Abstract
As sessile organisms, plants have evolved sophisticated ways to constantly gauge and adapt to changing environmental conditions including extremes that may be harmful to their growth and development and are thus perceived as stress. In nature, stressful events are often chronic or recurring and thus an initial stress may prime a plant to respond more efficiently to a subsequent stress event. An epigenetic basis of such stress memory was long postulated and in recent years it has been shown that this is indeed the case. High temperature stress has proven an excellent system to unpick the molecular basis of somatic stress memory, which includes histone modifications and nucleosome occupancy. This review discusses recent findings and pinpoints open questions in the field.
Current Opinion in Plant Biology 2021, 61:102007
This review comes from a themed issue on Epigenetics
Edited by Mary Gehring and François Roudier
For a complete overview see the Issue and the Editorial
Available online 8th February 2021
https://doi.org/10.1016/j.pbi.2021.102007
1369-5266/© 2021 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
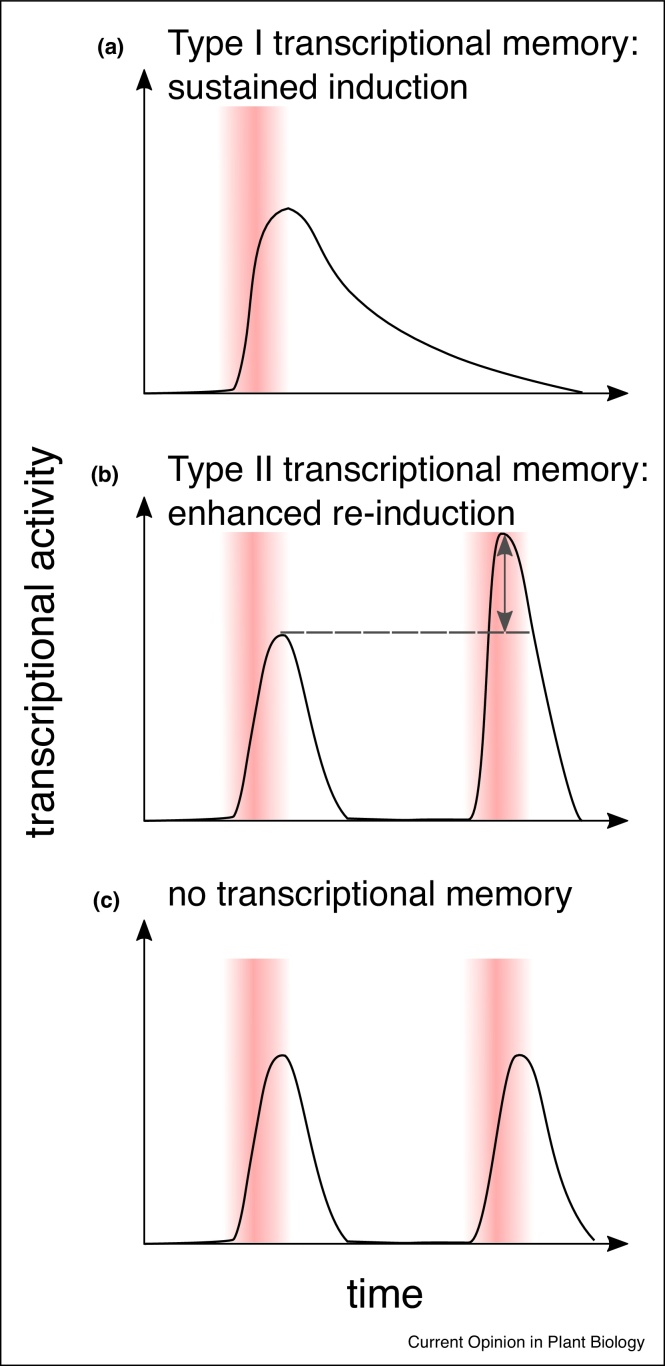
Stress in plants is defined as a condition that negatively interferes with growth and development and may severely damage or even kill the plant. Such stressful conditions include extreme temperatures, drought, salinity as well as pathogen and herbivore attacks. Plants are able to quickly adapt to stressful environments by changing their gene expression programme within minutes after the onset of a stress condition [1]. Exposure to stress is also able to prime a plant for a repeated exposure to the same or different stress (Table 1) [2, 3, 4, 5, 6, 7, 8, 9]. Depending on the duration of the memory relative to the life cycle of the organism, somatic, intergenerational and transgenerational memory are distinguished (Table 1). Somatic stress priming frequently involves transcriptional memory of which there are two types (Figure 1). One is a sustained activation (or repression) of a gene in response to a short stimulus that significantly exceeds the duration of that stimulus. The second type is a modified transcriptional response after a recurrent stress stimulus if a plant has been primed, compared to a naïve plant, for example an enhanced re-induction. Notably, during the stress-free memory phase, transcription of stress-induced genes returns to baseline levels in this second type of memory. As a subgroup of the second type it is also possible that the primary stress stimulus does not elicit a response but only the second stress does [4,10].
Table 1.
Terms describing different aspects of stress memory
| Priming | Process through which a stress stimulus prepares a plant for a future stress exposure; evidenced by a modified response of a primed plant to a repeated stress relative to a naïve plant’s response; described in plant immunity and in various abiotic stresses |
| Memory | Process of maintenance of the primed state over time, which may be apparent in altered gene activation patterns (→ transcriptional memory) |
| Transcriptional memory | Altered gene expression responses in a primed plant; either a sustained change after a short stimulus or a modified gene activation pattern upon a recurring stress. |
| Somatic (stress) memory | (Stress) memory that is limited in duration to the lifetime of an organism |
| Intergenerational (stress) memory | (Stress) memory that extends in duration into the next generation |
| Transgenerational (stress) memory | (Stress) memory that is maintained for at least two stress-free generations |
Figure 1.
Types of stress-induced transcription patterns.
(a) Transcriptional memory can manifest itself as stress-induced sustained induction (depicted here) or repression of gene expression that significantly exceeds the duration of the stress cue. (b) Another type of transcriptional memory describes modified gene expression upon repeated exposure to similar stress cues. Depicted here is enhanced re-induction where a gene is induced to higher levels upon the second compared to the first stress exposure. All four combinations of sequential induction/repression exist. (c) In non-memory transcription transient induction (depicted here) or repression of gene expression occurs regardless of a previous stress cue to a similar degree upon repeated stress exposure. Red bars represent stress cues.
Chromatin structure is an important determinant of gene regulation and is affected by nucleosome positioning, histone variants and posttranslational modifications of histones. Depending on their nature and position, these modifications can promote or repress transcription by altering chromatin accessibility or interaction with protein complexes. Histone acetylation is usually associated with active transcription and correlates with the rate of transcription [11]. Histone H3K4 can be mono-methylated, di-methylated or trimethylated. H3K4 trimethylation (me3) is closely associated with actively transcribed genes in several organisms including Arabidopsis thaliana [12,13]. In contrast, H3K4 dimethylation (me2) and monomethylation (me1) are not associated with transcriptional activation in A. thaliana [13]. Conversely, H3K27me3 is a repressive mark in genes that is deposited by the Polycomb repressive complex 2 [14].
In this review, we will use the transcriptional memory after HS in A. thaliana as an example of stress-induced somatic plant memory and will draw connections to other stresses where appropriate.
Transcriptional memory after heat stress (HS)
Priming and memory after exposure to high temperature stress, that is, heat stress (HS), has proven to be an efficient experimental system in which the underlying molecular mechanisms of priming and memory have started to be unravelled in the past few years [7,15, 16, 17, 18]. The two types of transcriptional memory were found to be activated after HS on overlapping sets of genes; however, not every gene shows both types of transcriptional memory [7,19•]. The sustained activation memory was observed after a two-step acclimation treatment that mimics the natural gradual increase in temperature. It typically lasts for a couple of days. By quantifying the level of unspliced transcripts, it was possible to show that the sustained accumulation of transcripts reflects ongoing transcriptional activity rather than transcript stability [15,16]. Several recent studies globally assessed transcript stability and found it to be generally in the range of minutes to hours [20,21••,22,23], which further supports the notion that the lasting accumulation of transcripts is caused by ongoing transcription. The enhanced re-induction memory is most prominently found on the ASCORBATE PEROXIDASE2 (APX2) gene [7,19•]. Recent work found that it lasts for at least six days after a moderate HS. Furthermore, the authors showed that it is most likely caused by a faster re-induction of transcription, suggesting that during the memory phase the locus remains in a state of elevated transcriptional competence, despite being transcriptionally inactive. Such a state could either be achieved by marking or shifting the nucleosomes at the locus, by accumulating stalled transcription initiation complexes, or by binding of transcription factors that are on their own not sufficient for activating transcription. In principle, these might be locus-specific activators or components of the general transcriptional machinery. A combination of different factors is also possible.
Interaction with the general transcriptional machinery
Controlling the elongation of RNA polymerase II has emerged as a major step in gene expression control in animals [24, 25, 26, 27]. In plants, stalled polymerase has been implicated in stress priming after dehydration stress by detecting elevated levels of phosphorylated RNA Pol II [2,28]. It was also implicated in gene expression control by premature termination of transcription [29••]. Furthermore, components of the Mediator co-activator complex have recently been shown to be involved in rapid gene induction after environmental stimuli [30,31••]. Similar observations have been made for transcriptional memory in yeast [32]. Thus, while the mechanisms may differ between plants and animals, it is conceivable that also in plants partial assembly of RNA PolII transcription complexes contributes to transcriptional memory.
Histone modifications
Stress-induced transcriptional memory was reported for recurring heat stress, drought stress, hyperosmotic stress and defense priming [2, 3, 4, 5, 6, 7]. In all cases, primed genes were associated with lasting changes in chromatin modifications, such as H3K4 hyper-methylation or loss of H3K27me3 (Figure 2). After a priming HS, histone H3K4me2 and H3K4me3 were elevated for up to five days compared to a plant in the naïve state [7,19•]. Interestingly, H3K4me3 started to decline earlier than H3K4me2, suggesting that the latter may be responsible for the long-term transcriptional memory. The accumulation of these modifications after HS is not a default consequence of HS-mediated transcriptional activation, as they did not accumulate at other HS-inducible genes that did not show memory-like gene expression patterns [7]. The accumulation of H3K4me2 and H3K4me3 was dependent on the HSFA2 transcription factor [7,33]. However, HSFA2 binds to these loci only transiently and dissociates from the locus before the histone methylation peaks, suggesting the involvement of other processes in the establishment and maintenance of these marks. These findings make H3K4me2/me3 good candidates for mediating the transcriptional memory in the absence of ongoing transcription. Previously, H3K4 hyper-methylation was proposed to act as a mark of recent transcriptional activity of a locus and may mediate a modified response following a recurrent stimulus in yeast [32,34]. Elevated H3K4me3 is also important for rapid (de-)activation of genes by environmental stimuli and may prevent gene silencing [34]. Notably, H3K4 hyper-methylation was also associated with somatic stress memory in response to dehydration stress and salt stress [2,3,35]. It remains an open question which enzymes are involved and what factors cause the locus-specific accumulation of H3K4 hyper-methylation.
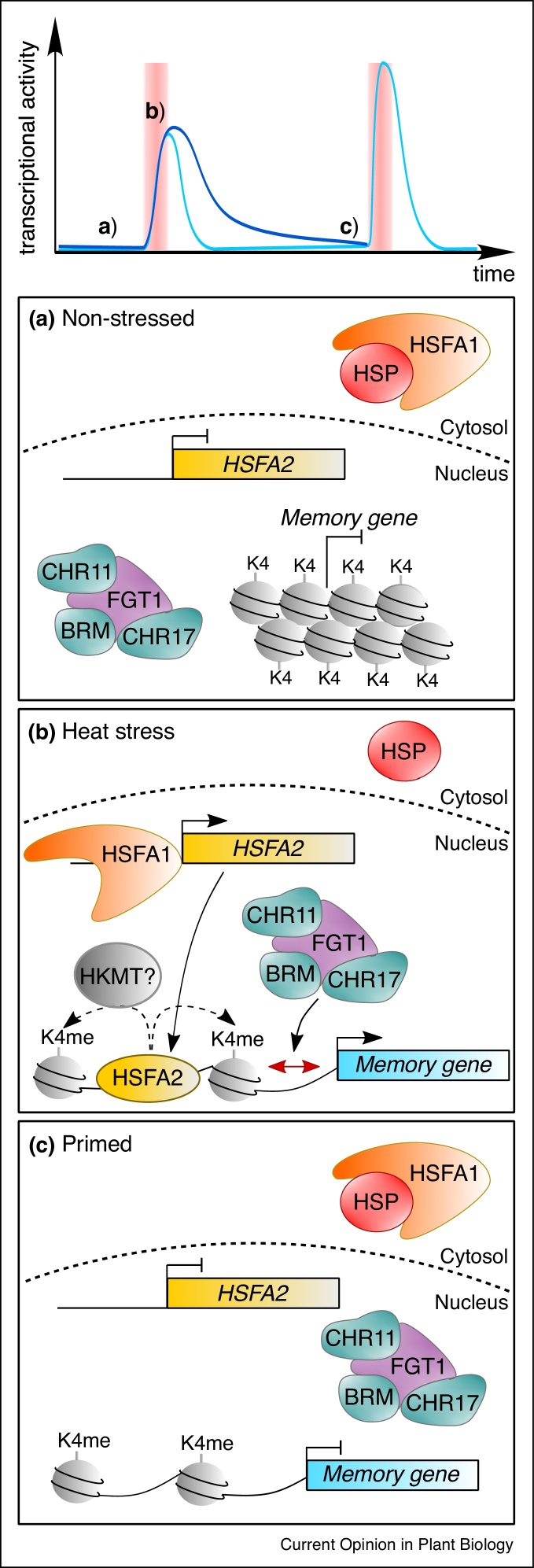
Figure 2.
Summary of mechanisms of heat stress memory.
The top panel illustrates the three stages detailed in (a)–(c). (a) Before HS, HSFA1 proteins are retained in the cytosol by chaperone proteins (HSP). Memory genes are inactive and the chromatin around them is dense in nucleosomes and poor in histone H3K4 methylation. (b) During HS, chaperone proteins release HSFA1 proteins. Free HSFA1 translocates to the nucleus and activates a set of heat stress-responsive genes, including HSFA2. HSFA2 binds the promoter regions of memory genes and sustains activation of their transcription. HSFA2 binding to memory genes also triggers H3K4 methylation of the neighbouring nucleosomes. Concomitantly, a chromatin remodeling complex that includes FGT1, BRM, CHR11 and CHR17 targets the TSS of memory genes to reduce nucleosome occupancy. (c) Up to several days after a HS, memory genes are primed. In absence of HSFA2 binding, the chromatin around memory genes is still enriched in H3K4 methylation, and shows low nucleosome occupancy. Thus, memory genes show a sustained expression and/or will be more quickly re-induced if another HS happens.
Interestingly, a positive feedback loop between HSFA2 and the H3K27me3 histone demethylase RELATIVE OF EARLY FLOWERING 6 (REF6) also contributes to a transgenerational memory of HS that drives early flowering while attenuating immunity [36•,37].
Nucleosome remodeling
The positioning and density of nucleosomes is an important determinant of the regulation of gene expression. Nucleosomes are obstacles to RNA PolII complexes. Thus, they have to be disassembled before RNA PolII can transcribe and are immediately reassembled after it has passed [38,39]. Thus, a lower nucleosome density at a given locus increases the ease with which it can be transcribed. Genetic analysis has identified a role for nucleosome remodeling in HS memory [16]. The FORGETTER1 (FGT1) protein is required for sustained induction of memory loci after HS [16]. The gene is highly conserved and contains two helicase domains and a plant homeodomain (PHD) that binds histones. FGT1 interacts directly with chromatin remodelers of the SWI/SNF and ISWI families and thereby mediates low nucleosome occupancy at memory loci throughout the memory phase [16]. Correspondingly, the SWI/SNF chromatin remodeler BRAHMA and the ISWI remodelers CHR11 and CHR17 were also found to be required for physiological HS memory. Thus, FGT1 may be acting as a hub to coordinate chromatin remodelers of different families to regulate nucleosome occupancy. Interestingly, FGT1 appears to be dispensable for cold stress memory [40].
The CHROMATIN ASSEMBLY FACTOR-1 (CAF-1) histone chaperone provides an additional link between priming and nucleosome positioning [41]. Mutants of CAF-1 show a constitutive priming response against pathogens and this was associated with low nucleosome occupancy and increased H3K4me3 at primed defense-response genes. CAF-1 is a histone chaperone that acts in depositing histone H3H4 tetramers into newly replicated DNA. This suggests that the regulated deposition of H3H4 tetramers onto replicated DNA is essential for priming responses and may hint to a role for the inheritance of histone modifications.
Open questions
How does the inheritance over mitoses work?
Enhanced re-induction memory is active for roughly a week [6,19•]. This is a relatively short period of time, compared to well-known repressive epigenetic responses that are often active throughout the whole life cycle of a plant or a large fraction of it. These repressive epigenetic phenomena include the silencing of transposons and repeats, as well as vernalization, and are in large part based on DNA methylation or polycomb-group mediated silencing [42, 43, 44, 45]. These mechanisms are well-known for being able to create sustained repressive chromatin states. In contrast, enhanced re-induction memory provides a sustained state of latent gene activation, and the maintenance of such a latent hyper-activation is mechanistically much less researched/understood. While we have reviewed factors that play a role in it, the mechanistic basis of its inheritance across replication and cell division is not clear. Based on the observed growth during the active memory period, it is obvious that the primed chromatin state needs to be inherited across cell divisions. An open question is whether this inheritance through cell division is a cis effect that is solely based on the chromatin conformation (histone modifications and nucleosome occupancy) at the time of replication, or whether it is a trans effect, that relies on soluble factors that mediate faithful copying of the chromatin conformation through replication. One candidate trans factor is the BRUSHY1/TONSOKU/MGOUN3 (BRU1/TSK/MGO3) protein [46,47]. BRU1 is required for the sustained activation of HS memory genes [48•]. Whether it is also required for enhanced re-induction memory remains to be investigated. Interestingly, BRU1 has previously been implicated in the faithful inheritance of chromatin states across DNA replication and cell division. It was originally identified as a factor that is implicated in the epigenetic inheritance of transcriptional silencing and the DNA damage response [46,47]. The mutant has similar phenotypes to that of the CAF-1 histone chaperone [47]. More recently, a BRU1 orthologue from mammals was found to bind to single-stranded DNA and newly incorporated nucleosomes after replication [49,50]. This raises the exciting hypothesis that BRU1 is directly involved in copying epigenetic marks onto new nucleosomes during replication and DNA damage repair.
Certain types of differentiated plant cells undergo several rounds of DNA replication without subsequent cell division, which is referred to as endoreduplication. It is an interesting question whether inheritance of chromatin modifications during DNA replication is in place in those cells.
Is the memory gradual or digital?
At the whole seedling level, the transcriptional memory appears to decrease gradually with the duration of the lag phase. One unresolved question is how many different states this apparently gradual decline is based on at the single cell level. For the FLC gene, which is epigenetically silenced during vernalization by H3K27me3 hyper-methylation, it has been shown that an individual gene locus can either adopt an ON state or an OFF state and that this state is faithfully inherited during replication [51,52]. This strongly indicates a cis-encoded memory. For the stress-induced re-induction memory, it remains to be investigated whether the apparent gradual decrease in the memory is caused by gradual reduction of the number of cells with active memory, or all cells exhibiting a weaker memory. Analyses of gene expression at single-cell resolution will yield answers to these exciting questions.
Why (and how) does the memory end?
For many stress memory phenomena with physiological benefit the duration was not systematically investigated, but is generally believed to be around one week [2,6,35]. For HS memory, physiological benefits last for at least three days while at the molecular level the enhanced re-induction was still detected after six days [15,19•]. The mechanisms that limit the duration of the memory are still unresolved. It is tempting to speculate that unlimited somatic stress memory may not be advantageous in evolutionary terms. In nature, a plant is likely exposed to a number of different stresses over a season and remembering them all may be more costly than the actual benefits from such stress priming. Accordingly, a pioneering study has shown that disease priming provides fitness advantages in a situation where the pathogen recurs, but not in a stress-free environment, where it was slightly disadvantageous [53].
How many mechanisms are there for stress memory?
Several epigenetic modifications have been implicated in the stress memory after priming, such as histone H3K4 hyper-methylation, histone H3K27 methylation, and nucleosome occupancy. Whether these are all organized into one linear pathway, or whether they are acting independently on the same locus, or whether different loci show preferences for different pathways, is currently poorly understood. To improve our understanding it will be necessary to investigate the interconnections between different pathways. It is, however, clear that different stresses trigger similar epigenetic signatures, although mechanisms are not necessarily conserved. For example, as mentioned before FGT1 does not appear to be involved in cold stress memory [40]. Although thermosensors are not directly involved in the stress memory, it is nevertheless interesting to note from recent work that several independent thermosensors have evolved [54,55,56••,57]. Analogously, multiple mechanisms may be active in stress memory.
The involvement of chromatin organization as a way to mark loci for altered re-induction upon a recurrent stress is intuitive and has been confirmed in several independent studies with independent stressors. However, the question remains whether there are chromatin-independent mechanisms of stress memory. Although beyond the topic of this review, recent work suggests that a component of HS memory is localized at the plasma membrane [58]. In a forward genetic screen the authors identified a protein phosphatase that interacts with phospholipase D and both proteins have a specific memory defect [58]. How diverse cellular mechanisms are integrated with regulation at the chromatin level (if at all) remains to be investigated.
Conclusion
Temporal dynamics are an important but under researched aspect of biological questions. Lifetimes of individual organic molecules are mostly short, yet they sustain processes that exceed the lifetime of any individual molecule by several orders of magnitude. Understanding the priming and memory of environmental stress responses will be a fruitful topic for such studies that will be much aided by new technologies such as super resolution technology and single cell-omics techniques. Insights into the molecular basis of priming and memory may also lead to novel approaches in breeding stress-resistant crop plants.
Conflict of interest statement
The authors declare that they have no competing interests.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the European Research Council (CoG 725295 CHROMADAPT).
References
- 1.Zhu J.-K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding Y., Fromm M., Avramova Z. Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat Commun. 2012;3:740. doi: 10.1038/ncomms1732. [DOI] [PubMed] [Google Scholar]
- 3.Sani E., Herzyk P., Perrella G., Colot V., Amtmann A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013;14:R59. doi: 10.1186/gb-2013-14-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaskiewicz M., Conrath U., Peterhänsel C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011;12:50–55. doi: 10.1038/embor.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lämke J., Bäurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017;18:124. doi: 10.1186/s13059-017-1263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh P., Yekondi S., Chen P.W., Tsai C.H., Yu C.W., Wu K., Zimmerli L. Environmental history modulates arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell. 2014;26:2676–2688. doi: 10.1105/tpc.114.123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lämke J., Brzezinka K., Altmann S., Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016;35:162–175. doi: 10.15252/embj.201592593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilker M., Schwachtje J., Baier M., Balazadeh S., Bäurle I., Geiselhardt S., Hincha D.K., Kunze R., Mueller-Roeber B., Rillig M.C. Priming and memory of stress responses in organisms lacking a nervous system. Biol Rev Cambridge Philos Soc. 2016;91:1118–1133. doi: 10.1111/brv.12215. [DOI] [PubMed] [Google Scholar]
- 9.Crisp P.A., Ganguly D., Eichten S.R., Borevitz J.O., Pogson B.J. Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv. 2016;2 doi: 10.1126/sciadv.1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N., Staswick P.E., Avramova Z. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ. 2016;39:2515–2529. doi: 10.1111/pce.12806. [DOI] [PubMed] [Google Scholar]
- 11.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 12.Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugues A., Jacobs C.S., Roudier F. Mitotic inheritance of PRC2-mediated silencing: mechanistic insights and developmental perspectives. Front Plant Sci. 2020;11:262. doi: 10.3389/fpls.2020.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stief A., Altmann S., Hoffmann K., Pant B.D., Scheible W.-R., Bäurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell. 2014;26:1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brzezinka K., Altmann S., Czesnick H., Nicolas P., Górka M., Benke E., Kabelitz T., Jähne F., Graf A., Kappel C. Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife. 2016;5 doi: 10.7554/eLife.17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sedaghatmehr M., Mueller-Roeber B., Balazadeh S. The plastid metalloprotease FtsH6 and small heat shock protein HSP21 jointly regulate thermomemory in Arabidopsis. Nat Commun. 2016;7:12439. doi: 10.1038/ncomms12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bäurle I., Trindade I. Chromatin regulation of somatic abiotic stress memory. J Exp Bot. 2020;71:5269–5279. doi: 10.1093/jxb/eraa098. [DOI] [PubMed] [Google Scholar]
- 19•.Liu H.C., Lamke J., Lin S.Y., Hung M.J., Liu K.M., Charng Y.Y., Baurle I. Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J. 2018;95:401–413. doi: 10.1111/tpj.13958. [DOI] [PubMed] [Google Scholar]; The authors characterize different aspects of the transcriptional memory after HS, such as its duration and its localization. They show that besides HSFA2, other HSF transcription factors are involved.
- 20.Szabo E.X., Reichert P., Lehniger M.-K., Ohmer M., de Francisco Amorim M., Gowik U., Schmitz-Linneweber C., Laubinger S. Metabolic labeling of RNAs uncovers hidden features and dynamics of the Arabidopsis transcriptome. Plant Cell. 2020;32:871–887. doi: 10.1105/tpc.19.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Chantarachot T., Sorenson R.S., Hummel M., Ke H., Kettenburg A.T., Chen D., Aiyetiwa K., Dehesh K., Eulgem T., Sieburth L.E. DHH1/DDX6-like RNA helicases maintain ephemeral half-lives of stress-response mRNAs. Nat Plants. 2020;6:675–685. doi: 10.1038/s41477-020-0681-8. [DOI] [PubMed] [Google Scholar]; Three RNA helicases are required to degrade short-lived stress-responsive mRNAs to ensure growth under normal conditions. The authors pinpoint a role for stress-granules in clearing unwanted transcripts, and nicely illustrate the fine balance between growth and defense.
- 22.Crisp P.A., Ganguly D.R., Smith A.B., Murray K.D., Estavillo G.M., Searle I., Ford E., Bogdanović O., Lister R., Borevitz J.O. Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell. 2017;29:1836–1863. doi: 10.1105/tpc.16.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hetzel J., Duttke S.H., Benner C., Chory J. Nascent RNA sequencing reveals distinct features in plant transcription. Proc Natl Acad Sci U S A. 2016;113:12316–12321. doi: 10.1073/pnas.1603217113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonkers I., Lis J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Core L., Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeitlinger J., Stark A., Kellis M., Hong J.-W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avramova Z. Transcriptional “memory” of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. Plant J. 2015;83:149–159. doi: 10.1111/tpj.12832. [DOI] [PubMed] [Google Scholar]
- 29••.Thomas Q.A., Ard R., Liu J., Li B., Wang J., Pelechano V., Marquardt S. Transcript isoform sequencing reveals widespread promoter-proximal transcriptional termination in Arabidopsis. Nat Commun. 2020;11:2589. doi: 10.1038/s41467-020-16390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; The role of elongation in the regulation of transcription in plants remains an unexplored topic. In this elegant study the authors discovered an additional point of plant transcriptional regulation by linking promoter proximal stalling of RNA PolII with premature termination of transcription. Whether elongation control globally provides a mechanism for priming remains to be investigated.
- 30.Crawford T., Karamat F., Lehotai N., Rentoft M., Blomberg J., Strand Å, Björklund S. Specific functions for Mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Sci Rep. 2020;10:5073. doi: 10.1038/s41598-020-61758-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Liu Q., Bischof S., Harris C.J., Zhong Z., Zhan L., Nguyen C., Rashoff A., Barshop W.D., Sun F., Feng S. The characterization of Mediator 12 and 13 as conditional positive gene regulators in Arabidopsis. Nat Commun. 2020;11:2798. doi: 10.1038/s41467-020-16651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Two subunits of the Mediator complex are implicated in gene activation in the context of inactive chromatin and also environmental signals (light and radiation). They were identified from a mutant screen. The Mediator complex bridges the interaction from sequence-specific transcription factors to the general transcription machinery. MED12 and MED13 are particularly important for the transition between activity states.
- 32.D’Urso A., Takahashi Y.H., Xiong B., Marone J., Coukos R., Randise-Hinchliff C., Wang J.P., Shilatifard A., Brickner J.H. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. eLife. 2016;5 doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charng Y.Y., Liu H.C., Liu N.Y., Chi W.T., Wang C.N., Chang S.H., Wang T.T. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng H.H., Robert F., Young R.A., Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 35.Feng X.J., Li J.R., Qi S.L., Lin Q.F., Jin J.B., Hua X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113:E8335–E8343. doi: 10.1073/pnas.1610670114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Liu J., Feng L., Gu X., Deng X., Qiu Q., Li Q., Zhang Y., Wang M., Deng Y., Wang E. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019;29:379–390. doi: 10.1038/s41422-019-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Besides the somatic thermomemory that is discussed in this review, HSFA2 also acts in transgenerational memory affecting flowering time and defense responses.
- 37.Lu F., Cui X., Zhang S., Jenuwein T., Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 38.Teves S.S., Weber C.M., Henikoff S. Transcribing through the nucleosome. Trends Biochem Sci. 2014;39:577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Lai W.K.M., Pugh B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. 2017;18:548–562. doi: 10.1038/nrm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leuendorf J.E., Frank M., Schmülling T. Acclimation, priming and memory in the response of Arabidopsis thaliana seedlings to cold stress. Sci Rep. 2020;10:689. doi: 10.1038/s41598-019-56797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mozgova I., Wildhaber T., Liu Q., Abou-Mansour E., L’Haridon F., Metraux J.P., Gruissem W., Hofius D., Hennig L. Chromatin assembly factor CAF-1 represses priming of plant defence response genes. Nature plants. 2015;1:15127. doi: 10.1038/nplants.2015.127. [DOI] [PubMed] [Google Scholar]
- 42.Berry S., Dean C. Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 2015;83:133–148. doi: 10.1111/tpj.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hepworth J., Dean C. Flowering Locus C’s lessons: conserved chromatin switches underpinning developmental timing and Adaptation. Plant Physiol. 2015;168:1237–1245. doi: 10.1104/pp.15.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matzke M.A., Mosher R.A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 45.Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T., Inagaki S., Nakajima S., Akashi T., Ohto M.A., Kobayashi M., Seki M., Shinozaki K., Kato T., Tabata S. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. 2004;38:673–684. doi: 10.1111/j.1365-313X.2004.02074.x. [DOI] [PubMed] [Google Scholar]
- 47.Takeda S., Tadele Z., Hofmann I., Probst A.V., Angelis K.J., Kaya H., Araki T., Mengiste T., Mittelsten Scheid O., Shibahara K. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. doi: 10.1101/gad.295404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Brzezinka K., Altmann S., Baurle I. BRUSHY1/TONSOKU/MGOUN3 is required for heat stress memory. Plant Cell Environ. 2019;42:765–775. doi: 10.1111/pce.13365. [DOI] [PubMed] [Google Scholar]; BRU1 has previously been implicated in the epigenetic inheritance of repressive chromatin states. This article raises the hypothesis that the role of BRU1 extends to active chromatin states. It provides a model of how HS memory is maintained across cell divisions.
- 49.Huang T.H., Fowler F., Chen C.C., Shen Z.J., Sleckman B., Tyler J.K. The histone chaperones ASF1 and CAF-1 promote MMS22L-TONSL-mediated Rad51 loading onto ssDNA during homologous recombination in human cells. Mol Cell. 2018;69:879–892.e5. doi: 10.1016/j.molcel.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saredi G., Huang H., Hammond C.M., Alabert C., Bekker-Jensen S., Forne I., Reveron-Gomez N., Foster B.M., Mlejnkova L., Bartke T. H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex. Nature. 2016;534:714–718. doi: 10.1038/nature18312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angel A., Song J., Yang H., Questa J.I., Dean C., Howard M. Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci U S A. 2015;112:4146–4151. doi: 10.1073/pnas.1503100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry S., Hartley M., Olsson T.S., Dean C., Howard M. Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. eLife. 2015;4 doi: 10.7554/eLife.07205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hulten M., Pelser M., van Loon L.C., Pieterse C.M., Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:5602–5607. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Legris M., Klose C., Burgie E.S., Rojas C.C.R., Neme M., Hiltbrunner A., Wigge P.A., Schäfer E., Vierstra R.D., Casal J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- 55.Jung J.-H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A.K., Box M.S., Charoensawan V., Cortijo S. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- 56••.Jung J.-H., Barbosa A.D., Hutin S., Kumita J.R., Gao M., Derwort D., Silva C.S., Lai X., Pierre E., Geng F. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585:256–260. doi: 10.1038/s41586-020-2644-7. [DOI] [PubMed] [Google Scholar]; The evening complex is a major signaling hub of the plant circadian clock. Temperature sensing is mediated by a prion-like polyQ domain in its ELF3 subunit. Depending on this domain and on temperature, ELF3 alternates between an active and an inactive state where it is included in liquid droplets. Interestingly, variation in the length of the polyQ domain fine-tunes temperature responses across natural variants.
- 57.Finka A., Cuendet A.F., Maathuis F.J., Saidi Y., Goloubinoff P. Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell. 2012;24:3333–3348. doi: 10.1105/tpc.112.095844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urrea Castellanos R., Friedrich T., Petrovic N., Altmann S., Brzezinka K., Gorka M., Graf A., Bäurle I. FORGETTER2 protein phosphatase and phospholipase D modulate heat stress memory in Arabidopsis. Plant J. 2020;104:7–17. doi: 10.1111/tpj.14927. [DOI] [PubMed] [Google Scholar]