Abstract
Working memory is a basic human cognitive function. However, the genetic signatures and their biological pathway remain poorly understood. In the present study, we tried to clarify this issue by exploring the potential associations and pathways among genetic variants, brain morphometry and working memory performance. We first carried out association analyses between 2‐back accuracy and 212 image‐derived phenotypes from 1141 Human Connectome Project (HCP) subjects using a linear mixed model (LMM). We found a significantly positive correlation between the left cuneus volume and 2‐back accuracy (T = 3.615, p = 3.150e−4, Cohen's d = 0.226, corrected using family‐wise error [FWE] method). Based on the LMM‐based genome‐wide association study (GWAS) on the HCP dataset and UK Biobank 33 k GWAS summary statistics, we identified eight independent single nucleotide polymorphisms (SNPs) that were reliably associated with left cuneus volume in both UKB and HCP dataset. Within the eight SNPs, we found a negative correlation between the rs76119478 polymorphism and 2‐back accuracy accuracy (T = −2.045, p = .041, Cohen's d = −0.129). Finally, an LMM‐based mediation analysis elucidated a significant effect of left cuneus volume in mediating rs76119478 polymorphism on the 2‐back accuracy (indirect effect = −0.007, 95% BCa CI = [−0.045, −0.003]). These results were also replicated in a subgroup of Caucasians in the HCP population. Further fine mapping demonstrated that rs76119478 maps on intergene CTD‐2315A10.2 adjacent to protein‐encoding gene DAAM1, and is significantly associated with L3HYPDH mRNA expression. Our study suggested this new variant rs76119478 may regulate the working memory through exerting influence on the left cuneus volume.
Keywords: brain morphometry, cuneus, genome‐wide association study, Human Connectome Project, mediation, n‐back, UK biobank, working memory
Working memory's genetic signatures and biological pathways remain poorly understood. Using the UK Biobank GWAS summary statistic and the HCP dataset, new variant rs76119478 may regulate the working memory through exerting influence on the left cuneus volume.
1. INTRODUCTION
Working memory (WM) plays an essential role in human cognition that stores and manipulates information temporarily while performing complex tasks such as reasoning, comprehension, and learning (Baddeley, 2010). Deficits in WM performance are associated with a variety of psychiatric and neurological disorders, including attention deficit hyperactivity disorder (Martinussen, Hayden, Hogg‐Johnson, & Tannock, 2005), schizophrenia (Van Snellenberg et al., 2016), multiple sclerosis (Bobholz & Rao, 2003), Alzheimer's disease (Jahn, 2013), and addiction (Livny et al., 2018). Based on the multicomponent model proposed by Baddeley et al., the WM system can be separated into at least four interacted components, a prefrontal central executed system, a parietal episodic buffer, an occipital visual‐spatial sketchpad, and a prefrontal‐temporal phonological loop (Baddeley, 2012; Chai, Abd Hamid, & Abdullah, 2018). Although the term “working memory” has been introduced for about 60 years (Miller, Galanter, & Pribram, 1960), the genetic signatures and their biological pathway are still poorly understood.
Some studies attempted to quantify the heritability of WM. They found that the estimates of heritability for WM range from 0.32 to 0.66 in either healthy individuals or clinical samples, indicating moderate to high heritability of WM (Ando, Ono, & Wright, 2001; Karlsgodt et al., 2010; Vogler et al., 2014). Besides, studies based on SNP also indicate the genetic association with WM. For example, a recent study reported an SNP rs1625579 at the coding gene MIR137 is associated with both neural activation and behavioral performance during a WM task (Zhang et al., 2018). Another study reported a significant SPON1 × APOE genotype interaction on WM performance (Liu et al., 2018). Besides, Gregory et al. reported that rs3918242 regulated the expression of the gene SLC12A5 and was associated with WM‐associated brain structure and activation (Gregory et al., 2019). In addition to candidate gene approaches, recent advances in high‐resolution genome‐wide association studies (GWAS) provide an opportunity to identify potential genomic loci that contribute to complicated cognitive processes without a priori assumption (Davies et al., 2016; Savage et al., 2018; Zhao et al., 2019). Several research groups tried to use the GWAS method to explore the potential loci that are associated with WM or WM‐related activation. Unfortunately, there were no loci that survived under the stringent GWAS threshold (p < 5e–8; Blokland et al., 2017; Cirulli et al., 2010; Need et al., 2009), except for one GWAS based on 905 individuals reported five SNPs were significantly associated with N‐back performance which mapped on two genes PAWR and OLFM3(Heck et al., 2014). Besides, gene set enrichment analysis demonstrated that gene sets related to voltage‐gated cation channel activity and neuronal excitability were robustly linked to WM performance and WM‐related brain activity (Heck et al., 2014). Although these studies have provided meaningful genetic associations of WM, little attention is paid to the link among genetic variation, brain structural organization, and WM performance.
Imaging genetics allows measuring the associations between genetic variation and intermediate neuroimaging phenotypes and is becoming an up‐and‐coming interdisciplinary tool for studying the biological pathway of genetics on behavior (Xu et al., 2020). As the world's largest neuroimaging genetic dataset, the UK Biobank (UKB) study team has collected and released magnetic resonance imaging (MRI) data from about 40,000 healthy participants (https://open.win.ox.ac.uk/ukbiobank/big40/; Elliott et al., 2018; Smith et al., 2020). Another informative database, Human Connectome Project (HCP) (Van Essen et al., 2013), provides state‐of‐art high‐quality multimodal MRI data, SNP‐array genotyping data, and multiple cognitive data for about 1,200 healthy young subjects. Early studies had identified many genetic loci associated with image‐derived phenotypes (IDPs) based on GWAS study (Satizabal et al., 2019; Zhao et al., 2019), and many morphometric IDPs were associated with the performance of WM (Ding, Qin, Jiang, Zhang, & Yu, 2012; Owen, McMillan, Laird, & Bullmore, 2005; Salat, Kaye, & Janowsky, 2002; Yeo et al., 2015). However, little attention was paid to the mediating pathways between genetics, brain morphometry, and WM.
In the present study, we tried to elucidate this issue by integrating the merits of the two imaging genetic datasets (UKB and HCP). We first carried out association analyses between 2‐back WM accuracy and morphometric IDPs using the HCP dataset. Based on the GWAS of the HCP dataset and UK Biobank 33 k discovery + validation GWAS summary statistics, we further tried to figure out SNPs that were reliably associated with the selected morphometric IDPs. Finally, we introduced a mediation analysis to explore the potential mediated relationships among the identified genetic loci, morphometric IDPs, and WM performance in the HCP dataset. It is worth noting that because the HCP cohort contains subjects from the same families and mixed population, we performed the above association and mediation analyses using a linear mixed model (LMM) to control for the population stratification caused by both ancestry differences and cryptic relatedness (Sul, Martin, & Eskin, 2018). Many studies indicated that LMM could handle nearly arbitrary genetic relationships between individuals and is an effective and powerful approach for human association studies to control for most forms of population stratification (Goodrich et al., 2016; Loh, Kichaev, Gazal, Schoech, & Price, 2018; Sul et al., 2018; W. Zhou et al., 2020; X. Zhou & Stephens, 2012). We further repeated the previous steps in only Caucasians of the HCP samples to validate the reliability of the main findings. The workflow of the study can be seen in Figure 1.
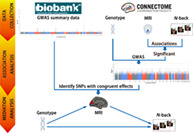
FIGURE 1.
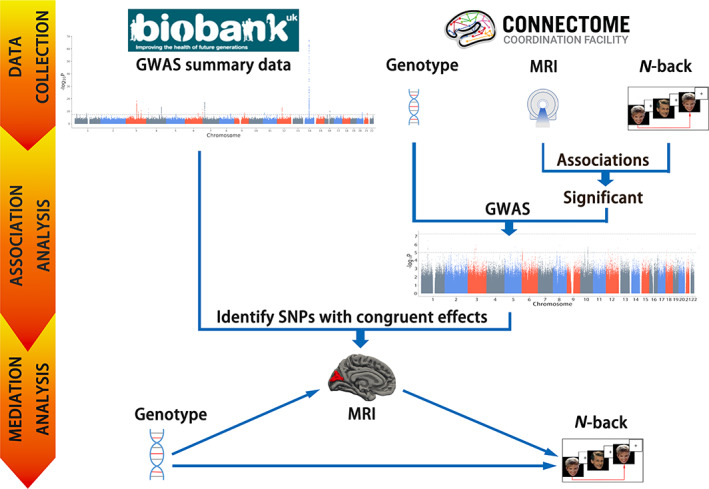
Flowchart of this study. GWAS, genome‐wide association study; MRI, magnetic resonance imaging
2. METHODS
2.1. Datasets
2.1.1. HCP dataset
One of the datasets used in the present study is HCP release S1200 (Van Essen et al., 2012). Participants were young adults without significant mental or neurological illness. Exclusion criteria included neurodevelopmental, neuropsychiatric, or neurological disorders, as well as severe health conditions, such as diabetes, multiple sclerosis, and cerebral palsy, and premature birth. Detailed information is available on the HCP website: https://humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Reference_Manual.pdf. In the present study, 1,141 participants with genotyping data were initially enrolled from 454 families (average age = 28.82 ± 3.69 years, ranging from 22 to 36 years, 54.16% female).
The DNA samples for the HCP dataset were extracted from the whole blood or saliva sample. All subjects were genotyped using the Illumina Multi‐Ethnic Global Array (MEGA) SNP‐array that included chip‐specific content from PsychChip and ImmunoChip and provided extended coverage of European, East Asian, and South Asian populations. Imputation was conducted by IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html). The genotyping data for 1,141 subjects were downloaded through the dbGAP repository (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001364.v1.p1).
The MRI data for 1,054 HCP subjects were collected using a 3T Siemens Connectome Skyra scanner (Siemens AG, Erlanger, Germany), with a customized SC72 gradient insert and a 32‐channel head coil. High‐resolution T1‐weighted structural images had been acquired with a resolution of 0.7 mm3 isotropic (field of view = 224 × 240, matrix = 320 × 320, 256 sagittal slices, repetition time = 2,400 ms, inversion time = 1,000 ms, and echo time = 2.14 ms). Morphometric analyses had been carried out using software HCP pipeline (https://github.com/Washington-University/HCPpipelines) with a standard preprocessing step (Glasser et al., 2013). Desikan_Killiany atlas had been used to extract the quantitative cortical volume, surface area, and thickness (Desikan et al., 2006). A total of 218 IDPs were initially selected for this study, including 14 IDPs for subcortical volume, 68 for cortical surface area, cortical volume, and cortical thickness, respectively. All the IDPs were downloaded directly from the website (https://db.humanconnectome.org/data/projects/HCP_1200).
WM performance was evaluated using a 2‐back task paradigm in which participants were presented with blocks of pictures that consisted of tools or faces. Participants were instructed to press their right index finger if the current image matches the one presented two before (2‐Back) and press the right middle finger if not. In the present study, we used 2‐back accuracy to represent WM performance, and 1,025 HCP subjects had valid WM data.
2.1.2. UK biobank dataset
Another dataset used was the imaging genetic GWAS summary data from UKB released in early 2020 (https://open.win.ox.ac.uk/ukbiobank/big40/). We use the most recent release version, which included the GWAS summary statistics of 22,138 discovery and 11,086 replication subjects (Alfaro‐Almagro et al., 2018; Smith et al., 2020). The process details of population selection, genotype quality control and imputation, and GWAS calculation were provided in Elliott's study (Elliott et al., 2018). For the UKB image data processing, an automated image processing pipeline was used to eliminate artifacts and to make images comparable across modalities and participants; it also generated thousands of IDPs. The genome‐wide genotype data were collected using specially designed genotyping arrays that consist of approximately 96 million genetic variants from nearly 500,000 participants. A custom quality control (QC), staging, and imputation pipeline was developed to address the challenges specific to the UKB experimental design, scale, and diversity (Bycroft et al., 2018). We only downloaded the UKB 33 k discovery + validation GWAS summary statistics with IDPs matching the HCP dataset for the latter analysis. Since the UKB GWAS summary data did not provide the cortical metric for the temporal pole (6 IDPs), we finally included 212 IDPs for later analysis. This work was approved by the ethics committee of Tianjin Medical University General Hospital.
2.2. Association analysis between morphometric IDPs and WM
We carried out imaging‐WM associations analysis with 212 morphometric IDPs as the dependent variables and 2‐back accuracy as the fixed‐effect independent variable using an LMM python package glimix‐core 3.1.11 (https://pypi.org/project/glimix‐core/), controlling for the random‐effects confounders kinship matrix and fixed‐effect confounders intracranial volume (ICV), sex, age, and education years. A family‐wise error (FWE) method was used to correct the effective independent comparisons (Hernandez‐Ferrer et al., 2019; Li, Yeung, Cherny, & Sham, 2012) (effective independent comparisons = 85.476, p < 5.850e−4 was considered statistically significant). The kinship coefficients matrix between each individual was computed using the Genome‐wide Efficient Mixed Model Association Analysis (GEMMA) toolbox, version 0.98.1 (X. Zhou & Stephens, 2012; https://github.com/genetics‐statistics/GEMMA/releases/download/0.98.1/gemma‐0.98.1‐linux‐static.gz).
2.3. GWAS analysis for morphometric IDPs in HCP dataset
The downloaded genotype data from HCP were first conducted QC with the same parameters as UKB GWAS: genome referencing using human genome assembly hg19, excluding SNPs with MAF <1%, genotype call rate <95%, info <0.3, and Hardy–Weinberg equilibrium <1e–7. Finally, 11,061,864 validated SNPs survived under the above QC steps. Considering the family structure and mixed population in the HCP samples, an LMM‐based GWAS was carried out between the identified morphometric IDPs and 11,061,864 qualified SNPs using an LMM‐based GWAS toolbox GEMMA 0.98.1 (https://github.com/genetics-statistics/GEMMA/releases/download/0.98.1/gemma-0.98.1-linux-static.gz) (Sul et al., 2018; X. Zhou & Stephens, 2012). Precisely, a kinship coefficients matrix representing each pair of individuals' genetic similarity was calculated using the preprocessed genotyping data. The additive associations between each SNP and each IDP were then performed using the LMM, controlling for the random‐effect confounds (kinship coefficients) and 20 fixed‐effect confounds identical with UKB GWAS.
2.4. Genomic risk loci characterization
Candidate SNPs with congruent effects in both UKB and HCP dataset were chosen from UKB 33 k discovery + validation GWAS summary statistics and HCP GWAS statistics based on the following three criteria: (a) SNPs survived under a threshold of p < 5e−8 in UKB 33 k GWAS summary statistics; (b) SNPs had nominal P‐value lower than 0.05 in HCP GWAS statistical results; and (c) SNPs had the same effect directions in both HCP and UKB datasets. Besides, we also validate the identified loci on UKB 11 k validation GWAS summary statistics to test if the genotype‐morphometric associations are still significant (p < 5e−8).
Then genomic risk loci were defined by the FUMA online platform version 1.3.6 (http://fuma.ctglab.nl/; Watanabe, Taskesen, van Bochoven, & Posthuma, 2017). FUMA identified independent significant variants with their p values less than a predefined criterion as shown above, and independent of other significant variants at linkage disequilibrium (LD) r2 <0.6. These variant LDs refer to the 1,000 Genome phase 3 European population Reference Panel. If the LD blocks of independently significant variants are close (<250 kb based on the block's closest boundary variant), they are combined into a single genomic locus. The regional views of independent significant variants of Manhattan plots were plotted in LocusZoom (Pruim et al., 2010). We also searched for candidate genes whose mRNA expression is associated with the independent SNPs based on the GTEx V8 eQTL database (GTEx, 2015).
2.5. Pathway analyses among genetic variants, morphometry and WM
An LMM‐based mediation analysis was introduced to explore the potential causal relationships among the identified genetic loci, morphometric IDPs, and WM performance. The LMM‐based mediation method is a modification from the classical equation (VanderWeele, 2016):
| (1) |
| (2) |
| (3) |
In which Y represents the outcome variable, X is the exposure variable, M is the mediator variable, C is the fixed‐effect covariates, Z is the random‐effect covariates, ∅ is an intercept, c is the total effect, c ′ is the direct effect, k and β represent coefficients for random‐ and fixed‐effects, respectively.
The indirect effect (IE), or named mediation effect, is defined according to the production method as following (VanderWeele, 2016):
| (4) |
To precisely estimate the significance of the IE, a bias‐corrected and accelerated (BCa) bootstrap interval was used, which can correct for bias and skewness in the distribution of bootstrap estimates (Bonett & Seier, 2006).
Before mediation analysis, we carried out association analyses between the identified independent SNPs and 2‐back accuracy using the LMM model to identify which variants were associated with the WM performance (p < .05, uncorrected) after controlling confounders as mentioned in imaging‐WM association analysis. During mediation analysis, the identified independent SNPs associated with the WM were considered the exposure variables; the identified morphometric IDP was defined as a mediator; two‐back accuracy was defined as the outcome variable. Besides, sex, age, ICV, and education were regarded as the fixed‐effect confounders, and kinship coefficients were determined as random‐effect confounders. The Bootstrap method with 1,000 resampling was used to estimate the bias‐correction parameter, and the jackknife method was used to calculate the acceleration (skewness) parameter for the BCa interval. IEs that fall within 95% confidence interval (IC) was considered statistically significant. The mediation analysis was carried out using a customized package based on MediationToolbox (https://github.com/canlab/MediationToolbox) and glimix‐core 3.1.11 (https://pypi.org/project/glimix-core/).
2.6. Validation on the Caucasian population in the HCP dataset
To test whether population stratification caused by races would influence our findings, we repeated the above association and mediation analyses on a subset of 753 Caucasians of the HCP samples with the same configurations.
3. RESULTS
3.1. Morphometry—WM association results
The association results between 2‐back accuracy and 212 morphometric IDPs were shown in Figure 2. We found a significantly positive correlation between the left cuneus volume and 2‐back accuracy (T = 3.615, p = 3.150e−4, Cohen's d = 0.226, corrected using FWE method), indicating greater gray volume of the left cuneus cortex predicts higher WM performance.
FIGURE 2.
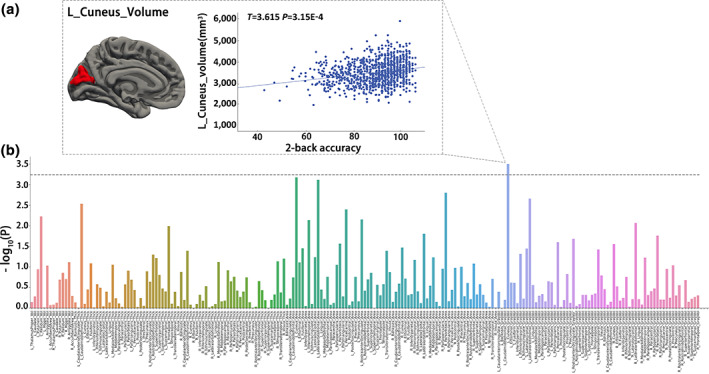
Associations between brain morphometry and working memory performance. A linear mixed model (LMM) was used to test the associations between 212 morphological imaging‐derived phenotypes (IDP) and 2‐back accuracy. A family‐wise error (FWE) method was used to correct the effective independent comparisons. (a) Identified left cuneus volume that is positively correlated with the 2‐back accuracy. (b) The x‐axis and y‐axis represent 212 phenotypes and ‐log10P of imaging‐working memory associations, respectively. The dashed horizontal line indicates the FWE threshold (p = 5.85e−4)
3.2. Morphometry—genome association results
The Manhattan and quantile‐quantile (QQ) graphs of left cuneus volume of UKB 33 k discovery+validation GWAS summary statistics were Figure S1. The LMM‐based GWAS plots of the HCP dataset with full samples and Caucasian samples were also shown in Figures S2 and S3. QQ plot demonstrated that the HCP GWAS results based on GEMMA showed no evidence of inflation in both full samples and Caucasian sub‐samples of the HCP dataset, indicating the efficiency of GEMMA in controlling the population stratification.
We screened 1,521 SNPs significantly associated with the identified left cuneus volume (p < 5e−8) based on the UKBB 33 k discovery+validation GWAS summary statistics. Furthermore, 108 candidate SNPs with congruent effects in both UKBB and HCP dataset were chosen based on the following three criteria: (a) SNPs survived under a threshold of p < 5e−8 in UKBB 33 k GWAS summary statistics; (b) SNPs had nominal P‐value lower than 0.05 in HCP GWAS statistical results; and (c) SNPs had the same effect directions in both HCP and UKBB datasets.
3.3. Independent SNPs identification
Eight significant independent variants of 108 SNPs with congruent effects in both the UKB and HCP were identified using FUMA (Table 1). One independent SNP was located on Chromosome 6 (rs74580701) and the other seven were located on Chromosome 14 (rs55633651, rs17095711, rs9323338, rs55643369, rs11158249, rs76119478, and rs140400396).
TABLE 1.
Identified independent significant SNPs that are associated with the left cuneus volume
| SNP | CHR | BP | A1/A2 | UKB_beta | UKB_P | HCP_beta | HCP_P |
|---|---|---|---|---|---|---|---|
| rs74580701 | 6 | 127000881 | G/A | −0.113 | 5.37e−9 | −0.198 | 0.048 |
| rs17095711 | 14 | 59580311 | A/G | −0.158 | 9.48e−23 | −0.149 | 0.040 |
| rs9323338 | 14 | 59588471 | C/A | −0.048 | 3.99e−9 | −0.100 | 0.014 |
| rs11158249 | 14 | 59605315 | C/G | −0.055 | 7.76e−9 | −0.100 | 0.033 |
| rs140400396 | 14 | 59864362 | T/C | −0.150 | 4.68e−19 | −0.190 | 0.035 |
| rs76119478 | 14 | 59622767 | C/A | −0.188 | 6.44e−24 | −0.236 | 0.026 |
| rs55633651 | 14 | 59551357 | A/G | −0.079 | 7.31e−14 | −0.107 | 0.040 |
| rs55643369 | 14 | 59595245 | C/T | −0.138 | 1.05e−34 | −0.160 | 0.008 |
Abbreviations: BP, base pair; CHR, chromosome; HCP, Human Connectome Project; SNP, single‐nucleotide polymorphism; UKB, UK biobank.
3.4. Genetic variants, morphometry, and WM pathway findings
LMM regression found a negative correlation between the rs76119478 polymorphism and 2‐back accuracy accuracy (T = −2.045, p = .041, Cohen's d = −.129; Table S1). Thus, an LMM‐based medication analysis was used to explore the potential pathway among rs76119478 polymorphism (exposure variable), left cuneus volume (mediator), and 2‐back accuracy (outcome variable). We found a significant effect of the left cuneus volume in mediating the regulation of locus rs76119478 (14q23.1) on 2‐back accuracy (IE = −0.007, 95% BCa CI = [−0.045, −0.003]; Figure 3b ).
FIGURE 3.
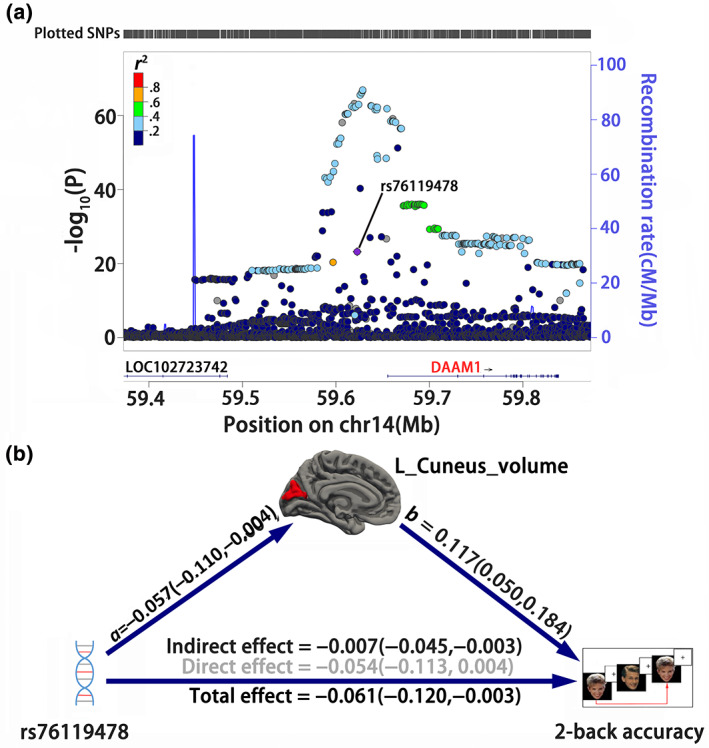
The mediation pathways among rs76119478, cuneus morphometry, and working memory. (a) Regional Manhattan plots of genome‐wide association study of left cuneus volume in UKB GWAS summary statistics are generated based on website http://locuszoom.org/ with linkage disequilibrium from 1000 Genomes Project Phase 3 EUR subjects. The identified variant rs76119478 is located at the intergene region and is nearest to the DAMM1 gene. (b) A linear mixed model‐based mediation analysis is used to test the mediation pathways from rs76119478 polymorphism, left cuneus volume, and 2‐back accuracy. A bias‐corrected and accelerated (BCa) bootstrap confidence interval (CI) is used to represent the statistical significance. Values in parentheses indicate the 95% BCa CI. Black values indicate significance under 95% BCa CI
3.5. Validation base on Caucasian sub‐samples of the HCP dataset
Association based on 753 Caucasian sub‐samples of the HCP dataset demonstrated that the left cuneus volume was still positively associated with the 2‐back accuracy (T = 3.218, p = 1.345e−3, Cohen's d = 0.235; Figure 4a ), whose effect size (Cohen's d) was comparable with the results using the full HCP samples (T = 3.615, p = 3.150e−4, Cohen's d = 0.226). On the association between imaging morphometry and genotypes, we also found a significantly negative association between the left cuneus volume and rs76119478 polymorphism in the Caucasians subset (T = −2.396, p = .017, Cohen's d = −0.175), whose effect size (Cohen's d) was also comparable with the results using the full HCP dataset (T = −2.232, p = .026, Cohen's d = −0.139). On the genotype ‐WM association, we only found a marginal negative association between rs76119478 polymorphism and 2‐back accuracy in the Caucasians subset (T = −1.322, p = .187, Cohen's d = −0.097), whose effect size was lower than the findings based on full HCP samples (T = −2.045, p = .041, Cohen's d = −0.129). Finally, the mediation analysis was repeated in the Caucasian population, and we also found a significant effect of left cuneus volume in mediating rs76119478 polymorphism on the 2‐back accuracy (IE = −0.009, 95% BCa CI = [−0.057, −0.003]; Figure 4b ).
FIGURE 4.
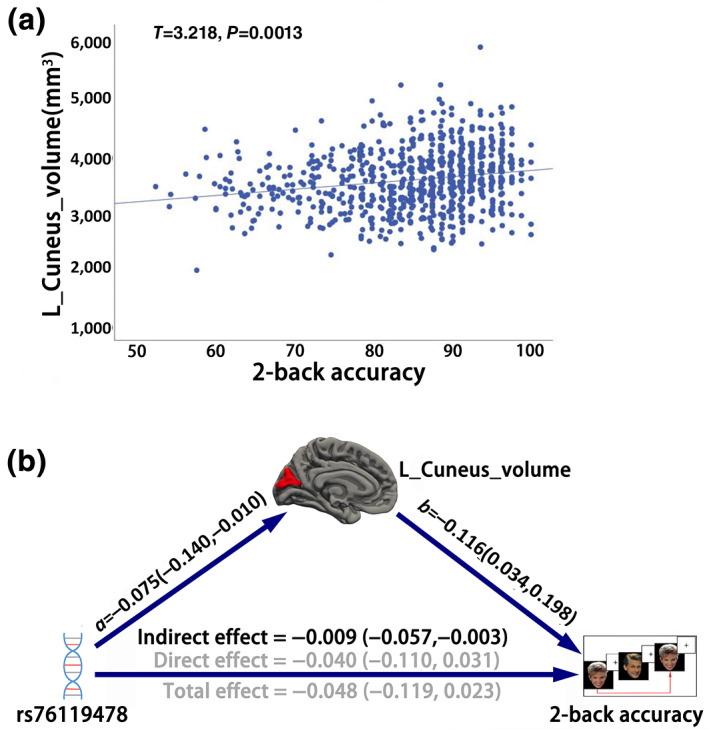
Association and mediation analysis based on Caucasian sub‐population of HCP dataset. (a) The scatter plot of the association between 2‐back accuracy and left cuneus volume using a linear mixed model (LMM). (b) LMM‐based mediation analysis is used to test the pathways among rs76119478 polymorphism, left cuneus volume, and working memory performance. A bias‐corrected and accelerated (BCa) bootstrap confidence interval (CI) is used to test the significance. Values in parentheses indicate the 95% BCa CI. Black values indicate significance under 95% BCa CI
3.6. Fine mapping of independent SNPs
In the previous steps, we found that rs76119478 may regulate WM via the left cuneus volume. We then used LocusZoom (Pruim et al., 2010) to locate the related gene based on the hg19 UCSC Genome Browser assembly and found its mapped gene within a ±250‐kilobase genomic region was a noncoding gene CTD‐2315A10.2 (ENSG00000258685), an intergene (or pseudogene) of AKR1B. Its nearest functional gene is DAAM1 (physical distance: 37524 bp; Figure 3a). We further searched for candidate genes whose mRNA expression is associated with the identified rs76119478 using the GTEx V8 eQTL database. We found this variant was significantly associated with the mRNA expression of gene L3HYPDH in the cerebellum (normalized effect size = −0.66, p = 9.5e−5; Figure S4).
4. DISCUSSION
In the present study, we aimed to explore the potential associations and pathways among genetic variants, brain morphometry and N‐back WM performance. We found that the left cuneus volume was significantly associated with 2‐back accuracy in the HCP samples. Based on the genome‐imaging GWAS, we identified eight independent SNPs congruently associated with left cuneus volume in both UKB and HCP datasets. Within the eight SNPs, we found a negative correlation between the rs76119478 polymorphism and 2‐back accuracy. Finally, mediation analysis elucidated a significant effect of left cuneus volume in mediating rs76119478 polymorphism on the 2‐back accuracy. These results were also replicated in Caucasian sub‐samples of HCP datasets. Our study provided a potential neurobiological pathway for WM.
The present study showed that left cuneus gray volume is significantly associated with WM performance. This morphometric phenotype was positively associated with 2‐back accuracy using a strict FWE correction method among the 212 morphometric phenotypes. The association was still replicated in a sub‐sample of the Caucasian population. Early studies have indicated that cuneus is the dorsal visual stream that participants in visual–spatial processing (Qin, Xuan, Liu, Jiang, & Yu, 2015), which is also a core hub of WM processing system named visual–spatial sketchpad (Baddeley, 2012; Chai et al., 2018). Our findings were consistent with two recent reports: one study reported that the activation of left cuneus was associated with both encoding and recall phases of a WM capacity task (reading span) (Bomyea, Taylor, Spadoni, & Simmons, 2018); and a second study reported a close association between the gray matter volume of left cuneus and 2‐back accuracy (Owens, Duda, Sweet, & MacKillop, 2018). The positive correlation between morphometry of the left cuneus and 2‐back accuracy implies a larger volume of this region contributes to better WM performance.
In the present study, we also found the left cuneus volume was congruently associated with eight independent SNPs in both UKB and HCP cohorts. This finding was consistent with early reports (Elliott et al., 2018; Hibar et al., 2015; Stein et al., 2012). Furthermore, among the eight independent SNPs, we found a new SNP, rs76119478, was significantly negatively associated with 2‐back accuracy, suggesting that this locus's mutation contributes to poor WM performance. Genome fine mapping demonstrated that this SNP is mapped on a nonencoding gene CTD‐2315A10.2, a pseudogene for AKR1B gene, whose functions are still unknown. An early study provided some indicative association of this gene with WM (Martins‐de‐Souza et al., 2012), which reported that AKR1B was differentially expressed in the occipital lobe in frontotemporal degeneration patients, whose WM performance are generally impaired (Libon et al., 2009; Stopford, Thompson, Neary, Richardson, & Snowden, 2012). The nearest protein‐encoding gene of rs76119478 is DAAM1. A recent study reported that this gene was significantly associated with the surface area of the cuneus (Grasby et al., 2020). DAMM1 had been indicated to be associated with synapse loss (Sellers et al., 2018). According to the GTEx database, we found that SNP rs76119478 is one trans‐eQTL of gene L3HYPDH in the brain tissue (Visser, Verhoeven‐Duif, & de Koning, 2012). The protein encoded by this gene is a dehydratase that converts trans‐3‐hydroxy‐l‐proline to delta (1)‐pyrroline‐2‐carboxylate. This enzyme may function to degrade dietary proteins containing trans‐3‐hydroxy‐l‐proline and other proteins such as collagen IV, which is involved in Arginine and proline metabolism (Kanehisa, Furumichi, Tanabe, Sato, & Morishima, 2017). A recent study reported that this gene was differentially methylated in the mitochondrial of autism spectrum disorder associated with Glutaryl‐CoA degradation (Stathopoulos et al., 2020). Although the functional indications of rs76119478 were sporadically reported, its exact molecular and cellular biological pathways on WM are still known.
The common genomic and WM association of left cuneus volume suggested this phenotype may bridge genetics and WM, which was supported by mediation pathway analysis. We reported a significant effect of left cuneus volume in mediating the regulation of rs76119478 polymorphism on 2‐back accuracy, and this medication effect can be validated in the Caucasian sub‐population in the HCP dataset. It should be noted that the direct pathway of rs76119478 on WM is not significant in the mediation analysis. Thus, rs76119478 may indirectly regulate the WM by exerting influence on the left cuneus volume. Specifically, the rs76119478 mutation might cause a reduction in left cuneus volume, and gray matter reduction in this region may lead to poorer WM accuracy.
Several metrological considerations strengthened the findings of the present studies. First, the identified IDPs and SNPs can be replicable in both UKB and HCP dataset with consensus effect direction and significance; second, the application of LMM in associations and medications can effectively control population stratification caused by both ancestry differences and cryptic relatedness (Sul et al., 2018; X. Zhou & Stephens, 2012), which thus made the statistics more powerful on HCP dataset. However, several limitations should not be neglected: The first issue is about the relatively small sample size of the HCP samples. HCP only contains 1,000+ genotyping imaging and cognition data; thus, it is difficult to identify significant genome‐WM and genome‐morphometric associations at a GWAS level directly. In the present study, we tried to increase the reliability of genome‐morphometric association by combining the UKB GWAS summary statistics with the HCP GWAS results. However, the genome‐WM association of rs76119478 is relatively weak and has not yet been replicated by large‐sample studies. Future studies with larger datasets are needed to test the genome‐imaging‐WM pathways. Second, it is unknown if racial differences exist on the associations among genome, brain morphometry, and WM. Thus, future studies based on larger race‐specific neuroimaging genetic datasets are promising. Fortunately, there are several big open‐shared neuroimaging genetic projects such as UK biobank (Bycroft et al., 2018), ENIGMA (Thompson et al., 2014), and CHIMGEN (Xu et al., 2020) that may reach this goal. Finally, although we found a potential statistical link among rs76119478, cuneus morphometry, and WM, the molecular and cellular experiments are necessary to elucidate and validate the biological pathways on WM.
5. CONCLUSIONS
In summary, this study not only validated previous studies on the relevance of brain morphology to genetics and WM, but also identified a new variant, rs76119478, which may indirectly modulate WM by exerting an effect on the morphology of the left cuneus.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of China (81771818, 81971599, 81701676, 82030053), Natural Science Foundation of Tianjin City (19JCYBJC25100), National Key Research and Development Program of China (2018YFC1314300), and Key Technology Research and Development Program of Tianjin (17ZXMFSY00090). The authors thank the individuals represented in the UKB, HCP datasets for their participation, and the research teams for their work in collecting, processing, and disseminating these datasets for analysis. HCP data were provided by the HCP, WU‐Minn Consortium (Principal Investigators: D. Van Essen and K. Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
He X, Li X, Fu J, et al. The morphometry of left cuneus mediating the genetic regulation on working memory. Hum Brain Mapp. 2021;42:3470–3480. 10.1002/hbm.25446
Xiaoxi He and Xi Li authors contributed equally to the manuscript.
Funding information Key Technology Research and Development Program of Tianjin, Grant/Award Number: 17ZXMFSY00090; National Key Research and Development Program of China, Grant/Award Number: 2018YFC1314300; Natural Science Foundation of China, Grant/Award Numbers: 81771818, 81971599, 81701676; 82030053; Natural Science Foundation of Tianjin City, Grant/Award Number: 19JCYBJC25100
Contributor Information
Zhaoxiang Ye, Email: zye@tmu.edu.cn.
Wen Qin, Email: wayne.wenqin@gmail.com.
DATA AVAILABILITY STATEMENT
All datasets used in this work were obtained from two publicly available datasets. The image‐derived phenotypes GWAS summary statistics of UKB can be found at the website (https://open.win.ox.ac.uk/ukbiobank/big40/). The HCP genotype was applied in dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001364.v1.p1); the image‐derived phenotypes were downloaded in the HCP website (https://db.humanconnectome.org/data/projects/HCP_1200).
REFERENCES
- Alfaro‐Almagro, F. , Jenkinson, M. , Bangerter, N. K. , Andersson, J. L. R. , Griffanti, L. , Douaud, G. , … Smith, S. M. (2018). Image processing and quality control for the first 10,000 brain imaging datasets from UKbiobank. NeuroImage, 166, 400–424. 10.1016/j.neuroimage.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, J. , Ono, Y. , & Wright, M. J. (2001). Genetic structure of spatial and verbal working memory. Behavior Genetics, 31(6), 615–624. 10.1023/a:1013353613591 [DOI] [PubMed] [Google Scholar]
- Baddeley, A. (2010). Working memory. Current Biology, 20(4), R136–R140. 10.1016/j.cub.2009.12.014 [DOI] [PubMed] [Google Scholar]
- Baddeley, A. (2012). Working memory: Theories, models, and controversies. Annual Review of Psychology, 63, 1–29. 10.1146/annurev-psych-120710-100422 [DOI] [PubMed] [Google Scholar]
- Blokland, G. A. M. , Wallace, A. K. , Hansell, N. K. , Thompson, P. M. , Hickie, I. B. , Montgomery, G. W. , … Wright, M. J. (2017). Genome‐wide association study of working memory brain activation. International Journal of Psychophysiology, 115, 98–111. 10.1016/j.ijpsycho.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobholz, J. A. , & Rao, S. M. (2003). Cognitive dysfunction in multiple sclerosis: A review of recent developments. Current Opinion in Neurology, 16(3), 283–288. 10.1097/01.wco.0000073928.19076.84 [DOI] [PubMed] [Google Scholar]
- Bomyea, J. , Taylor, C. T. , Spadoni, A. D. , & Simmons, A. N. (2018). Neural mechanisms of interference control in working memory capacity. Human Brain Mapping, 39(2), 772–782. 10.1002/hbm.23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonett, D. G. , & Seier, E. (2006). Confidence interval for a coefficient of dispersion in nonnormal distributions. Biometrical Journal, 48(1), 144–148. 10.1002/bimj.200410148 [DOI] [PubMed] [Google Scholar]
- Bycroft, C. , Freeman, C. , Petkova, D. , Band, G. , Elliott, L. T. , Sharp, K. , … Marchini, J. (2018). The UKbiobank resource with deep phenotyping and genomic data. Nature, 562(7726), 203–209. 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, W. J. , Abd Hamid, A. I. , & Abdullah, J. M. (2018). Working memory from the psychological and neurosciences perspectives: A review. Frontiers in Psychology, 9, 401. 10.3389/fpsyg.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli, E. T. , Kasperaviciute, D. , Attix, D. K. , Need, A. C. , Ge, D. , Gibson, G. , & Goldstein, D. B. (2010). Common genetic variation and performance on standardized cognitive tests. European Journal of Human Genetics, 18(7), 815–820. 10.1038/ejhg.2010.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, G. , Marioni, R. E. , Liewald, D. C. , Hill, W. D. , Hagenaars, S. P. , Harris, S. E. , … Deary, I. J. (2016). Genome‐wide association study of cognitive functions and educational attainment in UKbiobank (N=112 151). Molecular Psychiatry, 21(6), 758–767. 10.1038/mp.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R. S. , Segonne, F. , Fischl, B. , Quinn, B. T. , Dickerson, B. C. , Blacker, D. , … Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31(3), 968–980. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Ding, H. , Qin, W. , Jiang, T. , Zhang, Y. , & Yu, C. (2012). Volumetric variation in subregions of the cerebellum correlates with working memory performance. Neuroscience Letters, 508(1), 47–51. 10.1016/j.neulet.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Elliott, L. T. , Sharp, K. , Alfaro‐Almagro, F. , Shi, S. , Miller, K. L. , Douaud, G. , … Smith, S. M. (2018). Genome‐wide association studies of brain imaging phenotypes in UKbiobank. Nature, 562(7726), 210–216. 10.1038/s41586-018-0571-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Sotiropoulos, S. N. , Wilson, J. A. , Coalson, T. S. , Fischl, B. , Andersson, J. L. , … WU‐Minn HCP Consortium . (2013). The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage, 80, 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J. K. , Davenport, E. R. , Beaumont, M. , Jackson, M. A. , Knight, R. , Ober, C. , … Ley, R. E. (2016). Genetic determinants of the gut microbiome in UKtwins. Cell Host & Microbe, 19(5), 731–743. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby, K. L. , Jahanshad, N. , Painter, J. N. , Colodro‐Conde, L. , Bralten, J. , Hibar, D. P. , … Enhancing NeuroImaging Genetics through Meta‐Analysis Consortium (ENIGMA)—Genetics working group . (2020). The genetic architecture of the human cerebral cortex. Science, 367(6484), eaay6690. 10.1126/science.aay6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, M. D. , Kippenhan, J. S. , Callicott, J. H. , Rubinstein, D. Y. , Mattay, V. S. , Coppola, R. , & Berman, K. F. (2019). Sequence variation associated with SLC12A5 gene expression is linked to brain structure and function in healthy adults. Cerebral Cortex, 29(11), 4654–4661. 10.1093/cercor/bhy344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx . (2015). Human genomics. The genotype‐tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science, 348(6235), 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, A. , Fastenrath, M. , Ackermann, S. , Auschra, B. , Bickel, H. , Coynel, D. , … Papassotiropoulos, A. (2014). Converging genetic and functional brain imaging evidence links neuronal excitability to working memory, psychiatric disease, and brain activity. Neuron, 81(5), 1203–1213. 10.1016/j.neuron.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Ferrer, C. , Wellenius, G. A. , Tamayo, I. , Basagana, X. , Sunyer, J. , Vrijheid, M. , & Gonzalez, J. R. (2019). Comprehensive study of the exposome and omic data using rexposome bioconductor packages. Bioinformatics, 35(24), 5344–5345. 10.1093/bioinformatics/btz526 [DOI] [PubMed] [Google Scholar]
- Hibar, D. P. , Stein, J. L. , Renteria, M. E. , Arias‐Vasquez, A. , Desrivieres, S. , Jahanshad, N. , … Medland, S. E. (2015). Common genetic variants influence human subcortical brain structures. Nature, 520(7546), 224–229. 10.1038/nature14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, H. (2013). Memory loss in Alzheimer's disease. Dialogues in Clinical Neuroscience, 15(4), 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Furumichi, M. , Tanabe, M. , Sato, Y. , & Morishima, K. (2017). KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research, 45(D1), D353–D361. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt, K. H. , Kochunov, P. , Winkler, A. M. , Laird, A. R. , Almasy, L. , Duggirala, R. , … Glahn, D. C. (2010). A multimodal assessment of the genetic control over working memory. The Journal of Neuroscience, 30(24), 8197–8202. 10.1523/JNEUROSCI.0359-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. X. , Yeung, J. M. , Cherny, S. S. , & Sham, P. C. (2012). Evaluating the effective numbers of independent tests and significant p‐value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics, 131(5), 747–756. 10.1007/s00439-011-1118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libon, D. J. , McMillan, C. , Gunawardena, D. , Powers, C. , Massimo, L. , Khan, A. , … Grossman, M. (2009). Neurocognitive contributions to verbal fluency deficits in frontotemporal lobar degeneration. Neurology, 73(7), 535–542. 10.1212/WNL.0b013e3181b2a4f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Dai, X. , Tao, W. , Liu, H. , Li, H. , Yang, C. , … Zhang, Z. (2018). APOE influences working memory in non‐demented elderly through an interaction with SPON1 rs2618516. Human Brain Mapping, 39(7), 2859–2867. 10.1002/hbm.24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny, A. , Cohen, K. , Tik, N. , Tsarfaty, G. , Rosca, P. , & Weinstein, A. (2018). The effects of synthetic cannabinoids (SCs) on brain structure and function. European Neuropsychopharmacology, 28(9), 1047–1057. 10.1016/j.euroneuro.2018.07.095 [DOI] [PubMed] [Google Scholar]
- Loh, P. R. , Kichaev, G. , Gazal, S. , Schoech, A. P. , & Price, A. L. (2018). Mixed‐model association for biobank‐scale datasets. Nature Genetics, 50(7), 906–908. 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins‐de‐Souza, D. , Guest, P. C. , Mann, D. M. , Roeber, S. , Rahmoune, H. , Bauder, C. , … Bahn, S. (2012). Proteomic analysis identifies dysfunction in cellular transport, energy, and protein metabolism in different brain regions of atypical frontotemporal lobar degeneration. Journal of Proteome Research, 11(4), 2533–2543. 10.1021/pr2012279 [DOI] [PubMed] [Google Scholar]
- Martinussen, R. , Hayden, J. , Hogg‐Johnson, S. , & Tannock, R. (2005). A meta‐analysis of working memory impairments in children with attention‐deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry, 44(4), 377–384. 10.1097/01.chi.0000153228.72591.73 [DOI] [PubMed] [Google Scholar]
- Miller, G. , Galanter, E. , & Pribram, K. (1960). Plans and the structure of behavior. New York: Holt, Rinehart and Winston, Inc. [Google Scholar]
- Need, A. C. , Attix, D. K. , McEvoy, J. M. , Cirulli, E. T. , Linney, K. L. , Hunt, P. , … Goldstein, D. B. (2009). A genome‐wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Human Molecular Genetics, 18(23), 4650–4661. 10.1093/hmg/ddp413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, A. M. , McMillan, K. M. , Laird, A. R. , & Bullmore, E. (2005). N‐back working memory paradigm: A meta‐analysis of normative functional neuroimaging studies. Human Brain Mapping, 25(1), 46–59. 10.1002/hbm.20131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, M. M. , Duda, B. , Sweet, L. H. , & MacKillop, J. (2018). Distinct functional and structural neural underpinnings of working memory. NeuroImage, 174, 463–471. 10.1016/j.neuroimage.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim, R. J. , Welch, R. P. , Sanna, S. , Teslovich, T. M. , Chines, P. S. , Gliedt, T. P. , … Willer, C. J. (2010). LocusZoom: Regional visualization of genome‐wide association scan results. Bioinformatics, 26(18), 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, W. , Xuan, Y. , Liu, Y. , Jiang, T. , & Yu, C. (2015). Functional connectivity density in congenitally and late blind subjects. Cerebral Cortex, 25(9), 2507–2516. 10.1093/cercor/bhu051 [DOI] [PubMed] [Google Scholar]
- Salat, D. H. , Kaye, J. A. , & Janowsky, J. S. (2002). Greater orbital prefrontal volume selectively predicts worse working memory performance in older adults. Cerebral Cortex, 12(5), 494–505. 10.1093/cercor/12.5.494 [DOI] [PubMed] [Google Scholar]
- Satizabal, C. L. , Adams, H. H. H. , Hibar, D. P. , White, C. C. , Knol, M. J. , Stein, J. L. , … Ikram, M. A. (2019). Genetic architecture of subcortical brain structures in 38,851 individuals. Nature Genetics, 51(11), 1624–1636. 10.1038/s41588-019-0511-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J. E. , Jansen, P. R. , Stringer, S. , Watanabe, K. , Bryois, J. , de Leeuw, C. A. , … Posthuma, D. (2018). Genome‐wide association meta‐analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nature Genetics, 50(7), 912–919. 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers, K. J. , Elliott, C. , Jackson, J. , Ghosh, A. , Ribe, E. , Rojo, A. I. , … Killick, R. (2018). Amyloid beta synaptotoxicity is Wnt‐PCP dependent and blocked by fasudil. Alzheimers Dement, 14(3), 306–317. 10.1016/j.jalz.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Douaud, G. , Chen, W. , Hanayik, T. , Alfaro‐Almagro, F. , Sharp, K. , & Elliott, L. T. (2020). Enhanced brain imaging genetics in UKbiobank. bioRxiv . https://doi.org/10.1101/2020.07.27.223545
- Stathopoulos, S. , Gaujoux, R. , Lindeque, Z. , Mahony, C. , Van Der Colff, R. , Van Der Westhuizen, F. , & O'Ryan, C. (2020). DNA methylation associated with mitochondrial dysfunction in a south African autism Spectrum disorder cohort. Autism Research, 13(7), 1079–1093. 10.1002/aur.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J. L. , Medland, S. E. , Vasquez, A. A. , Hibar, D. P. , Senstad, R. E. , Winkler, A. M. , … Enhancing Neuro Imaging Genetics through Meta‐Analysis (ENIGMA) Consortium . (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44(5), 552–561. 10.1038/ng.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopford, C. L. , Thompson, J. C. , Neary, D. , Richardson, A. M. , & Snowden, J. S. (2012). Working memory, attention, and executive function in Alzheimer's disease and frontotemporal dementia. Cortex, 48(4), 429–446. 10.1016/j.cortex.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Sul, J. H. , Martin, L. S. , & Eskin, E. (2018). Population structure in genetic studies: Confounding factors and mixed models. PLoS Genetics, 14(12), e1007309. 10.1371/journal.pgen.1007309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Stein, J. L. , Medland, S. E. , Hibar, D. P. , Vasquez, A. A. , Renteria, M. E. , … Alzheimer's Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group . (2014). The ENIGMA Consortium: Large‐scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior, 8(2), 153–182. 10.1007/s11682-013-9269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. , Yacoub, E. , Ugurbil, K. , & WU‐Minn HCP Consortium . (2013). The WU‐Minn human Connectome project: An overview. NeuroImage, 80, 62–79. 10.1016/j.neuroimage.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen, D. C. , Ugurbil, K. , Auerbach, E. , Barch, D. , Behrens, T. E. , Bucholz, R. , … WU‐Minn HCP Consortium . (2012). The human Connectome project: A data acquisition perspective. NeuroImage, 62(4), 2222–2231. 10.1016/j.neuroimage.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Snellenberg, J. X. , Girgis, R. R. , Horga, G. , van de Giessen, E. , Slifstein, M. , Ojeil, N. , … Abi‐Dargham, A. (2016). Mechanisms of working memory impairment in schizophrenia. Biological Psychiatry, 80(8), 617–626. 10.1016/j.biopsych.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele, T. J. (2016). Mediation analysis: A Practitioner's guide. Annual Review of Public Health, 37, 17–32. 10.1146/annurev-publhealth-032315-021402 [DOI] [PubMed] [Google Scholar]
- Visser, W. F. , Verhoeven‐Duif, N. M. , & de Koning, T. J. (2012). Identification of a human trans‐3‐hydroxy‐L‐proline dehydratase, the first characterized member of a novel family of proline racemase‐like enzymes. The Journal of Biological Chemistry, 287(26), 21654–21662. 10.1074/jbc.M112.363218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler, C. , Gschwind, L. , Coynel, D. , Freytag, V. , Milnik, A. , Egli, T. , … Papassotiropoulos, A. (2014). Substantial SNP‐based heritability estimates for working memory performance. Translational Psychiatry, 4, e438. 10.1038/tp.2014.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Taskesen, E. , van Bochoven, A. , & Posthuma, D. (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Guo, L. , Cheng, J. , Wang, M. , Geng, Z. , Zhu, W. , … The CHIMGEN Consortium . (2020). CHIMGEN: A Chinese imaging genetics cohort to enhance cross‐ethnic and cross‐geographic brain research. Molecular Psychiatry, 25(3), 517–529. 10.1038/s41380-019-0627-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, B. T. , Krienen, F. M. , Eickhoff, S. B. , Yaakub, S. N. , Fox, P. T. , Buckner, R. L. , … Chee, M. W. (2015). Functional specialization and flexibility in human association cortex. Cerebral Cortex, 25(10), 3654–3672. 10.1093/cercor/bhu217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Yan, T. , Wang, Y. , Zhang, Q. , Zhao, W. , Chen, X. , … Li, J. (2018). Polymorphism in schizophrenia risk gene MIR137 is associated with the posterior cingulate cortex's activation and functional and structural connectivity in healthy controls. NeuroImage: Clinical, 19, 160–166. 10.1016/j.nicl.2018.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. , Luo, T. , Li, T. , Li, Y. , Zhang, J. , Shan, Y. , … Zhu, H. (2019). Genome‐wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co‐architecture with cognitive and mental health traits. Nature Genetics, 51(11), 1637–1644. 10.1038/s41588-019-0516-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W. , Zhao, Z. , Nielsen, J. B. , Fritsche, L. G. , LeFaive, J. , Gagliano Taliun, S. A. , … Lee, S. (2020). Scalable generalized linear mixed model for region‐based association tests in large biobanks and cohorts. Nature Genetics, 52(6), 634–639. 10.1038/s41588-020-0621-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , & Stephens, M. (2012). Genome‐wide efficient mixed‐model analysis for association studies. Nature Genetics, 44(7), 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
All datasets used in this work were obtained from two publicly available datasets. The image‐derived phenotypes GWAS summary statistics of UKB can be found at the website (https://open.win.ox.ac.uk/ukbiobank/big40/). The HCP genotype was applied in dbGAP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001364.v1.p1); the image‐derived phenotypes were downloaded in the HCP website (https://db.humanconnectome.org/data/projects/HCP_1200).


