Abstract
Antibody titers against SARS-CoV-2 slowly wane over time. Here, we examined how time affects antibody potency. To assess the impact of antibody maturation on durable neutralizing activity against original SARS-CoV-2 and emerging variants of concern (VOCs), we analyzed receptor binding domain (RBD)-specific IgG antibodies in convalescent plasma taken 1–10 months after SARS-CoV-2 infection. Longitudinal evaluation of total RBD IgG and neutralizing antibody revealed declining total antibody titers but improved neutralization potency per antibody to original SARS-CoV-2, indicative of antibody response maturation. Neutralization assays with authentic viruses revealed that early antibodies capable of neutralizing original SARS-CoV-2 had limited reactivity toward B.1.351 (501Y.V2) and P.1 (501Y.V3) variants. Antibodies from late convalescents exhibited increased neutralization potency to VOCs, suggesting persistence of cross-neutralizing antibodies in plasma. Thus, maturation of the antibody response to SARS-CoV-2 potentiates cross-neutralizing ability to circulating variants, suggesting that declining antibody titers may not be indicative of declining protection.
Keywords: SARS-CoV-2, SARS-CoV-2 variants of concern, neutralizing antibody, affinity maturation
Graphical abstract
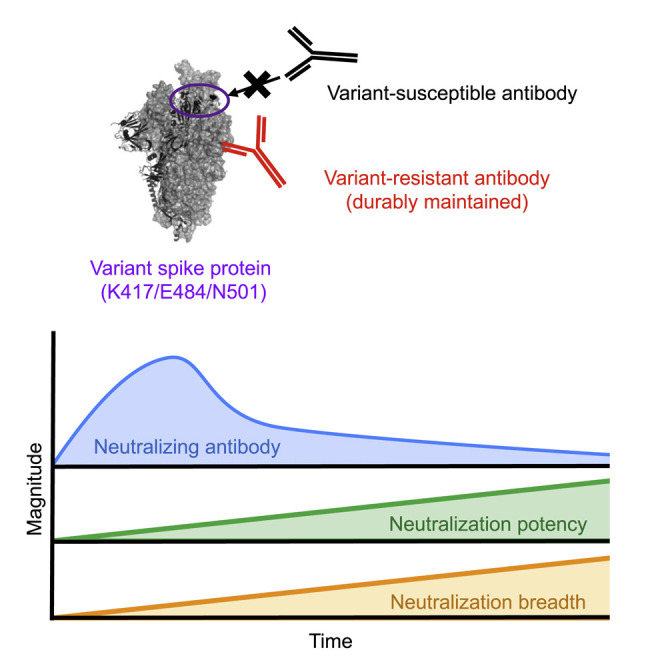
Antigenic drifts in SARS-CoV-2 variants permit escape from neutralizing antibody in COVID-19 convalescent plasma. Moriyama et al. reveal the evolution of serological immunity with time that counters SARS-CoV-2 variants via affinity maturation and durable elicitation of IgG antibodies that are resistant to viral escape.
Introduction
The novel coronavirus, SARS-CoV-2, first described in Wuhan, China, in December 2019, triggers multiple arms of acquired immunity, such as virus-binding antibodies, B cells, CD4+ T cells, and CD8+ T cells (Rydyznski Moderbacher et al., 2020). Coordinated induction and maintenance of these immune components is required to control COVID-19 pathogenesis (Rydyznski Moderbacher et al., 2020), among which neutralizing antibodies confer protection against reinfection in animal models (Baum et al., 2020; McMahan et al., 2021) and can be used as therapeutics in humans (Gottlieb et al., 2021; Weinreich et al., 2021). Neutralization activities of polyclonal antibodies to SARS-CoV-2 virus and its variants are the sum of two parameters of individual antibodies: neutralization potency that represents NT ability per virus-binding antibodies and neutralization breath that represents cross-neutralizing ability to variants per neutralizing antibodies.
Major epitopes of neutralizing antibodies reside in the receptor-binding domain (RBD) of the spike protein (Andreano et al., 2021; Piccoli et al., 2020; Rogers et al., 2020). RBD epitopes are further divided into at least four classes on the basis of the structure of the antigen-antibody complex (Barnes et al., 2020; Yuan et al., 2021). Among these epitopes, class 1 and 2 epitopes overlap with angiotensin-converting enzyme 2 (ACE2)-binding sites (receptor binding site) and are targeted by potent neutralizing antibodies (Barnes et al., 2020). Nevertheless, similar to other viral antigens, the receptor binding site epitopes on SARS-CoV-2 spike protein are functionally plastic (Greaney et al., 2021; Piccoli et al., 2020) and thus are highly susceptible to mutations.
Paradoxically, concentrations of RBD antibodies and, more specifically, neutralizing antibodies correlate with COVID-19 severity, with higher antibody titers observed in patients with severe relative to mild disease (Chen et al., 2020; Garcia-Beltran et al., 2021; Lynch et al., 2021; Piccoli et al., 2020; Rijkers et al., 2020). However, the results of these neutralization assays cannot discriminate whether high neutralization activities reflect the presence of highly neutralizing antibodies or a high abundance of less potent antibodies. To reconcile this paradox, an additional antibody parameter, termed the neutralization potency index (NPI), represents the sum of the neutralization potencies of individual antibodies (Garcia-Beltran et al., 2021). Compared with total neutralization activity, NPIs efficiently predict disease prognosis and survival in the case of severe disease (Garcia-Beltran et al., 2021). Therefore, it is important to quantify NPI in addition to neutralization activity in order to assess their possible impacts on clinical outcomes.
Emerging variants of concern (VOCs) with increased transmissibility and/or resistance to neutralizing antibodies elicited by parental virus infection or vaccination include those that emerged in the United Kingdom (B.1.1.7, 501Y.V1) (Volz et al., 2021), South Africa (B.1.351, 501Y.V2) (Tegally et al., 2021), and Brazil (P.1, 501Y.V3) (Faria et al., 2021). They all carry the N501Y mutation, which increases ACE2 binding (Starr et al., 2020). Moreover, 501Y.V2 and 501Y.V3 strains carry two additional RBD mutations (E484K and K417N/T); among these E484K has a greater impact on resistance to antibody neutralization (Chen et al., 2021; Wang et al., 2021a). Although all VOCs acquire resistance to neutralizing monoclonal antibodies, convalescent sera, and sera from vaccinees, levels of resistance differ among VOC strains, with strong, moderate, and weak resistance observed in 501Y.V2, 501Y.V3, and 501Y.V1 strains, respectively (Chen et al., 2021; Dejnirattisai et al., 2021; Hoffmann et al., 2021; Supasa et al., 2021; Wang et al., 2021a; Zhou et al., 2021). These antigenic characteristics are depicted via comparative analysis of total neutralization activities in parental strains compared with those in VOC strains; however, the effects of these antigenic drifts on other qualitative parameters including NPI remain unclear.
In the present study, we monitored the neutralizing activity and NPI of convalescent individuals longitudinally up to 10 months after symptom onset. Furthermore, an additional qualitative parameter, neutralization breadth index (NBI), which represents the relative neutralizing activities to parental strain versus those to VOC strain, was also quantitated. This parameter allowed us to visualize changes in neutralization breath independent of the magnitudes of neutralizing antibody titers to the parental virus strain. Despite the quantitative decay of antibody titers and total neutralizing activities, we demonstrated a time-dependent increase in NPI and NBI during the late convalescent phase beyond 3 months after symptom onset. The progressive increase in these qualitative parameters against circulating SARS-CoV-2 variants was highly correlated with the affinity maturation of IgG antibodies against RBD conserved sites that were durably maintained. Thus, the serological immunity elicited by SARS-CoV-2 infection evolves with time to counter SARS-CoV-2 variants through affinity maturation and durable elicitation of IgG antibodies to viral conserved sites.
Results
COVID-19 convalescent individuals mount durable, albeit declining, neutralizing antibody titers against ancestral SARS-CoV-2 RBD
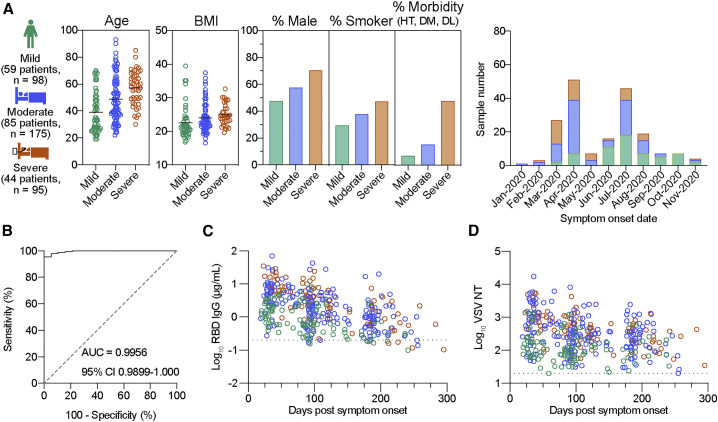
We collected 368 plasma samples between June 2020 and February 2021 from 188 PCR-confirmed COVID-19 patients who suffered from symptomatic infection and were seroconverted by anti-nucleocapsid antibodies. Symptoms were grouped into mild, moderate, and severe categories on the basis of severity. Mild cases were associated with signs and symptoms of COVID-19 without pneumonia, whereas moderate and severe cases were associated with pneumonia without and with the need for supplemental oxygen, respectively (moderate, 93% < oxygen saturation [SpO2] < 96%; severe, SpO2 ≤ 93%). This cohort comprised 59 patients with mild, 85 with moderate, and 44 with severe disease (Figure 1 A). Some subjects provided blood samples at a single time point for cross-sectional analysis, and 120 subjects provided blood samples two or three times at intervals of 2–3 months for longitudinal analysis. Participant demographics and clinical characteristics are summarized in Figure 1A. Subjects ranged in age from 19 to 93 years, with severe cases observed in the elderly group. Patients with severe COVID-19 exhibited higher BMI, were men and smokers, and manifested comorbidities at increased frequencies (Figure 1A), which is in accordance with the established clinical characteristics (www.cdc.gov/coronavirus/2019-ncov). The time period of symptom onset was between January and November 2020, prior to the initial detection of SARS-CoV-2 VOCs in Japan (December 2020; hCoV-19/Japan/IC-0446/2020).
Figure 1.
Durable neutralizing antibody response to original virus strain in COVID-19 convalescent individuals
(A) Participant demographics and clinical characteristics for cross-sectional blood samples are summarized: number of individuals in each severity group, age, body mass index (BMI), percentage of gender, smoking, and morbidity (HT, hypertension; DM, diabetes mellitus; DL, dyslipidemia), and date of symptom onset. All individuals were confirmed positive via PCR and seropositive via the detection of anti-nucleocapsid antibodies (Roche kit) and provided blood samples up to three times.
(B) ROC curve for RBD IgG titers of 368 convalescent plasma samples and 19 pre-pandemic negative samples. Area under the curve (AUC) and 95% confidence interval (CI) are indicated.
(C and D) Cross-sectional RBD IgG titers (C) and pseudovirus neutralizing antibody (VSV NT) titers (D) from COVID-19 convalescent plasma samples (n = 368). Dashed lines indicate the cut-off determined by ROC analysis.
As the major epitopes for potently neutralizing antibodies reside in the spike RBD (Andreano et al., 2021; Piccoli et al., 2020; Rogers et al., 2020), we focused on RBD antibodies. Moreover, IgG class was selected instead of IgM and IgA classes because of its abundance and persistence in convalescent plasma (Garcia-Beltran et al., 2021; Iyer et al., 2020; Piccoli et al., 2020). We quantitated the RBD IgG antibodies by ELISA using the reference monoclonal IgG, COVA1-18 (Brouwer et al., 2020). Receiver operating characteristic (ROC) curve analysis using plasma from convalescent and pre-pandemic healthy individuals (n = 19) was performed to determine the cut-off values that distinguished seropositive from seronegative samples (Figure 1B). Cross-sectional analysis of 368 plasma samples revealed a decline in RBD IgG antibodies over a period of 10 months in this study cohort (Figure 1C), thereby reproducing the gradual decay seen in other studies (Dan et al., 2021; Iyer et al., 2020; Piccoli et al., 2020; Pradenas et al., 2021). All samples were subjected to the vesicular stomatitis virus (VSV)-based pseudovirus neutralization assay to quantify the neutralizing (NT) antibody (Figure 1D). Similar to RBD IgG antibodies, NT antibodies declined over a period of 10 months, although they decayed more slowly than RBD IgG antibodies. During the follow-up period, seropositivity in this study cohort was 95.4% for RBD IgG and 99.5% for NT antibodies.
Neutralizing potency index of RBD antibodies increases with time in COVID-19 convalescent individuals
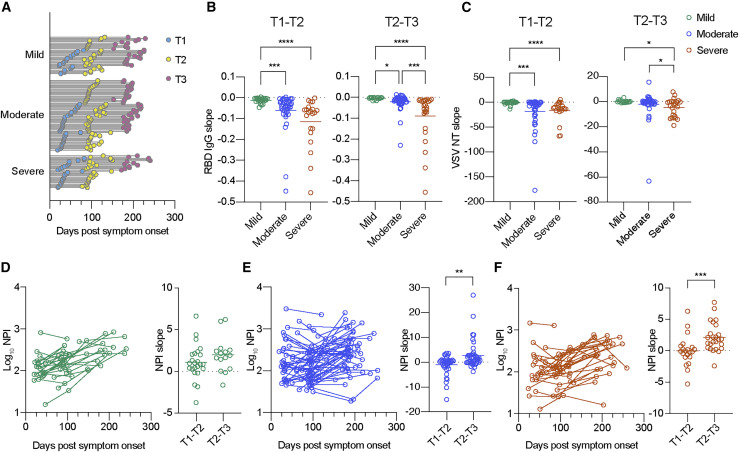
Blood samples from 120 subjects were assessed for longitudinal analysis to determine the decay slopes of RBD IgG and NT antibodies. Participant demographics and clinical characteristics for longitudinal analysis were similar to those observed for cross-sectional analysis (Figure S1A). Blood samples were collected from subjects (28 mild, 60 moderate, and 32 severe cases) at two or three time points, as illustrated in Figure 2 A. Time point 1 (T1), T2, and T3 were within the ranges of 1–2 months, 2–4 months, and 5–7 months after symptom onset, respectively. To visualize the levels of antibody decay, we calculated and plotted the decay slopes of RBD IgG (Figure 2B) and NT antibodies (Figure 2C) on the basis of the disease severity. From T1 to T2, RBD IgG antibodies decayed more rapidly in the severe group than in the mild group, presumably reflecting higher levels of RBD IgG antibodies at the initial T1 period in the severe group (Figure S1B) (Gaebler et al., 2021; Pradenas et al., 2021). This rapid decay in the severe group was reproduced in NT antibodies as well and continued in the late convalescent phase (T2–T3).
Figure 2.
Longitudinal decay in neutralizing antibody titers but increase in neutralizing potency index of RBD antibodies
(A) Timeline of sample collection for longitudinal analysis: mild (n = 67 from 28 individuals), moderate (n = 144 from 60 individuals), and severe (n = 78 from 32 individuals). Timing and number of samplings are indicated as line-connected circles; first (T1, light blue), second (T2, yellow), and third (T3, pink).
(B and C) Decay analysis of RBD IgG titers (B) and pseudovirus NT titers (C) from mild (green; n = 24 for T1–T2 and n = 15 for T2–T3), moderate (blue; n = 40 for T1–T2 and n = 44 for T2–T3), and severe (red; n = 21 for T1–T2 and n = 25 for T2–T3) groups. Bars represent the mean. Time points are within the following range of days after symptom onset: T1, 1–2 months; T2, 2–4 months; and T3, 5–7 months. Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (Kruskal-Wallis test).
(D–F) Longitudinal neutralization potency index (NPI) and decay analysis of mild (D), moderate (E), and severe (F) groups. Data from the same individual are connected with lines in left panels. Bars represent the mean in right panels. Statistical significance is indicated as follows: ∗∗p < 0.01 and ∗∗∗p < 0.001 (Mann-Whitney test).
The NPI was calculated by dividing the NT titer with RBD IgG titer, as previously described (Garcia-Beltran et al., 2021), and the temporal dynamics of this qualitative parameter were longitudinally analyzed. During the convalescent phase, we did not observe an apparent difference of NPI among the individuals who recovered from the varying severity, but we did observe a temporal increase of calculated NPI (Figures 2D–2F), inversely to decreasing amounts of antibodies (RBD IgG and NT) (Figures 2B and 2C). To compare the rate of increase between the T1/T2 and T2/T3 periods, the NPI slopes were calculated in each period and plotted separately (Figures 2D–2F). We observed higher increasing rates in the T2/T3 period compared with those in the T1/T2 period in the same subjects, although the difference in the mild group was not significant. Thus, longitudinal evaluation of convalescent plasma up to 10 months revealed improved neutralization potency per antibody to original SARS-CoV-2 during the late convalescent phase, despite declining total antibody titers.
Potently neutralizing monoclonal antibodies fail to neutralize SARS-CoV-2 variants 501Y.V2 and 501Y.V3
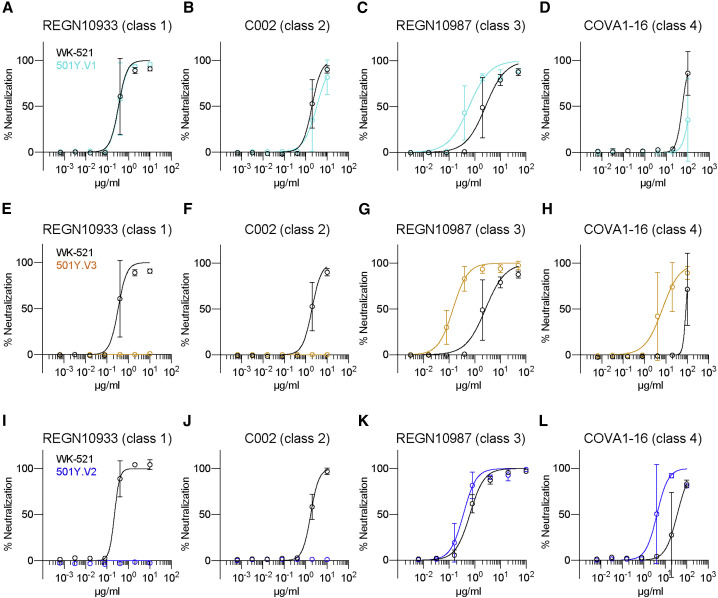
SARS-CoV-2 VOCs spreading worldwide include at least three lineages (501Y.V1, 501Y.V2, and 501Y.V3). The robust resistance of the 501Y.V2 strain to the potently neutralizing class 1 and 2 antibodies is, at least partly, attributed to K417N and E484K mutations (Chen et al., 2021; Wang et al., 2021a). The presence of K417T and E484K mutations in 501Y.V3 predict a similar resistance to antibody neutralization; however, this point is addressed by only a few studies using pseudotyped viruses (Hoffmann et al., 2021; Wang et al., 2021b) and authentic viruses (Dejnirattisai et al., 2021). The outcome of neutralization resistance varied depending on the use of authentic and pseudotyped virus systems (Chen et al., 2021). Therefore, we examined the resistance of VOC strains toward neutralizing antibodies using authentic viruses.
Furthermore, we selected four neutralizing monoclonal antibodies that recognize class 1–4 RBD epitopes: REGN10933 (class 1) (Hansen et al., 2020), C002 (class 2) (Barnes et al., 2020; Robbiani et al., 2020), REGN10987 (class 3) (Hansen et al., 2020), and COVA1-16 (class 4) (Liu et al., 2020). The neutralization activities of these antibodies to 501Y.V1 (QHN-002), 501Y.V2 (TY8-612), and 501Y.V3 (TY7-503) were compared with that of the Wuhan strain (WK-521). The positions of all mutations and deletions in the S proteins are illustrated in Figure S2A. In agreement with previous studies (Chen et al., 2021; Dejnirattisai et al., 2021; Wang et al., 2021a), the 501Y.V1 strain was relatively sensitive to all classes of neutralizing monoclonal antibodies examined; however, the 501Y.V2 and 501Y.V3 strains were resistant to class 1 and 2 antibodies (Figure 3 ). Thus, these two VOC lineages escape neutralization by class 1 and 2 antibodies, presumably by virtue of the E484K and K417N/T mutations that are shared among 501Y.V2 and 501Y.V3.
Figure 3.
Potently neutralizing monoclonal antibodies fail to neutralize VOCs 501Y.V2 and 501Y.V3
(A–L) Levels of neutralization by serially diluted neutralizing monoclonal antibodies were quantitated on the basis of the prevention of cytopathic effects. Resistance of 501Y.V1 (QHN002) (A–D), 501Y.V3 (TY7-503) (E–H), and 501Y.V2 (TY8-612) (I–L) was compared with that of the Wuhan strain. Data are plotted as mean ± SD with sigmoidal curves. Representative data from three independent experiments are shown.
501Y.V2 and 501Y.V3 escape from convalescent plasma antibodies
Cross-neutralizing activity of convalescent plasma against the 501Y.V2 and 501Y.V3 strains was determined as the inverse of the highest dilution preventing 50% of cytopathic effects. The neutralizing activities of polyclonal antibodies against different virus strains could be influenced by several factors other than the antigenicity of S proteins. In the present study, neutralizing activities against different virus strains were normalized in an individual plate using a reference antibody that equivalently neutralizes the SARS-CoV-2 variants. For this, we collected hyper-immune sera from rabbits that were repeatedly immunized with recombinant RBD. The rabbit serum exhibited neutralization activity equivalent to that of the Wuhan, 501Y.V2, and 501Y.V3 strains (Figure S2B) under similar conditions as those indicated in Figure 3. Likewise, the binding of rabbit serum to RBD proteins with K417N, E484K, N501Y, or all three mutations was similar to that observed with original RBD (Figure S2C). Therefore, we included rabbit serum as a reference antibody for all plates, and the NT titers of plasma samples were normalized to those of the rabbit serum in the following analyses. A significant correlation between NT titers determined using this method and plaque reduction neutralization titers (PRNT50) was observed (Figure S3A).
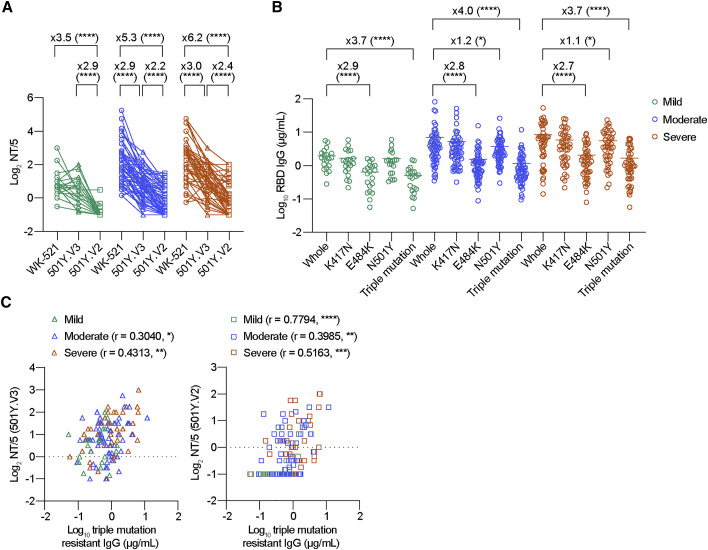
The plasma samples were subjected to NT assay using four different virus strains (Figure 4 A; Figure S3B). In agreement with the findings of previous studies using convalescent polyclonal antibodies (Chen et al., 2021; Dejnirattisai et al., 2021), the 501Y.V2 stain escaped neutralization from convalescent plasma (3.5-fold in mild, 5.3-fold in moderate, and 6.2-fold in severe cases), and the 501Y.V3 strain was modestly resistant in moderate and severe groups (2.9-fold in moderate and 3.0-fold in severe cases) that mounted higher NT titers to Wuhan strain (Figure 4A). However, no significant resistance was observed in the 501Y.V3 strain in the mild group, suggesting that the low NT titers in this group are insufficient to visualize the antigenic drifts of 501Y.V3 strain. The resistance of 501Y.V1 strain was weaker (1.3-fold) than that of 501Y.V2 and 501Y.V3 strains (Figure S3B).
Figure 4.
VOCs escape from convalescent antibodies through RBD mutations
(A) Neutralization antibody titers were determined as the inverse of the highest plasma dilution, preventing 50% of cytopathic effects, and plotted as cut-off dilution index. Convalescent plasmas comprised 20 mild (green), 59 moderate (blue), and 46 severe (red). Data from the same sample are connected with lines and depicted as circles (WK-521), triangles (501Y.V3), and squares (501Y.V2) symbols. Statistical significance and fold-reduction are indicated above columns; ∗∗∗∗p < 0.0001 (Friedman test). Pooled data from more than two independent experiments are shown.
(B) IgG titers against original RBD and RBD mutants carrying K417N, E484K, N501Y, or triple mutations are comparably plotted. Bars represent the mean. Statistical significance and fold reduction are indicated; ∗p < 0.05 and ∗∗∗∗p < 0.0001 (Friedman test).
(C) Correlation of mutation-resistant IgG titers with NT titers against VOCs is plotted. Spearman r and p values are indicated; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
We assessed the relative contribution of three RBD mutations to antibody resistance. RBD mutant proteins carrying either K417N, E484K, N501Y, or triple K417N/E484K/N501Y mutations were used as antigens in ELISA, and the binding IgG titers were quantitated by using the reference monoclonal IgG, CR3022 (Yuan et al., 2020). CR3022 recognizes the cryptic and conserved epitope in class 4 that does not overlap with the three mutations. Indeed, this epitope specificity resulted in comparable binding of CR3022 to RBD mutant proteins (Figure S3C). The quantitation of RBD IgG antibodies by ELISA revealed significant (2.7- to 2.9-fold) reduction in the binding IgG antibodies due to E484K mutation (Figure 4B). Furthermore, the N501Y mutation slightly reduced the binding by IgG antibodies in the moderate and severe groups (1.1- to 1.2-fold reduction). Compared with RBDs with any single mutation, the most profound reduction was observed in triple-mutant RBD (3.7- to 4.0-fold reduction), thus revealing the additive effects of K417N and/or N501Y mutations with respect to the effects of the E484K mutation. Considering the significant reduction in the RBD-binding IgG antibodies due to the presence of triple RBD mutations, we denoted IgG antibodies against the triple-mutant RBD protein as mutation-resistant IgG antibodies.
Correlation between the mutant-resistant IgG antibodies and the VOC NT titers was further assessed (Figure 4C). Except the NT titers to 501Y.V3 strain in the mild group, the mutant-resistant IgG antibodies correlated with the VOC NT titers, suggesting that the mutant-resistant IgG antibodies detected in our method are indeed relevant to the cross-neutralizing activities to VOCs. Therefore, we performed longitudinal analysis to determine the temporal dynamics of cross-neutralizing IgG antibodies in the following studies.
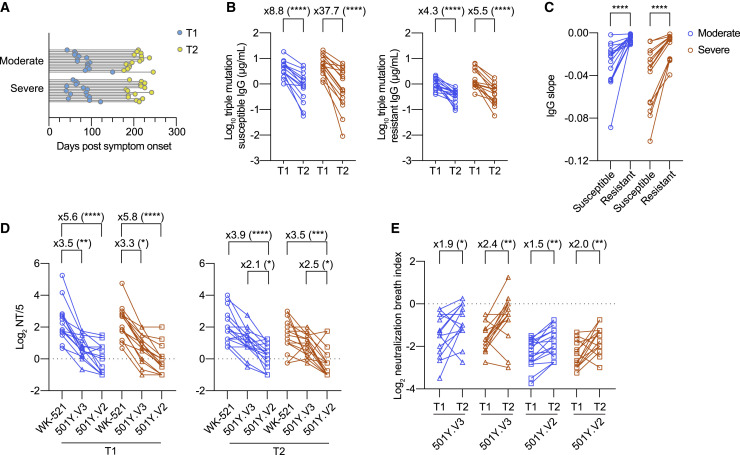
Neutralization breadth against VOCs increases over time in association with durable IgG response that is resistant to RBD mutations
Longitudinal analysis was performed during the convalescent phase to identify any temporal parameters that affected VOC cross-neutralizing activities. For detecting VOC cross-neutralizing activities above the detection limit (NT titer = 5), we used convalescent plasma that had >10 NT titers to Wuhan strain among the moderate and severe plasma panel used in Figure 4A. Also, the longitudinal plasma samples covering the T1 (day 79 ± 24) and T2 (day 207 ± 19) periods with >90 day intervals were selected for the following studies (Figure 5 A). We subtracted whole RBD-binding IgG titers by triple mutant-binding (mutation-resistant) IgG titers and defined the titers as mutation-susceptible IgG titers; that is, mutation-resistant IgG antibodies likely target relatively conserved epitopes among SARS-CoV-2 variants or target variable epitopes but overcome the mutations via high-affinity binding, whereas mutation-susceptible IgG antibodies lose the binding by the mutations at position 417, 484, or 501. IgG antibodies during the T1 period were dominated by the mutation-susceptible IgG antibodies (78% ± 12% in moderate and 77% ± 7% in severe cases) (Figure 5B). Both mutation-susceptible and mutation-resistant IgG titers declined from T1 to T2 in both groups (Figure 5B); however, the fold decrease was smaller in mutation-resistant IgG titers compared with mutation-susceptible IgG titers over the 3 month period (4.3 versus 8.8 in moderate cases and 5.5 versus 37.7 in severe cases). Indeed, quantitation of the decay slope revealed a significantly slower decline in mutation-resistant IgG titers relative to mutation-susceptible IgG titers (Figure 5C).
Figure 5.
Temporal increase of neutralization breadth against VOCs in association with durable IgG antibodies that are resistant to RBD mutations
(A) Timeline of sample collection for longitudinal analysis: moderate (n = 30 from 15 individuals) and severe (n = 30 from 15 individuals). Timing and number of samplings are indicated as line-connected circles: first (T1, light blue) and second (T2, yellow).
(B) Mutation-susceptible and mutation-resistant IgG titers from moderate (blue; n = 15) and severe (red; n = 15) groups were quantitated using ELISA. Data from the same individual are connected with lines. Statistical significance and fold reduction are indicated above columns; ∗∗∗∗p < 0.0001 (Wilcoxon test).
(C) Decay slopes of mutation-susceptible and mutation-resistant IgG titers from T1 to T2 period are plotted. Data from the same sample are connected with lines. Statistical significance is indicated above columns; ∗∗∗∗p < 0.0001 (Wilcoxon test).
(D) Cross-neutralizing titers to the indicated virus strains from moderate and severe groups are plotted (left, T1; right, T2). Data from the same sample are connected with lines (circles, WK-521; triangles, 501Y.V3; squares, 501Y.V2). Statistical significance and fold reduction are indicated above column as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 (Friedman test).
(E) Cross-neutralization breadth index in each plasma was expressed by dividing the VOC NT titers by the Wuhan NT titers. Data from the same individual are connected with lines (triangles, 501Y.V3; squares, 501Y.V2). Statistical significance and fold reduction are indicated above column as follows: ∗p < 0.05 and ∗∗p < 0.01 (Wilcoxon test).
To further assess the possible impact of differential IgG decay on VOC cross-neutralization activity, the 501Y.V2 NT and 501Y.V3 NT titers were determined in parallel to the Wuhan NT titers at the T1 and T2 periods (Figure 5D). Consistent with the data in Figure 4A, NT titers against both VOCs were significantly reduced in the T1 period; however, the fold decrease became smaller in 501Y.V2 from T1 (5.6-fold in moderate and 5.8-fold in severe cases) to T2 (3.9-fold in moderate and 3.5-fold in severe cases). Moreover, no significant reduction was observed in the T2 period for the 501Y.V3 NT titers.
Although the NT titers against SARS-CoV-2 variants represent the sum of broad neutralizing activities of polyclonal antibodies, this number cannot discriminate whether these activities reflect the total abundance or individual potency of neutralizing antibodies against SARS-CoV-2 variants. To quantify the neutralization breadth, we further calculated the NBI by dividing the VOC NT titers by the Wuhan NT titers. This parameter allows us to visualize the changes in neutralization breath independent of the amounts of NT titers to the original Wuhan strain. We observed that the level of NBI for the 501Y.V2 and 501Y.V3 strains was below log2 0 at T1, indicating that neutralization breadth to VOCs was limited during the early time point (Figure 5E). However, the NBI gradually increased toward T2 in all groups, resulting in a 1.5- to 2.4-fold increase from T1 to T2 (Figure 5E). These data indicate that the maturation of serological immunity not only improves neutralization potency to the originally infected viruses but also expands neutralization breadth to the drifted variants.
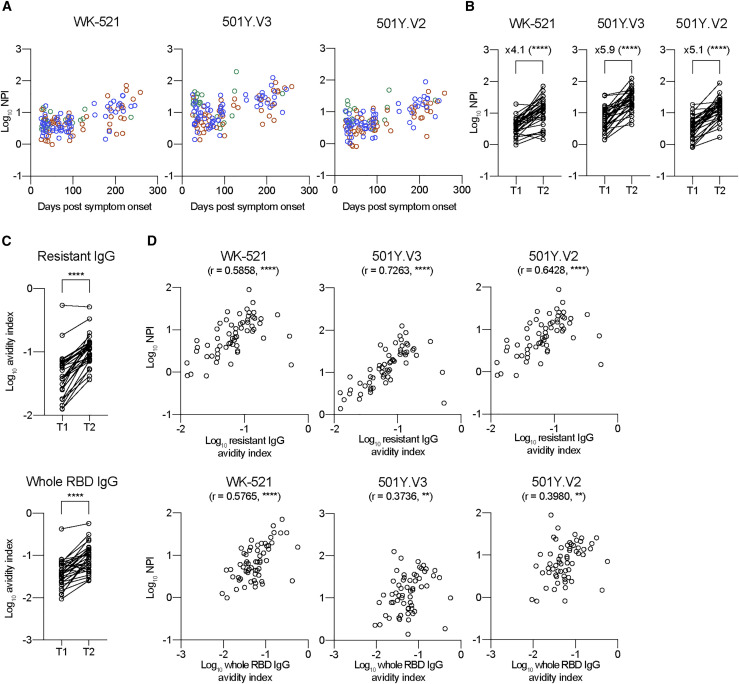
NPI to VOCs increases with time in correlation with an affinity maturation of mutation-resistant IgG antibodies
NPIs against the 501Y.V2 and 501Y.V3 strains were quantitated by dividing VOC NT titers with mutation-resistant IgG titers. We observed a gradual increase in the NPIs over time (Figure 6 A), resulting in 4.1- to 5.9-fold increase from T1 to T2 (Figure 6B). A progressive increase in NPI implies the possible relevance to an affinity maturation of antibody that is a hallmark of long-lasting serological immunity (Eisen and Siskind, 1964; Siskind and Benacerraf, 1969). Therefore, the average affinities of polyclonal plasma antibodies to RBD antigens were quantitated using a modified ELISA to dissociate plate-bound, low-affinity IgG selectively via 7 M urea treatment (de Souza et al., 2004; Onodera et al., 2016). The average affinities, indicated as avidity index, of mutation-resistant IgG antibodies increased from T1 to T2, and the avidity index of whole RBD-binding IgG antibodies that predominantly include mutation-susceptible IgG antibodies also increased at comparable levels (Figure 6C). The affinity maturation of antibodies is achieved through the decay of low-affinity antibodies and the supply of high-affinity antibodies with time. The comparable affinity maturation between the two types of IgG antibodies, regardless of the differential decline in the total amounts (Figures 5B and 5C), suggest that the progressive supply of high-affinity antibodies, presumably mediated by persistent viral antigen deposit (Gaebler et al., 2021), is the primary driving force for the affinity maturation during the convalescent period in COVID-19 patients.
Figure 6.
Temporal increase of NPIs to VOCs in correlation with affinity maturation of mutation-resistant IgG antibodies
(A) NPIs against WK-521, 501Y.V3, and 501Y.V2 strains were quantitated from mild (green, n = 20), moderate (blue, n = 59), and severe (red, n = 46) groups and plotted as log10.
(B) NPIs against the indicated strains are plotted as log10 in T1 and T2 periods (longitudinal samples; n = 31). Data from the same individual are connected with lines. Statistical significance and fold increase are indicated; ∗∗∗∗p < 0.0001 (Wilcoxon test).
(C) Avidity indexes of IgG to triple mutant (top; resistant IgG) and whole RBD (bottom; whole RBD-binding IgG) are plotted. Data from the same individual are connected with lines. ∗∗∗∗p < 0.0001 (Wilcoxon test).
(D) Correlations between avidity indexes of resistant IgG/whole RBD IgG and NPI against WK-521 (left), 501Y. V3 (middle), and 501Y.V2 (right) strains are plotted. Spearman r and p values are indicated; ∗∗p < 0.01 and and ∗∗∗∗p < 0.0001.
Correlation between VOC NPI and the affinity of IgG antibodies was further assessed (Figure 6D). For reference, correlation with Wuhan NPI was plotted in parallel. Wuhan NPI equally correlated with the affinity of mutation-resistant IgG and whole RBD-binding IgG antibodies. In contrast, we observed significant and clear correlations between VOC NPIs (501Y.V2/501Y.V3) and affinity of mutation-resistant IgG antibodies, whereas levels of correlation were reduced between the NPIs and affinity of whole RBD-binding IgG antibodies. These differential correlations highlight profound contribution of affinity-matured IgG antibodies to increasing VOC neutralization potency.
Discussion
Affinity maturation of serum antibodies, one of the fundamental phenomena in serological immunity, has attracted intense interest in research (Victora and Nussenzweig, 2012). Longitudinal analysis of RBD-binding monoclonal antibodies exhibited overall increase in binding affinities and neutralization potency and breadth in many, but not all, tested antibodies (Clark et al., 2021; Gaebler et al., 2021). Although these data support the contribution of affinity maturation on neutralization potency and breadth at the monoclonal level, the impacts of affinity maturation in polyclonal antibodies, which are a mixture of diverse clonality, remains unknown. Moreover, monoclonal antibodies have been recovered from memory B cells (Clark et al., 2021; Gaebler et al., 2021), but not from long-lived plasma cells, a major source of serum antibodies, because of their limited accessibility. As the extent of affinity maturation in memory B cells is not strictly correlated with those of serum antibodies (Smith et al., 1997), the gap in the cellular sources potentially affects the outcome. In this context, we showed a progressive increase in the neutralizing potency and breadth of convalescent plasma antibodies to SARS-CoV-2 variants. The increased levels were markedly correlated with affinity maturation of IgG antibodies that are resistant to RBD mutations. Thus, this study highlights the impacts of affinity maturation on VOC neutralization and the immune strategy to counteract against SARS-CoV-2 variants.
Continuous affinity maturation of plasma antibodies even after 3 months post-symptom onset resembles the evolution of memory B cell repertoires during the convalescent phase in COVID-19 patients (Gaebler et al., 2021; Sokal et al., 2021). Intriguingly, SARS-CoV-2 mRNA and protein were detected in the small intestinal epithelium in certain subjects, even months after symptom onset. The persistent viral antigen deposits could act as a driving force for continuous germinal center responses, whereby affinity-based selection occurs on the precursors for long-lived plasma cells (Kurosaki et al., 2015). The persistent viral antigens in COVID-19 patients are presumably insufficient to continuously recruit native B cells into plasma cells to sustain high levels of antibody, as observed in other chronic virus infections; however, they remarkably improve the affinity and neutralization potency and breadth of IgG antibodies that are resistant to RBD mutations. Therefore, we speculate that the mode of antigen persistence in COVID-19 patients is a key trigger for the selective increase in the numbers of memory B cells while reducing the amounts of serum/plasma antibodies over time (Abayasingam et al., 2021; Dan et al., 2021; Gaebler et al., 2021; Rodda et al., 2021; Sokal et al., 2021).
The quantities of RBD IgG antibodies declined in different slopes according to the resistance to RBD mutations (Figure 5C), with more slow decline in mutation-resistant IgG antibodies relative to mutation-susceptible IgG antibodies. The persistence of mutation-resistant IgG antibodies contributes to sustaining neutralizing activities to SARS-CoV-2 variants, additively with the qualitative improvement of IgG affinities over time. It is important to mention that the mutation-susceptible IgG antibodies dominated (77%–78% in Figure 5B) among all RBD-binding IgG antibodies; therefore, variable epitopes including position 417, 484, or 501 are immunodominant. Whereas immunodominant epitopes induce robust IgG responses, high titers of IgG antibodies mask the epitopes that are required for further stimulation of the epitope-binding B cells, a phenomenon known as antibody feedback (McNamara et al., 2020; Zhang et al., 2013). In light of the progressive accumulation of somatic hypermutations in memory B cells during the convalescent phase (Gaebler et al., 2021; Sokal et al., 2021), the unique mode of antigen persistence in COVID-19 patients probably stimulates virus-specific B cells for a longer period than we initially speculated. In this scenario, the epitopes targeted by the mutation-resistant IgG antibodies would be increasingly presented to B cells with time by antibody feedback, possibly accounting for the persistence of the IgG antibodies. This point is worthwhile to be further investigated using animal models.
Limitations of the study
A primary limitation of our study is the insufficient knowledge regarding the levels of neutralization activity and potency required for protection against reinfection. Therefore, we cannot estimate how the temporal increase of neutralization potency and breadth affect protection levels in vivo. An additional limitation is that there are poorly neutralizing in vitro but protective antibodies in vivo (Schäfer et al., 2021; Winkler et al., 2021). The heterogeneity in the protection pathways may generate the gaps between in vivo efficacy and in vitro neutralization potency and breadth detected in this study. Finally, we were unable to confirm the findings at monoclonal levels, because of difficulties in preparing monoclonal antibodies from long-lived plasma cells that are infrequently present in the blood during the convalescent phase.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| REGN10933 | Hansen et al., 2020 | N/A |
| C002 | Robbiani et al., 2020 | N/A |
| REGN10987 | Hansen et al., 2020 | N/A |
| COVA1-16 | Brouwer et al., 2020 | MT599835.1, MT599919.1 |
| COVA1-18 | Brouwer et al., 2020 | MT599837.1, MT599921.1 |
| CR3022 | Meulen et al., 2006 | DQ168569.1, DQ168570.1 |
| HPR-conjugated goat anti-human IgG | Southern Biotech | Cat#2040-05; RRID: AB_2795644 |
| Bacterial and virus strains | ||
| hCoV-19/Japan/TY-WK-521/2020 | National Institute of Infectious Diseases | EPI_ISL_408667 |
| hCoV-19/Japan/QHN002/2020 | National Institute of Infectious Diseases | EPI_ISL_804008 |
| hCoV-19/Japan/TY8-612/2021 | National Institute of Infectious Diseases | EPI_ISL_1123289 |
| hCoV-19/Japan/TY7-503/2021 | National Institute of Infectious Diseases | EPI_ISL_877769 |
| VSV pseudovirus bearing SARS-CoV-2 spike protein | Tani et al., 2021 | N/A |
| Biological samples | ||
| SARS-CoV-2-infected convalescent patient blood sample | Tokyo-Shinagawa Hospital, Yokohama Municipal Citizen’s Hospital, Kawakita General Hospital, and Tokyo Center Clinic | N/A |
| Pre-pandemic healthy individual blood sample | Japanese Red Cross | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RBD (amino acids: 331–529) | In-house | MN994467 |
| K417N mutant RBD | In-house | N/A |
| E484K mutant RBD | In-house | N/A |
| N501Y mutant RBD | In-house | N/A |
| K417N, E484K, N501Y triple mutant RBD | In-house | N/A |
| HiTrap Protein G HP Columns | Cytiva | 17-0404-01 |
| Bovine serum albumin | Sigma-Aldrich | A2153 |
| Tween-20 | Fujifilm | 167-11515 |
| Can Get Signal #2 | TOYOBO | NKB-301 |
| OPD substrate | Sigma-Aldrich | P8287 |
| D-MEM (Low Glucose) with L-Glutamine and Phenol Red | Fujifilm Wako Pure Chemicals | 041-29775 |
| D-MEM (High Glucose) with L-Glutamine and Phenol Red | Fujifilm Wako Pure Chemicals | 044-29765 |
| Fetal bovine serum | Biowest | S1780-500 |
| Geneticin | Thermo Fisher Scientific | 10131-027 |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15140-122 |
| Vacutainer CPT tube | BD Biosciences | 362761 |
| Formalin | Fujifilm Wako Pure Chemicals | 062-01661 |
| Crystal violet solution | Sigma-Aldrich | V5265 |
| Methylene blue | Fujifilm Wako Pure Chemicals | 137-06982 |
| Ni-NTA agarose | QIAGEN | 30230 |
| Critical commercial assays | ||
| Bright-Glo luciferase assay system | Promega | E2620 |
| Expi293 expression system | Thermo Fisher Scientific | A29133 |
| Elecsys Anti-SARS-CoV-2 | Roche Diagnostics | 518316181 |
| Experimental models: Cell lines | ||
| VeroE6/TMPRSS2 cells | JCRB Cell Bank | JCRB1819 |
| Software and algorithms | ||
| Graphpad Prism 9 | Graphpad | N/A |
| Other | ||
| iMark microplate reader | Bio-Rad | 168-1130J1 |
| Epoch2 | Biotek | EPOCH2NS |
| GroMax Navigator Microplate Luminometer | Promega | GM2000 |
| cobas e 411 plus | Roche Diagnostics | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yoshimasa Takahashi (ytakahas@niid.go.jp).
Materials availability
All resources generated in this study are available on request from the lead contact upon providing a completed Materials Transfer Agreement.
Data and code availability
All data needed to support the conclusion of this manuscript are included in the main text and supplementary materials.
Experimental model and subject details
Human subjects and sampling
SARS-CoV-2-infected individuals (confirmed using SARS-CoV-2 PCR on nasopharyngeal swab samples upon admission) were enrolled at Tokyo Shinagawa Hospital, Yokohama Municipal Citizen’s Hospital, Kawakita General Hospital, and Tokyo Center Clinic. Blood samples were collected over the course of 10 months at the intervals of 2–3 months for longitudinal analysis. The following information was collected for each patient: sex, age, BMI, date of symptom onset, severity, smoking history, and pre-existing medical conditions (hypertension, diabetes mellitus, and dyslipidemia). Blood samples were collected in Vacutainer CPT tubes (BD Biosciences), and plasma samples were isolated via centrifugation at 1800 × g for 20 minutes followed by additional centrifugation at 800 × g for 15 minutes after removing the cells. Thereafter, plasma was heat-inactivated at 56°C for 30 minutes before use for the assays. All samples were seropositive for nucleocapsid antibody by Elecsys Anti-SARS-CoV-2 (Roche). Pre-pandemic blood samples from healthy individuals were collected before the COVID-19 pandemic (from August 2018 to July 2019) at the Japanese Red Cross. All studies were approved by the Institutional Review Board of the National Institute of Infectious Diseases (#1237, #1132), #1197, and #913). This study was performed in accordance with the Declaration of Helsinki. All volunteers provided written informed consent prior to enrollment.
Method details
SARS-CoV-2 virus
SARS-CoV-2 virus strain (hCoV19/Japan/TY-WK-521/2020) and the variants (hCoV19/Japan/QHN002/2020, hCoV19/Japan/TY8-612/2021, and hCoV19/Japan/TY7-503/2021) were isolated at National Institute of Infectious Diseases in Japan.
Cells
Expi293F cells were maintained in Expi293 expression medium (Thermo Fisher Scientific). VeroE6/TMPRSS2 cells (JCRB1819, JCRB Cell Bank) were maintained in low glucose DMEM (Fujifilm) containing 10% heat-inactivated fetal bovine serum (FBS, biowest), 1 mg/mL geneticin (Thermo Fisher Scientific), and 100 U/mL penicillin/streptomycin (Thermo Fisher Scientific) at 37°C supplied with 5% CO2.
Recombinant RBD antigens
Human codon-optimized nucleotide sequence coding the spike protein of the SARS-CoV-2 isolate (GenBank: MN994467) was commercially synthesized (Eurofinsgenomics). RBD (amino acids: 331–529) with the signal peptide (amino acids 1–20; MIHSVFLLMFLLTPTESYVD) with a C-terminal histidine tag was cloned into the mammalian expression vector pCAGGS. RBD mutant proteins bearing K417N, E484K, N501Y, and triple mutations (K417N, E484K, and N501Y) were generated by site-directed mutagenesis. Recombinant proteins were produced using Expi293F cells, according to the manufacturer’s instruction (Thermo Fisher Scientific). The supernatant from transfected cells was harvested on day 5 post-transfection, and recombinant proteins were purified using Ni-NTA agarose (QIAGEN).
Monoclonal antibody generation
Recombinant monoclonal antibodies were prepared as previously described (Adachi et al., 2019; Tiller et al., 2008). Briefly, VH/VL genes of reference monoclonal antibodies (REGN10933, C002, REGN10987, COVA1-16, COVA1-18, and CR3022) were cloned into the expression vectors with human IgG1 heavy chain and kappa/lambda light chain. Pairs of heavy and light chain vectors were transfected into Expi293F cells according to the manufacturer’s instructions. Thereafter, antibodies were purified from the culture supernatant using a protein G column (Thermo Fisher Scientific) and subjected to further analysis after dialysis using PBS.
ELISA
F96 Maxisorp Nunc-Immuno plates (Thermo Fisher Scientific) were coated with 2 μg/mL of either original RBD or RBD mutants overnight at 4°C. After washing with PBS, the plates were blocked with 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 1.5 hour at room temperature. Heat-inactivated plasma and monoclonal antibodies were serially-diluted in PBS containing 1% BSA and 0.05% Tween-20 (eight 4-fold serial dilutions starting at 1:20 dilution for plasma, 1:10, 1:20, 1:80, and 1:320 dilutions for pre-pandemic plasma, or eight 4-fold serial dilutions starting at a 1 μg/mL for monoclonal antibodies), and then incubated for overnight at 4°C. The following day, the plates were washed with PBS containing 0.05% Tween-20. To determine the avidity index of RBD IgGs, serially diluted plasma samples were loaded in duplicate, and one set of wells was treated with 7 M urea for 30 minutes at room temperature before adding HPR-conjugated goat anti-human IgG (Southern Biotech). HRP-activity was visualized by the addition of OPD substrate (Sigma), and OD490 was measured using iMark microplate reader (Bio-Rad) and Epoch2 (Biotek). IgG titers were quantitated by using COVA1-18 or CR3022 as reference antibodies in each plate. The avidity index was calculated by dividing the amount of RBD-binding IgG with urea treatment by the amount of RBD-binding IgG without urea treatment. IgG slopes between two time points were calculated by dividing the difference in IgG titers by time difference.
Pseudovirus production
VSV pseudovirus bearing SARS-CoV-2 spike protein was generated as described previously (Tani et al., 2021). Briefly, the cDNA of SARS-CoV-2 spike protein was synthesized (Integrated DNA Technologies Inc.) and cloned into the pCAGGS expression vector. The plasmid (pCAG-SARS-CoV-2) comprising 19 aa truncation at the C terminus of the spike protein was constructed. The pCAG-SARS-CoV-2 expression vector was transfected into 293T cells on collagen-coated tissue culture plates. After 24 hours incubation, the cells were infected with G-complemented VSVΔG/Luc (Tani et al., 2010) at a multiplicity of infection of 0.5, and thereafter, the uninfected viruses were washed. After 24 hours of incubation, the culture supernatants with VSV pseudovirus were collected, centrifuged to remove cell debris, and then stored at −80°C until use for the virus neutralization assay.
Virus neutralization assay
For the pseudovirus neutralization assay, SARS-CoV-2 pseudovirus was incubated with an equal volume of serially diluted, heat-inactivated plasma (five 5-fold serial dilutions starting at 1:10 dilution) for 1 hour at 37°C. The mixture was inoculated with VeroE6/TMPRSS2 cells seeded in 96-well solid white flat-bottom plates (Corning), and then incubated for 24 hours at 37°C in a chamber supplied with 5% CO2. Luciferase activity in cultured cells was measured using the Bright-Glo luciferase assay system (Promega) with a GroMax Navigator Microplate Luminometer (Promega). Half-maximal inhibitory concentration (IC50) was calculated by Prism 9 (GraphPad) and was presented as neutralization titers.
For the authentic virus neutralization assay, a mixture of 100 TCID50 virus and serially diluted antibodies (2-fold serial dilutions starting at 1:5 dilution diluted with high glucose DMEM supplemented with 2% FBS and 100 U/mL penicillin/streptomycin) was incubated at 37°C for 1 hour before being placed on VeroE6/TMPRSS2 cells seeded in 96-well flat-bottom plates (TPP). After culturing for 4–6 days at 37°C supplied with 5% CO2, cells were fixed with 20% formalin (Fujifilm Wako Pure Chemicals), and stained with crystal violet solution (Sigma-Aldrich). Each sample was assayed in 4–6 wells and the average of cut-off dilution index with > 50% cytopathic effect was presented as neutralization titer. NT titer of the sample below the detection limit (1:5 dilution) was set as 2.5. For certain experiments, absorbance at 595 nm was measured using Epoch 2 area scanning mode (Biotek) and the neutralization percentage was calculated as follows; (sample signals – virus control signals)/(cell-only control signals – virus control signals) × 100 (Chi et al., 2020). Sigmoidal curves were calculated with Prism 9 (GraphPad).
For plaque reduction neutralization assay, heat-inactivated plasma was serially diluted with 2-fold from 1:2.5 to 1:40, mixed with equal volume of 2 × 103 PFU/mL of viruses, and then incubated for 1 hour at 37 °C. VeroE6/TMPRSS2 cells were plated in 24 well plates (Sumitomo Bakelite) and 50 μL of the virus/plasma mixtures were inoculated in duplicates after washing with DMEM containing 2% FBS and penicillin–streptomycin. After 1 hour incubation at 37°C, the inoculums were removed and cells were washed twice with media. The medium containing 1:100-diluted penicillin–streptomycin and 1% methylcellulose were overlayed and the plates were incubated at 37 °C in 5% CO2. After 4 days incubation, the cells were fixed with 10% formalin (Fujifilm Wako Pure Chemicals) at room temperature and stained with 0.037% of methylene blue (Wako Pure Chemicals). Plaques were counted and PRNT50 was determined by calculating the sigmoidal curves with Prism 9 (GraphPad).
Quantification and statistical analysis
Statistical analysis and ROC analysis were performed using Prism 9 (GraphPad). Statistical test for each analysis was described in each legend. Statistical significance was defined as p < 0.05. Error bars represent standard deviation (SD).
Acknowledgments
We thank Dr. Katsumi Maenaka at Hokkaido University for providing the S trimer graphic; Akira Dosaka, Eriko Izumiyama, Masataka Tokita, and Ryoko Itami at the National Institute of Infectious Diseases for their technical support; and the Japanese Red Cross Society for providing donated blood based on the "Guidelines on the use of donated blood in R&D, etc". This work was supported by Japan Agency for Medical Research and Development under grant numbers JP19fk0108104 (T. Suzuki and Y.T.) and JP20fk0108104 (T. Suzuki and Y.T.).
Author contributions
Conceptualization, Y.T.; Methodology, S.F., Y.A., K.M., and T. Suzuki; Investigation, S.M., Y.A., T. Sato, K. Tonouchi, L.S., S.F., S.Y., H.K., K.N., T.K., M.T., Y.K., P.E., T. Onodera, T. Matsumura, T.T., K. Terahara, M.I., A.N., A.K.-T., M.S., N.T., S.N., and T. Okai; Funding Acquisition, T. Suzuki, and Y.T.; Project Administration, T. Matano, T.F., K.M., M.O., T.W., T. Suzuki, and Y.T.; Supervision, M.O., T.W., T. Suzuki, and Y.T.; Writing – Original Draft, S.M., Y.A., and Y.T.; Writing – Review & Editing, S.M., Y.A., M.I., T. Matano, T.W., and Y.T.
Declaration of interests
The authors declare no competing interests.
Published: August 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.06.015.
Supplemental information
References
- Abayasingam A., Balachandran H., Agapiou D., Hammoud M., Rodrigo C., Keoshkerian E., Li H., Brasher N.A., Christ D., Rouet R., et al. COSIN Study Group Long-term persistence of RBD+ memory B cells encoding neutralizing antibodies in SARS-CoV-2 infection. Cell Rep Med. 2021;2:100228. doi: 10.1016/j.xcrm.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., Tonouchi K., Nithichanon A., Kuraoka M., Watanabe A., Shinnakasu R., Asanuma H., Ainai A., Ohmi Y., Yamamoto T., et al. Exposure of an occluded hemagglutinin epitope drives selection of a class of cross-protective influenza antibodies. Nat. Commun. 2019;10:3883. doi: 10.1038/s41467-019-11821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Nicastri E., Paciello I., Pileri P., Manganaro N., Piccini G., Manenti A., Pantano E., Kabanova A., Troisi M., et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell. 2021;184:1821–1835.e16. doi: 10.1016/j.cell.2021.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Pan Z., Yue S., Yu F., Zhang J., Yang Y., Li R., Liu B., Yang X., Gao L., et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct. Target. Ther. 2020;5:180. doi: 10.1038/s41392-020-00301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza V.A., Fernandes S., Araújo E.S., Tateno A.F., Oliveira O.M., Oliveira R.R., Pannuti C.S. Use of an immunoglobulin G avidity test to discriminate between primary and secondary dengue virus infections. J. Clin. Microbiol. 2004;42:1782–1784. doi: 10.1128/JCM.42.4.1782-1784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen H.N., Siskind G.W. Variations in affinities of antibodies during the immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., Hauser B.M., Caradonna T.M., Clayton K.L., Nitido A.D., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb R.L., Nirula A., Chen P., Boscia J., Heller B., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325:632–644. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5:eabe0367. doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Kometani K., Ise W. Memory B cells. Nat. Rev. Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K.L., Whitman J.D., Lacanienta N.P., Beckerdite E.W., Kastner S.A., Shy B.R., Goldgof G.M., Levine A.G., Bapat S.P., Stramer S.L., et al. Magnitude and kinetics of anti-severe acute respiratory syndrome coronavirus 2 antibody responses and their relationship to disease severity. Clin. Infect. Dis. 2021;72:301–308. doi: 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara H.A., Idris A.H., Sutton H.J., Vistein R., Flynn B.J., Cai Y., Wiehe K., Lyke K.E., Chatterjee D., Kc N., et al. Antibody feedback limits the expansion of B cell responses to malaria vaccination but drives diversification of the humoral response. Cell Host Microbe. 2020;28:572–585.e7. doi: 10.1016/j.chom.2020.07.001. [DOI] [PubMed] [Google Scholar]
- Onodera T., Hosono A., Odagiri T., Tashiro M., Kaminogawa S., Okuno Y., Kurosaki T., Ato M., Kobayashi K., Takahashi Y. Whole-virion influenza vaccine recalls an early burst of high-affinity memory B cell response through TLR signaling. J. Immunol. 2016;196:4172–4184. doi: 10.4049/jimmunol.1600046. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradenas E., Trinité B., Urrea V., Marfil S., Ávila-Nieto C., Rodríguez de la Concepción M.L., Tarrés-Freixas F., Pérez-Yanes S., Rovirosa C., Ainsua-Enrich E., et al. Stable neutralizing antibody levels 6 months after mild and severe COVID-19 episodes. Med (N Y) 2021;2:313–320.e4. doi: 10.1016/j.medj.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers G., Murk J.L., Wintermans B., van Looy B., van den Berge M., Veenemans J., Stohr J., Reusken C., van der Pol P., Reimerink J. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J. Infect. Dis. 2020;222:1265–1269. doi: 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siskind G.W., Benacerraf B. Cell selection by antigen in the immune response. Adv. Immunol. 1969;10:1–50. doi: 10.1016/s0065-2776(08)60414-9. [DOI] [PubMed] [Google Scholar]
- Smith K.G., Light A., Nossal G.J., Tarlinton D.M. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Kimura M., Tan L., Yoshida Y., Ozawa T., Kishi H., Fukushi S., Saijo M., Sano K., Suzuki T., et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein. Virol. J. 2021;18:16. doi: 10.1186/s12985-021-01490-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., Matsuura Y. Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 2010;84:2798–2807. doi: 10.1128/JVI.02499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. COVID-19 Genomics UK (COG-UK) consortium Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Wilson I.A. Recognition of the SARS-CoV-2 receptor binding domain by neutralizing antibodies. Biochem. Biophys. Res. Commun. 2021;538:192–203. doi: 10.1016/j.bbrc.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Meyer-Hermann M., George L.A., Figge M.T., Khan M., Goodall M., Young S.P., Reynolds A., Falciani F., Waisman A., et al. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to support the conclusion of this manuscript are included in the main text and supplementary materials.