Abstract
Campylobacter jejuni is one of the leading causes of gastrointestinal illness worldwide and is mainly transmitted from chicken through the food chain. Previous studies have provided increasing evidence that this pathogen can colonize and replicate in broiler chicken during its breeding; however, its temporal kinetics in laying hen are poorly understood. Considering the possible interaction between C. jejuni and gut microbiota, the current study was conducted to address the temporal dynamics of C. jejuni in the cecum of laying hen over 40 weeks, with possible alteration of the gut microbiota and fatty acid (FA) components. Following oral infection with C. jejuni 81-176, inocula were stably recovered from ceca for up to 8 weeks post-infection (p.i.). From 16 weeks p.i., most birds became negative for C. jejuni and remained negative up to 40 weeks p.i. 16S rRNA gene sequencing analyses revealed that most of the altered relative rRNA gene abundances occurred in the order Clostridiales, in which increased relative rRNA gene abundances were observed at >16 weeks p.i. in the families Clostridiaceae, Ruminococcaceae, Lachnospiraceae, and Peptococcaceae. Lipidome analyses revealed increased levels of sterols associated with bile acid metabolisms in the cecum at 16 and/or 24 weeks p.i. compared with those detected at 8 weeks p.i., suggesting that altered microbiota and bile acid metabolism might underlie the decreased colonization fitness of C. jejuni in the gut of laying hens.
Keywords: Campylobacter jejuni, laying hen, long-term breeding, gut microbiota, lipidome
Introduction
Campylobacter jejuni is one of the most reported foodborne pathogens to cause gastrointestinal illness worldwide (1, 2). Similar to the western countries, foodborne campylobacteriosis accounted for 27.0% of the total cases of foodborne gastrointestinal illness reported in 2019 in Japan (3). Several source attribution studies have provided increasing evidence that poultry and poultry products are among the main sources of human campylobacteriosis (4–6), which highlights the necessity to control this pathogen in poultry and poultry products.
The prevention of C. jejuni invasion and spread in poultry farms is recognized as one of the key issues for reducing the incidence of human campylobacteriosis because this pathogen can achieve stable and asymptomatic chicken colonization as commensal microbiota, thereby leading to bird-to-bird horizontal transmission after the invasion of farms (7). Despite the amount of research that has been performed on this subject, it is likely that the source and transmission routes of C. jejuni invasion in broiler farms have farm-to-farm variations (8). Comparatively, many attempts have been made to reduce this pathogen in the broilers during farm rearing using phage therapy (9), feed and/or water supplementation with short- or medium-chain fatty acids (FAs) (10–12), essential oils (13), organic acids (14), and probiotics (15), as practical control measures. These possible effectors are thought to have either direct or indirect bactericidal effects; however, further study would be required to clarify molecular basis of these approaches in terms of reducing Campylobacter in the field.
Chickens as a food source are largely divided into broiler chickens and laying hens. Broiler chickens are rapidly grown to slaughter weight, which is mainly affected by genetic background, digestive efficiency, and energy-use efficiency (16). During broiler chicken breeding, C. jejuni starts to colonize the gut at around 3–4 weeks of age, and then spread in the flocks, thereby becoming a burden at slaughter age (17–19). Similarly, in laying hens, which are used mainly for egg production, C. jejuni also exhibits increased colonization fitness between 0 and 4 weeks post-infection (p.i.) after experimental oral administration (20); moreover, both broilers and laying hens harbor C. jejuni at high percentages in the gut at slaughter age (21). Although laying hens are generally raised for longer periods compared with broiler chickens, the temporal dynamics of C. jejuni colonization in laying hens is not well-understood.
The recent advancements in the application of next-generation sequencing have allowed the investigation of the microbiome in specific organs of various animals. A recent study reported an association between Campylobacter burden and the microbiome in the cecum of broiler chickens (22); i.e., the authors reported a possible association between the decreased abundance of Lactobacillus spp. and high Campylobacter loads, raising questions pertaining to temporality and causation. In addition, Videnska and co-workers found different compositions of fecal microbiota between broiler chickens and laying hens at 30 or 61 weeks of age (23). However, they used chickens bred at different ages under different environments and using distinct feeds. It is likely that the fecal microbiota composition of laying hens varies according to their age; the gut microbiota of young hens are quite complex, whereas those of older hens are simpler and consist mainly of the phyla Bacteroidetes and Firmicutes (24). These observations suggest the alteration of the gut microbiota in laying hens during long-term growth, thus underscoring the need to monitor the temporal characteristics of gut microbiota in laying hens during breeding under similar environmental and feed conditions.
Based on this background, here we examined the dynamics of C. jejuni and microbiota compositions in the cecum of laying hens after experimental infection. After observing the time-to-time differences in the colonization fitness of C. jejuni and microbiota composition between 8 and 16 weeks p.i., we performed comparative lipidome analyses between these time points. Finally, we discussed their possible associations.
Materials and Methods
Bacterial Strain and Media
The C. jejuni 81-176 strain was employed as the inoculum in the chicken infection experiment. Bacteria were grown on Mueller-Hinton agar (MHA) or in Mueller-Hinton broth (MHB) (Merck, Darmstadt, Germany) at 42°C for 20 h under microaerophilic conditions using AnaeroPack-MicroAero system (Mitsubishi Gas Chemicals, Tokyo, Japan), unless otherwise indicated.
Chicken Infection Experiment
Two-week-old female specific-pathogen-free (SPF) white leghorn (Line-M) chickens (n = 38 in total) were obtained from Nisseiken (Yamanashi, Japan) and introduced into our animal facility at the ABSL2 level. Animals were fed in sterilized cages ad libitum with sterile water and antibiotic-free pellet diets (CR, Nisseiken) at 25°C with lighting from 9 a.m. to 5 p.m. in biosafety level 2 room. To prepare the bacterial inoculum, C. jejuni 81-176 was microaerobically grown in MHB at 42°C for 20 h using AnaeroPack-Microaero (Mitsubishi Gas Chemicals, Tokyo, Japan). The bacterial culture was then washed twice with PBS, and adjusted to 6.84 log CFU per 1 ml of PBS. One milliliter aliquots of the bacterial suspensions was orally inoculated into each bird via 18G-feeding gavage (Thermo Fisher Scientific, Waltham, MA, USA). At 0, 2, 8, 16, 24, 32, and 40 weeks p.i., five each animals were sacrificed per time point and samples of at least 1 g of cecum content were aseptically collected. Simultaneously, whole blood was collected from animals at 2, 8, 16, and 24 weeks p.i. (two each birds per time point), followed by centrifugation at 3,000 rpm for 5 min, to collect sera. For the control, three animals were fed for 2 weeks after their introduction, and their cecal and serum samples were collected in a similar manner. The numbers of C. jejuni from the cecum samples were enumerated according to the method of ISO 10272-2: 2017 (25).
Experiments utilizing animals were approved by the board of Animal Welfare and Ethical Committee of the National Institute of Health Science with the approval number of 680.
Enumeration of C. jejuni in Chicken Ceca
Campylobacter jejuni 81-176 was enumerated in chicken ceca essentially as described previously (20). Briefly, 1 g samples of fresh cecum were suspended in 9 ml of sterile buffered peptone water (BPW; Merck, Darmstadt, Germany); 1 ml aliquots of the BPW suspension and its serial dilutions were then spread on mCCDA agar plates (Oxoid, Hampshire, UK) and microaerobically incubated at 42°C for 48 h. The number of typical colonies was counted, and at least five suspected colonies per plate were subjected to real-time PCR to confirm C. jejuni, as described previously (26). Fisher's extract test was used to assess the statistical significance of the differences in the number of bacteria between the groups (2 and 8 vs. 16–40 weeks p.i.).
DNA Extraction
Three representative chicken cecum samples were selected at each time point, to exclude the samples with maximum and minimum bacterial counts, and subjected to 16S rRNA sequencing analysis. Aliquots of the BPW suspensions (1 ml) were centrifuged at 21,500 × g for 10 min at 4°C. The pellets were then resuspended in 400 μl of homogenization solution containing 2 μl of proteinase K (Promega, Madison, WI, USA). After incubation at 37°C for 10 min, the samples were vortexed for 5 min with Zirconia beads (ZircoPrep Mini; Nippon Genetics, Tokyo, Japan) on a Disruptor Genie instrument (Scientific Industries, Bohemia, NY, USA). After centrifugation at 11,000 × g for 5 min, 100 μl of each supernatant were transferred into 300 μl of lysis buffer (Promega). DNA extraction was then carried out using a Maxwell Blood DNA kit in a Maxwell RSC instrument (Promega). The concentration and quality of the extracted DNA were measured on a Tape Station 4150 system (Agilent Technologies, Santa Clara, CA, USA), and the samples were stored at −80°C until use.
16S rRNA Gene Sequencing
Barcoded semi-conductor sequencing analysis was performed essentially as described previously (27). Briefly, the 16S rRNA V5–V6 region sequences were amplified from 2 to 4 ng of DNA from each sample by PCR using the primers 799f and 1115r (27). The PCR amplicons were purified using E-gel Size Select 2% (Thermo Fisher Scientific) and Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA). After measuring DNA concentration using the Ion library quantification kit (Thermo Fisher Scientific), equal quantities of tagged amplicons were pooled. The pooled DNA samples (5 pM per sample) were then subjected to the Ion Chef and Ion PGM (400 bases) sequencing platform using a 318v2 chip (Thermo Fisher Scientific), according to the manufacturer's instructions.
Analysis of Microbiome Composition Data
FASTAQ files generated here were processed using the CLC Genomic Workbench ver. 20 (CLC-Qiagen, Aarhus, Denmark) to remove barcode sequences and low-quality sequences, which were defined as sequences with <275 bases, with ambiguous bases and homopolymers >6 bases, or without a barcode and a primer sequence. The 16S gene copy numbers were adjusted to 100,000 per a sample and taxonomical classification was carried out using the RDP pipeline (28) with an 80% confidence threshold. Operational taxonomy units (OTUs) were assigned using the average neighbor algorithm at 99% similarity on the RDP program, and the obtained OTUs which was then subjected to Permutational multivariate analysis of variance (PERMANOVA) test to calculate the statistical significance between three groups (group 1: 0 w p.i., group 2: 2 w and 8 w p.i., group 3: ≥16 w p.i.) by Bray-Curtis dissimilarity index under 10,000 times permutation using in-house program. Calculation of Shannon diversity indexes and Simpson indexes, and principle coordinate analysis (PCoA) were performed using Metagenome@KIN program (World Fusion, Tokyo, Japan) accordingly. All raw sequences were deposited into the DDBJ/GenBank database with accession number DRA009061 in BioProject PRJDB8861.
Cytokine Assay
Semi-quantitative cytokine assays were performed using the RayBio® C-Series Gallus (Chicken) Cytokine Array C1 kit (Raybiotech, Peachtree Corners, GA, USA), according to the manufacturer's instructions. For this assay, two representative serum samples collected from laying hens at 2, 8, 16, and 24 weeks p.i. were used in duplicate sets. Chemiluminescence detection was performed using an ImageQuant LAS 500 system (Cytiva, Marlborough, MA, USA). Densitometrical data analyses were performed according to the guidelines of the manufacturer.
Lipidome Analysis
Sample Preparation
Each pair of cecum samples collected at 8 weeks p.i. (samples 8-1 and 8-2), 16 weeks p.i. (16-1 and 16-2), and 24 weeks p.i. (24-1 and 24-2) (two samples per the time point) was subjected to a lipidome analysis. To extract lipids, 0.5 mg from each sample were sonicated in 300 μl of homogenization solution (CHCl3:methanol, = 1:2), followed by vortexing for 20 min at 20°C. The homogenate was then mixed with 20 μl of distilled water and vortexed again prior to centrifugation at 1,670 × g for 10 min at 20°C. The resultant supernatant was used as a sample in the subsequent analysis.
Analytical Equipment and Conditions
Three-microliter aliquots of the above-mentioned supernatants were injected for non-biased lipidome analysis using UPLC (Waters) in combination with Triple TOF 6600 (AB Sciex, Framingham, MA, USA) essentially as described previously (29). Mobile phase A consisted of 1:1:3 acetonitrile:methanol:water (v/v/v) with 5 mM ammonium acetate and 10 nM EDTA. Mobile phase B was 100% isopropanol with 5 mM ammonium acetate and 10 nM EDTA. The LC column was an Acquity UPLC Peptide BEH C18 column (50 × 2.1 mm; 1.7 μm; 130 Å). The gradient was 0 min, 0% B; 1 min, 0% B; 5 min, 40% B; 7.5 min, 64% B; 12.0 min, 64% B; 12.5 min, 82.5% B; 19 min, 85% B; 20 min, 95% B; 20.1 min, 0% B; and 25 min, 0% B. The column flow rate was 0.3 ml/min and the autosampler temperature was 5°C. The column temperature was 45°C. MS was performed on a TripleTOF 6600 system equipped with a DuoSpray ion source. All analyses were performed in the high sensitivity mode for both TOF–MS and product ion scanning. Data-dependent MS/MS acquisition (DDA) was used. The common parameters in both positive and negative ion mode were as follows: collision energy, 45 V; collision energy spread, 15 V; mass range, m/z 140–1,700; temperature, 300°C; and declustering potential, 80 V.
Statistical Analysis
The statistical differences of bacterial numbers among the different age groups (2, 8, 16, 24, 32, and 40 weeks p.i.) were calculated by Steel-Dwass test, and P < 0.05 were considered to be significant. To compare 16S rRNA gene DNA sequence data between 2/8 and 16/24/32/40 weeks p.i., relative abundances were comparatively analyzed by a non-parametric joint ranked Dunn test, and P < 0.05 were considered to be significant. The MS/MS spectra of each fragment ranged from 70 to 1,700 m/z obtained by lipidome analyses were analyzed using the MS-DIAL program and MS-FINDER software (30) to identify and classify lipids. The statistical significance of the differences among the different age groups (8, 16, and 24 weeks p.i.) was calculated by Bonferroni test and P-value of <0.05 were considered to be significant.
Results
Colonization Fitness of C. jejuni in the Cecum of Laying Hens Over a Period of 40 Weeks
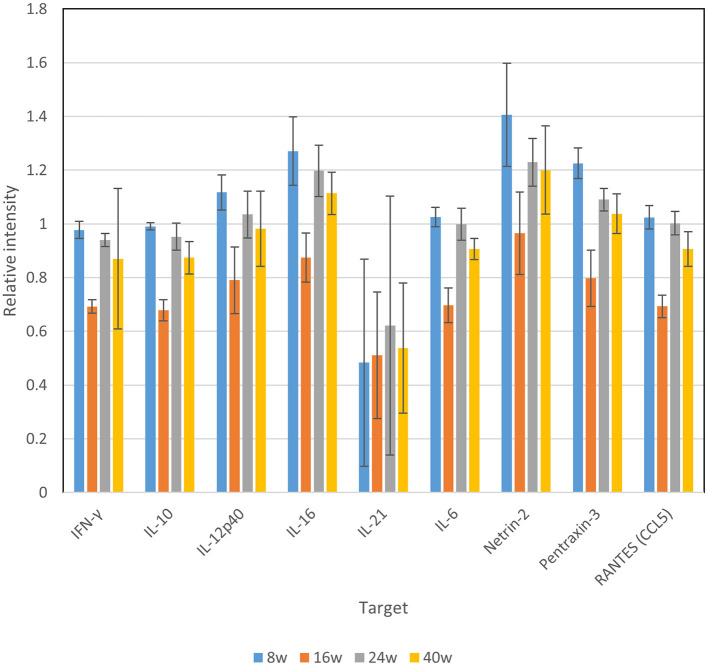
After oral infection with C. jejuni 81-176, the inocula were stably recovered from the cecum of laying hens for up to 8 weeks p.i.; the number of pathogens recovered was 7.08 and 6.90 log CFU/g at 2 and 8 weeks p.i., respectively (Figure 1). At 16 weeks p.i., C. jejuni were recovered only from two chickens (40%, 2/5 birds), with average means of 5.18 log CFU/g (Figure 1). At 24, 32, and 40 weeks p.i., C. jejuni was recovered from one out of five birds, at 5.02, 5.65, and 4.30 log CFU/g, respectively (Figure 1). Statistically, there was a significant difference (P = 0.0002) in the recovered bacterial burden between 2 and 8 weeks p.i. (defined as C. jejuni colonizer) and 16–40 weeks p.i. (C. jejuni excluser) (Figure 1). Thus, these data indicate that C. jejuni retained colonization at an early stage, but tended to have a reduced colonization fitness after 16 weeks p.i. in the gut of laying hens.
Figure 1.
Temporal colonization fitness of C. jejuni 81-176 in the caecum of laying hens. Each dot (gray) represents bacterial numbers detected per a gram of caecum. Dotted line represents median at each sampling time point. Statistical significance of the differences in the number of bacteria between the groups (2 and 8 weeks p.i. vs. 16–40 weeks p.i.) by Fisher's extract test.
Alteration of the Cecum Microbiome of the Laying Hens
After confirming the alteration in the colonization fitness of C. jejuni in the cecum of the SPF laying hens, their bacterial community structures (n = 3 each at 0, 2, 8, 16, 24, 32, and 40 weeks p.i.) were analyzed using a 16S rRNA gene sequencing approach. The Ion Torrent sequencer output 217,473–333,238 reads, and after filtering, 160,209–235,614 reads were remained (Supplementary Table 1). After normalization to 100,000 valid reads per a sample, a total of 446, 160, 76, 43, and 27 taxa were finally detected at the genus, family, order, class, and phylum levels, respectively, by RDP program. Shannon diversity index showed the increased trend at >16 weeks p.i. (Supplementary Table 1). In contrast, Simpson index resulted in the decreased trends in the means at >16 weeks p.i. (Supplementary Table 1). Permutational multivariate analysis of variance analysis showed the significant differences of the bacterial community between three groups (group 1: 0 w p.i., group 2: 2 and 8 w p.i., group 3: ≥16 w p.i.) at R2 of 0.375 and P-value of 0.0001.
Phylum Level Comparison
Overall, the main bacterial phyla detected in the cecum of laying hens were represented by Firmicutes, followed by Actinobacteria, Bacteroidetes, and Proteobacteria, with means ± SD of 99.05 ± 0.65, 0.69 ± 0.59, 0.14 ± 0.14, and 0.07 ± 0.07%, respectively (Figure 2A). Compared with 0 weeks p.i., the results obtained at >16 weeks p.i. showed significant differences in the relative abundance of Firmicutes (z = −2.36, P = 0.036) and Bacteroidetes (z = 2.96, P = 0.006), while the abundance of C. jejuni colonizers was not significantly different (Figure 2A), as assessed using the non-parametric Dunn test. The Firmicutes/Bacteroidetes (F/B) ratio, which is related to age in humans (31), was decreased in a time-dependent manner, from 0 weeks p.i. (mean, 12,104) to 16 weeks p.i. (340), and gradually increased thereafter (mean = 439, 535, and 648 at 24, 32, and 40 weeks p.i., respectively) (Figure 2B).
Figure 2.
Altered microbiome in the caecum of laying hens during infection experiments. (A) Time-dependent dynamics of representative bacterial phylum as detected in caecal samples of laying hens. (B) Time-dependent dynamics of the Firmicutes/Bacteroidetes (F/B) ratio of the caecal microbiomes. (C) Time-dependent dynamics of representative bacterial family as detected in caecal samples of laying hens. (D) Time-dependent dynamics of representative bacterial genera as detected in caecal samples of laying hens. (E) Principle coordinate analysis (PCoA) plot of representative bacterial phylum as detected in caecal samples of laying hens. Dots are shown in different colors by time points (0 weeks p.i., orange; 2 weeks and 8 weeks p.i., blue; >16 weeks p.i., green). (F) Fold changes in the relative abundances of representative bacterial genera between >16 weeks p.i. and <8 weeks p.i. Bacterial genera exhibiting the increased relative abundance at >16 weeks p.i. compared with 2/8 weeks p.i. are shown with green color, and those exhibiting the decreased relative abundances at >16 weeks p.i. compared with 2/8 weeks p.i. are shown with red color, respectively.
Family-Level Comparison
Throughout the experimental periods, the family Lachnospiraceae was predominant (41.20 ± 3.76%), followed by Ruminococcaceae (34.63 ± 4.99%), and Carnobacteriaceae (4.80 ± 3.77%) (Figure 2C and Supplementary Figure 1). According to time point, the family Lachnospiraceae showed a temporal decrease in relative abundance from 0 to 16 weeks p.i.; in turn, it increased thereafter, up to 32 weeks p.i. (Figure 2C and Supplementary Figure 1). The families Ruminococcaceae and Erysipelotrichaceae showed an increased relative abundance at 16 weeks p.i., as a plateau, thereby stably existing in these samples (Figure 2C and Supplementary Figure 1). In contrast, the families Carnobacteriaceae and Lactobacillaceae exhibited an initial (up to 8 weeks p.i.) increase in their relative abundance, to then decrease after 16 weeks p.i. (Figure 2C and Supplementary Figure 1). Finally, the families Peptostreptococcaceae and Clostridiales Incertae Sedis XII showed a time-dependent increase in relative abundance (Figure 2C and Supplementary Figure 1), whereas the family Enterococcaceae showed a time-dependent decrease in relative abundance (Figure 2C and Supplementary Figure 1).
Genus-Level Comparison
Among all tested samples, the genus Blautia was predominant (7.08 ± 1.16%), followed by Clostridium IV (5.57 ± 1.85%), Lactobacillus (4.80 ± 3.77%), and Gemmiger (4.72 ± 1.42%) (Table 1, Figure 2D). According to sampling time point, before C. jejuni infection (0 weeks p.i.), Blautia was the predominant genus (7.65 ± 0.13%), followed by Gemmiger (6.46 ± 1.10%), Weissella (4.63 ± 1.34%), and Faecalibacterium (4.52 ± 0.73%) (Table 1, Figure 2D, and Supplementary Figure 2). At 2 and 8 weeks p.i., the predominant genera were Lactobacillus (8.98 ± 4.25%), Blautia (8.30 ± 1.01%), Desemzia (5.76 ± 1.30%), and Gemmiger (5.57 ± 1.21%) (Table 1, Figure 2D, and Supplementary Figure 2). At >16 weeks p.i., Clostridium IV was the predominant genus (6.96 ± 1.04%), followed by Blautia (6.33 ± 0.68%), Ruminococcus (3.88 ± 0.69%), and Gemmiger (3.86 ± 0.87%) (Table 1, Figure 2D, and Supplementary Figure 2).
Table 1.
Relative abundance of bacterial genera (%) in the cecum of laying hens.
| Genus | 0 weeks p.i. | 2/8 weeks p.i. | 16/24/32/40 weeks p.i. | |||
|---|---|---|---|---|---|---|
| Mean ±SD (%) | Mean ±SD (%) | P-value to | Mean ±SD (%) | P-value to * | ||
| 0 w p.i. | 0 w p.i. | 2/8 w p.i. | ||||
| INCREASED RELATIVE ABUNDANCE AT >16 WEEKS P.I. COMPARED WITH 2/8 WEEKS P.I. | ||||||
| Clostridium IV | 3.91 ± 0.80 | 3.61 ± 0.37 | 1.0000 | 6.96 ± 1.04 | 0.0488 | 0.0017 |
| Sporobacter | 0.49 ± 0.35 | 0.58 ± 0.40 | 1.0000 | 3.66 ± 0.96 | 0.0239 | 0.0030 |
| Acetanaerobacterium | 0.83 ± 0.18 | 1.22 ± 0.26 | 1.0000 | 2.37 ± 0.40 | 0.0036 | 0.0107 |
| Ruminococcus | 0.89 ± 0.16 | 0.86 ± 0.13 | 1.0000 | 1.62 ± 0.21 | 0.0488 | 0.0017 |
| Saccharofermentans | 0.79 ± 0.13 | 0.89 ± 0.23 | 1.0000 | 1.44 ± 0.34 | 0.0255 | 0.0138 |
| Subdoligranulum | 1.08 ± 0.19 | 0.91 ± 0.17 | 0.8886 | 1.27 ± 0.25 | 0.9690 | 0.0127 |
| Shuttleworthia | 0.64 ± 0.09 | 0.76 ± 0.12 | 1.0000 | 1.11 ± 0.21 | 0.0085 | 0.0208 |
| Ethanoligenens | 0.53 ± 0.16 | 0.62 ± 0.16 | 1.0000 | 1.14 ± 0.35 | 0.0225 | 0.0265 |
| Anaerotruncus | 0.41 ± 0.08 | 0.39 ± 0.10 | 1.0000 | 1.03 ± 0.14 | 0.0488 | 0.0017 |
| Papillibacter | 0.40 ± 0.18 | 0.42 ± 0.06 | 1.0000 | 0.84 ± 0.17 | 0.0144 | 0.0044 |
| Dehalobacter | 0.16 ± 0.03 | 0.21 ± 0.08 | 1.0000 | 0.85 ± 0.29 | 0.0085 | 0.0063 |
| Guggenheimella | 0.15 ± 0.02 | 0.37 ± 0.31 | 1.0000 | 0.76 ± 0.13 | 0.0344 | 0.0452 |
| DECREASED RELATIVE ABUNDANCE AT >16 WEEKS P.I. COMPARED WITH 2/8 WEEKS P.I. | ||||||
| Blautia | 7.65 ± 0.13 | 8.30 ± 1.01 | 1.0000 | 6.33 ± 0.68 | 0.0989 | 0.0048 |
| Lactobacillus | 4.37 ± 3.03 | 8.98 ± 4.25 | 0.5700 | 2.81 ± 1.43 | 1.0000 | 0.0226 |
| Gemmiger | 6.46 ± 1.10 | 5.57 ± 1.21 | 1.0000 | 3.86 ± 0.87 | 0.0324 | 0.0361 |
| Desemzia | 4.19 ± 2.19 | 5.76 ± 1.30 | 0.4308 | 3.54 ± 1.16 | 1.0000 | 0.0265 |
| Fusicatenibacter | 3.60 ± 0.39 | 3.78 ± 0.54 | 1.0000 | 2.89 ± 0.41 | 0.1791 | 0.0095 |
| Clostridium XlVa | 2.31 ± 0.40 | 2.39 ± 0.39 | 1.0000 | 1.63 ± 0.24 | 0.0939 | 0.0036 |
| Oscillibacter | 2.30 ± 0.66 | 2.18 ± 0.34 | 1.0000 | 1.64 ± 0.26 | 0.1408 | 0.0286 |
| Bavariicoccus | 1.24 ± 0.30 | 1.00 ± 0.16 | 1.0000 | 0.45 ± 0.08 | 0.0111 | 0.0053 |
| Lacticigenium | 1.24 ± 0.32 | 0.79 ± 0.26 | 1.0000 | 0.25 ± 0.06 | 0.0085 | 0.0063 |
Bacterial genera exhibiting >0.50% of relative abundance on average in total are listed. p.i., post-infection.
P-values were calculated using a non-parametric joint ranked Dunn test.
Bold values represent P-values of <0.05.
Characterization of the Time-Dependent Dynamics of Cecal Microbiota
The principal coordinate analysis illustrated a distinct distribution of the samples at >16 weeks p.i. compared with those observed at 0, 2, and 8 weeks p.i. (Figure 2E). A comparison of the relative abundance of each bacterial genus between the groups (2 and 8 vs. >16 weeks p.i.) revealed a significant alteration in the relative abundance of several bacterial genera: a total of 12 and 9 genera exhibited a significant increase or decrease in their relative abundance between the groups, respectively (Table 1, Figure 2F). The 12 genera that showed an increased relative abundance at >16 weeks p.i. were Clostridium IV, Sporobacter (family Clostridiaceae), Acetanaerobacterium, Ruminococcus, Subdoligranulum, Ethanoligenens, Anaerotruncus, Papillibacter (family Ruminococcaceae), Shuttleworthia (family Lachnospiraceae), Dehalobacter (family Peptococcaceae), and Guggenheimella (family Clostridiales incertae sedis), all of which were classified in the order Clostridiales (Table 1, Figure 2F). In contrast, among the nine genera exhibiting a time-dependent decrease in relative abundance, five genera were in the order Clostridiales (Blautia, Gemmiger, Fusicatenibacter, Clostridium XIVa, and Oscillibacter), whereas the remaining genera (Lactobacillus, Desemzia, Bavariicoccus, and Lacticigenium) were in the order Lactobacillales (Figure 2F and Supplementary Figure 2). Thus, these data clearly suggest time-dependent alterations of the chicken gut microbiota composition throughout the experimental period.
Time-Course Dynamics of Cytokine Production in the Serum of Laying Hens
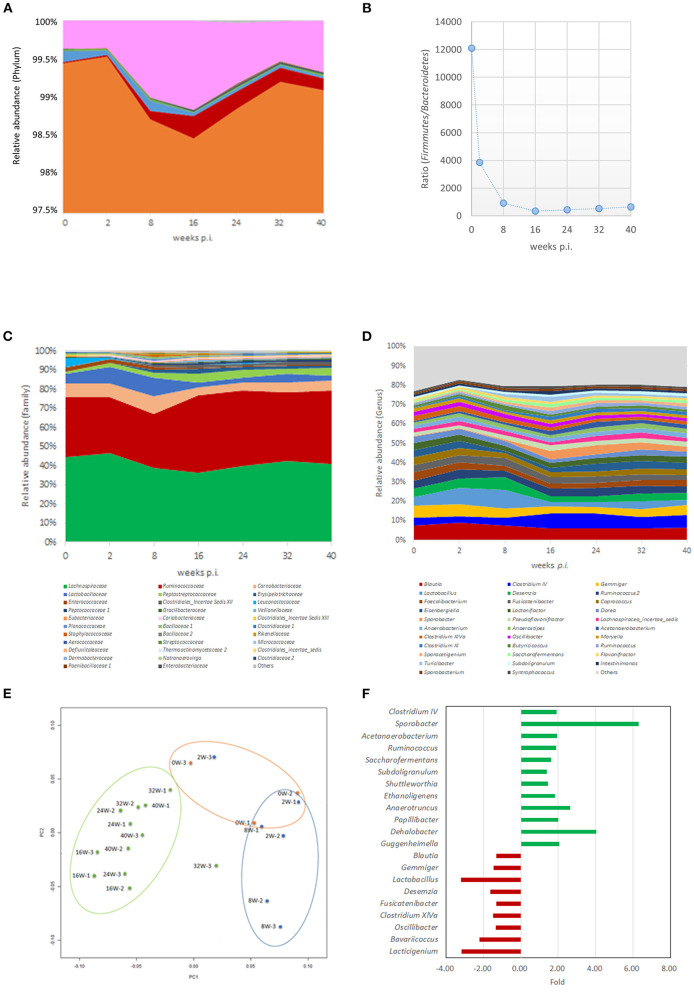
The levels of representative chicken cytokines [IFN-γ, IL-10, IL-12p40, IL-16, IL-21, IL-6, netlin-2, pentraxin-3 (PTX-3), and RANTES (CCL5)] in the serum samples were comparatively examined using a semi-quantitative membrane array. Compared with the serum samples collected at 8 weeks p.i., the samples obtained at 24 and 40 weeks p.i. exhibited no apparent differences in all target molecules (Figure 3). In contrast, the samples collected at 16 weeks p.i. exhibited a reduced level of cytokines, with the exception of IL-21 (Figure 3). Thus, these data indicate the presence of altered cytokine production at 16 weeks p.i. compared with the other time points of the experimental period.
Figure 3.
Cytokine production in the sera of laying hens after experimental C. jejuni infection.
Lipid Metabolic Profiles in the Cecum of Laying Hens
Untargeted LC-MS/MS analyses were conducted to comparatively measure chick cecum lipids and lipid metabolites in a total of six cecum samples collected at 8, 16, and 24 weeks p.i. These comparative analyses revealed that 22 or 36 lipids were significantly increased or decreased in the samples collected at 16/24 weeks p.i., respectively, compared with those obtained at 8 weeks p.i. (Table 2 and Supplementary Figure 2). These dynamics between the time courses were explained as follows.
Table 2.
Summary of altered lipids in the gut of laying hens between 8, 16, and 24 weeks p.i.
| Category | Lipid component1* | Average | P-value1,2 | |||||
|---|---|---|---|---|---|---|---|---|
| Blank | 8 w | 16 w | 24 w | 8 w vs. 16 w | 8 w vs. 24 w | 16 w vs. 24 w | ||
| INCREASED AT 16 WEEKS AND/OR 24 WEEKS P.I. (16 W/24 W) COMPARED WITH 8 WEEKS P.I. (8 W) | ||||||||
| Fatty acid metabolite | FA (27:1) | 21 | 2,905 | 11,031 | 12,297.5 | 0.123 | 0.003 | 0.594 |
| FA (28:0) | 64 | 1,220.5 | 4,682.5 | 4,289.5 | 0.012 | 0.026 | 0.186 | |
| AAHFA (5:0/26:0) | 59 | 560.5 | 2,532.5 | 4,867 | 0.426 | 0.032 | 0.371 | |
| AH2FA (4:0/25:0) | 22 | 713.5 | 1,605 | 2,449 | 0.411 | 0.036 | 0.432 | |
| CmpE (26:0) | 53 | 335.5 | 1,167 | 1,462 | 0.249 | 0.010 | 0.549 | |
| Glycerolipid | TG (45:1) | 203 | 336.5 | 800 | 798.5 | 0.370 | 0.049 | 0.997 |
| MGDG (18:0/20:2) | 11 | 439 | 1,652.5 | 596 | 0.041 | 0.316 | 0.044 | |
| Phospholipid | MLCL (16:0/16:0/18:2) | 0 | 237 | 892.5 | 452.5 | 0.006 | 0.069 | 0.038 |
| CL (e/e)(28:1e/30:1e) | 11 | 2,024.5 | 2,591.5 | 5,378.5 | 0.568 | 0.038 | 0.099 | |
| CL (e/e)(30:1e/33:3e) | 0 | 1,178.5 | 1,298.5 | 2,452.5 | 0.835 | 0.024 | 0.222 | |
| CL (30:1e/30:3e) | 0 | 125 | 219 | 296.5 | 0.140 | 0.013 | 0.198 | |
| DLCL (16:0/16:0) | 16 | 3,637.5 | 17,440 | 10,366 | 0.175 | 0.045 | 0.327 | |
| DLCL (16:0/18:2) | 0 | 1,853 | 5,436 | 2,487 | 0.002 | 0.628 | 0.200 | |
| HBMP (18:0/18:1/g18:1) | 11 | 100 | 237 | 331.5 | 0.017 | 0.240 | 0.496 | |
| HBMP (14:0/16:0/18:0) | 0 | 158.5 | 341.5 | 424.5 | 0.081 | 0.047 | 0.289 | |
| PG (18:2/18:2) | 16 | 1,353.5 | 2,083.5 | 2,179 | 0.007 | 0.620 | 0.950 | |
| Sphingolipid | CerPI (d44:0) | 0 | 158 | 700.5 | 990 | 0.056 | 0.039 | 0.060 |
| AcylCerPG (t18:0/16:0/15:0) | 0 | 361 | 2,758.5 | 2,101 | 0.052 | 0.041 | 0.324 | |
| Sterol lipids | StE (18:1) | 930 | 50,481.5 | 79,812.5 | 118,907.5 | 0.286 | 0.030 | 0.200 |
| StE (18:0) | 236 | 4,940 | 9,740.5 | 11,280 | 0.298 | 0.007 | 0.646 | |
| SISL G1 | 157 | 30,164.5 | 64,382 | 49,213 | 0.049 | 0.067 | 0.181 | |
| Stigmasterol G1 | 91 | 5,295 | 8,517 | 7,433 | 0.275 | 0.047 | 0.603 | |
| DECREASED AT 16 WEEKS AND/OR 24 WEEKS P.I. (16 W/24 W) COMPARED WITH 8 WEEKS P.I. (8 W) | ||||||||
| Fatty acid metabolite | FA (17:0) | 2,265 | 1,335,104 | 909,822.5 | 368,852.5 | 0.619 | 0.038 | 0.543 |
| FA (18:0) | 151,628 | 6,089,247 | 4,739,226.5 | 2,362,394.5 | 0.711 | 0.036 | 0.546 | |
| FA (19:1) | 0 | 555,886 | 95,071.5 | 64,656.5 | 0.018 | 0.004 | 0.246 | |
| FA (20:3) | 23 | 10,567.5 | 2,823.5 | 2,095 | 0.123 | 0.039 | 0.717 | |
| FA (24:1) | 348 | 246,110.5 | 127,115 | 94,823.5 | 0.088 | 0.009 | 0.309 | |
| FA (24:2) | 35 | 21,430 | 13,737.5 | 12,986 | 0.298 | 0.044 | 0.887 | |
| FA(O) (18:0) | 67 | 93,970 | 20,635 | 16,832 | 0.067 | 0.043 | 0.862 | |
| FA(aOH)(16:0) | 164 | 1,059,824.5 | 315,401 | 86,486 | 0.173 | 0.135 | 0.034 | |
| FA(aOH)(18:0) | 241 | 270,228 | 175,671 | 65,140.5 | 0.098 | 0.025 | 0.020 | |
| FA(aOH)(19:0) | 40 | 29,623.5 | 23,000 | 12,582.5 | 0.120 | 0.020 | 0.059 | |
| FA(aOH)(22:1) | 21 | 35,900.5 | 22,324.5 | 20,211.5 | 0.061 | 0.015 | 0.252 | |
| FA(aOH)(24:1) | 66 | 389,587.5 | 212,609.5 | 181,281 | 0.060 | 0.046 | 0.515 | |
| Glycerolipid | MGDG(sn2+O)(16:0/16:0) | 44 | 24,594.5 | 8,656.5 | 11,126 | 0.018 | 0.033 | 0.301 |
| AMGDGM(e) (16:1e/17:0/18:1) | 0 | 1,169.5 | 467 | 344.5 | 0.037 | 0.045 | 0.146 | |
| AMGDG(e)(18:1e/14:0/18:1) | 0 | 972.5 | 538.5 | 417.5 | 0.043 | 0.020 | 0.244 | |
| GPMGDG(e)(34:1e) | 34 | 23,957 | 11,863.5 | 10,900 | 0.189 | 0.044 | 0.870 | |
| GPMGDG(e)(34:3e) | 75 | 5,551 | 3,790 | 4,866 | 0.041 | 0.768 | 0.655 | |
| DGDG(16:0/18:2) | 21 | 1,161 | 1,267 | 728.5 | 0.705 | 0.023 | 0.228 | |
| DGDG(18:0/18:1) | 0 | 643.5 | 442 | 308 | 0.421 | 0.018 | 0.554 | |
| DGDG(e)(18:1e/18:1) | 0 | 6,650 | 2,918 | 1,610 | 0.244 | 0.015 | 0.563 | |
| DGDG(e)(17:1e/18:1) | 0 | 712.5 | 337.5 | 170.5 | 0.172 | 0.025 | 0.391 | |
| ADGDG(16:0/18:1/g16:0) | 0 | 746.5 | 630 | 415.5 | 0.457 | 0.048 | 0.255 | |
| TG(51:4) | 51 | 3,101 | 4,343.5 | 1,829 | 0.702 | 0.037 | 0.492 | |
| TG(60:4) | 0 | 1,959 | 1,898.5 | 1,046 | 0.934 | 0.035 | 0.372 | |
| DG (16:0/19:1) | 338 | 68,852.5 | 38,753.5 | 31,749 | 0.025 | 0.117 | 0.571 | |
| Phospholipid | PE(16:0/16:1) | 11 | 364.5 | 262.5 | 765.5 | 0.016 | 0.531 | 0.459 |
| Lyso PC (20:0) | 190 | 2,531.5 | 2,373.5 | 427 | 0.893 | 0.014 | 0.284 | |
| PE (18:0) | 134 | 1,619.5 | 1,485 | 373 | 0.844 | 0.014 | 0.286 | |
| Sphingolipid | AcylCer(phyto_aOH)(t18:1/24:0/2:0) | 0 | 4,412.5 | 2,732 | 708.5 | 0.281 | 0.010 | 0.226 |
| 3-O-AcylSM (d36:1/16:0) | 0 | 579 | 427 | 188 | 0.210 | 0.082 | 0.006 | |
| Sterol lipid | Taurin (19:1) | 16 | 962.5 | 532.5 | 157.5 | 0.049 | 0.041 | 0.133 |
| dehydro cholesterol | 856 | 2,425.5 | 3,853.5 | 1,408.5 | 0.212 | 0.007 | 0.127 | |
| Prenol lipid | Coenzyme Q9H2 | 155 | 78,620.5 | 11,985.5 | 14,275.5 | 0.049 | 0.031 | 0.747 |
| Coenzyme Q11 | 88 | 17,740 | 7,237.5 | 3,685 | 0.089 | 0.014 | 0.299 | |
| Coenzyme Q11H2 | 0 | 3,928.5 | 952.5 | 757.5 | 0.077 | 0.035 | 0.805 | |
| Others | Tocopherol | 76 | 16,937 | 6,859.5 | 5,661.5 | 0.003 | 0.040 | 0.459 |
p.i., post-infection.
*1 The abbreviation of each lipid component is defined in Supplementary Table 2.
*2 P < 0.05, indicated in bold.
Fatty Acids
Fatty acids (FAs), such as 27:1 and 28:0, as well as the FA ester of hydroxy fatty acid (AH2FA) 4:0/25:0 and acyl α-hydroxy fatty acid (AAHFA) 5:0/26:0, the latter of which was recently identified as a gut-microbiota-specific lipid (30), were present at higher levels at 24 weeks p.i. compared with 8 weeks p.i. (Table 2). In contrast, FAs 17:0, 18:0, 19:1, 20:3, 24:1, and 24:2, as well as α-hydroxy fatty acids [FA(aOH)] (18:0, 19:0, 24:1, 22:1, and 19:0), and FA(O) 18:0, showed decreased levels in the samples collected at 16 and/or 24 weeks p.i. compared with those obtained at 8 weeks p.i. (Table 2). Moreover, FAs 16:0 and 18:0 showed a continued decrease at 24 weeks p.i. compared with 16 weeks p.i. (Table 2).
Glycerolipids
Among the glycerolipids, triacylglycerolipid (TG) (C45:1), and monoglycosyl diacylglycerol (MGDG) (18:0/20:2) were increased at 24 or 16 weeks p.i. compared with 8 weeks p.i., although the increase of the latter was temporally detected only at 16 weeks p.i. (Table 2). Other glycerolipids, such as monogalactosyldiacylglycerol (MGDG) (sn2+O) (16:0/16:0), acylmonoglycosyl diacylglycerol (AMGDG) (16:1e/17:0/18:1), AMGDG (18:1e/14:0/18:1), glycerophosphomonoglycosyl monoacylglycerol (GPMGDG) (34:1e), GPMGDG (34:3e), diglycosyldiacylglycerol (DGDG) (16:0/18:2), DGDG (18:0/18:1), diglycosyl 1-alkyl, 2-acylglycerol [DGDG(e)] (18:1e/18:1), DGDG(e) (17:1e/18:1), TG (51:4), TG (60:4), and diacylglycerol (DG) (16:0/19:1), were decreased at 16 and/or 24 weeks p.i. compared with 8 weeks p.i. (Table 2).
Phospholipids
Among the phospholipids, monolysocardiolipin (MLCL) (16:0/16:0/18:2), cardiopin (CL) (28:1e/30:1e), CL (30:1e/33:3e), CL (30:1e/30:3e), dilysocardiolipin (DLCL) (16:0/16:0), DLCL (16:0/18:2), hemi bis (monoacylglycero) phosphate (HBMP) (18:0/18:1/g18:1), HBMP (14:0/16:0/18:0), and phosphatidyl glycerol (PG) (18:2/18:2) were increased at 16 or 24 weeks p.i. compared with 8 weeks p.i. (Table 2). Conversely, phosphatidyl ethanolamine (PE) (14:0/16:0/18:0), monoacylglycerophosphocholine (LysoPC) (20:0), and diacylglycerophosphoethanol (PE) (18:0) were decreased in the samples at 16 or 24 weeks p.i. compared with 8 weeks p.i. (Table 2).
Sphingolipids
Phytoceramide and acyl phosphoglycerol were increased at 24 weeks p.i. compared with 8 weeks p.i. (Table 2). In turn, acyl ceramide (AcylCer) (phyto_aOH) (t18:1/24:0/2:0) and N-palmitoyl-d-erythro-sphingosylphosphorylcholine were decreased at 24 weeks p.i. compared with 8 weeks p.i. (Table 2).
Sterol Lipids, Prenol Lipids, and Others
Sterol ester (StE) (18:0), StE (18:1), sitosterol (SISL) G1, and stigmasterol G1 were increased in the samples at 16 or 24 weeks p.i. compared with 8 weeks p.i. (Table 2). Conversely, 2-amino ethanesulfonic acid, taurine (19:1), and dehydro cholesterol were decreased in the samples at 16 and/or 24 weeks p.i. compared with 8 weeks p.i. (Table 2). Among the prenol lipids, coenzymes Q9H2, Q11, and Q11H2 exhibited decreased levels at 16 and/or 24 weeks p.i. compared with 8 weeks p.i. (Table 2). Finally, tocopherol was decreased at and 24 weeks p.i. compared with 8 weeks p.i. (Table 2).
Discussion
The current study investigated the temporal colonization fitness of C. jejuni in the cecum of laying hens after experimental infection. In parallel with the decreased bacterial colonization fitness observed after 16 weeks p.i., compositional changes in the gut microbiota and lipids were observed, suggesting their possible correlations.
After invasion, C. jejuni initializes and prolongs gut colonization in the gut of broiler chicken up to the slaughter age (generally <8 weeks) (17–21). At <2 weeks of age, this pathogen is rarely detected in commercial chicken flocks, regardless of the production system (32, 33), which implies that a biological mechanism to resist colonization may be present in young chicks. As a possible explanation for this phenomenon, maternal antibodies might be partly responsible for the absence of Campylobacter in young chicks (34).
It is noteworthy that C. jejuni could maintain the colonization for up to 8 weeks p.i., which is the general time point for the slaughter of broiler chickens; however, C. jejuni exhibited a decrease in its colonization ability thereafter and up to 40 weeks p.i. As the feed and water supplied in this study contained no antibiotics and no compositional changes, it could be considered that such a decreased colonization ability might be triggered by the maturation of the host immune response or certain interactions with gut microbiota occurring during the experimental period. Further studies would be required to clarify that all laying hens might exhibit similar trends for Campylobacter colonization, throughout the quantitative detection of this pathogen.
Our data revealed a temporal decrease in the production of IFN-γ, IL-10, IL-12p40, IL-16, IL-6, netrin-2, PTX-3, and RANTES (CCL5) at 16 weeks p.i., and constant production of IL-21 in the serum. This host immune response is considered to be one of the imperative factors affecting C. jejuni colonization, although it remains controversial; Pielsticker et al. reported that triggering an innate and acquired immune response, especially in the very early phase, affected bacterial colonization (35). However, in most experimental studies, contradictory data regarding the immune response in chickens following C. jejuni colonization were reported; one study contended that the chicken immune system is inefficiently activated, which might contribute to the persistent colonization of C. jejuni in the chicken gut (36, 37). In contrast, another study showed the presence of an inflammatory response following Campylobacter infection in chickens (38). The occurrence of such immune responses upon C. jejuni colonization might be due to the supposed genetic heterogeneity of both the chicken hosts and C. jejuni (39). Our data suggest that the laying hens used in this study might not represent an animal with a significant immunomodulatory response against C. jejuni infection during long-term grow-out. It remains unknown why the laying hens showed temporal decreases in the production of most cytokines at 16 weeks p.i. It is possible that, at this stage, certain physiological shifts occur in laying hens, as reflected in the visual observation of coloring of combs and male–female discrimination (data not shown). In contrast, IL-21, which is a T-cell-derived cytokine that modulates T cell, B cell, and natural killer cell responses and regulates the Th17/Treg balance in mice (40), exhibited no clear alteration at 16 weeks p.i. Further studies are required to evaluate the possible role of T-cell-mediated immunity in the colonization of C. jejuni in laying hens.
Gut microbiota play a pivotal role in conferring resistance to, or promoting, infection by pathogenic microorganisms (41). In fact, the administration of a large dose of streptomycin disrupted normal gut microbiota, thereby increasing susceptibility to Salmonella infection (42). Similarly, germ-free chickens were more susceptible to C. jejuni colonization compared with chickens possessing conventional intestinal microbiota (43). Campylobacter jejuni is likely to cooperate and compete with diverse commensal microbiota, thus becoming part of a well-balanced gut microbial community (44). Johansen and colleagues also found that C. jejuni colonization affected the development and complexity of the microbial communities in the ceca of chicken up to 17 days of age (45). A more recent study revealed that the experimental inoculation of C. jejuni into 1-day-old broiler chicks modulated the cecal microbial community structure, with a higher abundance of Firmicutes at the expense of the phylum Bacteroidetes and other taxa at 3–4 weeks p.i. (46). Accordingly, our data also showed that the phylum Firmicutes predominated at 0 weeks p.i. (2 weeks age), but was replaced thereafter with the representatives of Bacteroidetes at 8–16 weeks p.i. (10–18 weeks of age). At >16 weeks p.i., among the phylum Bacteroidetes, the genus Blautia showed negative associations with C. jejuni colonization. Including the genus Blautia, all genera in the phylum Bacteroidetes are likely to express enzymes for the biosynthesis of propionate, one of the main short-chain fatty acids (SCFAs) in the chicken cecum (47), which suggests a possible alteration of lipid metabolism in the cecum of laying hens during the experimental period. Referring to a recent study that demonstrated the age-dependent dynamics of cecal microbiota in laying hens (24), our data provided the idea that experimental infection with C. jejuni might not affect the age-dependent dynamics of cecal microbiota composition drastically during the experimental period, whereas age-dependent shifts in the gut microbiota might affect the C. jejuni colonization properties. To clarify this issue, our future study would be performed to include unchallenged control groups at different ages, in same animal lot.
Regarding the bacterium-to-bacterium interplay, a positive correlation between the relative abundance of the genus Clostridium and C. jejuni colonization in the gut of broiler chickens has been reported (48). This might be due to the fact that C. jejuni acts as a hydrogen sink, thus leading to improved growth conditions for some Clostridia through increased fermentation (49) and organic acid production, which can be used by C. jejuni as an energy source. As a consequence, C. jejuni infection affects the metabolic end products derived from the intestinal microbiota of chickens. In support of this notion, a recent study showed that butyrate, one of the SCFAs that are biosynthesized by a series of Clostridium species (50), is directly sensed by C. jejuni through the BumSR two-component signal transduction system (51).
It is likely that gut microbiota affect intestinal lipid metabolism, including microbiota-dependent changes in bile acid metabolism (52). To obtain further information on the altered microbiota dynamics and C. jejuni colonization fitness, we performed comparative lipidome analyses using samples collected at three different time points (8, 16, and 24 weeks p.i.).
Among the elevated lipids at >16 weeks p.i., we found increased levels of phytosterols, such as stigmasterol and sitosterol, which can reduce the reabsorption of bile acids and cholesterol in the gut, thereby increasing fecal lipid levels (53), at 16 weeks p.i. compared with 8 weeks p.i. Considering that bile acids are steroid acids that are synthesized in the liver and then conjugated with a taurine residue to give anions called bile salts (54), our data demonstrating the decreased levels of sterol lipids (i.e., taurine and dehydro cholesterol) and sphingolipids (i.e., phytoceramide), which are components of bile acids (55, 56), at 16/24 weeks p.i. compared with 8 weeks p.i. suggest that bile acid reabsorption might be altered at these time points. In cecal digesta of goats that were fed a high-grain diet, the level of stigmasterol was negatively correlated with the abundance of the genus Clostridium, Turicibacter, SMB53, and Pseudoramibacter (41). Together with our microbiome data, potential negative associations between phytosterols and Clostridium/C. jejuni colonization in laying hens should be considered. The temporal quantification of bile acids in the gut and gallbladder would clarify the kinetics of bile acid synthesis and absorption and provide a link with their impact on gut microbiota in a future study.
Among the glycerolipids, the cecum samples collected at 16/24 weeks p.i. showed increased levels of TG (45:1) and MGDG (18:0/20:2), while an additional 13 glycerolipids were decreased compared with those obtained at 8 weeks p.i. MGDG is metabolized by Streptococcus pneumoniae, with conversion between DGDG and MGDG (57). It could be considered that certain enzymatic reaction processes in S. pneumoniae might also be present in other bacterial genera; thus, lipid characterization in representative gut microbiota might contribute to the deciphering of the bacteria associated with the glycerolipid alteration observed here. Moreover, PE, which was decreased at 16/24 weeks p.i., was distributed in the representative human gut microbe Alistipes finegoldii in the phylum Bacteroidetes (58). This is not surprising because of the age-dependent decrease in F/B ratio observed.
Among other lipids, coenzymes (i.e., coenzyme Q9H2) showed decreased levels at 16 weeks p.i. compared with 8 weeks p.i. Considering the age-dependent reduction in plasma glucose detected in broiler chickens (59), the decreased levels of coenzymes might be part of the age-dependent dynamics.
In summary, we demonstrated that the long-term breeding of laying hens decreased C. jejuni colonization in the cecum after experimental infection. Comparative analyses of the alterations of gut microbiota and lipid components at 16 weeks p.i. or later unveiled possible negative associations between C. jejuni and several gut microbiota, such as those in the genera Blautia and Clostridium at younger or older age, respectively. It is likely that the chicken generally reaches maturity and starts laying eggs from 21 weeks old on average (60), which is close to the age at 16 weeks p.i. (18 weeks of age) when we observed the alterations in C. jejuni colonization, microbiota, and lipid compositions in the gut of laying hens. Thus, it could be considered that the altered phenomenon's observed in this study might be mainly due to certain host physiological change(s) accompanied with the host maturation. Our future study of the interplay between these gut microbiota and bile acid metabolism, as well as C. jejuni colonization, in laying hens is expected to improve our understanding of the possible interactions between these parameters, thereby leading to the discovery and establishment of control strategies for the reduction of C. jejuni intestinal carriage at poultry-production stages.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, DRA009061.
Ethics Statement
The animal study was reviewed and approved by Animal Welfare and Ethical Committee of the National Institute of Health Sciences.
Author Contributions
HA conceived and designed the study. HA, TN, SY, KI, and JK performed and analyzed the experiments, and HA wrote the manuscript. IK, JK, YT, and SM read and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The untargeted LC-MS/MS analyses were supported by the Kazusa DNA Research Institute (Japan). The authors sincerely thank the reviewers for their constructive comments.
Footnotes
Funding. This study was supported in part by the Food Safety Commission of Japan (grant number 1806), the Ministry of Health, Labour and Welfare of Japan (grant number 20KA1002), and the Japan Society for the Promotion of Science (JSPS) (grant number 19K06432).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.675570/full#supplementary-material
References
- 1.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) . The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. (2018) 16:e05500. 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geissler AL, Bustos Carrillo F, Swanson K, Patrick ME, Fullerton KE, Bennett C, et al. Increasing Campylobacter infections, outbreaks, and antimicrobial resistance in the United States, 2004-2012. Clin Infect Dis. (2017) 65:1624–31. 10.1093/cid/cix624 [DOI] [PubMed] [Google Scholar]
- 3.Ministry of Health Labor, and Welfare of Japan (MHLW) . Foodborne Statistics Report. (2020). Available online at: https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/kenkou_iryou/shokuhin/syokuchu/04.html (accessed February 22, 2021).
- 4.Cody AJ, Maiden MC, Strachan NJ, McCarthy ND. A systematic review of source attribution of human campylobacteriosis using multilocus sequence typing. Euro Surveill. (2019) 24:1800696. 10.2807/1560-7917.ES.2019.24.43.1800696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. (2015) 28:687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumagai Y, Pires SM, Kubota K, Asakura H. Attributing human foodborne diseases to food sources water in Japan using analysis of outbreak surveillance data. J Food Prot. (2020) 83:2087–94. 10.4315/JFP-20-151 [DOI] [PubMed] [Google Scholar]
- 7.Nauta M, Hill A, Rosenquist H, Brynestad S, Fetsch A, van der Logt P, et al. A comparison of risk assessments on Campylobacter in broiler meat. Int J Food Microbiol. (2009) 129:107–23. 10.1016/j.ijfoodmicro.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 8.Agunos A, Waddell L, Léger D, Taboada E. A systematic review characterizing on-farm sources of Campylobacter spp. for broiler chickens. PLoS ONE. (2014). 9:e104905. 10.1371/journal.pone.0104905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammerl JA, Jäckel C, Alter T, Janzcyk P, Stingl K, Knüver MT, et al. Reduction of Campylobacter jejuni in broiler chicken by successive application of group I and group II phages. PLoS ONE. (2014) 9:e114785. 10.1371/journal.pone.0114785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hovorková P, Skrivanová E. Use of caprylic acid in broiler chickens: effect on Campylobacter jejuni. Foodborne Pathog Dis. (2015) 12:712–8. 10.1089/fpd.2015.1978 [DOI] [PubMed] [Google Scholar]
- 11.Hankel J, Popp J, Meemken D, Zeiger K, Beyerbach M, Taube V, et al. Influence of lauric acid on the susceptibility of chickens to an experimental Campylobacter jejuni colonisation. PLoS ONE. (2018) 13:e0204483. 10.1371/journal.pone.0204483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Deun K, Haesebrouck F, van Immerseel F, Ducatelle R, Pasmans F. Short-chain fatty acids and L-lactate as feed additives to control Campylobacter jejuni infections in broilers. Avian Pathol. (2008). 37:379–83. 10.1080/03079450802216603 [DOI] [PubMed] [Google Scholar]
- 13.Micciche A, Rothrock MJ, Jr, Yang Y, Ricke SC. Essential oils as an intervention strategy to reduce Campylobacter in poultry production: a review. Front Microbiol. (2019) 10:1058. 10.3389/fmicb.2019.01058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beier RC, Byrd JA, Caldwell D, Andrews K, Crippen TL, Anderson RC, et al. Inhibition and interactions of Campylobacter jejuni from broiler chicken houses with organic acids. Microorganisms. (2019) 7:223. 10.3390/microorganisms7080223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabashsum Z, Peng M, Alvarado-Martinez Z, Aditya A, Bhatti J, Romo PB, et al. Competitive reduction of poultry-borne enteric bacterial pathogens in chicken gut with bioactive Lactobacillus casei. Sci Rep. (2020). 10:16259. 10.1038/s41598-020-73316-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tallentire CW, Leinonen I, Kyriazakis I. Breeding for efficiency in the broiler chicken: a review. Agron Sustain Dev. (2016) 36:66. 10.1007/s13593-016-0398-2 [DOI] [Google Scholar]
- 17.van Gerwe T, Miflin JK, Templeton JM, Bouma A, Wagenaar JA, Jacobs-Reitsma WF, et al. Quantifying transmission of Campylobacter jejuni in commercial broiler flocks. Appl Environ Microbiol. (2009) 75:625–8. 10.1128/AEM.01912-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yano S, Amano E, Katou A, Taneda I, Tsutsui T, Murase T. Intestinal carriage excretion of Campylobacter jejuni in chickens exposed at different ages. J Food Prot. (2014) 77:1184–7. 10.4315/0362-028X.JFP-14-061 [DOI] [PubMed] [Google Scholar]
- 19.Hermans D, Pasmans F, Messens W, Martel A, van Immerseel F, Rasschaert G, et al. Poultry as a host for the zoonotic pathogen Campylobacter jejuni. Vector Borne Zoonotic Dis. (2012) 12:89–98. 10.1089/vbz.2011.0676 [DOI] [PubMed] [Google Scholar]
- 20.Asakura H, Kawamoto K, Murakami S, Tachibana M, Kurazono H, Makino S, et al. Ex vivo proteomics of Campylobacter jejuni 81-176 reveal that FabG affects fatty acid composition to alter bacterial growth fitness in the chicken gut. Res Microbiol. (2016) 167:63–71. 10.1016/j.resmic.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 21.Rasschaert G, Houf K, van Hende J, de Zutter L. Investigation of the concurrent colonization with Campylobacter and Salmonella in poultry flocks and assessment of the sampling site for status determination at slaughter. Vet Microbiol. (2007) 123:104–9. 10.1016/j.vetmic.2007.03.011 [DOI] [PubMed] [Google Scholar]
- 22.Sakaridis I, Ellis RJ, Cawthraw SA, van Vliet AHM, Stekel DJ, Penell J, et al. Investigating the association between the caecal microbiomes of broilers and Campylobacter burden. Front Microbiol. (2018) 9:927. 10.3389/fmicb.2018.00927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Videnska P, Rahman MM, Faldynova M, Babak V, Matulova ME, Prukner-Radovcic E, et al. Characterization of egg laying hen and broiler fecal microbiota in poultry farms in Croatia, Czech Republic, Hungary and Slovenia. PLoS ONE. (2014) 9:e110076. 10.1371/journal.pone.0110076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Videnska P, Sedlar K, Lukac M, Faldynova M, Gerzova L, Cejkova D, et al. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS ONE. (2014) 9:e115142. 10.1371/journal.pone.0115142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.International Organization for Standardization (ISO) . Microbiology of the Food Chain — Horizontal Method for Detection and Enumeration of Campylobacter spp. — Part 2: Colony-Count Technique. ISO 10272-2:2017 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kawase J, Asakura H, Kurosaki M, Oshiro H, Etoh Y, Ikeda T, et al. Rapid accurate diagnosis based on real-time PCR cycle threshold value for the identification of Campylobacter jejuni, astA gene-positive Escherichia coli, eae gene-positive E. coli. Jpn J Infect Dis. (2018) 71:79–84. 10.7883/yoken.JJID.2017.151 [DOI] [PubMed] [Google Scholar]
- 27.Asakura H, Kawase J, Ikeda T, Honda M, Sasaki Y, Uema M, et al. Microbiological quality assessment of game meats at retail in Japan. J Food Prot. (2017) 80:2119–26. 10.4315/0362-028X.JFP-17-137 [DOI] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsugawa H, Ikeda K, Tanaka W, Senoo Y, Arita M, Arita M. Comprehensive identification of sphingolipid species by in silico retention time tandem mass spectral library. J Cheminform. (2017) 9:19. 10.1186/s13321-017-0205-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda S, Okahashi N, Tsugawa H, Ogata Y, Ikeda K, Suda W, et al. Elucidation of gut microbiota-associated lipids using LC-MS/MS and 16S rRNA sequence analyses. iScience. (2020) 23:101841. 10.1016/j.isci.2020.101841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. (2009) 9:123. 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conlan AJ, Coward C, Grant AJ, Maskell DJ, Gog JR. Campylobacter jejuni colonization transmission in broiler chickens: a modelling perspective. J Royal Soc Interface. (2007) 4:819–29. 10.1098/rsif.2007.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell DG, Fearnley C. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. (2003) 69:4343–51. 10.1128/aem.69.8.4343-4351.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cawthraw SA, Newell DG. Investigation of the presence and protective effects of maternal antibodies against Campylobacter jejuni in chickens. Avian Dis. (2010) 54:86–93. 10.1637/9004-072709-Reg.1 [DOI] [PubMed] [Google Scholar]
- 35.Pielsticker C, Glünder G, Rautenschlein S. Colonization properties of Campylobacter jejuni in chickens. Eur J Microbiol Immunol. (2012). 2:61–5. 10.1556/EuJMI.2.2012.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meade KG, Narciandi F, Cahalane S, Reiman C, Allan B, O'Farrelly C. Comparative in vivo infection models yield insights on early host immune response to Campylobacter in chickens. Immunogenetics. (2009) 61:101–10. 10.1007/s00251-008-0346-7 [DOI] [PubMed] [Google Scholar]
- 37.Hermans D, Pasmans F, Heyndrickx M, Van Immerseel F, Martel A, Van Deun K, et al. A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut. Crit Rev Microbiol. (2012) 38:17–29. 10.3109/1040841X.2011.615298 [DOI] [PubMed] [Google Scholar]
- 38.Li X, Swaggerty CL, Kogut MH, Chiang HI, Wang Y, Genovese KJ, et al. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS ONE. (2010) 5:e11827. 10.1371/journal.pone.0011827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey S, Chaloner G, Kemmett K, Davidson N, Williams N, Kipar A, et al. Campylobacter jejuni is not merely a commensal in commercial broiler chickens and affects bird welfare. mBio. (2014) 5:e01364-14. 10.1128/mBio.01364-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu JG, Lee J, Kim EK, Seo HB, Park JS, Lee SY, et al. Treatment of IL-21R-Fc control autoimmune arthritis via suppression of STAT3 signal pathway mediated regulation of the Th17/Treg balance and plasma B cells. Immunol Lett. (2015) 163:143–50. 10.1016/j.imlet.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 41.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. (2016). 535:85–93. 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. (1954) 86:132–37. 10.3181/00379727-86-21030 [DOI] [PubMed] [Google Scholar]
- 43.Han Z, Willer T, Li L, Pielsticker C, Rychlik I, Velge P, et al. Influence of the gut microbiota composition on Campylobacter jejuni colonization in chickens. Infect Immun. (2017) 85:e00380-17. 10.1128/IAI.00380-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Y, Kitamoto S, Kamada N. Microbial adaptation to the healthy and inflamed gut environments. Gut Microbes. (2020) 12:1857505. 10.1080/19490976.2020.1857505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansen CH, Bjerrum L, Finster K, Pedersen K. Effects of a Campylobacter jejuni infection on the development of the intestinal microflora of broiler chickens. Poultry Sci. (2006) 85:579–87. 10.1093/ps/85.4.579 [DOI] [PubMed] [Google Scholar]
- 46.Awad WA, Mann E, Dzieciol M, Hess C, Schmitz-Esser S, Wagner M, et al. Age-related differences in the luminal and mucosa-associated gut microbiome of broiler chickens and shifts associated with Campylobacter jejuni infection. Front Cell Infect Microbiol. (2016) 6:154. 10.3389/fcimb.2016.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. (2015) 82:1569–76. 10.1128/AEM.03473-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thibodeau A, Fravalo P, Yergeau É, Arsenault J, Lahaye L, Letellier A, et al. Chicken caecal microbiome modifications induced by Campylobacter jejuni colonization and by a non-antibiotic feed additive. PLoS ONE. (2015) 10:e0131978. 10.1371/journal.pone.0131978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaakoush NO, Sodhi N, Chenu JW, Cox JM, Riordan SM, Mitchell HM. The interplay between Campylobacter and Helicobacter species and other gastrointestinal microbiota of commercial broiler chickens. Gut Pathog. (2014) 6:18. 10.1186/1757-4749-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zu TNK, Liu S, Gerlach ES, Mojadedi W, Sund CJ. Co-feeding glucose with either gluconate or galacturonate during clostridial fermentations provides metabolic fine-tuning capabilities. Sci Rep. (2021) 11:29. 10.1038/s41598-020-76761-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodman KN, Powers MJ, Crofts AA, Trent MS, Hendrixson DR. Campylobacter jejuni BumSR directs a response to butyrate via sensor phosphatase activity to impact transcription and colonization. Proc Natl Acad Sci USA. (2020) 117:11715–26. 10.1073/pnas.1922719117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, Raka F, Adeli K. The role of the gut microbiota in lipid lipoprotein metabolism. J Clin Med. (2019) 8:2227. 10.3390/jcm8122227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng S, Dai Z, Liu AB, Huang J, Narsipur N, Guo G, et al. Intake of stigmasterol and beta-sitosterol alters lipid metabolism and alleviates NAFLD in mice fed a high-fat western-style diet. Biochem Biophys Acta Mol Cell Biol Lipids. (2018) 1863:1274–84. 10.1016/j.bbalip.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiang JYL. Bile acids: regulation of synthesis. J Lipid Res. (2009) 50:1955–66. 10.1194/jlr.R900010-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE. (2008) 3:e2945. 10.1371/journal.pone.0002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sofka D, Pfeifer A, Gleiß B, Paulsen P, Hilbert F. Changes within the intestinal flora of broilers by colonisation with Campylobacter jejuni. Berlr Münch Tierärzt Wochenschr. (2015) 128:104–10. 10.2376/0005-9366-128-104 [DOI] [PubMed] [Google Scholar]
- 57.Tatituri RV, Brenner MB, Turk J, Hsu FF. Structural elucidation of diglycosyl diacylglycerol monoglycosyl diacylglycerol from Streptococcus pneumoniae by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J Mass Spectrom. (2012) 47:115–23. 10.1002/jms.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radka CD, Frank MW, Rock CO, Yao J. Fatty acid activation and utilization by Alistipes finegoldii, a representative Bacteroidetes resident of the human gut microbiome. Mol Microbiol. (2020) 113:807–25. 10.1111/mmi.14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christensen K, McMurtry JP, Thaxton YV, Thaxton JP, Corzo A, McDaniel C, et al. Metabolic and hormonal responses of growing modern meat-type chickens to fasting. Brit Poultry Sci. (2013) 54:199–205. 10.1080/00071668.2013.772953 [DOI] [PubMed] [Google Scholar]
- 60.Zaheer K. An updated review on chicken eggs: production, consumption, management aspects and nutritional benefits to human health. Food Nutr Sci. (2015) 6:1208–20. 10.4236/fns.2015.613127 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, DRA009061.