Abstract
The rapid detection of novel pathogens including SARS-CoV-2 necessitates the development of easy-to-use diagnostic tests that can be readily adapted and utilized in both clinical laboratories and field settings. Delay in diagnosis has facilitated the rapid spread of this novel virus throughout the world resulting in global mortality that will surpass 2.5 million people. Development of point-of-care diagnostic assays that can be performed in rural or decentralized health care centers to expand testing capacity is needed. We developed a qualitative test based on recombinase-polymerase-amplification coupled with lateral flow reading (RPA-LF) for rapid detection of SARS-CoV-2. The RPA-LF detected SARS-CoV-2 with a limit of detection of 35.4 viral cDNA nucleocapsid (N) gene copies/μL. Additionally, the RPA-LF was able to detect 0.25–2.5 copies/μL of SARS-CoV-2 N gene containing plasmid. We evaluated 37 nasopharyngeal samples using CDC’s N3, N1 and N2 RT-real-time PCR assays for SARS-CoV-2 as reference test. We found a 100 % concordance between RPA-LF and RT-qPCR reference test as determined by 18/18 positive and 19/19 negative samples. All positive samples had Ct values between 19–37 by RT-qPCR. The RPA-LF primers and probe did not cross react with other relevant betacoronaviruses such as SARS and MERS. This is the first isothermal amplification test paired with lateral flow developed for qualitative detection of COVID-19 allowing rapid viral detection and with prospective applicability in resource limited and decentralized laboratories.
Keywords: Diagnostics, SARS-CoV-2, Lateral flow, RPA, Point-of-care, Coronavirus
1. Introduction
The development of accurate, rapid, and sensitive assays to detect novel pathogens has never been more important. The current pandemic acutely illustrates this need. The novel betacoronavirus, SARS-CoV-2, was identified as the etiological agent of an unknown pneumonia cluster occurring in Wuhan City, Hubei Province, China in early December 2019 (World Health Organization, 2020). The virus quickly spread throughout the world causing country-wide lockdowns and economic turmoil as countries struggled to develop testing protocols and treatments. At the time of this publication there has been 111.1 million confirmed cases and 2.5 million deaths globally with nearly 28 million cases and 500,000 deaths in the United States (World Health Organization, 2021). The U.S.’s response to the pandemic has lagged due to problems with reliable testing and insufficient production making up 25.2 % of the global infections and 20 % of the global deaths (Moore et al., 2020; Nalla et al., 2020).
Development of easy to use and relatively rapid detection assays are imperative to flatten the curve and return to “normal.” One such assay is isothermal recombinase-polymerase-amplification (RPA). Molecular tests based on isothermal RPA coupled with later flow reading (RPA-LF) allow for detection of pathogen nucleic acids with similar sensitivities as qualitative real-time PCR but without the need of expensive equipment (Saldarriaga et al., 2016; Castellanos-Gonzalez et al., 2015). To enable detection by lateral flow in the RPA-LF test, the reverse primer is biotinylated at the 5′ end and there is an internal fluorophore-labeled probe included. A positive test band is produced through an antibody-mediated interaction with amplification products. As the amplified DNA containing fluorophore labeled probe flow up the strip, it interacts with mouse anti-fluorophore antibody-labeled gold particles present in the strip. The products that contain the biotinylated reverse primer are immobilized by the anti-biotin antibodies on the LF strip, making the test line visible. Only amplified products containing both, biotinylated primers and fluorophore are visualized. The reaction is validated by the appearance of the control band in the upper part of the strip. This band appears upon the immobilization of excess free-gold particles (which are covered with mouse antibodies) by means of anti-mouse antibodies. To date, the RPA-LF assays for cutaneous and visceral leishmaniasis (Saldarriaga et al., 2016; Castellanos-Gonzalez et al., 2015), intestinal protozoa (Cryptosporidium and Giardia) (Crannell et al., 2016), Entamoeba histolytica (Nair et al., 2015) and Trypanosoma cruzi (Jimenez-Coello et al., 2018), have shown similar sensitivity and specificity as the respective real-time or conventional PCR assays.
In this study, we report the development of a two-step, RPA-LF assay that has similar sensitivity and specificity as the CDC 2019-nCoV rRT-PCR Panel (Control CoD, 2020; Lu et al., 2020). This assay does not require expensive real-time PCR equipment and would allow for testing of suspected COVID19 samples at rural or decentralized laboratories, with the capability to extract RNA and convert it to cDNA, reducing the time to diagnosis and potential treatment.
2. Results
2.1. Assay design
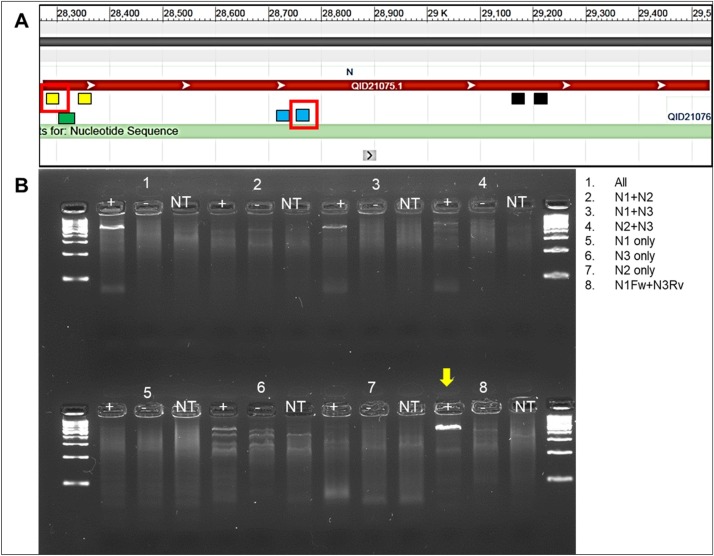
The nucleoprotein is one of the more conserved viral genes of SARS-CoV-2 according to the sequences currently available. The CDC 2019-nCoV rRT-PCR Panel was already available at the onset of our research. Consequently, we chose to assess the feasibility of using these primer sets under isothermal conditions. We ran the primers in mismatched forward and reversed pairs (i.e. N1 Fw with N3 RV, etc.) and in combinations (i.e. N1 pair plus N2 pair, etc.) to determine the best pairing that would work under our assay parameters (Fig. 1 A and B). Only combinations that contained N1FW and N3RV produced a significant band in the agarose gel under isothermal conditions (Fig. 1B). We did a blast search (NCBI) to ensure that this primer pairing would only target known N gene sequences of SARS-CoV-2 and would not cross-react with other betacoronaviruses. Only SARS-CoV-2 sequences returned in our searches.
Fig. 1.
Gene position and isothermal reaction results. A) Nucleotide schematic for N gene of SARS-CoV-2 with primer pair indicating relative positions of CDC N1-3. Selected primer pair for RPA-LF (N1 Fw and N3 Rv) are highlighted in red boxes. Green square depicts the position of RPA-LF probe. B) RPA-basic assessment of primer combinations at 40 °C.
2.2. SARS-CoV-2 RPA-LF analytical specificity
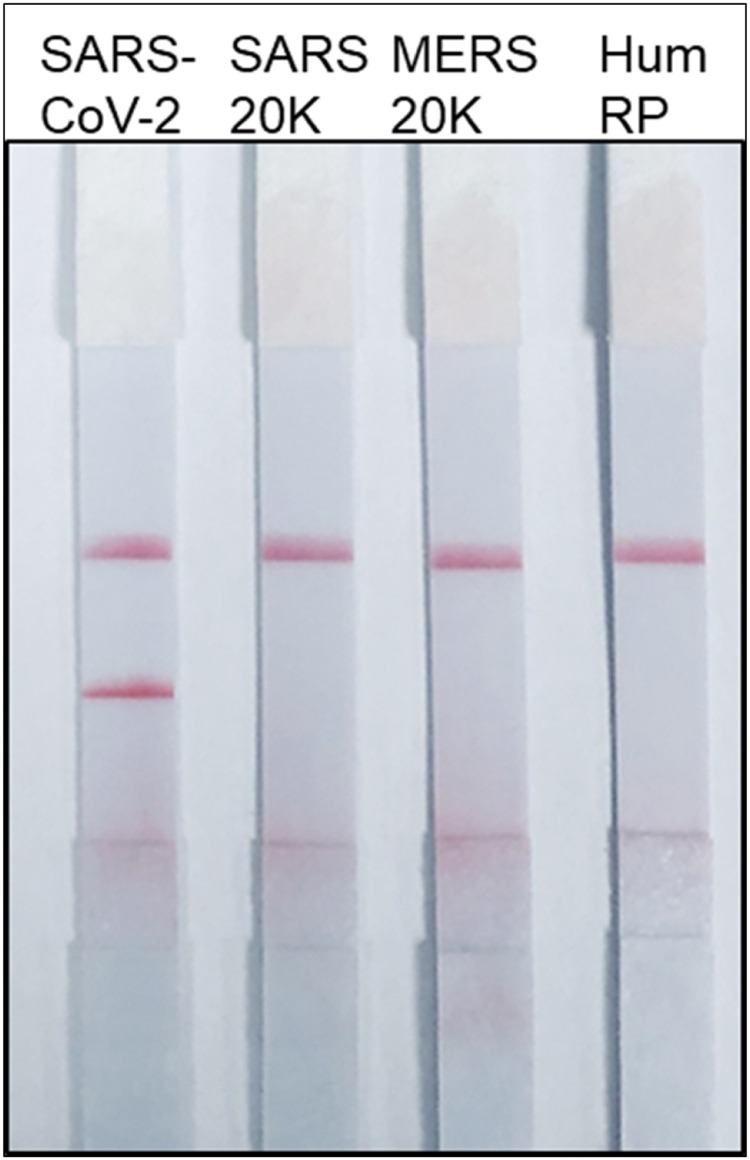
The specificity of the SARS-CoV-2 RPA-LF was tested against plasmids of the related betacoronaviruses, SARS and MERS. We confirmed that lack of RPA-LF cross-reactivity using ∼20,000 plasmid copies/reaction of SARS or MERS. The RPA-LF test was also negative for the human RNAse polymerase, indicating no cross reactivity with human DNA (n = 9 repetitions each, XLSTAT contingency analysis p < 0.0001) (Fig. 2 ).
Fig. 2.
Specificity of SARS-CoV-2 RPA-LF. Selected primer pair for RPA-LF (N1 Fw and N3 Rv) and N1 RPA-LF probe were assessed at 42 °C for 40 min. No cross-reactivity was observed using 20,000 genome copies/reactionof SARS or MERS.
2.3. Analytical sensitivity
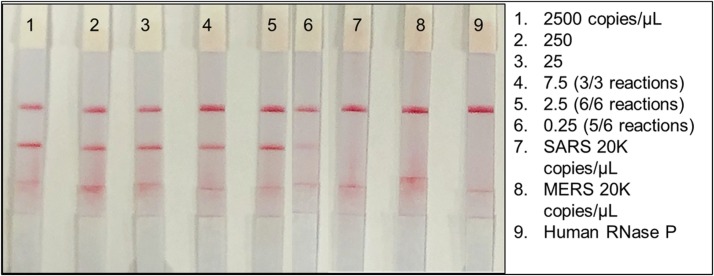
We initially assessed the limit of detection (LOD) of the RPA-LF using a plasmid containing the N gene, allowing the direct quantification of copies/μL/reaction. Known concentrations of SARS-CoV-2 plasmid DNA were serially diluted in water containing 5,000 copies/μL of Human RNase P (HRP) gene plasmid until it was no longer possible to detect bands in the RPA-LF test. The inclusion of HRP plasmid in each sample was to mimic clinical samples and to determine if excess of host DNA would interfere with viral gene detection. The assay, which included the (HRP) gene no virus control, was able to repeatedly detect diluted SARS-CoV-2 plasmid over a range of 2,500−0.25 copies/μL (Fig. 3 ), with 0.25 copies/μL being detected in 5 of 6 reactions. (p = 0.000197, indicating positive results are true). The assay was less consistent when evaluating dilutions equivalent to 0.125 copies/μL (n = 2/3 assays). Band intensity in the lateral flow strips was diminished but evident at this concentration (Fig. 3, Fig. 4).
Fig. 3.
Limit of detection using SARS-CoV-2 plasmid containing the nucleocapsid gene (copies/μL). Ten-fold serial dilutions of N gene plasmid compared to SARS and MERS plasmids that were used to test cross-reactivity of the RPA-LF and human RNase P plasmid (Hum RP) as the no virus control.
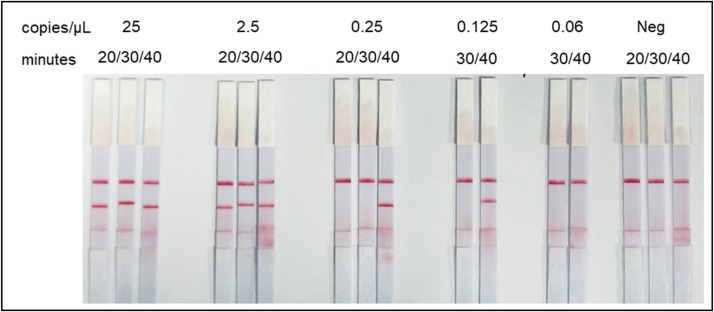
Fig. 4.
Sensitivity of RPA-LF according to the incubation time at 42 °C. The assay has a limit of detection equivalent to 0.25 copies/μL of SARS-coV-2 since it was able to consistently amplify this cDNA concentration (5/6 total repetitions). Concentrations of 0.125 copies/μL yielded positive reactions in 2/3 repetitions.
In addition to the limit of detection (LOD), we assessed the length of time and incubation temperature that more consistently delivered positive results. Using a range of concentrations, we determined the duration of incubation necessary to detect the lowest concentrations. We found that more concentrated samples could be detected as early as ten minutes post incubation (2,500 and 250 copies/μL data not shown), while lower concentrations required longer incubations, 20−40 min (Fig. 4 ).
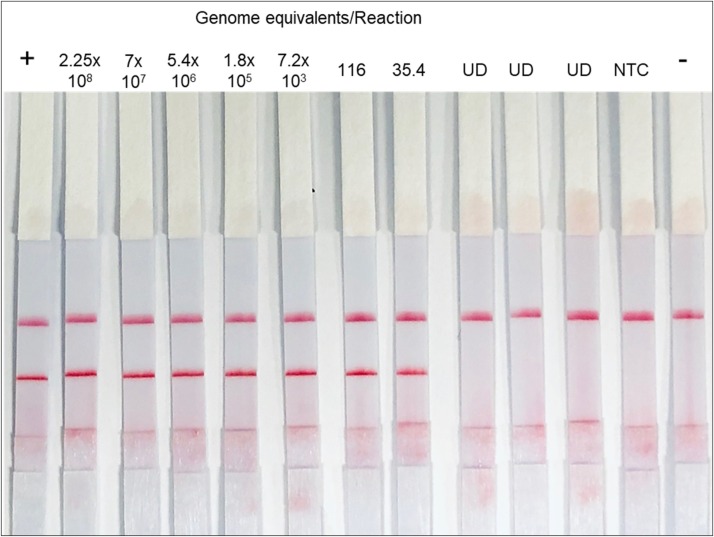
In addition to the N gene plasmid, we further tested the sensitivity of the RPA-LF assay by serially diluting viral RNA from cultured SARS-CoV-2provided by WRCEVA. The RNA was converted to cDNA and used in three replicate assays. Viral genome concentration was determined by the CDC’s nucleocapsid rRT-PCR for COVID-19 (Control CoD, 2020; Lu et al., 2020) using the 10-fold serial dilutions of known concentrations of control plasmids. The LOD of RPA-LF was 35.4 genome equivalents/reaction (n = 3) while all negative samples resulted negative (n = 6) (probit analysis, p = 0.0009) (Fig. 5 ); those samples dilutions that had CT values greater than 37 were labeled undetermined (UD). Therefore, in this limited number of samples the LOD of the RPA-LF was 100 % concordant with the rRT-PCR.
Fig. 5.
Genome equivalent limit of detection for SARS-CoV-2 RPA-LF. Concentration of genome equivalent was determined by standard curve of SARS-CoV-2 plasmid real-time PCR using the CDC N1 primer set. N1Fw and N3Rv were only able to detect 7.2 × 103 genome equivalents per reaction by real-time PCR. Values are for 2.5 μL of input cDNA. (UD, undetermined; NTC, no template control; +, positive control; -, no virus control/Human Rnase P).
2.4. Diagnostic efficacy of SARS-CoV-2 RPA-LF
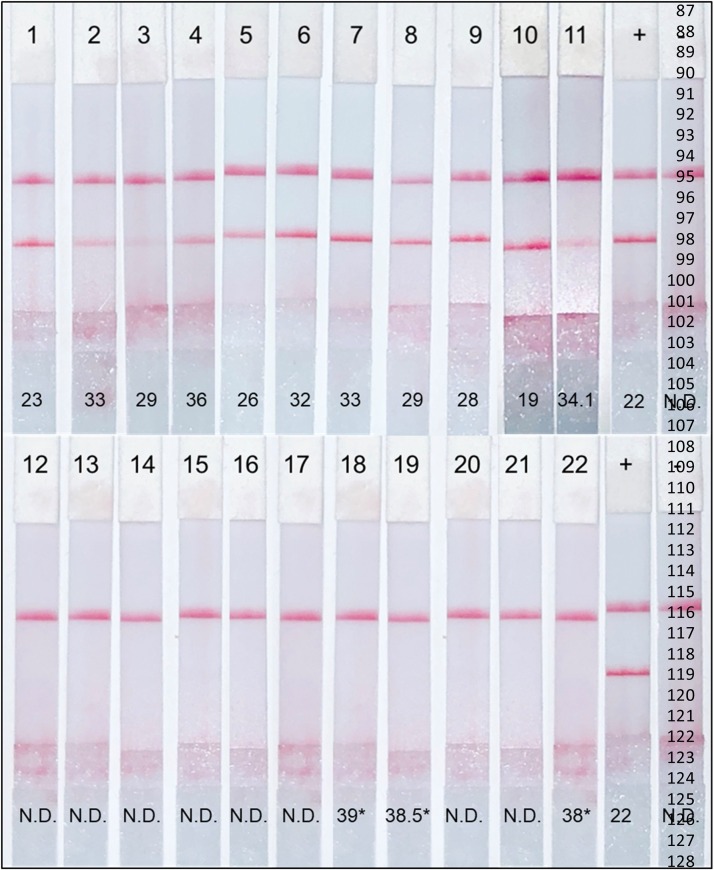
We evaluated the assay using 37 nasopharyngeal samples previously screened for COVID 19 by the University of Texas Medical Branch clinical laboratory. We extracted RNA from 100 μL of heat inactivated transport media and converted to cDNA or tested the samples again by rRT-PCR. RPA-LF detected 18 COVID-19 positive- and 19 negative samples. All positive tests had definitive bands in the lateral flow strips with varying intensities compared with the positive control (11 positive representative tests, Fig. 6 1-11), while the test band was absent in the negative samples (11 shown, Fig. 6 12-22). The results of the SARS-CoV-2 RPA-LF were 100 % concordant the CDC’s nucleocapsid rRT-PCR for COVID-19 (Control CoD, 2020; Lu et al., 2020) with Ct values ranging from 19 to 37 (Fig. 6, values).
Fig. 6.
Clinical efficacy of SARS-CoV-2 RPA-LF. Representative lateral flow strips of COVID-19 positive samples by RPA-LF, 1-11 and COVID-19 negative, 12-22. Numerical values represent corresponding Ct values for each sample; cut-off for positive samples is 37.5. (N.D., not detected; *, values greater than 37.5 are N.D.).
3. Discussion
Simplification of point-of-care diagnostics can decrease the time to diagnosis for clinics with limited equipment distant from urban settings. Available diagnostic methods for the detection of SARS-CoV-2 RNA focus on the use of real-time PCR which require technical expertise and expensive equipment. These requirements force small or rural clinics to ship suspected COVID-19 samples to centralized laboratories to be processed, leading to delays in treatment as well as public health interventions such as patient isolation and contact tracing. Additional issues arise with shipping the samples to centralized laboratories. Li et al. demonstrated that prolonged inactivation times and storage conditions, i.e. transit to testing laboratories, can decrease sensitivity of widely used RT real-time PCRs for SARS-CoV-2 leading to at least 10.2 % false negative results (Li et al., 2020). This data supports the need for the development of simplified assays, not to replace existing assays but to complement them, which can be done on site with minimal equipment and expertise.
In the current study, we demonstrated the development of a rapid diagnostic test for the detection of SARS-CoV-2. In its current state, the assay is a two-step reaction, requiring a separate cDNA synthesis step prior to the RPA reaction and lateral flow readout. The SARS-CoV-2 RPA-LF had 100 % specificity and was 100 % concordant with the CDC’s nucleocapsid rRT-PCR for COVID-19 (Control CoD, 2020; Lu et al., 2020) for 37 nasopharyngeal samples tested. We were able to determine the LOD of this assay using both N gene plasmid and whole viral RNA. The assay was able to detect between 0.125−0.25 plasmid copies/μL and 35.4 genome equivalents/reaction. The LOD for viral genome detection was identical to the CDC’s nucleocapsid rRT-PCR for COVID-19 (Fig. 5). As the pandemic persists, new variants have emerged with mutations in the viral spike gene (Plante et al., 2021). As our assay uses primers that target the N gene, these variants should be detected by RPA-LF. To verify this, BLASTn analysis using our primers detected all currently published sequences of SARS-CoV-2.
Several rapid and laboratory-based antibody detection assays were developed to determine if individuals have been exposed to the virus or if they are currently infected utilizing antigen capture LFA or ELISA. However, none of these methods have reached the sensitivity or wide spread use of real-time PCR (Yüce et al., 2021). The antigen capture assay has a wide range of sensitivity (0–94 %, average 56.2 %) depending on the subjects’ course of infection and the source of antigen (Dinnes et al., 2020). Recent LAMP assays have focused on minimalizing sample processing but have shown mixed results. Direct testing of saliva without any processing resulted in 47 % positivity compared to 97 % samples with minimal processing (Fowler et al., 2021; Howson et al., 2021; Taki et al., 2021). We are currently working on protocols that shorten the RNA extraction process, further reducing the need for specialized equipment or reagents as well as converting the assay into a one-step, reverse transcriptase RPA-LF. These optimizations will convert SARS-CoV-2 RPA-LF into a true POC test that can be used in rural clinics or decentralized laboratories allowing for rapid diagnosis, treatment, and isolation of infectious individuals to reduce transmission.
4. Materials and methods
4.1. SARS-CoV-2
Viral RNA used in this study was provided by the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at UTMB. SARS-CoV-2 strain USA_WA1/2020 was provided at a reported concentration of 6 × 104 PFU/μl or approximately 6 × 107 genomic equivalents/μl of extracted RNA.
4.2. Patient samples
De-identified samples that were suspected to contain SARS-CoV-2 genomic material were collected at UTMB for further laboratory analysis under UTMB IRB protocol #95-111. Briefly, nasopharyngeal swabs were collected and placed in transport media per clinic protocol for COVID screening. The transport media potentially containing active virus was heat inactivated at 95 °C for 65 min by the UTMB clinical laboratories prior to processing for RNA extraction and cDNA synthesis as described as below.
4.3. Human sample Ethics statement
All data acquisition and experimental procedures were performed in accordance with relevant guidelines and regulations. All clinical samples were obtained as de-identified patient samples and considered medical waste under protocol #95-111. This protocol was approved by the Institutional Review Board (IRB) of the University of Texas Medical Branch at Galveston, TX. All procedures performed within have been approved by the Institutional Biosafety Committee (IBC) under Notification of Use (NOU) #2020113.
4.4. Viral RNA isolation and cDNA synthesis
The virus was inactivated using Trizol followed by modified chloroform separation and RNA isolation using the Qiagen RNeasy Mini Kit (Qiagen). The cDNA synthesis was carried out using the iScript Select cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s protocol. For cDNA synthesis we used 10 μL of RNA and 3 μL of random primers (Bio-Rad) as per the manufacturer instructions.
4.5. Quantitative reverse transcriptase Real time PCR for SARS-CoV-2
For detection of clinical samples and quantification viral RNA (provided by WRECEVA) we used the CDC’s nucleocapsid (N1, N2, and N3 primers as described) rRT-PCR for COVID-19 (Control CoD, 2020; Lu et al., 2020). To determine the limit of detection by RPA-LF, we performed 10-fold serial dilutions of the N gene plasmid provided in the kit and compared it to serially diluted cDNA transcribed from SARS-CoV-2 RNA.
4.6. SARS-CoV-2 primer and probe design and position
Primers were selected from the commercially available CDC real-time Rt PCR for detection 2019-Novel Coronavirus (Control CoD, 2020; Lu et al., 2020). The assay consists of three primer pairs targeting the nucleocapsid gene (Fig. 1A). We used the primers in multiple combinations at 40 °C (Fig. 1B) to determine the pair that would work the best under isothermal conditions. Once we identified which primer pair functioned more efficiently under these conditions (N1FW and N3RV), several probes were designed based on the assay’s existing probes. The modifications of the RPA-LF probes required at least 50 base pairs and include FAM (5′-carboxy fluorescein amidite) at the 5′ end, an internal dSpacer that acts as an exonuclease cut-site, and a C3-Spacer at the 3′ end, as suggested by the manufacturer (TwistDx, UK). Results are shown for the CDC N1 probe (2019-nCoV_N1-P) (IDT-Petaluma, CA) modified to meet the above criteria (Fig. 1A). All primers and probe were BLASTed (NCBI GenBank) to ensure no cross-reactivity with other organisms, only SARS-CoV-2 sequences returned. To enable for detection by lateral flow, the reverse primer was biotinylated at the 5′ end. Table 1 shows the primers and probe sequences used in the RPA-LF assay.
Table 1.
qPCR and RPA-LF primers and Probes.
| 5′-3′ | Purpose | |
|---|---|---|
| 2019-nCoV_N1-F | GAC CCC AAA ATC AGC GAA AT | RT-qPCR |
| 2019-nCoV_N1-R | TCT GGT TAC TGC CAG TTG AAT CTG | RT-qPCR |
| 2019-nCoV_N1-P | FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1 | RT-qPCR |
| 2019-nCoV_N2-F | TTA CAA ACA TTG GCC GCA AA | RT-qPCR |
| 2019-nCoV_N2-R | GCG CGA CAT TCC GAA GAA | RT-qPCR |
| 2019-nCoV_N2-P | FAM-ACA ATT TGC CCC CAG CGC TTC AG-BHQ1 | RT-qPCR |
| 2019-nCoV_N3-F | GGG AGC CTT GAA TAC ACC AAA A | RT-qPCR |
| 2019-nCoV_N3-R | TGT AGC ACG ATT GCA GCA TTG | RT-qPCR |
| 2019-nCoV_N3-P | FAM-AYC ACA TTG GCA CCC GCA ATC CTG-BHQ1 | RT-qPCR |
| RP-F | AGA TTT GGA CCT GCG AGC G | RT-qPCR |
| RP-R | GAG CGG CTG TCT CCA CAA GT | RT-qPCR |
| RP-P | FAM – TTC TGA CCT GAA GGC TCT GCG CG – BHQ | RT-qPCR |
| RV2019-nCoV_N3 Biot | /5BiosG/TGTAGCACGATTGCAGCATTG | RPA-LF |
| FW2019-nCoV_N1 | GAC CCC AAA ATC AGC GAA AT | RPA-LF |
| Probe2019-nCoV_N1ext | /56-FAM/ACCCCGCATTACGTTTGGTGGACCCTCAGAT/idSp/CAACTGGCAGTAACCAGA/3SpC3/ | RPA-LF |
4.7. RPA reaction and LF reading
The amplification mixture was comprised of: 1) forward primer (4.8 μL, 5μM), 2) biotinylated reverse primer (4.8 μL, 5μM), 3) FAM-labeled probe (0.6 μL, 5 μM), 4) magnesium acetate (2.5 μL, 288 mM), 5) 6.6 μL of water), and 6) the rehydrated cocktail (Twist amp nfo RPA kit -TwistDx, UK). Viral cDNA or control plasmid (2.5 μL) was immediately added to the mixture and subjected to amplification at 42.0 °C for 40 min using a dry bath incubator (VWR). The RPA product was diluted 1:50 in 100 μL of dipstick assay buffer in a 1.5 μL Eppendorf tube. The bottom tip of the lateral flow strip (Ustar Biotechnologies, Hangzhou) was then immersed in the sample making the amplification product run upwards by capillarity. Viral cDNA amplification was confirmed with the naked eye after 5 min by the appearance of the test band in the lower part of the strip in addition to the control band.
4.8. Statistical analysis
Probit analysis was conducted for the lowest concentrations detected of N gene plasmid and viral genome cDNA samples. Statistical analysis was done using free software from https://astatsa.com/Logit_Probit or XLSTAT.
Author Disclosure Statement
We declare that the authors have no competing interests as defined by JCV, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
CRediT authorship contribution statement
Thomas R. Shelite, Bruno L. Travi: Conception or design of the work. Thomas R. Shelite, Alejandro Castellanos, Ashanti C. Uscanga-Palomeque: Data collection. WRECEVA: Provision of viral stock and reagents. Thomas R. Shelite, Alejandro Castellanos, Peter C. Melby, Bruno L. Travi: Data analysis and interpretation. Thomas R. Shelite, Bruno L. Travi: Drafting the article. Thomas R. Shelite, Alejandro Castellanos, Bruno L. Travi: Critical revision of the article. Thomas R. Shelite, Alejandro Castellanos, Ashanti C. Uscanga-Palomeque, Bruno L. Travi: Final approval of the version to be published.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank the WRCEVA at UTMB for providing viral samples to test in our assay, and the Sealy & Smith Foundation and Dr. Scott Weaver for obtaining clinical samples. This work would not have been possible without the support of the Division of Infectious Diseases, Department of Internal Medicine at UTMB.
References
- Castellanos-Gonzalez A., Saldarriaga O.A., Tartaglino L., Gacek R., Temple E., Sparks H., et al. A novel molecular test to diagnose canine visceral leishmaniasis at the point of care. Am. J. Trop. Med. Hyg. 2015;93(5):970–975. doi: 10.4269/ajtmh.15-0145. Epub 2015/08/03. PubMed PMID: 26240156; PubMed Central PMCID: PMC4703275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Control CoD . 2020. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primers and Probes.https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf?fbclid=IwAR3rqUhcAfzGLTVsYOdFN5Y10I_G-AZesQ8g4kc9Vhlub-uwjsK_seAnRec [cited 2020]. Available from: [Google Scholar]
- Crannell Z., Castellanos-Gonzalez A., Nair G., Mejia R., White A.C., Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal. Chem. 2016;88(3):1610–1616. doi: 10.1021/acs.analchem.5b03267. Epub 2016/01/12. PubMed PMID: 26669715. [DOI] [PubMed] [Google Scholar]
- Dinnes J., Deeks J.J., Adriano A., Berhane S., Davenport C., Dittrich S., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2020;8 doi: 10.1002/14651858.CD013705. Epub 2020/08/26. PubMed PMID: 32845525; PubMed Central PMCID: PMC8078202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler V.L., Armson B., Gonzales J.L., Wise E.L., Howson E.L.A., Vincent-Mistiaen Z., et al. A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection. J. Infect. 2021;82(1):117–125. doi: 10.1016/j.jinf.2020.10.039. Epub 2020/11/30. PubMed PMID: 33271166; PubMed Central PMCID: PMC7703389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howson E.L.A., Kidd S.P., Armson B., Goring A., Sawyer J., Cassar C., et al. Preliminary optimisation of a simplified sample preparation method to permit direct detection of SARS-CoV-2 within saliva samples using reverse-transcription loop-mediated isothermal amplification (RT-LAMP) J. Virol. Methods. 2021;289 doi: 10.1016/j.jviromet.2020.114048. Epub 2020/12/20. PubMed PMID: 33358911; PubMed Central PMCID: PMC7750029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Coello M., Shelite T., Castellanos-Gonzalez A., Saldarriaga O., Rivero R., Ortega-Pacheco A., et al. Efficacy of recombinase polymerase amplification to diagnose Trypanosoma cruzi infection in dogs with cardiac alterations from an endemic area of Mexico. Vector Borne Zoonotic Dis. 2018;18(8):417–423. doi: 10.1089/vbz.2017.2258. Epub 2018/05/16. PubMed PMID: 29768103. [DOI] [PubMed] [Google Scholar]
- Li L., Li X., Guo Z., Wang Z., Zhang K., Li C., et al. Influence of storage conditions on SARS-CoV-2 nucleic acid detection in throat swabs. J. Infect. Dis. 2020;222(2):203–205. doi: 10.1093/infdis/jiaa272. PubMed PMID: 32427340; PubMed Central PMCID: PMC7313924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect Dis. 2020;26(8) doi: 10.3201/eid2608.201246. Epub 2020/05/15. PubMed PMID: 32396505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore N.M., Li H., Schejbal D., Lindsley J., Hayden M.K. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV reverse transcriptase PCR assay for the detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00938-20. Epub 2020/07/23. PubMed PMID: 32461287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G., Rebolledo M., White A.C., Crannell Z., Richards-Kortum R.R., Pinilla A.E., et al. Detection of entamoeba histolytica by recombinase polymerase amplification. Am. J. Trop. Med. Hyg. 2015;93(3):591–595. doi: 10.4269/ajtmh.15-0276. Epub 2015/06/29. PubMed PMID: 26123960; PubMed Central PMCID: PMC4559702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalla A.K., Casto A.M., Huang M.W., Perchetti G.A., Sampoleo R., Shrestha L., et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J. Clin. Microbiol. 2020;58(6) doi: 10.1128/JCM.00557-20. Epub 2020/05/26. PubMed PMID: 32269100; PubMed Central PMCID: PMC7269385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Mitchell B.M., Plante K.S., Debbink K., Weaver S.C., Menachery V.D. The variant gambit: COVID-19’s next move. Cell Host Microbe. 2021;29(4):508–515. doi: 10.1016/j.chom.2021.02.020. Epub 2021/03/01. PubMed PMID: 33789086; PubMed Central PMCID: PMC7919536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldarriaga O.A., Castellanos-Gonzalez A., Porrozzi R., Baldeviano G.C., Lescano A.G., de Los Santos M.B., et al. An Innovative field-applicable molecular test to diagnose cutaneous leishmania Viannia spp. infections. PLoS Negl. Trop. Dis. 2016;10(4) doi: 10.1371/journal.pntd.0004638. Epub 2016/04/26.,PubMed PMID: 27115155; PubMed Central PMCID: PMC4845993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki K., Yokota I., Fukumoto T., Iwasaki S., Fujisawa S., Takahashi M., et al. SARS-CoV-2 detection by fluorescence loop-mediated isothermal amplification with and without RNA extraction. J. Infect. Chemother. 2021;27(2):410–412. doi: 10.1016/j.jiac.2020.10.029. Epub 2020/10/31. PubMed PMID: 33214073; PubMed Central PMCID: PMC7604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva, Switzerland: 2020. Novel Coronavirus (2019-nCoV) Situation Report–1. [Google Scholar]
- World Health Organization . 2021. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ [cited 2021 2/22/2021]. Available from: [Google Scholar]
- Yüce M., Filiztekin E., Özkaya K.G. COVID-19 diagnosis -a review of current methods. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112752. Epub 2020/10/24. PubMed PMID: 33126180; PubMed Central PMCID: PMC7584564. [DOI] [PMC free article] [PubMed] [Google Scholar]