Summary
The emergence of SARS-CoV-2 variants with evidence of antibody escape highlight the importance of addressing whether the total CD4+ and CD8+ T cell recognition is also affected. Here, we compare SARS-CoV-2-specific CD4+ and CD8+ T cells against the B.1.1.7, B.1.351, P.1, and CAL.20C lineages in COVID-19 convalescents and in recipients of the Moderna (mRNA-1273) or Pfizer/BioNTech (BNT162b2) COVID-19 vaccines. The total reactivity against SARS-CoV-2 variants is similar in terms of magnitude and frequency of response, with decreases in the 10%–22% range observed in some assay/VOC combinations. A total of 7% and 3% of previously identified CD4+ and CD8+ T cell epitopes, respectively, are affected by mutations in the various VOCs. Thus, the SARS-CoV-2 variants analyzed here do not significantly disrupt the total SARS-CoV-2 T cell reactivity; however, the decreases observed highlight the importance for active monitoring of T cell reactivity in the context of SARS-CoV-2 evolution.
Keywords: SARS-CoV-2, VOCs, T cells, CD4, CD8, COVID-19, vaccines
Graphical abstract
Highlights
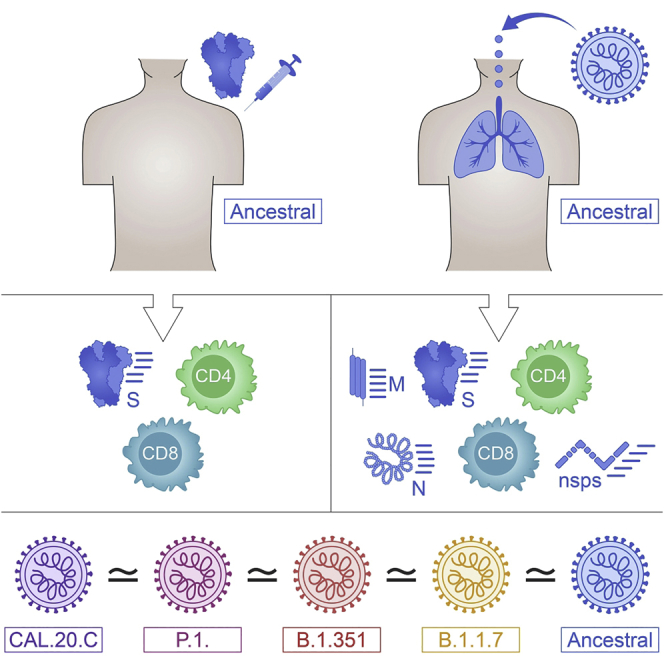
T cells of exposed donors or vaccinees effectively recognize SARS-CoV-2 variants
Effective recognition in AIM and FluoroSPOT assays, for spike and other proteins
93% and 97% of CD4 and CD8 epitopes are 100% conserved across variants
Tarke et al. show that SARS-CoV-2-specific memory CD4 and CD8 T cells exposed to the ancestral strain by infection or vaccination effectively recognize the variants B.1.1.7, B.1.351, P.1, and CAL.20C. The majority of T cell epitopes are unaffected by mutations in these variant strains.
Introduction
The emergence of several severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) variants of concern (VOC) with multiple amino acid replacements has implications for the future control of the coronavirus disease 2019 (COVID-19) pandemic.1, 2, 3, 4 VOCs include the United Kingdom (UK) variant 501Y.V1 lineage B.1.1.7,3 the South Africa (SA) variant 501Y.V2 lineage B.1.351,4 the BR (Brazilian) variant 501Y.V3 lineage P.1,5,6 and the California (CA) variant CAL.20C lineages B.1.427–429.7,8 The B.1.1.7 variant is associated with increased transmissibility,9,10 and similar epidemiological observations have been reported for the SA and BR variants.4,5
The mutations of greatest concern are present in the viral spike (S) protein and include notable mutations in the receptor-binding domain (RBD), the N-terminal domain (NTD), and the furin cleavage site region. Several of the RBD mutations directly affect angiotensin-converting enzyme 2 (ACE2) receptor-binding affinity, which may affect infectivity, viral load, or transmissibility.11, 12, 13, 14 Multiple mutations were also noted in regions bound by neutralizing antibodies, so it is crucial to address the extent to which mutations associated with the variants may affect immunity induced by either SARS-CoV-2 infection or vaccination.
Numerous reports address the effect of variant spike (S) mutations on antibody binding and function.11,14, 15, 16, 17, 18, 19, 20, 21, 22, 23 In general, the impact of B.1.1.7 mutations on neutralizing antibody titers is moderate.15, 16, 17, 18,20,24 In contrast, mutations in B.1.351 and P.1 variants are associated with the more pronounced loss of neutralizing capacity.15,16,22,23,25 Concerning vaccination responses, the AstraZeneca ChAdOx1 vaccine has been associated with a partial loss of neutralizing antibody activity against B.1.1.715 and a large loss of neutralizing activity against B.1.351.26 ChAdOx1 maintains efficacy against B.1.1.724,27 but has a major loss in efficacy against mild COVID-19 with the B.1.351 variant.26 Current epidemiological evidence is that the BNT162b2 Pfizer/BioNTech COVID-19 vaccine retains its efficacy against B.1.1.7 in the UK24 and in reports from Israel.28,29 The Novavax COVID-19 vaccine (NVX-CoV2373) has differential protective immunity against the parental strain, B.1.1.7, and B.1.351 in clinical trials (96%, 86%, and 60%, respectively),30 whereas the Janssen Ad26.COV2.S 1-dose COVID-19 vaccine has relatively similar protection for moderate COVID-19 against both the ancestral strain and B.1.351 (72% and 64%, respectively).31,32
Several lines of evidence suggest that CD4+ and CD8+ T cell responses play important roles in the resolution of SARS-CoV-2 infection and COVID-19,33 including modulating disease severity in humans34,35 and reducing viral loads in non-human primates.36 Furthermore, individuals with agammaglobulinemia or pharmaceutical depletion of B cells generally experience an uncomplicated COVID-19 disease course.33,37,38 Robust CD4+ and CD8+ T cell memory is induced after COVID-19,22,39, 40, 41 and multiple COVID-19 vaccines elicit CD4+ and CD8+ T cell responses.26,42, 43, 44, 45 It is therefore key to address the potential impact of mutations associated with SARS-CoV-2 variants on T cell reactivity; however, few data are available on this topic.46
Here, we take a combined experimental and bioinformatics approach to address T cell reactivity to SARS-CoV-2 VOCs. We directly assess T cell responses from individuals recovered from COVID-19 and T cell responses from recent Moderna mRNA-1273 or Pfizer/BioNTech BNT162b2 vaccinees for their capacity to recognize peptides derived from the ancestral reference sequence and the B.1.1.7, B1.351, P.1, and CAL.20C variants. As a complementary approach, bioinformatics analyses were used to predict the impact of mutations in the VOCs with sets of previously reported CD4+ and CD8+ T cell epitopes derived from the ancestral reference sequence.
Results
Sequence analysis and peptide pool generation
As a first step, we mapped the specific mutations (amino acid replacements and deletions) of the main current VOCs, including B.1.1.7, B1.351, P.1, and CAL.20C, as compared to the SARS-CoV-2 Wuhan ancestral sequence (NCBI: NC_045512.2) (Table S1). Then, we synthesized the corresponding peptides associated with the different variants and generated new peptide pools (megapools [MPs]) spanning the full genome sequences of the ancestral Wuhan strain and the respective B.1.1.7, B1.351, P.1, and CAL.20C variants (Table S2). As described below, the resulting peptide pools were assessed for their capacity to be recognized by memory T cell responses derived from natural infection in convalescents and vaccinees, and responses to the variant and ancestral genome antigen-specific pools were compared.
Cohorts of COVID-19 convalescent, vaccinees, and unexposed controls
We selected three donor cohorts to investigate T cell reactivity against VOCs. Our convalescent donors were adults ranging from 20 to 67 years of age (median 38); 43% were male and 57% were female (Table 1). SARS-CoV-2 infection in these donors was determined by PCR-based testing during the acute phase of their infection, if available (46% of the cases), and/or seropositivity determined by plasma SARS-CoV-2 S protein RBD immunoglobulin G (IgG) ELISA (Figure S1A). From these donors, peripheral blood mononuclear cell (PBMC) samples were collected between May and October 2020, a period when none of the VOCs analyzed were prevalent in the San Diego, California, area, where the donations from convalescent donors were obtained. The convalescent donor samples reflect the local ethnic demographics (81% non-Hispanic white). The peak of COVID-19 severity was representative of the distribution observed in the general population, with a prevalence of mild cases (86% of the cohort analyzed).
Table 1.
Characteristics of donor cohorts
| COVID-19 (n = 28) | Vaccinees (n = 29) | Unexposed (n = 23) | |
|---|---|---|---|
| Age, y | 20–67 (median = 38, IQR = 32) | 23–67 (median = 47, IQR = 36) | 21–82 (median = 34, IQR = 24) |
| Gender, % | |||
| Male | 43 (12/28) | 31 (9/29) | 35 (8/23) |
| Female | 57 (16/28) | 69 (20/29) | 56 (13/23) |
| Unknown | 0 (0/28) | 0 (0/29) | 9 (2/23) |
| Sample collection date | May–October 2020 | January–March 2021 | May 2014–March 2018 (13/23) |
| March–May 2020 (10/23) | |||
| SARS-CoV-2 PCR, % | positive 92 (12/13) | N/A | N/A |
| not tested 54 (15/28) | |||
| S RBD IgG+ (%) | 96 (27/28) | 100 (29/29) | 0 (10/10, collected in 2020) |
|
Peak disease severity, %a | |||
| Mild | 86 (24/28) | N/A |
N/A |
| Moderate | 14 (4/28) | ||
| Severe | 0 (0/28) | ||
| Critical | 0 (0/28) | ||
| Race-ethnicity, % | |||
| White—not Hispanic or Latino | 81 (23/28) | 52 (15/29) | 43 (10/23) |
| Hispanic or Latino | 4 (1/28) | 10 (3/29) | 9 (2/23) |
| Asian | 7 (2/28) | 38 (11/29) | 26 (6/23) |
| American Indian/Alaska Native | 0 (0/28) | 0 (0/29) | 0 (0/23) |
| Black or African American | 0 (0/28) | 0 (0/29) | 9 (2/23) |
| >1 race | 4 (1/28) | 0 (0/29) | 4 (1/23) |
| Not reported | 4 (1/28) | 0 (0/29) | 9 (2/23) |
| Days at collection | 38–163 (28/28) (median = 78, IQR = 50)b | 13–30 (14/29) Pfizer | N/A |
| 12–16 (15/29) Moderna (median = 14, IQR = 14)c | |||
According to World Health Organization criteria.
Post-symptom onset.
2nd dose of vaccine.
From the vaccinated donors, we collected PBMCs after recent vaccination with the Moderna mRNA-1273 or the Pfizer/BioNTech BNT162b2 vaccines, ∼14 days following second dose administration (Table 1). These donors ranged in age from 23 to 67 years (median 47); 31% were male and 69% were female. All of the vaccinees had RBD IgG titers indicative of vaccination (Figure S1A).
PBMCs from unexposed donors were collected between May 2014 and March 2018 or in the March–May 2020 period, and were seronegative for RBD IgG. The unexposed donors ranged from 21 to 82 years old (median 34); 35% were male and 56% were female, while 9% were unknown (Table 1).
CD4+ and CD8+ T cell reactivity against ancestral S
We previously described the use of activation-induced marker (AIM) assays to measure CD4+ and CD8+ T cell responses to pools of overlapping peptides spanning SARS-CoV-2 antigens.34,39,47,48 Here, we used the same AIM techniques using OX40+CD137+ and CD69+CD137+ markers for CD4+ and CD8+ T cell reactivity, respectively.
We compared the CD4+ and CD8+ T cell reactivity of the three cohorts against SARS-CoV-2 S by AIM and FluoroSPOT assays. The T cell responses to the ancestral S MP were significantly higher than the DMSO controls in unexposed (p < 0.0001 for CD4, p = 0.0063 for CD8, and p = 0.0036 for spot-forming cells [SFCs]/106 PBMCs by the Wilcoxon test), convalescent (p < 0.0001 for CD4, p < 0.0001 for CD8, and p = 0.0008 for SFC/106 PBMCs by the Wilcoxon test), and vaccinated donors (p < 0.0001 for CD4, p < 0.0001 for CD8, and p < 0.0001 for SFC/106 PBMCs by the Wilcoxon test) (Figures S1B–S1D).
When the AIM responses in the different donor cohorts were compared, we found significantly higher reactivity in COVID-19 convalescents compared to unexposed individuals (CD4: p < 0.0001; CD8: p < 0.0001 by the Mann-Whitney test) (Figures S1E and S1F), with 93% and 39% of donors positive (responses above the threshold of positivity as described in Method details) for CD4+ and CD8+ T cell responses, respectively. These rates of positivity are similar to what was observed in a previous study that analyzed responses to peptides spanning the whole SARS-CoV-2 proteome over a similar range of post-symptom onset (PSO) days,39 in which 93% and 50% positivity rates were observed for CD4+ and CD8+ T cells, respectively. Likewise, the responses observed in the S MP in vaccinated donors were significantly higher than in unexposed donors (CD4: p < 0.0001; CD8: p < 0.0001 by Mann-Whitney test), with 100% and 48% of donors positive for CD4 and CD8 responses, respectively (Figures S1E and S1F).
With an interferon-γ (IFN-γ) FluoroSPOT assay, IFN-γ T cell responses were detected in 17%, 50%, and 76% of unexposed, convalescent, and vaccinated donors, respectively (unexposed versus convalescent: p = 0.019; unexposed versus vaccinees: p < 0.0001 by the Mann-Whitney test) (Figure S1G). These donor T cell responses provided the benchmarks for subsequent assessment of the impact of VOC mutations.
CD4+ and CD8+ T cell reactivity against VOC S protein
We measured the CD4+ and CD8+ T cell responses to S MPs derived from the ancestral strain and corresponding MPs representing the B.1.1.7, B1.351, P.1, and CAL.20C variants. As shown in Figures 1A–1C, good CD4+ and CD8+ T cell reactivity was observed in convalescent donors with peptides spanning the S protein of the ancestral Wuhan sequence and the corresponding variant S peptides. Geomean S-specific responses ranged from 0.07 to 0.08 for CD4+ T cells and 0.08 to 0.10 for CD8+ T cells. No significant difference was observed between the ancestral and VOC S peptide pools (CD4: B.1.1.7 p = 0.17; B.1.351 p = 0.30; P.1 p = 0.21; CAL.20C p = 0.06 and CD8: B.1.1.7 p = 0.15; B.1.351 p = 0.25; P.1 p = 0.47; CAL.20C p = 0.17 by Wilcoxon test) (Figures 1A–1C). These values (here and in subsequent graphs) are not corrected for multiple comparisons, as a correction would decrease the statistical power for detecting significant differences; therefore, not performing multiple comparison corrections is the more conservative and stringent test.
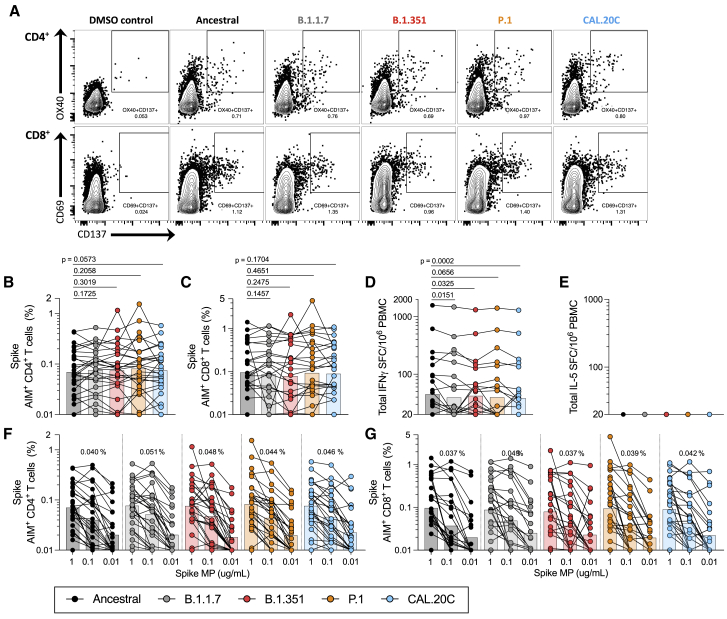
Figure 1.
T cell responses of COVID-19 convalescent individuals against ancestral and variant SARS-CoV-2 spike (S)
PBMCs of COVID-19 convalescent individuals (n = 28) were stimulated with the S MPs corresponding to the ancestral reference strain (black) and the B.1.1.7 (gray), B.1.351 (red), P.1 (orange), and CAL.20C (light blue) SARS-CoV-2 variants.
(A) The gating strategy for the AIM assay is illustrated by representative graphs defining S-specific CD4+ or CD8+ T cells by expression of OX40+CD137+ and CD69+CD137+, respectively. These graphs depict 1 of the COVID-19 convalescent donors from this study tested with the S MPs corresponding to each of the VOCs tested.
(B) Percentages of AIM+ (OX40+CD137+) CD4+ T cells.
(C) Percentages of AIM+ (CD69+CD137+) CD8+ T cells.
(D) IFN-γ spot-forming cells (SFCs) per million PBMCs.
(E) IL-5 SFCs per million PBMCs.
(F and G) The data shown in (B) and (C) are plotted to show the titration of the S MPs (1 μg/mL, 0.1 μg/mL, and 0.01 μg/mL) for CD4+ (F) and CD8+ (G) T cells for each SARS-CoV-2 variant. The geometric mean of the 0.1 μg/mL condition is listed above each titration.
Paired comparisons of ancestral S MPs versus each of the variants were performed by 1-tailed Wilcoxon test and are indicated by the p values in (B)–(D). In all of the panels, the bars represent the geometric mean. See also Figures S1, S2, S4, and S5 and Tables S1–S3.
To further test for potential differences in the recognition of VOCs by these T cells, we calculated fold change values per individual donor (Figures S2A and S2B). We then performed a non-inferiority test on one sample Wilcoxon signed-rank test compared to the lower bound fold change threshold calculated based on technical repeats (Table S3). No significant differences were observed, demonstrating that the decreases observed were within the range expected from technical repeat variation.
The VOC T cell analyses were extended using FluoroSPOT to measure the capacity of the various SARS-CoV-2 peptide pools to elicit functional responses in terms of secretion of IFN-γ and interleukin-5 (IL-5) cytokines (Figures 1D, 1E, and S2C). The results from the FluoroSPOT assay (Figures 1D and 1E) showed ex vivo IFN-γ reactivity using whole PBMCs, with a geomean of 45 SFCs/106 PBMCs (range 20–1,578) for the ancestral strain peptides. This overlaps with the 0–800 SFC range (median 110) detected in a previous study describing S reactivity in symptomatic donors49 and with the 20–110 range for 2–5 months’ symptomatic donors reported in a separate study.50
VOC reactivity was observed for the S MPs in convalescent donors, with geomean IFN-γ SFCs per million PBMCs ranging from 38 to 45 (Figure 1D). Compared to the ancestral strain, there were significant decreases of 12%, 6%, and 14% for the B.1.1.7, B.1.351, and CAL.20C variant pools (B.1.1.7 p = 0.02; B.1.351 p = 0.03; P.1 p = 0.07, and CAL.20C p < 0.01 by Wilcoxon test), while no difference was observed for P.1 (Figure 1D). No significant differences were observed by fold change analysis, suggesting that the decreases observed were still within the technical fluctuation range (Figure S2C). No IL-5 reactivity was observed for any of the pools (Figure 1E).
To further assess the functionality of T cell recognition of these variants, we considered the variant peptide dose-response curves of the CD4+ and CD8+ T cells. As shown in Figures 1F and S2D, peptide concentration sensitivity of CD4+ T cell responses to the ancestral and four variant pools was similar. The same pattern was also observed for CD8+ T cell responses to S (Figures 1G and S2E).
CD4+ and CD8+ T cell total reactivity against VOCs
As shown in Table S1, mutations found in the variants studied herein were not limited to the S protein, but also occurred in several additional antigens encoded in the SARS-CoV-2 genome. To address the potential impact of non-S variant mutations on overall proteome-wide CD4+ and CD8+ T cell reactivity, we tested overlapping peptide MPs spanning the entire proteome of the ancestral Wuhan sequence in comparison with corresponding MPs representing the different variants.
Overall, reactivity to the peptide pools spanning the variant genomes was found to be similar to that against the ancestral Wuhan strain (Figures 2 and S2). When the sum total of reactivity throughout the genome was considered, no decreases in reactivity compared to the ancestral were noted for the variant pools (Figures 2A–2C, S2F, and S2G).
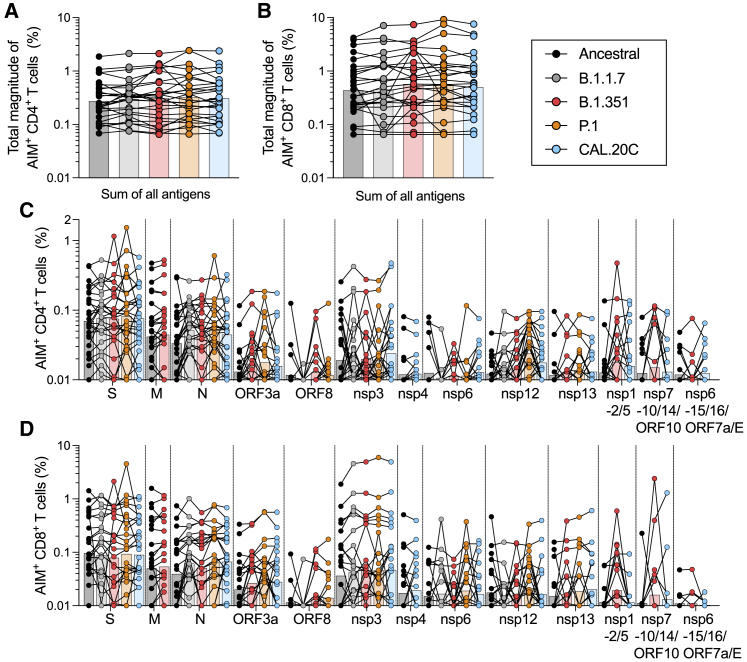
Figure 2.
T cell responses of COVID-19 convalescent individuals against ancestral and variant SARS-CoV-2 proteomes
PBMCs of COVID-19 convalescent individuals (n = 28) were stimulated with MPs for the entire viral proteome corresponding to the ancestral reference strain (black) and the B.1.1.7 (gray), B.1.351 (red), P.1 (orange), and CAL.20C (light blue) SARS-CoV-2 variants.
(A) Percentages of AIM+ (OX40+CD137+) CD4+ T cells for the total reactivity.
(B) Percentages of AIM+ (CD69+CD137+) CD8+ T cells for the total reactivity.
(C) Percentages of AIM+ (OX40+CD137+) CD4+ T cells for each MP.
(D) Percentages of AIM+ (CD69+CD137+) CD8+ T cells for each MP.
All of the bars represent the geometric mean. See also Figure S1, S2, S4, and S5 and Tables S1–S3.
We previously showed in COVID-19 convalescent subjects that a set of 10 different antigens (nsp3, nsp4, nsp6, nsp12, nsp13, S, ORF3a, M, ORF8, and N) account for 83% and 81% of the total CD4+ and CD8+ T cell response, respectively.51 Here, a similar overall pattern of dominant antigens was observed (Figures 2C and 2D). It is worth noting that this specific comparison is for illustration purposes only, as this study is not powered to rule out differences in individual antigens.
For comparison purposes, unexposed donors were also tested in the AIM assay, with MPs encompassing the ancestral and variant strains (Figures S2H–S2M). As expected, some T cell reactivity against the SARS-CoV-2 MPs was observed in unexposed donors, possibly due to previous exposure to common cold coronaviruses47,48,52,53 or to other pathogens or autoantigens. The magnitude of the responses was lower than in COVID-19 convalescents (unexposed versus convalescent: CD4 p < 0.0001; CD8 p < 0.0001 by the Mann-Whitney test, comparison not shown in the graphs). Similar to the COVID-19 convalescents, no decrease in the geomean of AIM+ CD4+ or CD8+ T cell responses to the sum of all of the antigens of individual MPs tested was observed in unexposed donors (Figures S2H–S2K). These experiments suggest that CD4+ and CD8+ T cells from individuals infected with the ancestral SARS-CoV-2 strain recognize VOCs with similar efficiency.
CD4+ and CD8+ T cell reactivity against VOCs by vaccinees
We also studied T cell responses by individuals who received US Food and Drug Administration (FDA)-authorized COVID-19 mRNA vaccines. We focused our analysis on T cell reactivity to the S antigen of the ancestral strain, which is the basis of the presently used vaccines.
For both CD4+ and CD8+ T cell reactivity, the magnitude of responses to pools encompassing the sequences from the ancestral Wuhan genome and the VOCs ranged from a geomean of 0.15–0.17 for CD4+ T cells and a geomean of 0.10–0.15 for CD8+ T cells (Figures 3A–3C). Comparison of the variant pools to the ancestral sequence showed no significant differences for CD4+ and CD8+ T cell reactivity in the AIM assay for B.1.1.7 and P.1 (CD4: B.1.1.7 p = 0.41, P.1 p = 0.29; CD8: B.1.1.7 p = 0.10, P.1 p = 0.09 by Wilcoxon test). Decreases of 14% and 22%, respectively, were observed with the B.1.351 pools for CD4+ and CD8+ T cells (B.1.351: p < 0.01 for both comparisons), and a 10% decrease with the CAL.20C pool for CD8+ T cells (p = 0.04) (Figures 3A–3C). The FluoroSPOT assay (Figures 3D–3F and S3C) showed IFN-γ reactivity, with geomeans ranging from 58 to the 74 SFCs per million PBMCs (Figures 3C and S3C). Given the number of comparisons made and that the reductions observed were on the low side (and others showed increases), we wanted to put these numbers into context by comparing them to what is expected based on the observed fold changes in repeat measurements of the same samples on different days for both AIM and FluoroSPOT assays, and asked whether the observed decreases were significantly higher (non-inferiority analysis; see Method details). No significant differences were observed, indicating that the magnitude of decreases for some strains was within the expected technical assay variability (Figures S3A–S3C). Minimal IL-5 responses were observed, with geomean reactivity ranging from 22 to 23 SFCs/106, which is slightly above the limit of detection (Figure 3E). On a per-donor basis, the IFN-γ response was found to account for >80% of the total response, on average (range 84%–89%), irrespective of whether the ancestral strain or VOCs were considered (Figure 3F).
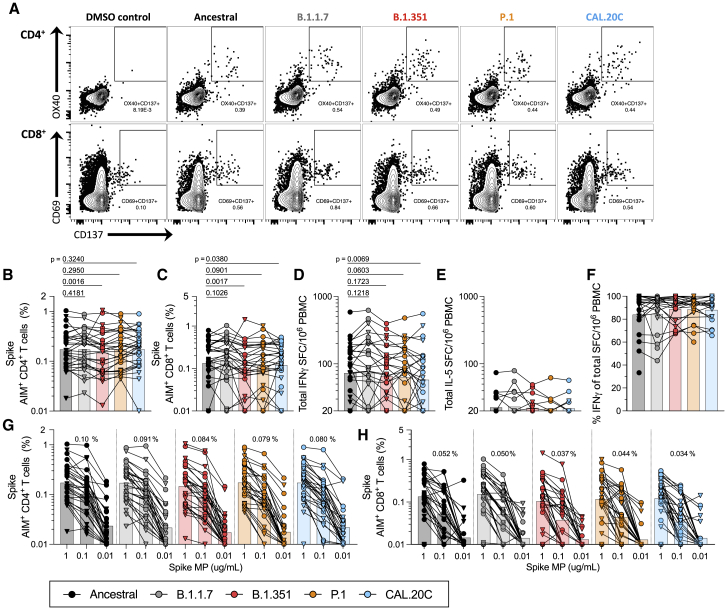
Figure 3.
T cell responses of COVID-19 vaccinees against ancestral and variant SARS-CoV-2 S
PBMCs of Pfizer/BioNTech BNT162b2 (n = 14, triangles) and Moderna mRNA-1273 COVID-19 vaccinees (n = 15, circles) were stimulated with the S MPs corresponding to the ancestral reference strain (black) and the B.1.1.7 (gray), B.1.351 (red), P.1 (orange), and CAL.20C (light blue) SARS-CoV-2 variants.
(A) The gating strategy for the AIM assay is illustrated by representative graphs defining S-specific CD4+ or CD8+ T cells by the expression of OX40+CD137+ and CD69+CD137+, respectively. These graphs depict one of the COVID-19 vaccinated donors from this study tested with the S MPs corresponding to each of the VOCs tested.
(B) Percentages of AIM+ (OX40+CD137+) CD4+ T cells.
(C) Percentages of AIM+ (CD69+CD137+) CD8+ T cells.
(D) IFN-γ SFCs per million PBMCs.
(E) IL-5 SFCs per million PBMCs.
(F) Percentages of IFN-γ were calculated from the total IFN-γ and IL-5 SFCs per million PBMCs.
(G and H) The data shown in (B) and (C) are also plotted showing the S MPs titration (1, 0.1, and 0.01 μg/mL) for CD4+ (G) and CD8+ (H) T cells with each SARS-CoV-2 variant. The geometric mean of the 0.1 μg/mL condition is listed above each titration.
Paired comparisons of the ancestral reference strain-based S MP versus each of the variants were performed by one-tailed Wilcoxon test and are indicated by the p values in (B)–(D). In all of the panels, the bars represent the geometric mean. See also Figures S1 and S3–S5 and Tables S1–S3.
Similar to the experiments in convalescent donors, we also examined T cell dose responses in vaccinated donors. As shown in Figures 3G, 3H, S3D, and S3E, CD4+ and CD8+ T cell dose responses to the ancestral and VOC S pools were similar.
Phenotypes of CD4+ and CD8+ T cells
We further assessed the quality of AIM+ T cell responses by phenotyping S-specific CD4+ and CD8+ AIM+ T cell responses. Representative gating strategy is shown in Figure S5. Consistent with previous observations,39,48 AIM+ T cell responses in COVID-19 convalescent donors above the threshold of positivity (CD4 >0.016% and CD8 >0.16%, as described in Method details; Figure S4), irrespective of the variant analyzed, had a memory phenotype, preferentially enriched for central (Tcm) and effector memory (Tem) phenotypes for CD4+ T cells (CD4+Tcm CD45RA+CCR7− and CD4+Tem CD45RA−CCR7−) (Figure S4A) and Tem phenotypes for CD8+ T cells (CD8+Temra CD45RA+CCR7−) (Figure S4B). Likewise, AIM+ T cell responses in COVID-19 vaccinees had a memory phenotype regardless of the origin of the SARS-CoV-2 sequences analyzed, with preferential enrichment for Tcm and Tem for CD4 (Figure S4C) and Tem and Temra for CD8 (Figure S4D). This provides additional evidence that donors primed by the ancestral strain S protein, by either infection or vaccination, mount a memory T cell response able to cross-recognize the SARS-CoV-2 VOCs.
Conservation analysis of sets of defined CD4+ and CD8+ T cell epitopes
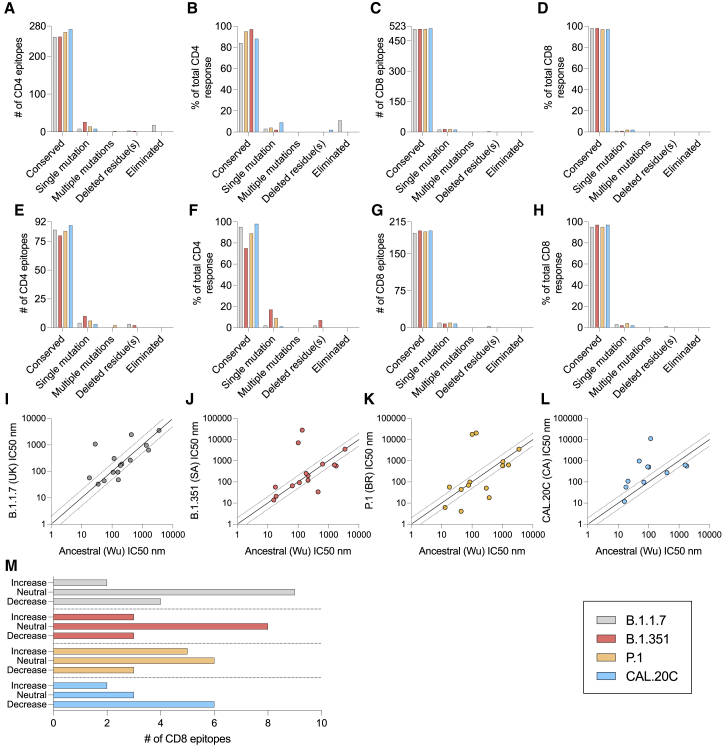
An analysis of experimentally defined CD4 and CD8 epitopes is presented in Figure 4 and examines both the number of epitopes and the experimentally determined magnitude of responses associated with the epitopes. Specifically, we recently reported a comprehensive study of epitopes recognized in convalescent subjects, leading to the identification of 280 different CD4+ T cell epitopes.51 Here, we analyzed how many of those epitopes would be affected by mutations in the different variants. As shown in Figure 4A, we found that 89.6%, 90%, 94.3%, and 97.1% (average 93%) of the CD4+ T cell epitopes are conserved in the B.1.1.7, B1.351, P.1, and CAL.20C variants. A similar pattern is observed when the magnitude of T cell responses associated with the various epitopes is considered, rather than the simple number of epitopes. Fully conserved CD4+ T cell epitopes can be inferred to account for 84.4%, 88.1%, 95.7%, and 97.8% (average 91.5%) of the recognition of the B.1.1.7, B.1.351, P.1, and CAL.20C variants, respectively, based on the ancestral sequence (Figure 4B).
Figure 4.
SARS-CoV-2 T cell epitope sequences affected by the variants
CD4+ and CD8+ T cell epitopes of the ancestral strain identified in a previous study51 were analyzed as a function of the number and percentage of responses that are or are not conserved across the B.1.1.7 (gray), B.1.351 (red), P.1 (orange), and CAL.20C (light blue) SARS-CoV-2 variants.
(A–D) The SARS-CoV-2 epitopes for the most immunodominant SARS-CoV-2 proteins in terms of numbers and percentage of response are shown for CD4+ (A and B) and CD8+ (C and D) T cells.
(E–H) The SARS-CoV-2 epitopes for the S protein only in terms of numbers and percentage of response are shown for CD4+ (E and F) and CD8+ (G and H) T cells.
(I–M) The peptide-binding algorithm data of mutated versus wild-type epitopes are shown for CD8+ (I–L) and for the effect of the mutations on CD8+ T cell epitopes, in which each instance was categorized as a function of whether the binding capability of the mutated peptide was increased (>2-fold), neutral, or decreased (<2-fold) (M).
See also Table S4.
That same study also reported 523 CD8+ T cell epitopes.51 Performing a similar analysis, 508 (97.1%) of these 523 CD8+ T cell epitopes are totally conserved within the B.1.1.7 variant, 509 (97.3%) within the B.1.351 and P.1 variants, and 512 (97.9%) within the CAL.20C variant (Figure 4C). Similarly, in terms of magnitude of the CD8+ T cell responses associated with the various epitopes, fully conserved CD8+ T cell epitopes can be inferred to account for 98.3%, 98.4%, 97.9%, and 97.8% of the recognition of the B.1.1.7, B.1.351, P.1, and CAL.20C variants, respectively, based on the ancestral sequence (Figure 4D, average of 98.1%).
Finally, we analyzed the degree of CD4+ and CD8+ T cell epitope conservation with an analysis restricted to epitopes in the S antigen. The number of S-derived epitopes conserved at 100% sequence identity was, on average, 84.5% for the CD4+ T cell epitopes (Figure 4E) and 95.3% for the CD8+ T cell epitopes (Figure 4G). Similarly, in terms of magnitude of CD4+ T cell responses to S epitopes, fully conserved CD4+ T cell epitopes can be inferred to account for 95.5%, 75.3%, 89.8%, and 98.3% of the recognition of B.1.1.7, B.1.351, P.1, and CAL.20C variants, respectively, with an average of 89.7% (Figure 4F). For CD8+ T cell responses, fully conserved epitopes can be inferred to account for 95.2%, 97.6%, 95.4%, and 97.3% of the recognition of B.1.1.7, B.1.351, P.1, and CAL.20C variants, respectively, with an average of 96.4% (Figure 4H).
While human leukocyte antigen (HLA) restriction of the class II epitopes51 could not be unequivocally assigned, the restriction of the class I epitopes was implicitly inferred based on HLA allele-specific predictions and testing in HLA-matched donors. Accordingly, we analyzed the predicted binding affinity for each epitope and matching variant for the corresponding putative HLA class I restriction element. The predicted binding affinity for each ancestral epitope/matching variant for B.1.1.7, B.1.351, P.1, and CAL.20C variants is shown in Figures 4I–4L, tabulated in Table S4, and summarized in Figure 4M. The predicted binding capacity was determined using the NetMHCpan BA 4.1 tool provided by the Immune Epitope Database’s (IEDB’s) analysis resource.54,55 In the case of the B.1.1.7, B.1.351, P.1, and CAL.20C variants, the number of mutations associated with binding capacity decreases (conservatively defined as a 2-fold reduction) was 4 out of 15, 3 out of 14, 3 out of 14, and 6 out of 11, respectively.
In conclusion, the analyses suggest that the vast majority of CD4+ and CD8+ T cell epitopes are unaffected by mutations found in all of the different variants. The corresponding mutations are also predicted to have minor effects on the total T cell response, thus providing a molecular basis for the overall impact on T cell reactivity by COVID-19 convalescent subjects and recipients of COVID-19 mRNA vaccines.
Discussion
The present study addresses a key knowledge gap pertaining to the potential of emergent SARS-CoV-2 variants to evade overall recognition by human immune responses. We focused on T cell responses elicited by either natural infection or vaccination with the Pfizer/BioNTech and Moderna COVID-19 mRNA vaccines. We found similar total CD4+ or CD8+ T cell reactivity against the four variants investigated herein, B.1.1.7, B.1.351, P.1, and CAL.20C lineages found initially in the United Kingdom, South Africa, Brazil, and California, respectively. To assess the overall total T cell functionalities, the comparison between the original Wuhan isolate and the variants was performed using different T cell methodologies, such as the AIM assay (quantifying T cells with a range of functionalities), and the FluoroSPOT assay (quantifying cells with specific cytokine-secreting activity). We also tested whether any of the variant sequences may be associated with an altered cytokine polarization.
The data herein provide some positive news in light of justified concern over the impact of SARS-CoV-2 VOCs on the efforts to control and eliminate the present pandemic. Multiples of the VOCs are associated with increased transmissibility, and several have been associated with decreased susceptibility to neutralizing antibodies from infected or vaccinated individuals. In contrast, the data presented here suggest that the total T cell responses are not significantly disrupted by the VOCs. While it is not anticipated that circulating memory T cells would be effective in preventing SARS-CoV-2 infection, it is plausible that they can contribute in reducing COVID-19 severity.33,56 Several lines of evidence support this notion, such as observations that early SARS-CoV-2 T cell responses are associated with milder COVID-19.34,35 Thus, the T cell response may contribute to limiting COVID-19 severity induced by VOCs that partially or largely escape neutralizing antibodies. This is consistent with T cell-mediated immunity observed in humans against a different respiratory pathogen, influenza, for which heterologous immunity against diverse influenza strains is associated with memory T cells to conserved epitopes.57, 58, 59
Our data also provide insights on the predicted impact of the mutations associated with the variants analyzed on T cell responses in the context of the T cell epitopes recognized. Prior reports have identified a large number of T cell epitopes recognized throughout the SARS-CoV-2 proteome, including S.40,53,60, 61, 62, 63 We furthered this point by an analysis of the Tarke et al. dataset, showing that 93% of CD4+ and 97% of CD8+ T cell epitopes in SARS-CoV-2 are completely conserved in the variants. Furthermore, we found that even in the epitopes affected by single mutations, no negative effect on the predicted HLA-binding capacity in the majority of cases is expected. Overall, it is plausible to hypothesize that single amino acid substitutions or deletions across large peptidomes do not significantly affect a polyclonal memory T cell response. Nevertheless, the methodology applied in this study is not powered to reveal whether T cells specific to amino acid sequences that are changed in variant strains still retain functionality. The apparent higher conservation of CD8+ T cell epitopes is to be expected based on the shorter length of HLA class I binding peptides (usually 9–10 amino acids) as compared to their class II counterparts (13–17 amino acids). This effect is counterbalanced by CD8+ T cells being generally less tolerant of amino substitutions as compared to CD4+ T cells.47,64 Marginal IL-5 production was detected in all of the conditions tested. This is relevant, since it was reported that single amino acid replacements in an epitope sequence can lead to a change in the cytokines produced,65,66 and a Th2-like response pattern was initially hypothesized to be linked to adverse outcomes in SARS-specific responses.67 Overall, we observed that VOC mutations do not significantly disrupt the total CD4+ and CD8+ T cell responses.
VOC mutations could be reflective of adaptation in terms of optimizing replication or binding to ACE2, but also reflective of adaptation to escape immune recognition by antibodies.13,14,22,68,69 Higher viral binding to a cellular receptor can be a mechanism of compensatory viral evolution in the presence of neutralizing antibodies.70 In that respect, while mutations to escape antibody binding have been well documented for influenza71, 72, 73 and SARS-CoV-2, immune escape at the level of T cell responses in human populations has not been reported for acute respiratory infections. Because of HLA polymorphism, the epitope repertoire recognized is likely to be substantially different from one individual to the next, greatly decreasing the likelihood of immune escape by an acute virus. An advantage conferred to the virus by a mutation in one person would not be linked to an immune response escape advantage in a non-HLA-matched individual. For SARS-CoV-2, this property of T cell recognition is further enhanced by the fact that the T cell responses against SARS-CoV-2 are highly multi-antigenic and multi-specific, with tens of different epitopes recognized by CD4+ and CD8+ T cells in a given individual.51,52,60,62 Nevertheless, our data do not rule out that certain individuals could be strongly affected by the mutations in specific VOCs.
These results here have potential implications for engineering coronavirus vaccines with broader protective immunity against VOCs. Clearly, the most straightforward path is to update the current vaccines to target a variant S, given how highly successful several COVID-19 vaccines have proven to be against the parental SARS-CoV-2 strain. Our results suggest that a parallel alternative approach could involve the inclusion of additional antigens and T cell epitopes, perhaps selected on the basis of low mutational propensity,74 to ensure that neutralizing antibodies are complemented with T cell responses to minimize COVID-19 morbidity and mortality.
Limitations of the study
The present study did not assess decreases in antibody reactivity, as several other studies have already investigated this matter.11,14, 15, 16, 17, 18, 19, 20,22,23 Our studies used overlapping peptide pools. As such, we cannot exclude that some of the mutations may involve alterations in terms of antigen processing for either class I or class II, which would be undetected by using pools of “preprocessed” peptides. While we have no reason to suspect that substantial differences may exist between the epitope specificity of responses elicited by different vaccines, our study did not address this point.
The statistical power of this study does not allow for the sensitivity needed to detect the loss of small populations of T cell clones that may be affected by variant sequences when sampled in the presence of the majority of conserved peptides. Alterations in major histocompatibility complex (MHC) binding do not necessarily confer changes in T cell receptor (TCR) affinity, as often several MHC-binding residues can be changed before changing TCR signaling, so we cannot exclude impaired T cell recognition of those specific mutated peptides. An additional limitation of the study in this respect is that one amino acid difference could influence TCR signal strength and memory T cell reactivation. However, each epitope can be recognized by multiple TCRs, and calculating the affinity would require cloning the multiple TCRs recognizing each of the epitopes seen in a polyclonal setting.
Our study was designed to test for differences in total T cell response against the different variants and was not powered or designed to investigate differences between the mRNA vaccines or at the single-peptide level. Furthermore, in our study, the sequences of the infecting virus were not determined, and therefore we cannot exclude that some of the donors may have been exposed to variants. To minimize this issue, the tested samples from convalescent donors were selected to be infected before October 2020; thus, it is unlikely that any of the donors would have been infected by any of the VOCs, as this date precedes their diffusion to an appreciable degree in the United States in general, and California in particular. While we could not clearly define the infecting strain in the COVID-19 convalescent, we showed similar results in vaccinated subjects in whom the response was induced by the ancestral S sequence. As also mentioned in the Results section, the cohorts investigated were predominantly White, reflective of the patient population available for recruitment.
Finally, we have only investigated whether the total T cell responses induced by the ancestral reference sequence are able to cross-recognize variant sequences, as this is relevant to the present situation. We have not examined whether responses induced by an infection with a variant sequence will be able to cross-recognize the ancestral reference sequence present in the approved vaccines.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| M5E2 (V500) [anti-CD14] | BD Biosciences | 561391 (RRID:AB_10611856) |
| HIB19 (V500) [anti-CD19] | BD Biosciences | 561121 (RRID:AB_10562391) |
| RPA-T4 (BV605) [anti-CD4] | BD Biosciences | 562989 (RRID:AB_2737935) |
| RPA-T8 (BV650) [anti-CD8] | BioLegend | 301042 (RRID:AB_2563505) |
| RPA-T8 (BUV496) [anti-CD8] | BD Biosciences | 612942 (RRID:AB_2870223) |
| FN50 (PE) [anti-CD69] | BD Biosciences | 555531 (RRID:AB_2737680) |
| Ber-ACT35 (PE-Cy7) [anti-OX40] | Biolegend | 350012 (RRID:AB_10901161) |
| 4B4-1 (APC) [anti-CD137] | BioLegend | 309810 (RRID:AB_830672) |
| OKT3 (AF700) [anti-CD3] | Biolegend | 317340 (RRID:AB_2563408) |
| UCHT1 (BUV805) [anti-CD3] | BD Biosciences | 612895 (RRID:AB_2870183) |
| HI100 (BV421) [anti-CD45RA] | Biolegend | 304130 (RRID:AB_10965547) |
| G043H7 (FITC) [anti-CCR7] | Biolegend | 353216 (RRID:AB_10916386) |
| Biological samples | ||
| Convalescent donor blood samples | UC San Diego Health | https://health.ucsd.edu/Pages/default.aspx |
| Convalescent donor blood samples | La Jolla Institute | https://www.lji.org |
| Convalescent donor blood samples | BioIVT | https://bioivt.com |
| COVID-19 vaccinee donor blood samples | La Jolla Institute | https://www.lji.org |
| Unexposed donor blood samples | San Diego Blood Bank | https://www.sandiegobloodbank.org |
| Unexposed donor blood samples | La Jolla Institute | https://www.lji.org |
| Unexposed donor blood samples | Carter Blood Care | https://www.carterbloodcare.org |
| Chemicals, peptides, and recombinant proteins | ||
| Synthetic peptides | TC Peptide Lab | https://tcpeptidelab.com |
| SARS-CoV-2 Receptor Binding Domain (RBD) protein | Stadlbauer et al.75 | N/A |
| Deposited data | ||
| Ancestral | https://www.ncbi.nlm.nih.gov | NC_045512.2 |
| B.1.1.7 | https://www.GISAID.org | EPI_ISL_601443 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_660629 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736930 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736932 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736944 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736971 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736966 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_736973 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_825104 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_825120 |
| B.1.351 | https://www.GISAID.org | EPI_ISL_825131 |
| P.1 | https://www.GISAID.org | EPI_ISL_804823 |
| CAL.20C | https://www.GISAID.org | EPI_ISL_847619 |
| CAL.20C | https://www.GISAID.org | EPI_ISL_847621 |
| CAL.20C | https://www.GISAID.org | EPI_ISL_847643 |
| Software and algorithms | ||
| IEDB | Vita et al.76 | https://www.iedb.org |
| IEDB-AR (analysis resource) | Reynisson et al.55 | http://tools.iedb.org/main |
| NetMHCpan BA 4.1 | Jurtz et al.77 | http://tools.iedb.org/mhci/ |
| VIGOR4 | Pickett et al.78 | https://www.viprbrc.org//brc/home.spg?decorator=vipr |
| FlowJo 10 | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism 8.4 | GraphPad | https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Alessandro Sette (alex@lji.org).
Materials availability
Aliquots of synthesized sets of peptides utilized in this study will be made available upon request. There are restrictions to the availability of the peptide reagents due to cost and limited quantity.
Data and code availability
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental materials.
Experimental model and subject details
Convalescent COVID-19 donors
Convalescent donors were enrolled at either a UC San Diego Health clinic under the approved IRB protocols of the University of California, San Diego (UCSD; 200236X), or at the La Jolla Institute (LJI; VD-214). All donors were California residents and samples were collected from May to October 2020, before any of the SARS-CoV-2 variants described herein had been detected in California. These donors were referred to the study by a health care provider or were self-referred. The CRO BioIVT provided additional cohorts of COVID-19 convalescent donors who had been confirmed positive for COVID-19 by PCR following the resolution of symptoms. The total cohort of convalescent donors represented both genders (43% male, 57% female) and ranged from 20 to 67 years of age (median 38 years). All samples were either PCR positive or seropositive against SARS-CoV-2 by ELISA, as described below. Details of this convalescent COVID-19 cohort are listed in Table 1. All convalescent COVID-19 donors provided informed consent to participate in the present and future studies at the time of enrollment.
COVID-19 vaccinees
The La Jolla Institute recruited 29 healthy adults who had received the first and second dose of the Pfizer/BioNTech BNT162b2 (n = 14) or Moderna mRNA-1273 COVID-19 vaccines (n = 15). Blood draws took place under IRB approved protocols (LJI; VD-214) two to four weeks after the second dose of the vaccine was administered. All donors had their SARS-CoV-2 antibody titers measured by ELISA, as described below. The cohort of vaccinees ranged from 23 to 67 years of age (median 47 years) and represented both genders (31% male, 69% female). At the time of enrollment in the study, all donors gave informed consent.
Unexposed donors
PBMCs from 23 healthy unexposed donors were used as controls. Healthy donors were recruited by the La Jolla Institute under IRB approved protocols (LJI; VD-214) or Carter Blood Care, where according to the blood center’s criteria for eligibility the donors were also eligible for our study. All healthy donors were collected from May 2014 to March 2018 (n = 13). Additional healthy donors were recruited from March to May 2020 by the San Diego Blood Bank (SDBB (n = 10) and were confirmed seronegative for SARS-CoV-2 by ELISA. This cohort was 35% male, 56% female, and 9% unknown and ranged from 21 to 82 years of age (median 34). All donors gave informed consent and permission for their samples to be used for future studies.
Method details
Isolation of peripheral blood mononuclear cells (PBMCs) and plasma
Collection and processing of blood samples was performed as previously described.39,51 Briefly, whole blood was collected in heparin coated blood bags or in ACD tubes and centrifuged for 15 minutes at 1850 rpm to separate the cellular fraction from the plasma. The plasma was then removed and stored at −20°C. The cellular fraction next underwent density-gradient sedimentation using Ficoll-Paque (Lymphoprep, Nycomed Pharma, Oslo, Norway) to separate the PBMCs as previously described.64 Isolated PBMCs were cryopreserved in cell recovery media containing 10% DMSO (GIBCO), supplemented with 90% heat inactivated fetal bovine serum (FBS; Hyclone Laboratories, Logan UT) and stored in liquid nitrogen until used in the assays.
SARS-CoV-2 RBD ELISA
Serology to SARS-CoV-2 was determined for all donor cohorts as previously described.34 Briefly, 96-well half-area plates (ThermoFisher 3690) were coated with 1 ug/mL SARS-CoV-2 Spike (S) Receptor Binding Domain (RBD) and incubated at 4°C overnight. The next day, plates were blocked at room temperature for 2 hours with 3% milk in phosphate buffered saline (PBS) containing 0.05% Tween-20. Heat-inactivated plasma was added to the plates for an additional 90-minute incubation at room temperature followed by incubation with the conjugated secondary antibody, detection, and subsequent data analysis by reading the plates on Spectramax Plate Reader at 450 nm using the SoftMax Pro. The limit of detection (LOD) was defined as 1:3. Limit of sensitivity (LOS) for SARS-CoV-2 infected individuals was established based on uninfected subjects, using plasma from normal healthy donors not exposed to SARS-CoV-2.
Mutation analysis of SARS-CoV-2 B.1.1.7, B.1.351, P.1 and CAL.20C variants
Genome sequences for the variant viruses were downloaded from GISAID. These sequences were screened to select those without ambiguous residues and generated from Illumina sequencing technologies using an in-house sequence QC script. The selected genomic sequences were then translated into protein amino acid sequences using the VIGOR4 tool available on the Virus Pathogen Resource (ViPR).78 Sequence variations in the variant viruses were derived by comparison with Wuhan-1 (NC_045512.2). One or more representative sequences were considered for the B.1.1.7 (EPI_ISL_601443), B.1.351 (EPI_ISL_660629; EPI_ISL_736930; EPI_ISL_736932; EPI_ISL_736944; EPI_ISL_736966; EPI_ISL_736971; EPI_ISL_736973; EPI_ISL_825104; EPI_ISL_825120; EPI_ISL_825131), P.1 (EPI_ISL_804823), and CAL.20C (EPI_ISL_847619; EPI_ISL_847621; EPI_ISL_847643) variants. A summary of all the amino acids mutated in the different variants considered in this study with respect to the Wuhan sequence and is available in Table S1.
SARS-CoV-2 Wuhan and variant peptide synthesis and pooling
Peptides were synthesized that spanned entire SARS-CoV-2 proteins and corresponded to the ancestral Wuhan sequence or the B.1.1.7, B.1.351, P.1 and CAL.20C SARS-CoV-2 variants. Peptides were 15-mers overlapping by 10 amino acids and were synthesized as crude material (TC Peptide Lab, San Diego, CA). All peptides were individually resuspended in dimethyl sulfoxide (DMSO) at a concentration of 10–20 mg/mL. Megapools (MP) for each antigen were created by pooling aliquots of these individual peptides, undergoing another lyophilization, and resuspending in DMSO at 1 mg/mL.
Bioinformatic analysis of T cell epitopes
The binding capacity of SARS-CoV-2 T cell epitopes, and their corresponding variant-derived peptides, for their putative HLA class I restricting allele(s) was determined utilizing the NetMHCpan BA 4.1 algorithm,55 as implemented by the IEDB’s analysis resource.54,76 Predicted binding is expressed in terms of IC50 nM. For each epitope-variant pair a ratio of affinities (variant IC50 nM/WT IC50 nM) was determined. Ratios > 2, indicating a 2-fold or greater decrease in affinity due to the mutation, were accordingly categorized as a decrease in binding capacity, and a ratio < 0.5 as an increase; ratios between 0.5 and 2 were designated as neutral.
Flow cytometry-based AIM assay
Activation induced cell marker (AIM) assay has previously been described in detail elsewhere.39,79,80 In summary, PBMCs were cultured in the presence of SARS-CoV-2-specific (ancestral or VOC) MPs [1 μg/ml] in 96-well U-bottom plates at a concentration of 1x106 PBMC per well. As a negative control, an equimolar amount of DMSO was used to stimulate the cells in triplicate wells and as positive controls phytohemagglutinin (PHA, Roche, 1 μg/ml) and a cytomegalovirus MP (CMV, combining CD4 and CD8 MPs, 1 μg/ml) were also included. After incubation for 20–24 hours at 37°C, 5% CO2, the cells were stained with CD3 BUV805 or CD3 AF700 (4:100 or 4:100; BD Biosciences Cat# 612895 or Life Technologies Cat# 56-0038-42, respectively), CD4 BV605 (4:100; BD Biosciences Cat# 562658), CD8 BUV496 or BV650 (2:100 or 4:100; BD Biosciences Cat# 612942 or Biolegend Cat# 301042), CD14 V500 (2:100; BD Biosciences Cat# 561391), CD19 V500 (2:100; BD Biosciences Cat# 561121), and Live/Dead eFluor506 (5:1000; eBioscience Cat# 65-0866-14). Cells were also stained to measure activation with the following markers: CD137 APC (4:100; Biolegend Cat# 309810), OX40 PE-Cy7 (2:100; Biolegend Cat#350012), and CD69 PE (10:100; BD Biosciences Cat# 555531). The memory phenotyping was analyzed by staining with CD45RA (1:100; Biolegend Cat# 304130) and CCR7 (2:100; Biolegend Cat# 353216). All samples were acquired on a ZE5 5-laser or 4-laser cell analyzer (Bio-rad laboratories) and analyzed with FlowJo software (Tree Star). In the resulting data generated from the AIM assays, the background was removed from the data by subtracting the average of the % of AIM+ cells plated in triplicate wells stimulated with DMSO. The Stimulation Index (SI) was calculated by dividing the % of AIM+ cells after SARS-CoV-2 stimulation with the average % of AIM+ cells in the negative DMSO control. An SI greater than 2 and a minimum of 0.016% or 0.16% AIM+ CD4+ or CD8+ cells, respectively, after background subtraction was considered to be a positive response based on the median twofold standard deviation of all negative controls measured in the study (> 80), as previously reported.39 The gates for AIM+ cells were drawn relative to the negative and positive controls for each donor. A representative example of the gating strategy is depicted in Figure S5. Specifically, lymphocytes were gated, followed by single cells determination. T cells were considered to be positive for CD3 and negative for a Dump channel including in the same colors CD14, CD19 and Live/Dead staining. The CD3+CD4+ and CD3+CD8+ were further gated based on OX40+CD137+ and CD69+CD137+ AIM markers, respectively. Memory phenotype was gated based on the expression of CD45RA and CCR7 markers on the CD3+CD4+ and CD3+CD8+ T cells and the AIM+ T cells.
FluoroSPOT assay
96-well FluoroSpot plates were coated with anti-cytokine antibodies for IFNγ and IL-5 (mAbs 1-D1K and TRFK5, respectively; Mabtech, Stockholm, Sweden) at a concentration of 10 μg/mL. PBMCs were stimulated in triplicate at a density of 200x103 cells/well with S MPs corresponding to each of the SARS-CoV-2 variants analyzed (1 μg/mL), PHA (1 μg/mL), and DMSO (0.1%), as positive and negative controls respectively. After 20 hours of incubation at 37°C, 5% CO2, cells were discarded and plates were washed before the addition of cytokine antibodies (mAbs 7-B6-1-BAM and 5A10-WASP; Mabtech, Stockholm, Sweden). After a 2-hour incubation, plates were washed again with PBS/0.05% Tween20 and incubated for 1 hour with fluorophore-conjugated antibodies (Anti-BAM-490 and Anti-WASP-640). An AID iSPOT FluoroSpot reader (AIS-diagnostika, Germany) was used to count the fluorescent spots that resulted from cells secreting IFNγ and IL-5. Each peptide MP was considered positive compared to the DMSO negative control based on the following criteria: 20 or more spot forming cells (SFC) per 106 PBMC after background subtraction, a stimulation index (S.I.) greater than 2, and a p value < 0.05 by either a Poisson or t test calculated between the triplicates of the MP and the relative negative control.
Quantification and statistical analysis
Data and statistical analyses were performed in FlowJo 10 and GraphPad Prism 8.4, unless otherwise stated. Statistical details of the experiments are provided in the respective figure legends and in each methods section pertaining to the specific technique applied. Data plotted in logarithmic scales are expressed as geometric mean. Statistical analyses were performed using Wilcoxon matched-pairs signed rank test for paired comparisons. Multi-hypothesis testing corrections (MHTC) have not been applied in the study by design. The study is not designed or powered to address differences across different proteins. The primary hypothesis is that no significant differences are observed across the different variants, and this is more stringently addressed avoiding to correct for MHTC, since a difference that is not significant would remain so even after corrections. Therefore, reporting the data without applying MHTC is a more stringent criterion which is appropriately being applied in this case to avoid false negatives. Details pertaining to significance are also noted in the respective figure legends.
To determine the expected variability of the AIM assays, we also analyzed 23 donors for which CD4+ and CD8+ T cell responses to Spike, M and N megapools were evaluated in up to 4 repeat measurements. A total of 208 measurements were made for these donors for both CD4 and CD8, resulting in 69 independent assessments of repeat measures (comparing the initial measurement to a repeat in the same donor) (Table S3). We evaluated the variability in these repeat measures using a corrected log2 fold-change calculated as:
where LOD is the level of detection threshold, which is 0.014% for CD4 and 0.06% for CD8 based on the twofold geometric 95%CI of all negative controls measured (> 80). The addition of the LOD ensures that variations between measurements close to- or below the level of detection do not result in inflated fold-change estimates. We evaluated the standard deviation of these corrected log-fold change values of repeat measurements (at an individual level), and used this value as a threshold for what would constitute a significant, non-random change at the population level. Similarly, FluoroSPOT assay variability was evaluated by analyzing 38 donors and a total of 301 measurements considering an LOD of 20 SFC/106 cells. For CD4-AIM assays, that corresponded to a corrected fold-change of 2.3, for CD8-AIM of 2.4 and for FluoroSPOT of 1.6 as shown in Table S3. Next, we performed a test of non-inferiority for the observed AIM assay or FluoroSPOT reactivities for any of the variants tested compared to the ancestral strain. A non-inferiority test determines if the observed values are significantly above what would be considered a biological meaningful reduction in responses, based on one sample Wilcoxon Signed Rank test compared to the lower bound fold change threshold. The test results in a p value expressing effectiveness of the comparison and a p value corresponding to the discrepancy. Confidence intervals for discrepancy are shown in all the graphs where the comparison was considered statistically significant (p < 0.05). The non-inferiority test found that all CD4 and CD8 responses or IFNγ responses to any of the variant peptide pools were significantly above the lower bound threshold. Therefore, by this approach at the population level, there is no biological meaningful reduction in the T cell response to the variant pools compared to the ancestral strain. Details pertaining to significance are also noted in the respective figure legends.
Acknowledgments
This study has been funded by the NIH NIAID (SARS-CoV-2 Assessment of Viral Evolution program, award no. AI142742 to S.C. and A. Sette, contract no. 75N9301900065 to A. Sette and D.W., contract no. 75N93019C00001 to A. Sette and B.P., NIH grant U01 CA260541-01 to D.W., K08 award AI135078 to J.M.D., AI036214 to D.M.S., and HHSN75N93019C00076 to R.H.S.). Additional support has been provided by UCSD T32s (AI007036 and AI007384 to S.A.R. and S.I.R.) and the Jonathan and Mary Tu Foundation (to D.M.S.). A.T. was supported by a PhD student fellowship through the Clinical and Experimental Immunology Course at the University of Genoa, Italy. We thank Gina Levi and the LJI clinical core for assistance in sample coordination and blood processing. We gratefully thank the authors from the originating laboratories responsible for obtaining the specimens, as well as the submitting laboratories where the genome data were generated and shared via GISAID and on which this research is based. We would like to thank Vamseedhar Rayaprolu and Erica Ollmann Saphire for providing the recombinant SARS-CoV-2 RBD protein used in the ELISA assay.
Author contributions
Conceptualization, A.T., A.G., S.C., and A. Sette.; data curation and bioinformatic analysis, Y.Z., R.H.S., and B.P.; formal analysis, A.T., J.S., and A.G.; funding acquisition, S.C., A.S., D.W., S.I.R., S.A.R., and J.M.D.; investigation, A.T., N.M., A. Sutherland, B.G., J.S., E.D.Y., E.W., D.W., A. Sette, and A.G.; project administration, A.F.; resources, S.I.R., S.A.R., and J.M.D.; supervision, J.S., S.C., D.W., A. Sette, R.d.S.A., and A.G.; writing, A.T., D.W., S.C., A. Sette, and A.G.
Declaration of interests
A. Sette is a consultant for Gritstone, Flow Pharma, CellCarta, Arcturus, Oxfordimmunotech, and Avalia. S.C. is a consultant for Avalia. All of the other authors declare no competing interests. LJI has filed for patent protection for various aspects of vaccine design and identification of specific epitopes.
Published: July 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100355.
Contributor Information
Alba Grifoni, Email: agrifoni@lji.org.
Alessandro Sette, Email: alex@lji.org.
Supplemental information
References
- 1.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., COVID-19 Genomics UK (COG-UK) consortium Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 3.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., CMMID COVID-19 Working Group. COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 5.Voloch C.M., da Silva Francisco R., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.C., Mariani D., da Costa R.M., Ferreira O.C., Jr., Covid19-UFRJ Workgroup, LNCC Workgroup, Adriana Cony Cavalcanti Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95 doi: 10.1128/JVI.00119-21. e00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021 doi: 10.1016/j.cell.2021.04.025. 2021.2003.2007.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., COVID-19 Genomics Consortium UK (CoG-UK . 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- 10.Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Barrett K.M.S., Larsen B.B., Anderson C., White S., Cassens T. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv. 2021 doi: 10.1101/2021.2002.2006.21251159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahradník J., Marciano S., Shemesh M., Zoler E., Chiaravalli J., Meyer B., Dym O., Elad N., Schreiber G. SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor. bioRxiv. 2021 doi: 10.1101/2021.2001.2006.425392. [DOI] [Google Scholar]
- 14.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 15.Skelly, D.T., Harding, A.C., Gilbert-Jaramillo, J., Knight, M.L., Longet, S., Brown, A., Adele, S., Adland, E., and Brown, H., et al.; Medawar Laboratory Team (2021). Vaccine-induced immunity provides more robust heterotypic immunity than natural infection to emerging SARS-CoV-2 variants of concern. Research Square, 10.21203/rs.3.rs-226857/v2.
- 16.Wu K., Werner A.P., Koch M., Choi A., Narayanan E., Stewart-Jones G.B.E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edara V.V., Norwood C., Floyd K., Lai L., Davis-Gardner M.E., Hudson W.H., Mantus G., Nyhoff L.E., Adelman M.W., Fineman R. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 2021;29:516–521.e3. doi: 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B.F., Sanders K.O. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29:529–539.e3. doi: 10.1016/j.chom.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 24.Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., COVID-19 Genomics UK Consortium. AMPHEUS Project. Oxford COVID-19 Vaccine Trial Group Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cele S., Gazy I., Jackson L., Hwa S.H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall V.J., Foulkes S., Saei A., Andrews N., Oguti B., Charlett A., Wellington E., Stowe J., Gillson N., Atti A. Effectiveness of BNT162b2 mRNA Vaccine Against Infection and COVID-19 Vaccine Coverage in Healthcare Workers in England, Multicentre Prospective Cohort Study (the SIREN Study) Lancet. 2021;397:1725–1735. doi: 10.1016/S0140-6736(21)00790-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amit S., Regev-Yochay G., Afek A., Kreiss Y., Leshem E. Early rate reductions of SARS-CoV-2 infection and COVID-19 in BNT162b2 vaccine recipients. Lancet. 2021;397:875–877. doi: 10.1016/S0140-6736(21)00448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novavax Inc . 2021. Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial.https://ir.novavax.com/2021-01-28-Novavax-COVID-19-Vaccine-Demonstrates-89-3-Efficacy-in-UK-Phase-3-Trial [Google Scholar]
- 31.US Food and Drug Administration . 2021. Vaccines and Related Biological Products Advisory Committee February 26, 2021. Sponsor Briefing Document Addendum.https://www.fda.gov/media/146218/download [Google Scholar]
- 32.US Food and Drug Administration . 2021. Vaccines and Related Biological Products Advisory Committee February 26, 2021 FDA Briefing Document—Addendum--Sponsor.https://www.fda.gov/media/146218/download [Google Scholar]
- 33.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Foca E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker D., Roberts C.A.K., Pryce G., Kang A.S., Marta M., Reyes S., Schmierer K., Giovannoni G., Amor S. COVID-19 vaccine-readiness for anti-CD20-depleting therapy in autoimmune diseases. Clin. Exp. Immunol. 2020;202:149–161. doi: 10.1111/cei.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breton G., Mendoza P., Hägglöf T., Oliveira T.Y., Schaefer-Babajew D., Gaebler C., Turroja M., Hurley A., Caskey M., Nussenzweig M.C. Persistent cellular immunity to SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202515. doi: 10.1084/jem.20202515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dowd S.M., Zalta A.K., Burgess H.J., Adkins E.C., Valdespino-Hayden Z., Pollack M.H. Double-blind randomized controlled study of the efficacy, safety and tolerability of eszopiclone vs placebo for the treatment of patients with post-traumatic stress disorder and insomnia. World J. Psychiatry. 2020;10:21–28. doi: 10.5498/wjp.v10.i3.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redd A.D., Nardin A., Kared H., Bloch E.M., Pekosz A., Laeyendecker O., Abel B., Fehlings M., Quinn T.C., Tobian A.A.R. CD8+ T cell responses in COVID-19 convalescent individuals target conserved epitopes from multiple prominent SARS-CoV-2 circulating variants. medRxiv. 2021 doi: 10.1101/2021.02.11.21251585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo J., Dowell A.C., Pearce H., Verma K., Long H.M., Begum J., Aiano F., Amin-Chowdhury Z., Hoschler K., Brooks T. Robust SARS-CoV-2-specific T cell immunity is maintained at 6 months following primary infection. Nat. Immunol. 2021;22:620–626. doi: 10.1038/s41590-021-00902-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218:e20202617. doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 53.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 54.Dhanda S.K., Mahajan S., Paul S., Yan Z., Kim H., Jespersen M.C., Jurtz V., Andreatta M., Greenbaum J.A., Marcatili P. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res. 2019;47(W1):W502–W506. doi: 10.1093/nar/gkz452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenbaum J.A., Kotturi M.F., Kim Y., Oseroff C., Vaughan K., Salimi N., Vita R., Ponomarenko J., Scheuermann R.H., Sette A., Peters B. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. USA. 2009;106:20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., Bean T., Barclay W., Deeks J.J., Lalvani A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013;19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson T.M., Li C.K., Chui C.S., Huang A.K., Perkins M., Liebner J.C., Lambkin-Williams R., Gilbert A., Oxford J., Nicholas B. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 60.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., Noakes M.N. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv. 2020 doi: 10.1101/2020.07.31.20165647. [DOI] [Google Scholar]
- 62.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lübke M., Bauer J., Rieth J., Wacker M. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 63.Keller M.D., Harris K.M., Jensen-Wachspress M.A., Kankate V.V., Lang H., Lazarski C.A., Durkee-Shock J., Lee P.H., Chaudhry K., Webber K. SARS-CoV-2-specific T cells are rapidly expanded for therapeutic use and target conserved regions of the membrane protein. Blood. 2020;136:2905–2917. doi: 10.1182/blood.2020008488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiskopf D., Angelo M.A., Sidney J., Peters B., Shresta S., Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J. Virol. 2014;88:11383–11394. doi: 10.1128/JVI.01108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evavold B.D., Allen P.M. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- 66.Sloan-Lancaster J., Allen P.M. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 67.Peeples L. News Feature: avoiding pitfalls in the pursuit of a COVID-19 vaccine. Proc. Natl. Acad. Sci. USA. 2020;117:8218–8221. doi: 10.1073/pnas.2005456117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A. SARS-CoV-2 escape < em > in vitro < /em > from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hensley S.E., Das S.R., Bailey A.L., Schmidt L.M., Hickman H.D., Jayaraman A., Viswanathan K., Raman R., Sasisekharan R., Bennink J.R., Yewdell J.W. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doud M.B., Lee J.M., Bloom J.D. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 2018;9:1386. doi: 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andrews S.F., Huang Y., Kaur K., Popova L.I., Ho I.Y., Pauli N.T., Henry Dunand C.J., Taylor W.M., Lim S., Huang M. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krammer F., García-Sastre A., Palese P. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Potential Target Antigens and Critical Aspects for a Universal Influenza Vaccine. Cold Spring Harb. Perspect. Biol. 2018;10:a028845. doi: 10.1101/cshperspect.a028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaiha G.D., Rossin E.J., Urbach J., Landeros C., Collins D.R., Nwonu C., Muzhingi I., Anahtar M.N., Waring O.M., Piechocka-Trocha A. Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science. 2019;364:480–484. doi: 10.1126/science.aav5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr Protoc Microbiol. 2020;57 doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R., Wheeler D.K., Sette A., Peters B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.P., Havenar-Daughton C. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.da Silva Antunes R., Pallikkuth S., Williams E., Esther D.Y., Mateus J., Quiambao L., Wang E., Rawlings S.A., Stadlbauer D., Jiang K. Differential T cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J. Infect. Dis. 2021:jiab176. doi: 10.1093/infdis/jiab176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data generated or analyzed during this study, and summarized in the accompanying tables, figures and supplemental materials.