Abstract
The SARS-CoV-2 N protein binds several cell host proteins including 14-3-3γ, a well-characterized regulatory protein. However, the biological function of this interaction is not completely understood. We analyzed the variability of ∼90 000 sequences of the SARS-CoV-2 N protein, particularly, its mutations in disordered regions containing binding motifs for 14-3-3 proteins. We studied how these mutations affect the binding energy to 14-3-3γ and found that changes positively affecting the predicted interaction with 14-3-3γ are the most successfully spread, with the highest prevalence in the phylogenetic tree. Although most residues are highly conserved within the 14-3-3 binding site, compensatory mutations to maintain the interaction energy of N-14-3-3γ were found, including half of the current variants of concern and interest. Our results suggest that binding of N to 14-3-3γ is beneficial for the virus, thus targeting this viral-host protein-protein interaction seems an attractive approach to explore antiviral strategies.
Keywords: Coronavirus, SARS-CoV-2, N protein, 14-3-3, Interaction, Viral success
Introduction
The 14-3-3s constitute a family of highly conserved eukaryotic proteins with more than 300 known binding partners, and 3000 predicted. Their binding to specific phosphorylated motifs modifies the activity, stability, and/or subcellular localization of the target proteins [1].
14-3-3 proteins contribute positively in the pathogenesis and progression of viral infections. Viral interactions with 14-3-3 proteins may change the original distribution and functions of this host family. For example, some viruses associate with and inhibit 14-3-3s from inducing apoptosis [2].
The Severe Acute Respiratory Syndrome SARS Coronavirus (SARS-CoV) Nucleocapsid (N) protein is frequently phosphorylated on several Ser/Thr residues in host cells [3], where it is mainly cytoplasmic [4]. It binds to nucleic acids, controlling viral transcription, replication, and assembly [5].
Phosphorylated N protein binds to 14-3-3 [4,6], which regulates its subcellular localization. Decreasing the levels of 14-3-3 using siRNA causes N protein nuclear concentration [4].
Sequence mutations in the Serine-Arginine (SR)-rich region (amino acids 180 to 210) were previously analyzed [7]. Tung and Limtung proposed that only the SARS-CoV-2 N protein monomer can bind the 14-3-3 dimer and that the phosphorylation of the N protein and its sequestration by 14-3-3 is a cellular defense mechanism to control the SARS-CoV-2 through hindering its replication, transcription, and packaging [7]. Nevertheless, Sluchanko and coll [6]. proposed that the N-14-3-3 interaction may sequester 14-3-3 proteins and impedes them to exert their cellular functions, suggesting that this interaction could be beneficial for the virus. They demonstrated that the conserved Ser197 in N protein is essential for the binding to the 14-3-3, in particular to 14-3-3γ paralog [6].
Here, we analyzed the variability of ∼90 000 SARS-CoV-2 N protein sequences, and studied how these mutations affected the ΔG of binding to 14-3-3γ. We analyzed if the variants bearing those mutations were successfully spread, and their implications in the discussion of the beneficial or detrimental role of N protein-14-3-3 interaction in SARS-CoV-2 life cycle. We compared the data with the consensus sequences of current circulating variants of concern and interest. This could be relevant to the planning of novel antiviral strategies.
Methods
Sequences
All SARS-CoV-2 N protein sequences analyzed in the present report (93,246) were downloaded from GISAID [8] (www.gisaid.org) in August 2020. Low quality or incomplete sequences were excluded from the analysis.
Multiple sequence alignment (MSA)
The 86,168 N protein sequences were aligned using the algorithm Clustal Omega v1.2.4 with parameters in their default setting. Five sequences containing amino acid insertions of more than 10 amino acids were removed, leaving 86,163 sequences. Mutation frequencies were computed using the Balcony package in Rstudio, and the score used in Fig. 2 (A, B) was calculated using the BLOSUM62 (BLOcks SUbstitution Matrix) matrix [9]. In our color scale, white represents conservative changes (score 0, as expected by chance) or no change, blue (positive score) indicates that the change was found in the database more, and red (negative score) less often than expected by chance respectively.
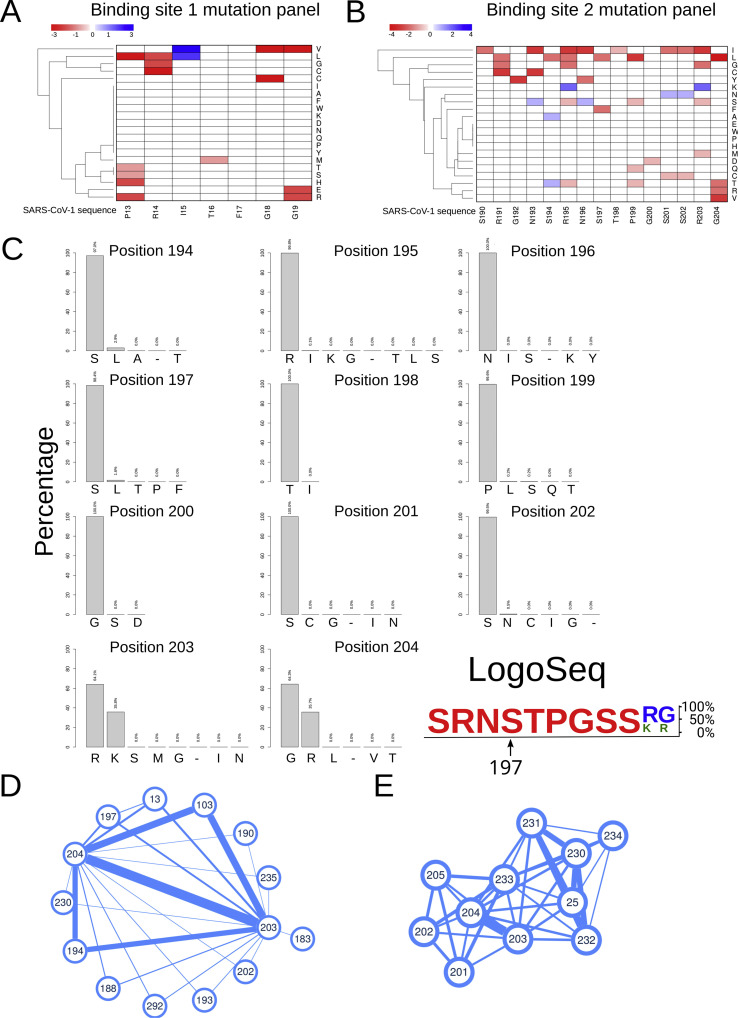
Fig. 2.
SARS-CoV-2 N protein variations in disordered regions containing 14-3-3 binding sites and co-evolution analysis. A, B) Heat maps showing amino acid mutations in two selected disordered regions of the SARS-CoV-2 N protein containing putative 14-3-3 binding sites. C) Percentages of amino acid variations in sites surrounding S197, the most important in 14-3-3 binding. D, E) Co-evolution network of N protein amino acids calculated by OMES (D), and DCA method (E). Lines thickness are proportional to the co-evolution score (minimum = 0, maximum = 1).
Computer Programming, Statistics, Intrinsic Structural Disorder Calculation, and Eukaryotic Linear Motif (ELM) Identification
Scripts for sequence analyses were programmed in bash. All statistical analyses were carried out using the Rstudio. Disorder predictions were done by using the software Espritz [10] (http://protein. bio.unipd.it/espritz/) as previously done [11]. Briefly, the SARS-CoV-2 N protein sequences were identified and translated from the downloaded SARS-CoV-2 genomes, and then Espritz was run on the sequences, resulting files were post-processed using bash scripting. ELMs for the binding of 14-3-3 proteins on the reference sequence of SARS-CoV-2 N protein were identified using the server http://elm.eu.org/
Entropy calculation
To compare the variability of each position in the MSA we scored the position entropy based on Shannon entropy (S), given by:
where i is the MSA interest position, x is any amino acid and p x is the frequency of x at i. The calculations were done in R using the package bios2cor [12] and plots were done with the package ggplot2 [13].
Protein-protein affinity calculation
The effects of single-point mutations on 14-3-3γ/peptide interactions were evaluated in the following 3D structures from the RBSD PDB (2B05, 4J6S, 4O46, 5D3E, 6FEL, 6GKF, 6GKG, 6S9K, 6SAD) using the mcsm-ppi2 algorithm [14] and Foldx [15]. Each 3D structure corresponded to 14-3-3γ in complex to a phosphorylated peptide. The point mutations of the amino acids in the crystal to the corresponding amino acids in SARS-CoV-2 N protein were done using the commands PositionScan, Optimize, and AnalyseComplex in FoldX or by using the PDB file and a list of point mutations in mcsm-ppi2.
Amino acid Co-evolution
SARS-CoV-2 N protein amino acid co-variation was evaluated by different methods, Observed Minus Expected Squared (OMES [12],), which is based on the χ2 test and the Direct Coupling Analysis (DCA [16],) that infers co-evolution between pairs of residues that are spatially close.
The OMES score was computed as:
where N (i, j) is the number of aligned sequences without gaps, at positions i and j, Nobs xy (i,j) and Nex xy (i,j) are the number of times each pair of amino acids (x, y) is observed and expected at (i,j), respectively.
The DCA score was computed as:
The sequence S = (S1, …, SL) is aligned to the MSA of length L; J and h are known parameters from a seed alignment. The energy H (DCA) is considered as a “score” (lower energy corresponds to higher score) for sequence S.
Results
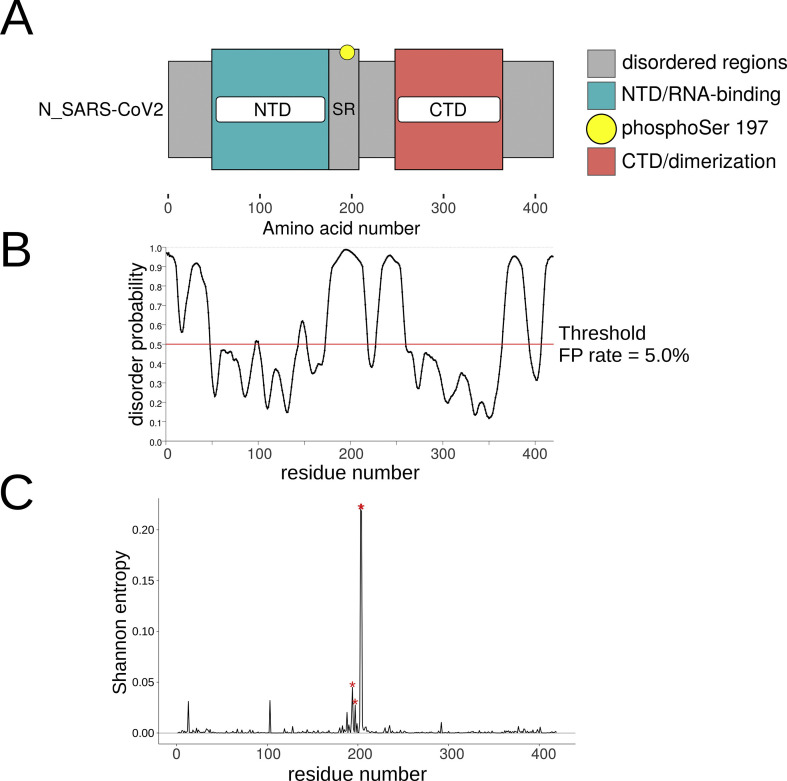
The host expressed SARS-CoV-2 N protein contains multiple phosphorylation sites [6]. Its most phosphorylated portion is the SR-rich region, located downstream to the N-terminal domain (NTD, Fig. 1 A). Based on reports about the earlier SARS-CoV-1 N protein interaction with host 14-3-3 proteins [4], and recent in silico data suggesting the interaction has been conserved in SARS-CoV-2 [7], we were interested in studying 14-3-3 interaction with SARS-CoV-2 N protein and the amino acidic mutations occurring in its 14-3-3 binding site/s. Because we have previously shown that the intrinsic disorder is crucial for the binding to 14-3-3s [17], we first analyzed the disorder probability in a SARS-CoV-2 reference amino acidic sequence (EPI_ISL_412977, Wuhan, China, Fig. 1B). We identified 4 disordered regions, one in the N-terminal domain (NTD), two nearby regions linking the NTD and the C-terminal Domain (CTD), and one in the CTD (Fig. 1B). We downloaded 93,246 SARS-CoV-2 N protein sequences available from GISAID [8] (www.gisaid.org) up to the end of August 2020. Low quality (containing undetermined amino acids, X) or incomplete sequences were excluded. The final number of complete sequences analyzed was 86,163. These were globally aligned using Clustal Omega v1.2.4; the resulting MSA is available at our lab page (https://dbustoslab.github.io/). Then, we calculated the Shannon Entropy to condense the MSA information in a plot (Fig. 1C). The graph showed the second disordered region as highly variable, making it especially attractive to study protein sequence variations, their effects on 14-3-3 binding and viral success. Within this region, the most prominent peak in the entropy plot of the SARS-CoV-2 N protein corresponded to sites 203 and 204, which are among the most variable of the complete SARS-CoV-2 proteome, which can be observed at the GISAID website (GISAID - NextStrain). These sites, together with S194 and S197 were marked with asterisks (Fig. 1C) because they are critical residues that will be discussed below.
Fig. 1.
The SARS-CoV-2 Nucleocapside (N) protein organization, intrinsic disorder and variability. SARS-CoV-2 N protein domains and organization (A), disorder prediction (B), and amino acidic sequence Shannon Entropy prediction (C).
The first and second disordered regions contained putative 14-3-3 binding sites (one and three sites respectively). These sites were identified using the SARS-CoV-2 reference amino acidic sequence (EPI_ISL_412977) as input in the ELM resource (http://elm.eu.org/), and through visual inspection. Another possible 14-3-3 binding site, covering amino acids 259 to 268, was located inside the structured CTD domain (Fig. 1), and because the binding to 14-3-3 requires structural intrinsic disorder [17], we excluded it from our analysis. Hence, we analyzed sequence variations on the two first disordered regions, specifically in residues contained or nearby the putative 14-3-3 binding sites. These corresponded to amino acids 13 to 19 in the first disordered region and 190 to 204 in the second and were labeled site 1 and 2 respectively (Fig. 2A and B). The latter fragment was located within the SR-rich region previously studied [6,7] and contained three possible concentrically arranged 14-3-3 binding sites of different lengths, the shortest inside the middle-sized and this one inside the longest. This fact, and visual comparison with optimal motifs [1], made us hypothesize that the second site could be the most important in the binding of N protein to 14-3-3 proteins. Indeed, a recent publication [6] demonstrated the critical role of pS197, contained in the SR-rich region (Fig. 1A and B) of the second disordered region, thus we later focused more specifically on mutations of this residue and its neighbors. Remarkably, the S197 showed lower levels of Shannon Entropy as compared to other residues in the SR-rich region (Fig. 1C).
Fig. 2A, B shows amino acid variations in the above-mentioned sites 1 and 2 of SARS-CoV-2 N protein in comparison to the homologous sequence in SARS-CoV1 (GenBank accession number AY274119.3 [3]). Each site including variations was assigned the value 1, which was multiplied by the corresponding score in the BLOSUM62 matrix. We observed that most of the non-conservative mutations were on the red side of the scale, meaning that the changes were naturally unexpected. Although only one site in these two regions was found strictly conserved (F17, Fig. 2A and B), the percentage of strains bearing each mutation is low (less than 3%), except for sites 203 and 204, which showed a significant percentage (36%) of strains bearing both mutations, R203K and G204R (Fig. 2C). Except for these two sites, the analyzed regions were highly conserved. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2D and E shows the co-evolution of the SARS-CoV-2 N protein amino acids, computed using two alternative methods (OMES and DCA). The OMES measures the co-variation in the composition of the residues in any two columns in the MSA. The DCA method additionally infers spatial proximity between residues. Both methods identified co-evolving amino acids within the SARS-CoV-2 N protein. However, only OMES found co-evolution involving one of the residues in the known binding motif of N protein to 14-3-3: the S197, critical in this interaction [6], is evolutionarily linked to R203 and G204. The DCA method recognized a strong co-evolution between R203 and G204 but not with S197. Based on these methods characteristics, the existence of a functional relationship (but not structural constrain) between residues S197 and R203/G204 is possible.
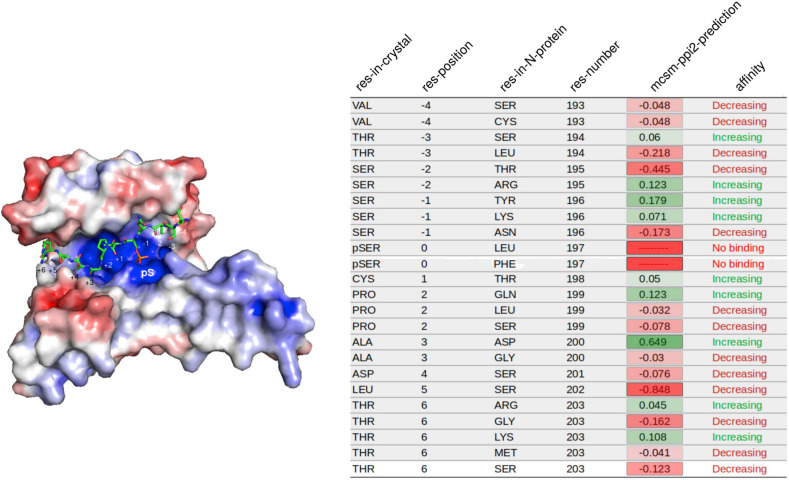
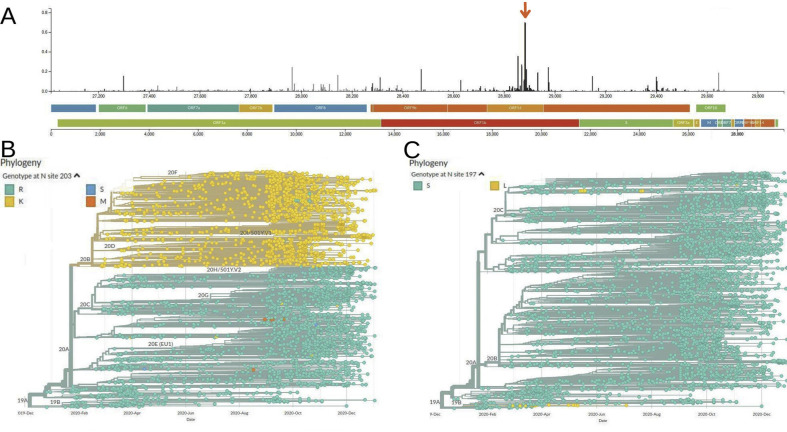
Although several structures of the N protein NTD and CTD are available (https://www.uniprot.org/uniprot/P59595), there is no data about the SR-rich region including the demonstrated 14-3-3 binding site, probably because of its highly disordered nature. Thus, to study the effect of different mutations on the binding affinity to 14-3-3, we used crystallographic structures of binding peptides with 14-3-3γ, the isoform with the highest binding affinity to the SARS-CoV-2 N protein [6]. Fig. 3 shows this structure and the mscm-ppi2 prediction for the replacement of each position of the 14-3-3 binding peptide by the residues present in that position in SARS-CoV-2 N protein sequences. We observed that those changes positively affecting the predicted interaction between the peptide and 14-3-3 were the ones with the highest prevalence in the phylogenetic tree. In other words, the strains bearing them were the most successfully spread. One of the most interesting examples was observed for mutations in the residue 203, which shows the maximum entropy within the N protein, and even the highest considering the complete SARS-CoV-2 genome (Fig. 4 A). The presence of this residue, R203 in the earlier SARS-CoV-2 sequences (Fig. 4B), and also shared with the former SARS-CoV-1, increases the binding to 14-3-3 compared to the T in position +6 in the 14-3-3 binding peptide (Fig. 3). Substitutions to amino acids G, K, M, and S were observed in SARS-CoV-2 N protein amino acid sequences in this site. Among these, only the change to K had a positive effect on the N-protein-14-3-3 interaction, compared to the original T on the 14-3-3 binding peptide. This mutation is the one that has spread most successfully, as it can be observed by its prevalence in the phylogenetic tree (Fig. 4B). The mutation in the contiguous site, G204R, showed a similar pattern on the phylogenetic tree (not shown). Indeed, we found strong evidence of co-evolution between these two sites 203 and 204 (Fig. 2D and E). Using FoldX [15], we observed that either of these two mutations occurring separately (R203K or G204R) destabilized 14-3-3-N protein interaction (R203K, interaction energy 30.08 kcal/mol, and G204R, 30.36 KCal/mol, compared to the wild-type, R203/G204, 35.35 kcal/mol). Intriguingly, the double mutant R203K G204R had compensatory effects, with an interaction energy of 32.22 kcal/mol. This double mutation was rapidly spread after its first appearance (Fig. 4B), and we found that it is present in half of the concern/interest variants, Alpha, Gamma, Zeta, Theta and Lambda. In contrast, the replacement of pS197 by L or F as occurred in a few SARS-CoV-2 variants (Fig. 2) led to impaired binding to 14-3-3 proteins (Fig. 3), and these strains have not been efficiently spread, as it can be observed in Fig. 4C.
Fig. 3.
Mutations on the SR-rich region of N protein affect the binding potential to 14-3-3 of SARS-CoV-2 variants. Effect of S197 and surrounding amino acids mutations in N protein on the binding potential to host 14-3-3γ. The crystal Structure (PDBID: 2B05) of 14-3-3γ in complex with a phosphoserine peptide was single mutated to each amino acid of SARS-CoV-2 N protein in the corresponding position. The binding energy was calculated for each change, and determined if it was stabilizing or destabilizing by comparison with the original peptide in the crystal structure.
Fig. 4.
General variability of the SARS-CoV-2 genome and phylogenetic distribution of sites 203 and 197. A) Bars graph (GISAID, https://www.gisaid.org) showing the entropy of the different residues along the SARS-CoV-2 genome. The bars corresponding to residues 203 and 204 were indicated with an arrow. B) Phylogeny (GISAID) showing mutations of site 203 to different amino acids with different levels of dissemination within the tree. C) Same as in B except that site 197 phylogenetic distribution is represented.
Discussion
Our data showed that although the N protein of SARS-CoV-2 is variable along its sequence (Fig. 1C), the region containing the S197 residue, critical for 14-3-3 binding [6] is relatively conserved, except for residues 203 and 204. These two appeared conjointly during December 2019 (https://www.gisaid.org/phylodynamics/global/nextstrain/), which correlates with our result of co-evolution between them (Fig. 2C and D). Interestingly, both mutations seem to drive the phylogenetic tree (amino acids at site 203 were mapped on the tree of Fig. 4B, the same for site 204 was identical, not shown) and match one of the current main clades at the GISAID site, named GR or 20B. For information about how the different clades are defined and named see Ref. [29]. According to previous studies, both mutations drop protein stability [19,20] while favoring torsion [19] or decrease overall structural flexibility [20]. More recently, Vahed et al. [21] found that the mutant type (K203, R204) showed higher free energy at these residues, suggesting enhanced structural flexibility. Besides these observations, we found that any of these two mutations occurring separately destabilized the interaction between 14-3-3 and the N protein, while the double mutant R203K G204R showed compensatory effects. This linked functionally these residues to the S197, the most important residue for the binding to 14-3-3 proteins [6]. In the 14-3-3-N protein interaction, the 203 residue accommodates near the border of the 14-3-3 binding grove (position +6, Fig. 3). Although 204 residue stays outside the 14-3-3 binding grove, we have previously identified essential residues for 14-3-3 interactions with this feature [22]. The SARS-CoV-2 N protein R203K/G204R double mutant has successfully spread, whereas single R203K or G204R mutants did not spread at all. As explained earlier, most strains bearing the R203K mutation also include the G204R mutation.
The current SARS-CoV-2 variants that raise the risk to public health have been recently classified (Tracking SARS-CoV-2 variants (who. int)) as either of concern (VOC): Alpha (B1.1.7, first documented in the UK, Sept. 2020), Beta (B1.351, SA, May 2020), Gamma (P1, Brazil, Nov. 2020), and Delta (B1.617.2, India, Oct. 2020), or of interest (VOI): Epsilon (B1.427/9, USA, Mar. 2020), Zeta (P2, Brazil, Apr. 2020), Eta (B1.525, multiple countries, Dec. 2020), Theta (P3, Philippines, Jan. 2021), Iota (B1.526, USA, Nov. 2020), Kappa (B1.617.1, India, Oct. 2020), and Lambda (C37, Peru, Aug. 2020). Thus, we compared the data presented with the consensus sequences of these circulating variants. We found that half of them (Alpha, Gamma, Zeta, Theta, and Lambda) include the mutation R203K, always linked to the G204R mutation. Only two of them (Delta and Kappa) contain in this site a different mutation, R203 M, however, this modification is not linked to any variation in position 204, which remains as glycine. This may be related to the fact that the change to K in position +6 in the crystal (which corresponds to residue 203 in N protein) increases affinity to 14-3-3 proteins, with a positive mscm-ppi2-prediction value of 0.108 ( Fig. 3 ). In contrast, the change to M is slightly negative (Figs. 3, -0.041), thus it decreases the binding affinity. As expected, the VOCs and VOIs do not show other mutations in the 14-3-3 binding groove, from residues 193 to 203. This agrees with our results, and again supports the idea that this region of the N protein is highly conserved, and that the interaction N protein-14-3-3 is an appealing target for the development of antiviral tools.
Besides, to investigate the main groove amino acid variations of the 14-3-3 protein family in different human ethnic groups we used the information from the gnomAD database [23]. Any possible loss-of-function analysis requires large sample sizes. The gnomAD database contains 125,748 exomes and 15,708 genomes from healthy humans of different ethnics backgrounds around the world. No 14-3-3 isoforms, including gamma isoform, which is the one with the strongest interaction with the N protein [6], accumulate mutations in the important residues for the binding to their partners.
Many clues suggest the biological importance of 14-3-3-N protein interaction. First, 14-3-3 isoforms bind the N protein with different affinities, with 14-3-3γ being the strongest [8]. We have previously studied the interaction networks of all human 14-3-3 isoforms and found that the 14-3-3γ interactome is specifically enriched in RNA processing proteins [11]. This isoform has been reported in P-bodies, structures specialized in translational repression, mRNA surveillance, and degradation. Its depletion by siRNA showed 14-3-3γ as a regulator of P-body formation [24]. The N protein is well known to form liquid condensates through binding specific sequences from its genome [25]. Liquid-Liquid Phase Separation (LLPS) induced by viral proteins are considered “viral factories”, where viral replication and assembly take place [26]. We propose that the SARS-CoV-2 uses 14-3-3 as part of its life cycle, for replication and/or assembly, through the interaction of N with 14-3-3γ proteins within liquid condensates. Another possibility is that LLPSs are not associated with viral replication and assembly, and rather interfere with host cell functions through alteration of gene transcription or interaction with cellular proteins [26]. If this is the case for SARS-CoV-2, the N protein could be “sequestering” 14-3-3s and disrupting their numerous functions, as previously suggested [6]. Among other examples, 14-3-3s are involved in autophagy [1], and other host cell defense mechanisms [6]. A third hypothesis is that 14-3-3 binding may regulate N protein shuttling, as seen previously for SARS-CoV-1 N protein [4]. In favor of this are the reported enhancement of nuclear localization and export Signals in Coronavirus associated with high case fatality reports, including SARS-CoV-2 [27].
Phosphorylation regulates the state and function of the N protein. The unmodified protein would be in a gel state for the nucleocapsid, while the phosphorylated version would be in a liquid state to process the viral genome [28]. This is in agreement with our results, which suggest that binding of N protein to 14-3-3 is beneficial for the virus and supports the hypothesis that either the virus uses 14-3-3 proteins to complete its life cycle, or it arrests them to impede their normal functions in the host cell. In any case, targeting this N protein-14-3-3 viral-host protein-protein interaction seems an attractive approach for exploring antiviral strategies. Future studies should be oriented in that direction.
Author contributions
DMB and MU conceived and supervised the study, analyzed data and wrote the manuscript; SDV, LR, DMB, and MU designed and performed experiments; SDV and LR contributed equally.
Role of the funding source
The funding institution had no role in study design; collection, analysis, and interpretation of data; in writing the report; and in the decision to publish.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgments
SDV and LR hold Ph.D. scholarships from CONICET, Argentina; DMB and MU are CONICET Researchers. This work was supported by ANPCYT, Argentina (grant PICT′17–1984). We thank all the researches that submitted the sequences used.
References
- 1.Sluchanko N.N., Bustos D.M. Danc. Protein Clouds Intrinsically Disord. Proteins Norm Pathol. First Ed. Elsevier Inc.; 2019. Intrinsic disorder associated with 14-3-3 proteins and their partners; pp. 19–61. [DOI] [PubMed] [Google Scholar]
- 2.Nathan K.G., Lal S.K. The multifarious role of 14-3-3 family of proteins in viral replication. Viruses. 2020;12:436. doi: 10.3390/v12040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marra M.A., Jones S.J.M., Astell C.R., et al. The Genome Sequence of the SARS-Associated Coronavirus. Science. 2003;300:1399–1405. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 4.Surjit M., Kumar R., Mishra R.N., et al. The Severe Acute respiratory Syndrome Coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/jvi.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong Y., Ulasli M., Schepers H., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2019;94 doi: 10.1128/jvi.01925-19. e01925–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tugaeva K.V., Hawkins D.E.D.P., Smith J.L., et al. The mechanism of SARS-CoV-2 nucleocapsid recognition by the human 14-3-3 proteins. J. Mol. Biol. 2021;433:166. doi: 10.1016/j.jmb.2021.166875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tung H.Y.L., Limtung P. Mutations in the phosphorylation sites of SARS-CoV-2 encoded nucleocapsid protein and structure model of sequestration by protein 14-3-3. Biochem. Biophys. Res. Commun. 2020;532:134–138. doi: 10.1016/j.bbrc.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastien O., Roy S., Maréchal É. Construction of non-symmetric substitution matrices derived from proteomes with biased amino acid distributions. Comptes Rendus Biol. 2005;328:445–453. doi: 10.1016/j.crvi.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Walsh I., Martin A.J.M., Di domenico T., et al. Espritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28:503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 11.Uhart M., Bustos D.M. Human 14-3-3 paralogs differences uncovered by cross-talk of phosphorylation and lysine acetylation. PloS One. 2013;8:e55703. doi: 10.1371/journal.pone.0055703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelé J., Taddese B., Deniaud M., et al. Co-variation approaches to the evolution of protein families. Adv. Tech. Biol. Med. 2017;5:250–254. doi: 10.4172/2379-1764.1000250. [DOI] [Google Scholar]
- 13.Valero-Mora P.M. ggplot2: elegant graphics for data analysis. J. Stat. Software. 2010;35:1–3. doi: 10.18637/jss.v035.b01. [DOI] [Google Scholar]
- 14.Pires D.E.V., Ascher D.B., Blundell T.L. MCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delgado J., Radusky L.G., Cianferoni D., et al. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35:4168–4169. doi: 10.1093/bioinformatics/btz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerihun M.B., Pucci F., Peter E.K., et al. Pydca v1. 0: a comprehensive software for Direct coupling analysis of RNA and protein sequences. Bioinformatics. 2020;36:2264–2265. doi: 10.1093/bioinformatics/btz892. [DOI] [PubMed] [Google Scholar]
- 17.Bustos D.M., Iglesias A.A. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins Struct. Funct. Genet. 2006;63:35–42. doi: 10.1002/prot.20888. [DOI] [PubMed] [Google Scholar]
- 19.Benvenuto D., Demir A.B., Giovanetti M., et al. Evidence for mutations in SARS-CoV-2 Italian isolates potentially affecting virus transmission. J. Med. Virol. 2020;92:2232–2237. doi: 10.1002/jmv.26104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman M.S., Islam M.R., Alam A.S.M.R.U., et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2020;93:2177–2195. doi: 10.1002/jmv.26626. [DOI] [PubMed] [Google Scholar]
- 21.Vahed M., Calcagno T.M., Quinonez E., et al. Impacts of 203/204: RG>KR mutation in the N protein of SARS-CoV-2 1. BioRxiv. 2021 doi: 10.1101/2021.01.14.426726. [DOI] [Google Scholar]
- 22.Uhart M., Iglesias A.A., Bustos D.M. Structurally constrained residues outside the binding motif are essential in the interaction of 14-3-3 and phosphorylated partner. J. Mol. Biol. 2011;406:552–557. doi: 10.1016/j.jmb.2010.12.043. [DOI] [PubMed] [Google Scholar]
- 23.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada N., Yabuta N., Suzuki H., et al. A novel Chk1/2–Lats2–14-3-3 signaling pathway regulates P-body formation in response to UV damage. J. Cell Sci. 2010;124:57–67. doi: 10.1242/jcs.072918. [DOI] [PubMed] [Google Scholar]
- 25.Iserman C., Roden C.A., Boerneke M.A., et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80:1078–1091. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brocca S., Grandori R., Longhi S., et al. Liquid–liquid phase separation by intrinsically disordered protein regions of viruses: roles in viral life cycle and control of virus–host interactions. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21239045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gussow A.B., Auslander N., Faure G., et al. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 2020;117:15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson C.R., Asfaha J.B., Ghent C.M., et al. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80:1092–1103. doi: 10.1016/j.molcel.2020.11.025. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.2020. https://nextstrain.org/blog/2020-06-02-SARSCoV2-clade-naming