contacting a. This is likely the case for different affinity sites noted here for the dual effects of venturicidins. For MsFOF1 incubated with excess BDQ, cryo-EM did not show decoupled complexes but ATP was absent, consistent with our finding that decoupling is ATP-dependent. A likely scenario is that decoupling of BDQ-saturated FOF1 involves the added stress of partial rotation driven by ATP binding on F1.
Methods
Venturicidin A was from BioViotica, and venturicidin B was from Cayman Chemical. Lactate dehydrogenase (salt-free, lyophilized) and Pyruvate kinase (type II) were both rabbit muscle enzymes obtained from Sigma-Aldrich. Before use, pyruvate kinase was desalted by column centrifugation53.
Plasmid pXH302S, which encodes the WT atpC gene and expresses near haploid levels of the ε subunit54, was subjected to fusion PCR mutagenesis to construct plasmid pMBε1, with an N-terminal affinity tag (MHHHHHHGH) added prior to the initial Met residue. To express the ε88-stop subunit, an AfeI-ScaI restriction fragment of WT pMBε1 was replaced by the analogous fragment from pH6ε88-stop46. To express haploid levels of EcFOF1 containing WT-ε or ε88-stop, the appropriate version of pMBε1 was transformed into E. coli strain XH1, which has a chromosomal deletion of the atpC gene for ε54. For these transformed strains, cultures were grown and inverted membrane vesicles (IMV) were prepared according to 55. WT ε, with the same N-terminal His6-tag, was over-expressed from pH6ε and purified as described46.
P. aeruginosa strain PAO1 was grown in LB medium at 37 °C as described56. For preparation of IMV, final cultures used 1 L of LB per 2L baffled flask and were inoculated from an overnight culture at an initial OD600 ~ 0.025. Cultures were shaken at 250 rpm and monitored until they reached mid- to late-logarithmic growth phase (OD600, 1–1.4). Cultures were chilled and subsequent steps for cell lysis and isolation of IMV were done at 4ºC. Cells were sedimented by centrifugation (3,700 × g, 30 min). Cells from 1 to 2 L of culture (~ 2–3 g wet cells/L) were resuspended in 25–40 ml of STEM buffer (TEM buffer plus sucrose55), transferred to a 50 ml Oak Ridge tube, sedimented again (11,617 × g, 10 min), resuspended in 20–40 ml of STEM and frozen at -80 °C. Thawed cells were lysed by two sequential passages through an SLM-Aminco French pressure cell at ~ 16,000 psi, essentially as described for E. coli55; the pressure cell was enclosed in a clear biohazard bag (MiniGrip, IP2024B3T) during use to contain any aerosols formed. Unlysed cells and cell wall debris were removed by centrifugation (50 ml Oak Ridge tubes, 16,743 × g, 25 min): the supernate was diluted with MTGM7.5 buffer (50 mM MOPS-TRIS, 10%(v/v) glycerol, 5 mM Mg(CH3COO)2, pH 7.5) and centrifugation was repeated at least twice, recovering the supernate each time. The final supernatant lysate was stored at -80 °C. To isolate IMV, the cleared lysate was thawed quickly and diluted with MTGM7.5 as needed to fill 1 or 2 polycarbonate tubes for use in a Beckman Ti70 rotor. The IMV (membranes) were sedimented by ultracentrifugation (250,000 × g max) for 1 h, the membranes were resuspended in MTGM7.5 buffer to fill one Ti70 tube and ultracentrifugation was repeated for 45 min. The membranes were resuspended in a small volume of MTGM7.5 buffer (typically ≥ 10 mg membrane protein/ml) and homogenized manually (Potter–Elvehjem type). Small aliquots of the final membrane sample were quick-frozen in liquid nitrogen and stored at − 80 °C.
ATP hydrolysis by membrane vesicles was assayed spectrophotometrically57 at 30 °C using an open-chamber, diode array spectrophotometer (Hewlett Packard 8453 UV–Vis) as described in29. The assay medium (1 ml per assay) contained 20 mM Mops/Tris, pH 8.0, 0.2 mM EDTA, 10 mM CH3COOK, 1 mM ATP, 3.2 mM Mg(CH3COO)2, 1 mM phosphoenolpyruvate, at least 0.3 mM NADH, 0.1 mg/ml pyruvate kinase, 0.1 mg/ml lactate dehydrogenase, 5 mM KCN, and 5 μM carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone. As noted before29, the rate of ATP hydrolysis by membranes increases to a small degree during assays. This is due, at least in part, to dissociation of a small fraction of EcF1 and/or ε from the membranes, since the increase is inhibited by added ε (e.g., Fig. 5). To avoid interference of this increase with the changes of ATPase activity induced by venturicidins, unless indicated otherwise, measurement of ATPase activities were started 60 min after addition of membranes to the complete assay medium. This minimizes further increases in ATPase rate during control assays to < 15%. To keep absorbance between 1.5 and 2 units after the 60-min preincubation, up to 0.2 mM additional NADH was added to the assay medium. In figures displaying activity traces, some traces are shifted vertically for visual clarity.
Protein was measured by a modified Lowry procedure58.
Supplementary Information
Acknowledgements
We thank Ms. Mariam Bhatti (now M.D.) for help with cloning, cell cultures, and membrane preparations. We acknowledge Vladimir V. Bulygin, PhD (deceased) and Marcus L. Hutcheon (now at Bristol Myers Squibb, East Syracuse, NY) for preparations of purified ε subunit. We thank Benjamin Lundgren, PhD and Professor Christopher T. Nomura of the SUNY College of Environmental Science and Forestry for P. aeruginosa strain PAO1 and initial advice in working with it. We acknowledge Upstate Medical University and the Department of Biochemistry and Molecular Biology for financial and collegial support.
Author contributions
Y.M.M. and T.M.D. designed the research. T.M.D. was responsible for preparation of bacterial cultures and membranes. Y.M.M. conducted and analyzed all enzymological experiments, and prepared all data figures. T.M.D. prepared the schematic Fig. 7. Both authors wrote and revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-93098-8.
References
- 1.Boyer PD. The ATP synthase–a splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 3.Kühlbrandt W. Structure and mechanisms of F-Type ATP synthases. Annu. Rev. Biochem. 2019;88:515–549. doi: 10.1146/annurev-biochem-013118-110903. [DOI] [PubMed] [Google Scholar]
- 4.Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Micro. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glasser NR, Kern SE, Newman DK. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol. Microbiol. 2014;92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hards K, Cook GM. Targeting bacterial energetics to produce new antimicrobials. Drug Resist. Update. 2018;36:1–12. doi: 10.1016/j.drup.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Rapid Communication: Key changes to treatment of drug-resistant tuberculosis (WHO/CDS/TB/2019.26). (2019).
- 8.Koul A, et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 2007;3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 9.Preiss L, et al. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. SCIENCE ADVANCES. 2015;1(4):E1500106. doi: 10.1126/sciadv.1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koul A, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hards K, et al. Ionophoric effects of the antitubercular drug bedaquiline. Proc. Natl. Acad. Sci. USA. 2018;115:7326–7331. doi: 10.1073/pnas.1803723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stokes JM, Lopatkin AJ, Lobritz MA, Collins JJ. Bacterial metabolism and antibiotic efficacy. Cell Metab. 2019;30:251–259. doi: 10.1016/j.cmet.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamontagne Boulet Maxime, et al. Tomatidine is a lead antibiotic molecule that targets Staphylococcus aureus ATP synthase subunit C. Antimicrobial Agents and Chemotherapy. 2018;62(6):e02197–17. doi: 10.1128/AAC.02197-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright GD. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Liu A, et al. Antibiotic sensitivity profiles determined with an escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vestergaard Martin, et al. Inhibition of the ATP Synthase Eliminates the Intrinsic Resistance of Staphylococcus aureus towards Polymyxins. mBio. 2017;8(5):e01114–17. doi: 10.1128/mBio.01114-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Molec. Biol. Rev. 2008;72:590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Symersky J, Osowski D, Walters DE, Mueller DM. Oligomycin frames a common drug-binding site in the ATP synthase. Proc. Natl. Acad. Sci. USA. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes A, Fantes KH, Boothroyd B, McGonagle MP, Crosse R. Venturicidin: a new antifungal antibiotic of potential use in agriculture. Nature. 1961;192:952–954. doi: 10.1038/192952a0. [DOI] [PubMed] [Google Scholar]
- 20.Masamune S, Sehgal JM, van Tamelen EE, Strong FM, Peterson WH. Separation and preliminary characterization of oligomycins A, B and C1. J. Am. Chem. Soc. 1958;80:6092–6095. doi: 10.1021/ja01555a049. [DOI] [Google Scholar]
- 21.Shaaban Khaled, et al. Venturicidin C, a new 20-membered macrolide produced by Streptomyces sp. TS-2–2. The Journal of Antibiotics. 2014;67:223–230. doi: 10.1038/ja.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yarlagadda V, Medina R, Wright GD. A membrane-active natural product inhibitor of atp synthase potentiates aminoglycoside antibiotics. Sci. Rep. 2020;10:8134. doi: 10.1038/s41598-020-64756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlin DS, Latchney LR, Senior AE. Inhibition of Escherichia coli H+-ATPase by venturicidin, oligomycin and ossamycin. Biochim. Biophys. Acta. 1985;807:238–244. doi: 10.1016/0005-2728(85)90254-3. [DOI] [PubMed] [Google Scholar]
- 24.Bosi E, et al. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc. Nat. Acad. Sci. USA. 2016;113:E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halsey Cortney, et al. Amino Acid Catabolism in Staphylococcus aureus and the Function of Carbon Catabolite Repression. mBio. 2017;8(1):e01434–16. doi: 10.1128/mBio.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosser Melinda, et al. Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLOS Pathogens. 2018;14(3):e1006907. doi: 10.1371/journal.ppat.1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacheco-Moises F, Garcia JJ, Rodriguez-Zavala JS, Moreno-Sanchez R. Sulfite and membrane energization induce two different active states of the Paracoccus denitrificans FOF1-ATPase. Eur. J. Biochem. 2000;267:993–1000. doi: 10.1046/j.1432-1327.2000.01088.x. [DOI] [PubMed] [Google Scholar]
- 28.Fillingame RH, Oldenburg M, Fraga D. Mutation of alanine 24 to serine in subunit c of the Escherichia coli F1FO-ATP synthase reduces reactivity of aspartyl 61 with dicyclohexylcarbodiimide. J. Biol. Chem. 1991;266:20934–20939. doi: 10.1016/S0021-9258(18)54800-6. [DOI] [PubMed] [Google Scholar]
- 29.Milgrom Yakov, Duncan Thomas. F-ATPase of Escherichia coli membranes: The ubiquitous MgADP-inhibited state and the inhibited state induced by the ε–subunit's C-terminal domain are mutually exclusive. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2020;1861(7):148189. doi: 10.1016/j.bbabio.2020.148189. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P, Moyle J. Activation and inhibition of mitochondrial adenosine triphosphatase by various anions and other agents. J Bioenerg. 1971;2:1–11. doi: 10.1007/BF01521319. [DOI] [PubMed] [Google Scholar]
- 31.Ebel RE, Lardy HA. Stimulation of rat liver mitochondrial adenosine triphosphatase by anions. J. Biol. Chem. 1975;250:191–196. doi: 10.1016/S0021-9258(19)41999-6. [DOI] [PubMed] [Google Scholar]
- 32.Dunn SD, Zadorozny VD, Tozer RG, Orr LE. ε subunit of Escherichia coli F1-ATPase: effects on affinity for aurovertin and inhibition of product release in unisite ATP hydrolysis. Biochemistry. 1987;26:4488–4493. doi: 10.1021/bi00388a047. [DOI] [PubMed] [Google Scholar]
- 33.Bulygin VV, Milgrom YM. A bi-site mechanism for Escherichia coli F1-ATPase accounts for the observed positive catalytic cooperativity. Biochim. Biophys. Acta. 2009;1787:1016–1023. doi: 10.1016/j.bbabio.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 34.Vasilyeva EA, Minkov IB, Fitin AF, Vinogradov AD. Kinetic mechanism of mitochondrial adenosine triphosphatase. Inhibition by azide and activation by sulphite. Biochem. J. 1982;202:15–23. doi: 10.1042/bj2020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murataliev MB, Boyer PD. The mechanism of stimulation of MgATPase activity of chloroplast F1-ATPase by non-catalytic adenine-nucleotide binding. Acceleration of the ATP-dependent release of inhibitory ADP from a catalytic site. Eur. J. Biochem. 1992;209:681–687. doi: 10.1111/j.1432-1033.1992.tb17336.x. [DOI] [PubMed] [Google Scholar]
- 36.Milgrom YM, Cross RL. Nucleotide binding sites on beef heart mitochondrial F1-ATPase. Cooperative interactions between sites and specificity of noncatalytic sites. J. Biol. Chem. 1993;268:23179–23185. doi: 10.1016/S0021-9258(19)49444-1. [DOI] [PubMed] [Google Scholar]
- 37.Murataliev MB, Milgrom YM, Boyer PD. Characteristics of the combination of inhibitory Mg2+ and azide with the F1 ATPase from chloroplasts. Biochemistry. 1991;30:8305–8310. doi: 10.1021/bi00098a004. [DOI] [PubMed] [Google Scholar]
- 38.Hyndman DJ, Milgrom YM, Bramhall EA, Cross RL. Nucleotide-binding sites on Escherichia coli F1-ATPase. Specificity of noncatalytic sites and inhibition at catalytic sites by MgADP. J. Biol. Chem. 1994;269:28871–28877. doi: 10.1016/S0021-9258(19)61988-5. [DOI] [PubMed] [Google Scholar]
- 39.Feniouk BA, Suzuki T, Yoshida M. The role of subunit ε in the catalysis and regulation of FOF1-ATP synthase. Biochim. Biophys. Acta. 2006;1757:326–338. doi: 10.1016/j.bbabio.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Sielaff H, Duncan TM, Börsch M. The regulatory subunit ε in Escherichia coli FOF1-ATP synthase. Biochim. Biophys. Acta Bioenerg. 2018;1859:775–788. doi: 10.1016/j.bbabio.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebald W, Hoppe J. On the structure and genetics of the proeteolipid subunit of the ATP synthase complex. Curr. Top. Bioenerg. 1981;12:1–64. doi: 10.1016/B978-0-12-152512-5.50007-5. [DOI] [Google Scholar]
- 42.Hutcheon ML, Duncan TM, Ngai H, Cross RL. Energy-driven subunit rotation at the interface between subunit a and the c oligomer in the FO sector of Escherichia coli ATP synthase. Proc. Natl. Acad. Sci. USA. 2001;98:8519–8524. doi: 10.1073/pnas.151236798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sternweis PC. The epsilon subunit of Escherichia coli coupling factor 1 is required for its binding to the cytoplasmic membrane. J. Biol. Chem. 1978;253:3123–3128. doi: 10.1016/S0021-9258(17)40811-8. [DOI] [PubMed] [Google Scholar]
- 44.Sternweis PC, Smith JB. Characterization of the inhibitory ε subunit of the proton-translocating adenosine triphosphatase from Escherichia coli. Biochemistry. 1980;19:526–531. doi: 10.1021/bi00544a021. [DOI] [PubMed] [Google Scholar]
- 45.Smith JB, Sternweis PC. Purification of membrane attachment and inhibitory subunits of the proton translocating adenosine triphosphatase from Escherichia coli. Biochemistry. 1977;16:306–311. doi: 10.1021/bi00621a023. [DOI] [PubMed] [Google Scholar]
- 46.Shah NB, Hutcheon ML, Haarer BK, Duncan TM. F1-ATPase of Escherichia coli: the ε-inhibited state forms after ATP hydrolysis, is distinct from the ADP-inhibited state, and responds dynamically to catalytic-site ligands. J. Biol. Chem. 2013;288:9383–9395. doi: 10.1074/jbc.M113.451583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klionsky DJ, Brusilow WS, Simoni RD. In vivo evidence for the role of the ε subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 1984;160:1055–1060. doi: 10.1128/jb.160.3.1055-1060.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarco-Zavala M, Mendoza-Hoffmann F, García-Trejo JJ. Unidirectional regulation of the F1FO-ATP synthase nanomotor by the ζ pawl-ratchet inhibitor protein of Paracoccus denitrificans and related α-proteobacteria. Biochim. Biophys. Acta Bioenerg. 2018;1859:762–774. doi: 10.1016/j.bbabio.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Guo H, et al. Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline. Nature. 2020;589:143–147. doi: 10.1038/s41586-020-3004-3. [DOI] [PubMed] [Google Scholar]
- 50.Armitage JP, Evans MCW. The motile and tactic behaviour of Pseudomonas aeruginosa in anaerobic environments. FEBS Lett. 1983;156:113–118. doi: 10.1016/0014-5793(83)80259-2. [DOI] [PubMed] [Google Scholar]
- 51.Linnett PE, Mitchell AD, Beechey RB. Changes in inhibitor sensitivity of the mitochondrial ATPase activity after detergent solubilisation. FEBS Lett. 1975;53:180–183. doi: 10.1016/0014-5793(75)80014-7. [DOI] [PubMed] [Google Scholar]
- 52.Sarathy JP, et al. TBAJ-876 retains Bedaquiline’s activity against subunit c and ϵ of Mycobacterium tuberculosis F-ATP synthase. Antimicrob. Agents Chemother. 2019;63:e01191–e1219. doi: 10.1128/AAC.01191-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penefsky HS. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J. Biol. Chem. 1977;252:2891–2899. doi: 10.1016/S0021-9258(17)40446-7. [DOI] [PubMed] [Google Scholar]
- 54.Xiong H, Vik SB. Construction and plasmid-borne complementation of strains lacking the ε subunit of the Escherichia coli F1FO ATP synthase. J. Bacteriol. 1995;177:851–853. doi: 10.1128/jb.177.3.851-853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wise JG. Site-directed mutagenesis of the conserved β subunit tyrosine 331 of Escherichia coli ATP synthase yields catalytically active enzymes. J. Biol. Chem. 1990;265:10403–10409. doi: 10.1016/S0021-9258(18)86960-5. [DOI] [PubMed] [Google Scholar]
- 56.Sarwar Zaara, et al. GcsR, a TyrR-Like Enhancer-Binding Protein, Regulates Expression of the Glycine Cleavage System in Pseudomonas aeruginosa PAO1. mSphere. 2016;1(2):e00020–16. doi: 10.1128/mSphere.00020-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pullman ME, Penefsky HS, Datta A, Racker E. Partial resolution of the enzymes catalysing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenyl-stimulated adonosine triphophatase. J. Biol. Chem. 1960;235:3322–3329. doi: 10.1016/S0021-9258(20)81361-1. [DOI] [PubMed] [Google Scholar]
- 58.Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 59.Sobti M, et al. Cryo-EM structures provide insight into how E. coli F1FO ATP synthase accommodates symmetry mismatch. Nat. Commun. 2020;11:2615. doi: 10.1038/s41467-020-16387-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.