Abstract
An excessive, non‐resolving inflammatory response underlies severe COVID‐19 that may have fatal outcomes. Therefore, the investigation of endogenous pathways leading to resolution of inflammation is of interest to uncover strategies for mitigating inflammation in people with SARS‐CoV‐2 infection. This becomes particularly urgent in individuals with preexisting pathologies characterized by chronic respiratory inflammation and prone to bacterial infection, such as cystic fibrosis (CF). Here, we analyzed the immune responses to SARS‐CoV‐2 virion spike 1 glycoprotein (S1) of macrophages (MΦ) from volunteers with and without CF and tested the efficacy of resolvins (Rv) D1 and D2 in regulating the inflammatory and antimicrobial functions of MΦ exposed to S1. S1 significantly increased chemokine release, including interleukin (IL)‐8, in CF and non‐CF MΦ, while it enhanced IL‐6 and tumor necrosis factor (TNF)‐α in non‐CF MΦ, but not in CF cells. S1 also triggered the biosynthesis of RvD1 and modulated microRNAs miR‐16, miR‐29a, and miR‐103, known to control the inflammatory responses. RvD1 and RvD2 treatment abated S1‐induced inflammatory responses in CF and non‐CF MΦ, significantly reducing the release of select chemokines and cytokines including IL‐8 and TNF‐α. RvD1 and RvD2 both restored the expression of miR‐16 and miR‐29a, while selectively increasing miR‐223 and miR‐125a, which are involved in NF‐κB activation and MΦ inflammatory polarization. During Pseudomonas aeruginosa infection, S1 stimulated the MΦ phagocytic activity that was further enhanced by RvD1 and RvD2. These results provide a map of molecular responses to SARS‐CoV‐2 in MΦ, key determinants of COVID‐19‐related inflammation, unveiling some peculiarity in the response of cells from individuals with CF. They also demonstrate beneficial, regulatory actions of RvD1 and RvD2 on SARS‐CoV‐2‐induced inflammation.
Keywords: ω‐3 fatty acids, COVID‐19, resolution, respiratory viruses, specialized proresolving lipid mediators
Abbreviations
- 17‐HDHA
17‐hydroxy‐DHA
- ACE2
angiotensin‐converting enzyme 2
- CFTR
cystic fibrosis transmembrane conductance regulator
- FBS
fetal bovine serum
- GM‐CSF
granulocyte‐monocyte colony stimulating factor
- IgG
immunoglobulin
- IL
interleukin
- IFN
interferon
- MΦ
macrophages
- NF‐κB
nuclear factor κB
- RANTES
regulated upon activation, normal T cell expressed and presumably secreted
- RPMI
Roswell Park Memorial Institute growth medium
- SARS
severe acute respiratory syndrome
- SPM
specialized pro‐resolving lipid mediators
- TMPRSS2
transmembrane Ser‐protease 2
1. INTRODUCTION
Acute inflammation is an innate protective response that is evolved to eliminate invading organisms. It should ideally be self‐limited and lead to complete resolution and return to homeostasis. 1 , 2 Resolution of inflammation is an active process introduced by the timely biosynthesis of specialized pro‐resolving mediators (SPM), which include lipoxins (LX), resolvins (Rv), protectins, and maresins. 3 SPM are evolutionarily conserved potent chemical signals with major roles in innate and adaptive immunity. Following inflammatory and infectious insults, they prevent excessive leukocyte infiltration and activation, balance inflammatory cytokines and chemokines, and regulate macrophage (MΦ) phenotype skewing. SPM also act on lymphocyte maturation, T cell differentiation, and IgG switch and enhance antimicrobial responses promoting resolution of infections, including bacterial and viral pneumonia. 4 , 5 , 6 , 7 , 8 In vivo, they counter cytokine storm, protect inflamed organs from damage, and enhance antimicrobial responses promoting resolution of infections. 5 , 6
The elucidation of SPM biology and actions has contributed to clarify the pathobiology of human diseases and has opened new trajectories in human pharmacology. An imbalance in SPM biosynthesis contributes to the pathogenesis of several diseases including atherosclerosis, 9 arthritis, 10 diabetes, 10 and sickle cell anemia. 11 Furthermore, SPM have proved safety and effectiveness in many clinical studies. 12 , 13
The coronavirus disease 2019 (COVID‐19) pandemic caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2) constitutes an unprecedented global health threat. Although extraordinary measures have been taken to restrain the spreading, this new virus has infected ~ 70,000,000 of people and claimed > 1,500,000 lives worldwide. 14 It is clear now that SARS‐CoV‐2 infection presents with variable severity, with many people developing pneumonia that progresses toward respiratory distress syndrome, sepsis, and multiple organ dysfunction, whereas others show a mild flu‐like illness that resolves in a few days. Of interest, evidence indicates that the viral load is not strictly correlated with disease severity and clinical evolution, whereas cytokine storm, increase in inflammatory mediators, and an imbalance in innate and adaptive immunity are associated with a poor prognosis. 15 In clinical settings, patients that received organ transplant or suffering from comorbidities linked to chronic inflammation (eg, chronic lung diseases, diabetes, and gastrointestinal disease) have worse outcomes and higher fatality rates among people with COVID‐19, 16 signifying that a defective resolution of inflammation can play key roles in clinical fate of COVID‐19. Hence, the role of pro‐resolution mechanisms and SPM in SARS‐CoV‐2‐mediated inflammatory responses are of considerable interest.
Cystic fibrosis (CF) is caused by mutations in the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene and is among the most common fatal genetic diseases worldwide. CF is a multiorgan condition characterized by chronic lung disease and a local and systemic inflammatory status that does not resolves and is marked by high serum concentrations of inflammatory mediators like interleukins (IL)‐6, C‐reactive protein, and ferritin. 17 , 18 , 19 Despite enormous strides have been made in the management of CF with the introduction of highly effective modulator therapies, unresolved inflammation and chronic infections remain constitutive in people with CF, as demonstrated by several longitudinal studies with patients taking CFTR modulators. 20 , 21 , 22 , 23 Since the early phases of the SARS‐CoV‐2 outbreak, people with CF have been considered at high risk for severe COVID‐19 12 because of their overzealous inflammatory response characterized by MΦ tonic activation, cytokine/chemokine overproduction, and chronic inflammation in organs such as lungs and gut that are targets of SARS‐CoV‐2. 17 , 18 , 19 Furthermore, immune system dysfunctions and defective pro‐resolution mechanisms, which are hallmarks of CF, can determine an overshooting inflammatory reaction with detrimental consequences for health and life of people with CF infected by SARS‐CoV‐2. 17 , 18 , 19 , 20 A recent report by the Cystic Fibrosis Registry Global Harmonization Group confirmed that COVID‐19 can cause severe illnesses in people with CF particularly in those who had lung transplant or had a severe lung disease. 24 Therefore, pathological mechanisms of SARS‐CoV‐2 in CF and actions of SPM in SARS‐CoV‐2‐induced inflammation are of outmost importance.
Here, we characterized immune responses to SARS‐CoV‐2 by human macrophages (MΦ) from study participants with and without CF, provided evidence for SPM biosynthesis during SARS‐CoV‐2 infection, and demonstrated the effects of RvD1 and RvD2 in mitigating MΦ inflammatory responses to this new virus.
Some of these results have been previously reported in the form of a preprint. 25
2. MATERIALS AND METHODS
2.1. Chemicals
SARS‐CoV‐2 S1, S2, and N recombinant proteins were purchased from RayBiotech (Peachtree Corners, GA). RvD1, RvD2, and RvD1 EIA were purchased from Cayman Chemicals. RvD1 and RvD2 were stored and prepared before each experiment as previously published. 6 Gibco cell culture media, fetal bovine serum (FBS), and supplements were purchased from Thermo Fisher Scientific. IL‐8 ELISA kits were purchased from PeproTech (London).
2.2. MΦ culture and phagocytosis
Peripheral blood was obtained from volunteers with CF (age ≥ 18 yrs; FEV1% ≥ 70) that did not have exacerbations in the 4 weeks prior to blood collection, as well as from age‐ and gender‐matched healthy volunteers. Study participants signed an informed consent form, and the protocol was approved by the Regional Ethics Committee (Prot. 1984/2019, Study Name RECCHI19). Monocytes were grown onto plastic surfaces (Eppendorf, Milan) 7‐10 days in 10% FBS RMPI medium plus GM‐CSF (10 ng/mL) inducing a phenotype close to that of lung MΦ. 26 A minimum of 24 hours washout period was carried out to remove the residual GM‐CSF effects and cells were maintained in FBS‐free or 1% FBS medium throughout the experiments.
In phagocytosis experiments the Pseudomonas aeruginosa RP73 strain was used and grown as recently reported. 27 Bacteria were collected at mid‐log phase, killed at 60°C for 40 minutes, and labeled (30 minutes, r.t., in the dark) with 0.5 mg/mL of FITC (Alfa Aesar, Thermo Fisher Scientific) in 100 mM of NaCl/50 mM of Na2CO3 buffer. After 3‐5 thorough washes, FITC‐RP73 was suspended in PBS at an OD600 nm = 1.
To assess phagocytosis, MΦ were gently collected (TrypLE, Gibco) and plated (150,000 cells) on a 24‐well Cell Imaging Plate (Eppendorf, Milan) in 10% of FBS RMPI medium 24‐48 hours prior to the experiments. MΦ were washed twice with PBS+/+ and treated or not with SARS‐CoV‐2 S1 recombinant protein for 3h in RPMI medium without FBS. Cells were then treated with RvD1 and RvD2 (10 or 100 nM) or vehicle (0.01% EtOH) for 15 minutes, infected by FITC‐RP73 (OD600 = 0.5, 15 µL/well) and incubated at 37°C, 5% CO2. After 45 minutes, infections were stopped, cells were washed with cold PBS+/+ and Trypan Blue (0.03% in PBS) was used to quench the fluorescent signal of non‐engulfed bacteria. MΦ were fixed with 3% formalin and fluorescence was determined using a plate reader (Synergy, BioTek) (Ex 530 nm/Abs 590 nm).
2.3. Luminex, RNA, and miRNA analyses
A Milliplex Magnetic Bead array kit (Merck Millipore, Milan) was used for measuring 30 cytokines, chemokines, interferons, and growth factors in cell‐free media from experiments with MΦ.
Total and small RNA (containing microRNA) were extracted from MΦ using the Roche High Pure miRNA Isolation kit (Roche, Milan) or the Quick‐RNA MicroPrep from Zymo Research (Irvine, CA) and quantified using a UV nanophotometer. cDNA was reverse transcribed from 100 to 150 ng of total RNA using the SuperScript VILO Master Mix (Thermo Fisher Scientific) with ezDNAse treatment and used (1‐5 ng/reaction) to assess the gene expression with real‐time PCR as in ref. 27 , 28 Primer pairs for ALX (Gene Name: FPR2; Ensembl: ENSG00000171049; Assay ID: qHsaCED0037673), DRV1 (Gene Name: GPR32 Ensemle: ENSG00000142511; Assay ID: qHsaCED0019486), and DRV2 (Gene Name GPR18; Ensembl: ENSG00000125245; Assay ID qHsaCED0036491) were purchased from Bio‐Rad (Segrate, Italy) and used as recommended. microRNAs were determined as previously published 27 from 100 pg of cDNA synthesized with the miScript RT kit (Qiagen, Milan).
Real‐time PCR were analyzed using the relative quantification method previously described calculated using β2 microglobulin, SNORD68 and SNORD95 as housekeeping mRNA or miRNAs for loading control, respectively. 22
2.4. Statistics
Results are presented as mean ± SEM. Differences between groups were assessed using One‐Way ANOVA and Holm‐Sidak or Dunn’s post hoc test depending on variances among groups or Student’s t test. The criterion for statistical significance was P < .05.
3. RESULTS
3.1. Characterization of SARS‐CoV‐2 triggered responses by MΦ
The SARS‐CoV‐2 is an enveloped virus whose virion is composed of a phospholipid bilayer, covered by spike (S) proteins, which encloses the nucleocapsid made of a single‐stranded RNA and phosphorylated nucleocapsid (N) proteins. The S protein has the function of conveying binding of SARS‐Cov‐2 to target cells (eg, MΦ and mucosal cells) through the interaction to ACE2 receptors on host cell surfaces. Once bound to ACE2, the S protein is cleaved by the transmembrane Ser‐protease 2 (TMPRSS2), which is essential for its priming, into the S1 and S2 subunits resulting in viral‐host membrane fusion and virus endocytosis. 29
In contrast, the N protein binds to the virus RNA to ensure maintenance of a “beads‐on‐a‐string” conformation and is, hence, essential for viral replication. 30
To characterize host responses to SARS‐CoV‐2 in the CF and non‐CF population, monocyte‐derived MΦ, which are key immune cells that contribute to the pathophysiology of COVID‐19 and contribute to chronic inflammation in CF, 31 , 32 were treated with the CoV‐2 S1 protein as surrogate of viral infection and the release of cytokines, chemokines, IFN, and growth factors was determined. Treatment with the glycoprotein S1 3 hours resulted in a significant increase in IL‐8 release by MΦ derived from volunteers with CF. Other chemokines involved in leukocyte recruitment, namely monocyte chemoattractant protein‐ (MCP)‐1, macrophage inflammatory protein (MIP)‐1α and 1β, and RANTES, were also significantly enhanced by S1, although at a much lower extent. In contrast, IL‐6, tumor necrosis factor (TNF)‐α, IL1‐β, and IFN‐α and γ were not modified at this time point and IL‐1 receptor antagonist (IL1RA) was the only protein reduced by S1 in CF MΦ. In keeping with results from CF cells, the S1 protein (3 h) gave a significant increase in IL‐8, MCP‐1, MIP‐1α and β, and RANTES, whereas IL‐1β and IFN‐α and γ were slightly, but not significantly, enhanced. In sharp contrast, there was a significant increase in IL‐6 and TNF‐α protein released by MΦ from volunteers without CF and secretion of IL1RA was not reduced by S1 (Figure 1A and Table 1). Moreover, the amount of MCP‐1 and MIP‐1β released by non‐CF MΦ in response to S1 was significantly higher compared to those from CF‐MΦ, whereas the secreted IL‐8 and MIP‐1α were comparable between MΦ from volunteers with and without CF.
FIGURE 1.
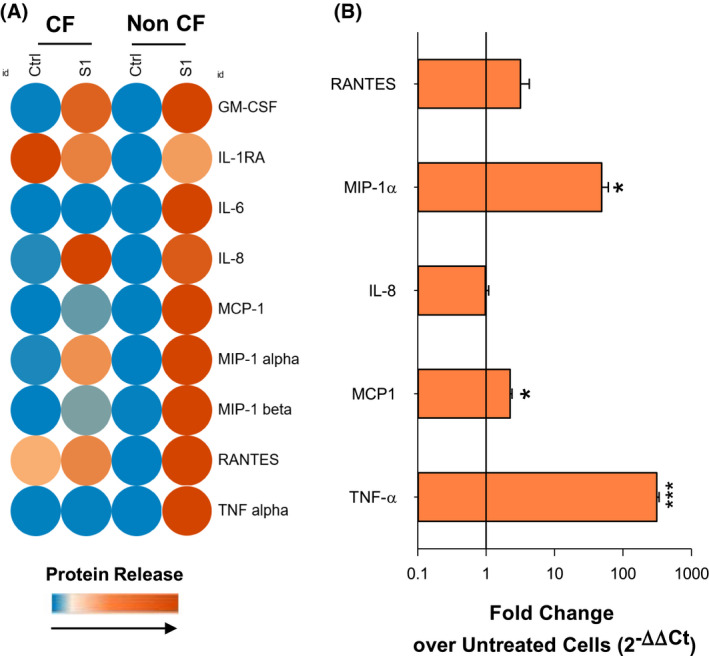
Inflammatory mediators regulated in MΦ in response to SARS‐CoV‐2 proteins. A, Protein concentrations of mediators involved in innate and adaptive immunity determined in monocyte‐derived MΦ collected from volunteers with and without CF stimulated with S1 (10 µg/mL, 3 hours) using a multiplex Luminex array kit. The heat map with relative protein concentrations was generated using the Morpheus software of the BROAD Institute (Cambridge, MA) (https://software.broadinstitute.org/morpheus) B, Real‐time PCR analysis of mRNA expression levels of inflammatory genes in CF MΦ treated (3 hours) with SARS‐CoV‐2 S1 protein (10 μg/mL). Results are mean ± SE of experiments with cells from four different donors. *P < .05; **P < .01; ***P < .001 vs untreated cells (One‐Way ANOVA)
TABLE 1.
Mediators of innate and adaptive immunity released by MΦ in response to CoV‐2
| CF | Non‐CF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl | S1 | Ctrl | S1 | ||||||||
| Mean | SE | Mean | SE | P vs Ctrl | Mean | SE | Mean | SE | P vs Ctrl | P CF +S1 vs Non‐CF +S1 | |
| IL‐8 | 2.1 | 0.7 | 1516.0 | 919.4 | * | 19.74 | 5.35 | 1272.28 | 323.68 | * | ns |
| IL‐1RA | 3092.8 | 353.4 | 2234.3 | 52.7 | * | 283.78 | 45.69 | 300.93 | 53.10 | ns | ** |
| GM‐CSF | 0.0 | 0.0 | 6.2 | 3.2 | * | 0.0 | 0.0 | 7.5 | 2.5 | * | ns |
| MCP‐1 | 10.2 | 4.5 | 107.6 | 24.5 | * | 11.00 | 1.36 | 2757.40 | 550.44 | * | *** |
| MIP‐1 alpha | 14.6 | 10.4 | 532.0 | 58.8 | * | 9.94 | 3.79 | 501.58 | 121.59 | * | ns |
| MIP‐1 beta | 4.4 | 4.1 | 292.4 | 21.6 | *** | 7.84 | 2.25 | 1843.81 | 635.47 | * | * |
| RANTES | 10.1 | 0.5 | 16.7 | 5.0 | * | 5.54 | 0.93 | 26.86 | 15.54 | ns | ns |
| TNF alpha | nd | ‒ | nd | ‒ | ‒ | 0.0 | 0.0 | 1011.68 | 130.42 | * | ns |
| VEGF | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| EGF | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| EOTAXIN | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‐ | ‒ | ‒ |
| FGF‐2 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‐ | ‒ |
| G‐CSF | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| HGF | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IFN alpha | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IFN gamma | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐1 beta | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐10 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐12 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐13 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐15 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐17A | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐2 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐2R | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐4 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐5 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‐6 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IL‒7 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| IP‒10 | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
| MIG | nd | ‒ | nd | ‒ | ‒ | nd | ‒ | nd | ‒ | ‒ | ‒ |
Concentrations of proteins involved in innate and adaptive immune response released by monocyte‐derived MΦ isolated from volunteers with and without CF upon treatment (3 h, 37 °C) with recombinant CoV‐2 spike proteins subunit 1 (S1, 10 µg/mL). Protein concentrations were measured using a multiplex Luminex array kit. Results are reported as pg/mL and are of experiments will cells from four different donors. nd, not detected (below limit). ns, not significant; *P < .05; **P < .01; ***P < .001 (One‐Way ANOVA).
Gene expression analyses revealed that mRNA levels of MIP‐1α, MCP‐1, and TNF‐α were significantly and strongly (~ 100‐ to 300‐fold vs baseline in untreated cells) upregulated, while IL‐8 mRNA was unchanged compared to untreated cells and RANTES was increased without reaching significance (Figure 1B).
These findings indicate that the increase in MIP‐1α, MCP‐1, and TNF‐α production involves activation of mRNA expression, whereas the enhancement of IL‐8 does not occur at the transcription level.
Collectively, these results indicate that the CoV‐2 S1 protein has a direct, infection‐independent pro‐inflammatory action on MΦ from volunteers with CF and subjects carrying a functional wild‐type CFTR.
3.2. SARS‐CoV‐2 activates SPM biosynthesis
SPM like RvD1 are tonically biosynthesized from polyunsaturated fatty acids by leukocytes in sterile inflammation and their production rapidly increases following infectious stimuli. 6 , 8 , 22 To determine if SARS‐CoV‐2 triggered SPM biosynthesis, we measured RvD1 released by MΦ following treatment with S1. As shown (Figure 2), unstimulated MΦ from CF volunteers produced significantly lower amounts of RvD1 compared to MΦ from donors without CF, confirming a defective tonic production of SPM in CF MΦ. 5 , 33 After a 3 hours treatment with S1, RvD1 concentrations significantly increased in CF MΦ but not in non‐CF cells (Figure 2). We also determined whether the expression of specific receptors for RvD1 and RvD2 was modulated by S1 in CF and non‐CF MΦ. Despite a variability in its reductive effect, S1 gave a decrease in ALX, DRV1, and DRV2 gene expression in MΦ from volunteers with and without CF, which reached the significance for DRV1. Of interest, RvD1 and RvD2 reverted the suppressive action of S1 on these receptors (Figure 2B).
FIGURE 2.
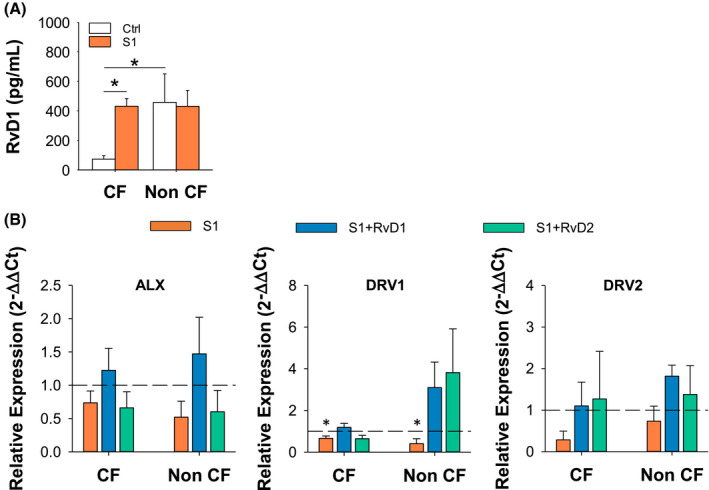
SARS‐CoV‐2 stimulates RvD1 biosynthesis. RvD1 concentrations in MΦ cell supernatants following stimulation (3 hours) with CoV‐2 proteins (10 µg/mL) and in unstimulated cells used as a control (Ctrl). RvD1 was measured using a validated EIA procedure. 5 Results are mean ± SE from experiments with cells from four different donors. *P < .05 (One‐Way ANOVA). B, Real‐time PCR analysis of ALX, DRV1, and DRV2 receptors in CF and non‐CF MΦ treated (3 hours, 37°C) with S1 (10 μg/mL) plus RvD1 (10 nM), RvD2 (10 nM) or Veh (0.01 % EtOH). Results are mean ± SE of experiments from three different donors. Gene expression was determined as a duplicate for each test condition. *P < .05 (One‐Way ANOVA). ALX, lipoxin A4 receptor; DRV1, RvD1 receptor; DRV2, RvD2 receptor
These results gave clues for the activation of SPM biosynthesis, modulation of their receptors, and SPM bioactions during SARS‐CoV‐2 infection.
3.3. RvD1 and RvD2 reduce MΦ inflammatory responses to SARS‐CoV‐2
To verify this hypothesis, we tested RvD1 and RvD2 bioactions on inflammatory responses triggered in MΦ by SARS‐CoV‐2 S1. RvD1 and RvD2 treatment gave a significant reduction in IL‐8, MCP‐1, and MIP‐1β in S1‐stimulated MΦ from volunteers with and without CF, whereas RvD2 reduced MIP‐1α secretion selectively in CF MΦ. Moreover, RvD1 and RvD2 abated the S1‐induced increase in TNF‐α and IL‐6 in CFTR competent MΦ and countered the inhibitory effect of S1 on IL1RA in CF MΦ (Figure 3).
FIGURE 3.
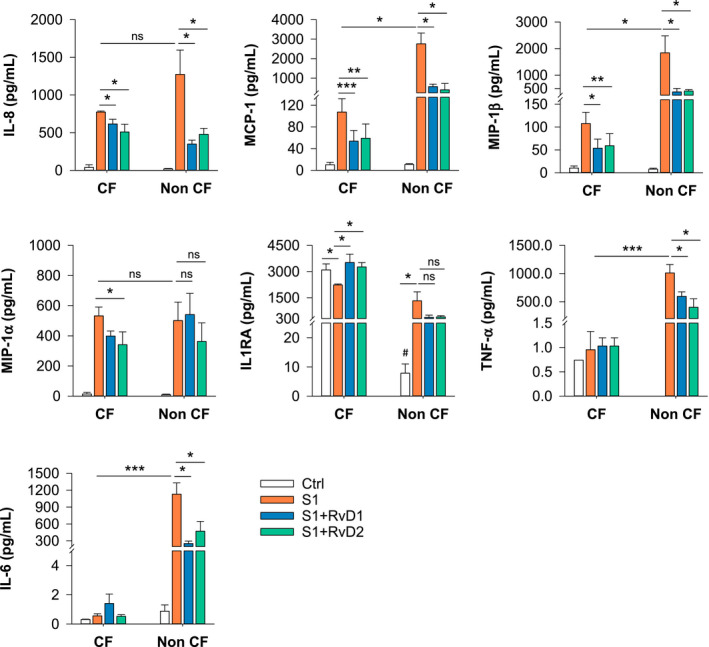
Rvd1 and RvD2 actions on SARS‐CoV‐2‐triggered inflammatory mediators. Inflammatory mediators were determined in supernatants from monocyte‐derived MΦ from volunteers with CF or without CF treated (3 hours, 37°C) with RvD1, RvD2 (10 nM), or Veh (0.01 % EtOH) plus S1 (10 µg/mL). Results are mean ± SE from experiments with cells from four different donors. *P < .05; **P < .01. ns, not significant 05 (One‐Way ANOVA)
To determine the time‐course of responses to S1 and effects of RvD1 and RvD2, MΦ were stimulated with the spike protein for 3 hours and treated thereafter with RvD1, RvD2, or vehicle control. Concentrations of cytokines and chemokines were measured in supernatants collected 24 hours post S1 stimulation.
As shown in Figure 4, IL‐8 and MIP‐1α steadily increased in a time‐dependent manner from 3 to 24 hours after stimulation with S1 of CF MΦ, while they plateaued after 3 hours in non‐CF MΦ. TNF‐α, which was not induced in CF MΦ by S1 at 3 hours, showed a time‐dependent increase in both MΦ from healthy and CF volunteers, whereas, consistent with 3 hours results, IL‐6 was enhanced only in non‐CF‐MΦ at 24 hours. Secretion of MIP‐1β showed opposite trends in the two cell types following a 24 hours stimulation with S1, with a slight increase S1‐stimulated CF MΦ and a decrease in non‐CF MΦ compared to cells treated with the SARS‐CoV‐2 protein for 3 hours. Notably, the production of IL‐8 and MIP‐1α, was significantly higher in S1‐activated CF MΦ at 24 hours compared to non‐CF counterparts, while TNF‐α, and IL‐6 were significantly more abundant in supernatants from non‐CF MΦ (Figure 4). Finally, IL‐1β, IL‐10, IL1RA, MCP‐1, and INF‐α and γ secretion was not increased by S1 in both cell types at 24 hours (Supplementary Figure S1).
FIGURE 4.
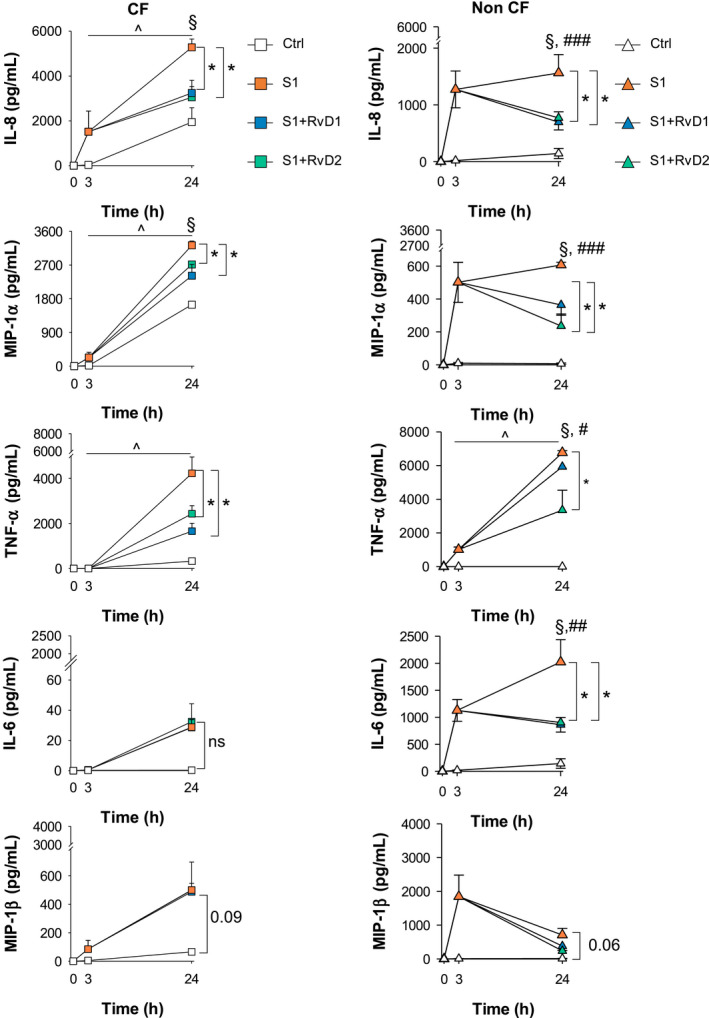
Time‐course of S1‐triggered responses and effects of RvD1 and RvD2. Cytokines and chemokines released by CF and non‐CF MΦ at the indicated time points post stimulation with S1 (10 µg/mL, 3 hours) followed by treatment with RvD1, RvD2 (10 nM), or Veh (0.01% EtOH). Cells that were not stimulated with S1 were used as a control. Supernatants were collected 24 hours after the initial stimulation. Results are mean ± SE from experiments with cells from four different donors *P < .05 vs S1‐treated cells; §P < .05 vs control cells at 24 hours; ^P < .05 vs 3 hours S1 stimulated cells; #P < .05 vs S1‐strimulated CF MΦ at 24 hours; ##P < .01 vs S1‐strimulated CF MΦ at 24 hours; ###P < .001 vs S1‐strimulated CF MΦ at 24 hours (One‐Way ANOVA)
Collectively, these results corroborate the evidence of differences in the magnitude and kinetics of the responses to S1 depending on the presence in MΦ of a fully competent or a mutated CFTR protein.
Regardless of the different responses observed in CF and non‐CF MΦ, RvD1 and RvD2 significantly dampened by ~ 25‐50 % the increase in IL‐8, MIP‐1α, and TNF‐α elicited by S1, demonstrating similar regulatory actions on these cytokines and chemokines, whereas they had selective reductive effects IL‐6 in non‐CF cells (Figure 4). There was a statistically significant (P = .021) difference in the reduction of MIP‐1α by RvD2 in non‐CF MΦ (61.18 ± 12.13% reduction) compared with CF MΦ (15.86 ± 2.42% reduction), which could be determined by the higher induction of this chemokine by S1 in CFTR mutated MΦ. The reducing effect of RvD1 and RvD2 on IL‐8 and TNF‐α was comparable between CF and non‐CF MΦ (not shown).
These results signify that SARS‐CoV‐2 can upregulate leukocyte chemotactic signals such as IL‐8 and MIP‐1α, but not cytokines like IL‐1β and IFN, involved in MΦ activation in both CF and non‐CF MΦ.
Molecular mechanisms of action of SPM encompass regulation of microRNAs (miRNAs) that are important controller of inflammation. 34 , 35 Therefore, we analyzed whether RvD1 and RvD2 modified miRNA in MΦ in response to SARS‐CoV‐2 S1 protein. Real‐time PCR analysis revealed that S1 significantly decreased the expression of miR‐29a, miR‐16, and miR‐103 in MΦ from volunteers with and without CF, increased miR‐197 selectively in CF MΦ, and regulated oppositely the microRNA let‐7b in CF and non‐CF cells (Figure 5A, B). RvD1 and RvD2 restored the expression of miR‐29a in CF and non‐CF MΦ and of miR‐16 in CF cells. Moreover, RvD1, but not RvD2, significantly upregulated miR‐223 in CF and non‐CF MΦ, whereas miR‐125a was selectively increased by RvD2 in these cells, indicating distinct regulatory actions of RvD1 and RvD2 on MΦ responses to S1 (Figure 5C).
FIGURE 5.
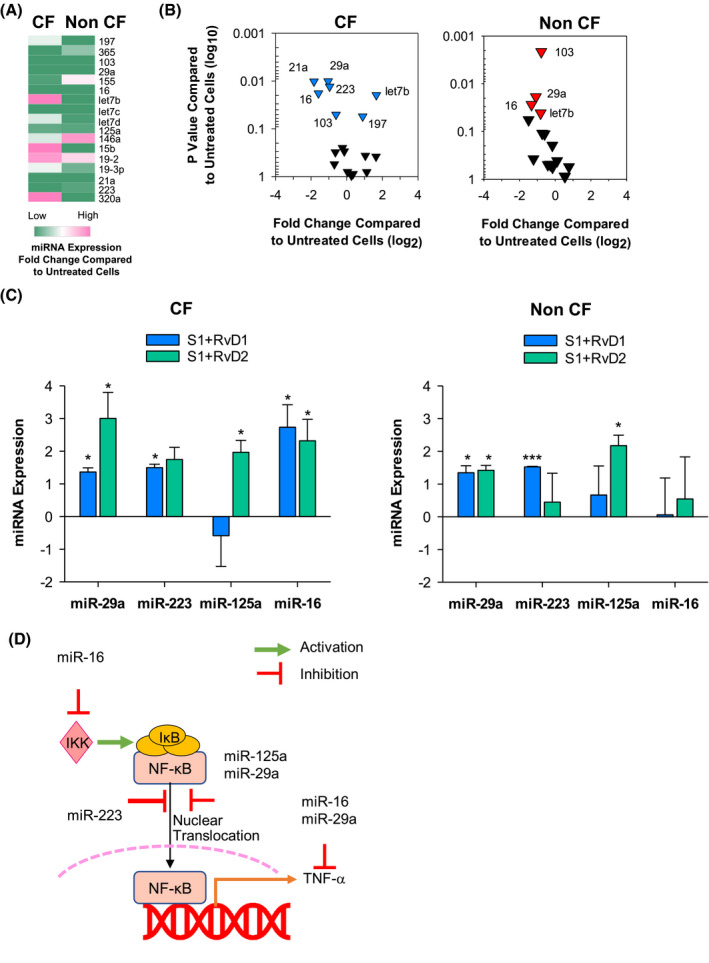
RvD1 and RvD2 regulates miRNAs expression in CF MΦ during CoV‐2 response. Heat map A, and Volcano plots B, showing relative expression miRNA determined in peripheral blood monocytes from volunteers with or without CF stimulated with S1 protein (10 µg/mL, 3 hours). The heat map was rendered using the algorithm of the Morpheus software (https://software.broadinstitute.org/morpheus). C, microRNAs relative expression levels in CF and non‐CF MΦ treated with (3 hours, 37°C) with RvD1, RvD2 (10 nM), or Veh (0.01 % EtOH) plus S1 (10 µg/mL). miRNA expression is reported as fold change over S1‐treated MΦ. Results are mean ± SE from experiments with cells from four different donors. *P < .05; ***P < .001 b. D, Biological roles of miRNAs modulated by S1 and regulated by RvD1 and RvD2 in CF and non‐CF MΦ (see text and ref. 35 for further details)
Bioinformatic analysis revealed that miR‐29a and miR‐16, upregulated by RvD1 and RvD2 and reduced by S1, and miR‐223 and miR‐125a, which were increased by RvD1 or RvD2, respectively, reduce IKK/NF‐κB activation and downstream cytokines and chemokines 36 (Figure 5D). In contrast, let‐7b amplifies cytokine signaling and MΦ pro‐inflammatory phenotype, 37 while miR‐103 induces MΦ anti‐inflammatory phenotype 38 and the role of miR‐197 in MΦ is debated as it can either enhance or reduce inflammation. 39 , 40 Hence, miRNAs modulated by S1 and targeted by RvD1 and RvD2 play pivot roles in regulating induction of cytokines and chemokines in response to SARS‐CoV‐2.
Collectively, these results characterize and unveil shared and common reactions activated in MΦ from individuals with or without CF by SARS‐CoV‐2. They also indicate that RvD1 and RvD2 regulate mediators and microRNA related to inflammatory responses triggered by SARS‐CoV‐2 in MΦ.
3.4. RvD1 and RvD2 enhance MΦ phagocytosis during P. aeruginosa infection
Pseudomonas aeruginosa infections are persistent in patients with CF 41 , 42 and are well documented in COVID‐19. 43 , 44 Since enhancement of MΦ bacterial phagocytosis are defining the functions of SPM including RvD1 and RvD2, 5 , 6 it was of interest to determine the effect of S1 and RvD1 and RvD2 on this essential host defensive MΦ task. To this end, phagocytosis was determined by treating MΦ from participants with and without CF with S1 and RvD1, RvD2, or vehicle control prior to feeding with FITC‐RP73. As shown (Figure 6) S1, per se, significantly stimulated the bacterial phagocytosis by both CF and non‐CF MΦ. RvD1 and RvD2 further increased (~ 2.5‐ to 3‐fold) the MΦ phagocytic capacity, as determined by the amount of intracellular FITC‐P. aeruginosa (Figure 6).
FIGURE 6.
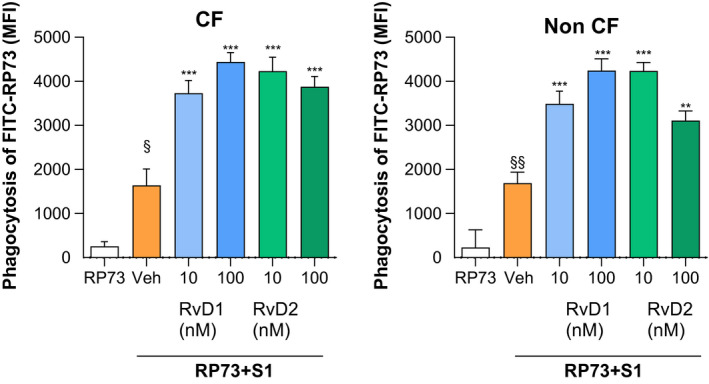
RvD1 and RvD2 enhance phagocytic activity of S1‐treated MΦ. Phagocytic activity was determined in MΦ from CF and healthy volunteers incubated with S1 (10 µg/mL, 3 hours, 37°C). RvD1, RvD2 (10 or 100 nM), or Veh (0.01 % EtOH) were added for 15 minutes and cells were infected with RP73‐FITC (45 minutes, 37°C, 5% CO2). Results are mean ± SE from experiments with cells from three different donors. Each condition was tested as a triplicate or quadruplicate. MFI, mean fluorescence intensity. *P < .05; **P < .01; ***P < .001
These results demonstrate that RvD1 and RvD2 retain antimicrobial proresolving actions in bacterial and viral co‐infection.
4. DISCUSSION
Here, we report the characterization of molecular and cell responses induced by SARS‐CoV‐2 in MΦ from healthy subjects and individuals with CF. We also provide the first evidence for SPM biosynthesis and regulatory activities of RvD1 and RvD2 on inflammatory, immune, and anti‐microbial responses of MΦ to SARS‐CoV‐2.
SARS‐CoV‐2 is a pleiotropic, highly contagious virus that can lead to an uncontrolled multi‐organ inflammatory syndrome especially in individuals with preexisting chronic diseases like CF. MΦ are sentinel of host defense and key players in inflammation, resolution, and innate and adaptive immunity. Therefore, we chose MΦ as a SARS‐CoV‐2 cell target for characterizing inflammatory pathways activated by this new virus and determine if cells from people with CF have different responses to SARS‐CoV‐2 compared to those from individuals that are not affected by this genetic disease.
While infection with whole virions is important to assess overall viral replication and host responses, the use of single proteins represents a useful strategy for discriminating the contribution of each viral component to inflammation in target cells. The innate immune response recognizing SARS‐CoV‐2 culminate with the activation of two general host defense programs, that is, the production of NF‐κB‐dependent cytokines and chemokines that recruit neutrophils and other leukocytes to the infected site and the induction of IFN and IFN‐dependent genes.
Here, we found that S1 stimulates the release of chemokines (IL‐8, MIP‐1α, and β, MCP‐1) that are downstream the NF‐κB axis, but it did not induce the IFN or inflammasome response (Figures 1, 3, 4). Recent studies 45 indicate that, unlike other respiratory viruses, SARS‐CoV‐2, does not induce IFN‐α/β in infected cells implying that this could underlie some clinical manifestations of COVID‐19 since a deficiency in type I IFN immunity is associated with severe symptoms of COVID‐19. 46 , 47 It is possible that multiple signals (eg, co‐stimulation of cell‐membrane and phagosome‐associated PRR) occurring in vivo are required to induce IFN secretion.
We also identified a clear difference in the induction of TNF‐α and IL‐6 as well as in the regulation of IL1RA between CFTR mutated and competent MΦ (Figures 1, 3, 4). CF MΦ have an hyper‐reactive behavior, with an increased production of cytokines and chemokines when exposed to phlogistic stimuli. 31 To the best of our knowledge, this is the first evidence that MΦ from individuals with CF have a delayed secretion of cytokines in response to viral proteins compared to MΦ from subjects without CF. IL1RA plays important roles in curbing IL‐1 actions that contributes to CF lung disease and persistent inflammation. 48 Consistent with this, concentrations of IL1RA and IL‐1 are abundant in lung secretions from CF patients and are considered indicative of an attempt of an homeostatic response to restrain the effects of IL‐1 on CF lungs. 17
The difference observed in IL‐6 and TNF‐α production is of interest since these cytokines promote systemic inflammation and cytokine storm that is considered a major driver of severe symptoms of COVID‐19. 49 Overall, these findings unveil that cells from individuals with CF have some biological differences in the underlying immune response to SARS‐CoV‐2. With large biological samples being collected during the COVI‐19 pandemic, it will be crucial to carry out serological and blood cell analyses to assess these differences in SARS‐CoV‐2 positive CF patients compared to the general population and their impact on the clinical manifestations of this virus, along with longitudinal studies to determine if these are biomarkers of COVID‐19 severity in people with or without CF.
SPM are rapidly biosynthesized from polyunsaturated fatty acids in sterile and infectious inflammation. Here were report that SARS‐CoV‐2 proteins trigger the biosynthesis of RvD1, which is paradigmatic of activation SPM biosynthetic pathway, in MΦ from volunteers with CF but not in non‐CF cells, which constitutively released a higher amount of RvD1 (Figure 2). These results are consistent with other studies demonstrating defective biosynthesis of LX and SPM by cells from subjects with CF 50 , 51 and suggest that SARS‐CoV‐2 may normalize the SPM‐biosynthetic machinery in patients with CF. Precursors, including 17‐HDHA (the precursors RvD1 and RvD2) have been identified in lipidomic analyses from mouse airway lavage fluids and human nasal washes during H1N1 influenza. 52 Moreover, 17‐HDHA and its derivative protectin D1 proved to have anti‐viral and vaccine‐adjuvant activities. 4 , 8 In this work, we found that DHA‐derived RvD1 and RvD2 have counter‐regulatory actions on SARS‐CoV‐2‐induced inflammatory responses in CF and non‐CF MΦ, including reduction of select inflammatory chemokines and cytokines (Figures 3, 4, 5).
SPM are potent regulator of neutrophil chemotaxis and recruitment in inflamed tissues. For instance, we recently demonstrated that RvD1 reduces IL‐8 in MΦ from volunteers with CF and KC (the murine IL‐8homolog) in CF mice infected with P. aeruginosa, 5 whereas RvD2 curbs chemokine storm in septic mice. 7
Therefore, these results unveil new actions of RvD1 and RvD2 in reducing virus‐triggered inflammatory responses.
miRNAs are intracellular regulatory mechanisms of MΦ functions and many of them are modulated by SPM to reduce inflammation and promote resolution. We defined here the first comparative signature of microRNA response to SARS‐CoV‐2 in MΦ from CF and non‐CF individuals, identifying miRNAs that were equally (miR‐29a, miR‐103, miR‐16, and miR‐125a) or distinctly (miR‐21, miR‐223, and let‐7b) modified by S1 and regulated by RvD1 and RvD2 in CF and non‐CF MΦ. These miRNAs block the NF‐κB pathway, control inflammatory proteins, and dictate MΦ polarization. 35 , 36 , 37 , 39 , 40 Furthermore, miR‐21 has been identified as one of the most downregulated microRNA by SARS‐CoV‐2 in peripheral blood leukocytes from patients with COVID‐19 53 and an in silico prediction suggested that miR‐16 can reduce SARS‐CoV‐2 replication by targeting its RNAs. 34 Regulation of NF‐κB and NF‐κB‐associated miRNAs is a central mechanism of action of SPM in resolution. RvD1 blocks NF‐κB nuclear translocation, 34 , 54 upregulates miR‐146b in human MΦ that results in downregulation of inflammatory cytokines and chemokines. 34 In vivo, RvD1 dampens NF‐κB activation and downstream genes in lungs and kidneys from sickle cell mice undergoing hypoxia‐reoxygenation injury, 15 enhances miR‐219 in mouse peritoneal leukocytes leading to increase in IL‐10, 33 and increases miR‐21 and miR‐155 that interact with the TLR‐NF‐κB axis in lung MΦ from mice bearing chronic P. aeruginosa infection. 21 RvD2 induces miR‐146a in human monocytes stimulated with LPS attenuating TLR‐mediated inflammation. 55 Of interest, reduced expression of the RvD1‐regulated miR‐219 has been associated with impaired resolution of inflammation, 56 corroborating the biological importance of SPM‐regulated miRNAs in the regulation of inflammation. Hence, results shown here provide the first evidence for roles of miRNAs in SPM‐mediated regulation of inflammatory responses to viral stimuli.
P. aeruginosa establishes chronic lung infections in 20‐60% of patients with CF 42 and is a primary opportunistic pathogens that can cause severe pneumonia in COVID‐19 patients under assisted ventilation or in intensive care unit. 43 , 44 Here, using a cell model that mimics viral and bacterial co‐infection frequently observed in the clinical setting of CF, we found that S1 activated MΦ phagocytic activity against P. aeruginosa (Figure 6). The influence of viruses on MΦ anti‐bacterial functionalities is complex. SARS and human immunodeficiency virus 1 proteins reduce phagocytosis, 57 , 58 whereas influenza A virus neuraminidase stimulates phagocytic function. 59 Although the exact mechanisms by which SARS‐CoV‐2 increases MΦ phagocytosis of P. aeruginosa require further investigations, results from this study indicate that RvD1 and RvD2 retain antimicrobial proresolving actions in viral and bacterial infections.
In summary, here we show that SARS‐CoV‐2 triggers common and distinct phlogistic responses in MΦ from volunteers with CF and from healthy individuals and that RvD1 and D2 reduce MΦ‐driven inflammation while potentiating their host defensive, phagocytic functions, thus suggesting host directed action of SPM regulating resolution of COVID‐19 and expanding the spectrum of their protective role in CF 5 (Figure 7) and expanding the spectrum of their protective role in CF. 5
FIGURE 7.
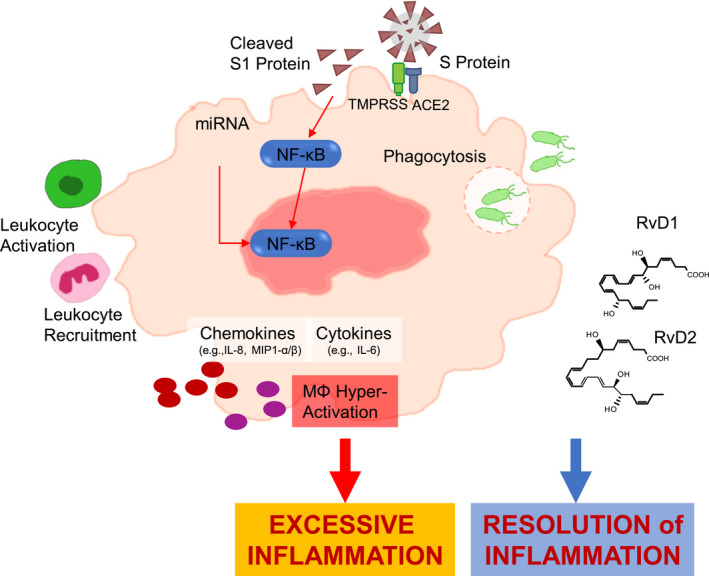
S1‐triggered responses in MΦ and actions of RvD1 and RvD2. S1 from SARS‐CoV‐2 triggers phlogistic responses in CF and non‐CF MΦ encompassing activation of microRNAs and NF‐κB‐regulated chemokines and cytokines that drive further leukocyte functions. An unremitting MΦ activation can lead to excessive inflammation in patients with COVID‐19. RvD1 and D2 RvD1 and D2 regulate select cytokines, chemokines, and microRNAs while enhancing MΦ phagocytosis of bacteria, thus proving important actions in SARS‐CoV‐2‐driven inflammation and its resolution
CONFLICT OF INTEREST
The authors declare that no conflict of interest exist that may affect the objectivity of this manuscript. A patent application related to this work has been submitted.
AUTHOR CONTRIBUTIONS
A. Recchiuti conceived the overall research, carried out the experiments, analyzed the results, and wrote the manuscript. S. Patruno, D. Mattoscio, E. Isopi, A. Pomilio, and R. Pecce carried out the experiments and analyzed the results. M. Romano revised the final version of the manuscript. All the authors contributed to the final version of the manuscript
Supporting information
Fig S1
ACKNOWLEDGMENTS
This study was supported by the Cystic Fibrosis Foundation (CFF) Grant RECCHI19I0 and Italian Ministry of University and Research (MIUR) grant ex 60% 2019 and 2020 (to A.R.) and grant L.548/93 from the Ministry of Health (to M.R.). AR was awarded a grant (“COVID‐19 Pathophysiology and Proresolving Therapies in Cystic Fibrosis” Grant # RECCHI20G1) from the CFF while the work reported here was accomplished. The authors thank the staff Center for Cystic Fibrosis San Liberatore (Atri, TE) for the recruitment of study participants. We are indebted to all the participants to this study.
Recchiuti A, Patruno S, Mattoscio D, et al. Resolvin D1 and D2 reduce SARS‐CoV‐2‐induced inflammatory responses in cystic fibrosis macrophages. The FASEB Journal. 2021;35:e21441. 10.1096/fj.202001952R
References
- 1. Perretti M, Norling LV. Actions of SPM in regulating host responses in arthritis. Mol Aspects Med. 2017;58:57‐64. [DOI] [PubMed] [Google Scholar]
- 2. Gilroy DW, Bishop‐Bailey D. Lipid mediators in immune regulation and resolution. Br J Pharmacol. 2019;176:1009‐1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spite M, Clària J, Serhan CN. Resolvins, specialized pro‐resolving lipid mediators and their potential roles in metabolic diseases. Cell Metab. 2014;19:21‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morita M, Kuba K, Ichikawa A, et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153:112‐125. [DOI] [PubMed] [Google Scholar]
- 5. Recchiuti A, Mattoscio D, Isopi E. Roles, actions, and therapeutic potential of specialized pro‐resolving lipid mediators for the treatment of inflammation in cystic fibrosis. Front Pharmacol. 2019;10:1‐18. 10.3389/fphar.2019.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spite M, Norling L, Summers L, et al. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 2009;461:1287‐1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiang N, Fredman G, Bäckhed F, et al. Infection regulates pro‐resolving mediators that lower antibiotic requirements. Nature. 2012;484:524‐528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramon S, Baker SF, Sahler JM, et al. The specialized proresolving mediator 17‐HDHA enhances the antibody‐mediated immune response against influenza virus: a new class of adjuvant? J. Immunol. 2014;193:6031‐6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fredman G, Hellmann J, Proto JD, et al. An imbalance between specialized pro‐resolving lipid mediators and pro‐inflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat Commun. 2016;7:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Norling L, Headland SE, Dalli J, Arnardottir HH. Pro‐resolving and cartilage‐protective actions of Resolvin D1 in inflammatory arthritis. JCI Insight. 2016;1:e85922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gromovsky AD, Schugar RC, Brown AL, et al. Δ‐5 fatty acid desaturase fads1 impacts metabolic disease by balancing proinflammatory and proresolving lipid mediators. Arterioscler Thromb Vasc Biol. 2018;38:218‐231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Motwani MP, Colas RA, George MJ, et al. Pro‐resolving mediators promote resolution in a human skin model of UV‐killed Escherichia coli‐driven acute inflammation. JCI insight. 2018;3:e94463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu S‐H, Chen X‐Q, Liu B, Wu H‐J, Dong L. Efficacy and safety of 15(R/S)‐methyl‐lipoxin A(4) in topical treatment of infantile eczema. Br J Dermatol. 2013;168:172‐178. [DOI] [PubMed] [Google Scholar]
- 14. World Health Organization (WHO) . Coronavirus disease (COVID‐19) outbreak situation.
- 15. Matte A, Recchiuti A, Federti E, et al. Resolution of sickle cell disease‐associated inflammation and tissue damage with 17%3cem%3eR%3c/em%3e‐resolvin D1. Blood. 2019;133:252‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Centers for Disease Control and Prevention . People Who Are at Higher Risk for Severe Illness.
- 17. Bonfield TL, Panuska JR, Konstan MW, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med. 1995;152:2111‐2118. [DOI] [PubMed] [Google Scholar]
- 18. Roesch EA, Nichols DP, Chmiel JF. Inflammation in cystic fibrosis: an update. Pediatr Pulmonol. 2018;53:S30‐S50. [DOI] [PubMed] [Google Scholar]
- 19. Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992‐2001. [DOI] [PubMed] [Google Scholar]
- 20. Heltshe SL, Mayer‐Hamblett N, Burns JL, et al. Pseudomonas aeruginosa in cystic fibrosis patients with G551D‐CFTR treated with ivacaftor. Clin Infect Dis. 2015;60:703‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hisert KB, Heltshe SL, Pope C, et al. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am J Respir Crit Care Med. 2017;195:1617‐1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rowe SM, Heltshe SL, Gonska T, et al. Clinical mechanism of the cystic fibrosis transmembrane conductance regulator potentiator ivacaftor in G551D‐mediated cystic fibrosis. Am J Respir Crit Care Med. 2014;190:175‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hisert KB, Birkland TP, Schoenfelt KQ, et al. CFTR modulator therapy enhances peripheral blood monocyte contributions to immune responses in people with cystic fibrosis. Front Pharmacol. 2020;11:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McClenaghan E, Cosgriff R, Brownlee K, et al. The global impact of SARS‐CoV‐2 in 181 people with cystic fibrosis. J Cystic Fibrosis. 2020;S1569199320308778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Recchiuti A, Patruno S, Mattoscio D, et al. Resolvin D1 and D2 reduce SARS‐Cov‐2‐induced inflammation in cystic fibrosis macrophages. bioRxiv. 2020:255463. 10.1101/2020.08.28.255463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lescoat A, Ballerie A, Augagneur Y, et al. Distinct properties of human M‐CSF and GM‐CSF monocyte‐derived macrophages to simulate pathological lung conditions in vitro: application to systemic and inflammatory disorders with pulmonary involvement. Int J Mol Sci. 2018;19:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Codagnone M, Cianci E, Lamolinara A, et al. Resolvin D1 enhances the resolution of lung inflammation caused by long‐term Pseudomonas aeruginosa infection. Mucosal Immunol. 2018;11:35‐49. [DOI] [PubMed] [Google Scholar]
- 28. Recchiuti A, Codagnone M, Pierdomenico AM, et al. Immunoresolving actions of oral resolvin D1 include selective regulation of the transcription machinery in resolution‐phase mouse macrophages. FASEB J. 2014;28:3090‐3102. [DOI] [PubMed] [Google Scholar]
- 29. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McBride R, Van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991‐3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bruscia EM, Bonfield TL. CF lung immunity: the role of the macrophage. J Innate Immun. 2016;8:550‐563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020;26:842‐844. [DOI] [PubMed] [Google Scholar]
- 33. Eickmeier O, Fussbroich D, Mueller K, et al. Pro‐resolving lipid mediator resolvin D1 serves as a marker of lung disease in cystic fibrosis. PLoS ONE. 2017;12:e0171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1‐miRNA circuits. FASEB J. 2011;25:544‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curtale G, Rubino M, Locati M. MicroRNAs as molecular switches in macrophage activation. Front Immunol. 2019;10:799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li T, Morgan MJ, Choksi S, Zhang Y, Kim Y‐S, Liu Z. MicroRNAs modulate the noncanonical transcription factor NF‐κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat Immunol. 2010;11:799‐805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Z, Xu L, Hu Y, et al. miRNA let‐7b modulates macrophage polarization and enhances tumor‐associated macrophages to promote angiogenesis and mobility in prostate cancer. Sci Rep. 2016;6:25602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu Y‐L, Hung J‐Y, Chang W‐A, et al. Hypoxic lung‐cancer‐derived extracellular vesicle Microrna‐103a increases the oncogenic effects of macrophages by targeting PTEN. Mol Ther. 2018;26:568‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H, Su X, Yang M, et al. Reciprocal control of miR‐197 and IL‐6/STAT3 pathway reveals miR‐197 as potential therapeutic target for hepatocellular carcinoma. Oncoimmunology. 2015;4:e1031440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akkaya‐Ulum YZ, Akbaba TH, Tavukcuoglu Z, et al. Familial Mediterranean fever‐related miR‐197‐3p targets IL1R1 gene and modulates inflammation in monocytes and synovial fibroblasts. Sci Rep. 2021;11:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cantin AM, Hartl D, Konstan MW, Chmiel JF. Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J Cyst Fibros. 2015;14:419‐430. [DOI] [PubMed] [Google Scholar]
- 42. Stefani S, Campana S, Cariani L, et al. Relevance of multidrug‐resistant Pseudomonas aeruginosa infections in cystic fibrosis. Int J Med Microbiol: IJMM. 2017;307:353‐362. [DOI] [PubMed] [Google Scholar]
- 43. Fattorini L, Creti R, Palma C, Pantosti A. Bacterial coinfections in COVID‐19: an underestimated adversary. Ann Ist Super Sanita. 2020;56:359‐364. [DOI] [PubMed] [Google Scholar]
- 44. Torres A, Niederman MS, Chastre J, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital‐acquired pneumonia and ventilator‐associated pneumonia: guidelines for the management of hospital‐acquired pneumonia (HAP)/ventilator‐associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J. 2017;50:1700582‐1‐1700582‐26. [DOI] [PubMed] [Google Scholar]
- 45. Blanco‐Melo D, Nilsson‐Payant BE, Liu W‐C, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181:1036‐1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science. 2020;370:eabd4570‐1‐eabd4570‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bastard P, Rosen LB, Zhang Q, et al. Auto‐antibodies against type I IFNs in patients with life‐threatening COVID‐19. Science. 2020;370:eabd4585‐1‐eabd4585‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Montgomery ST, Dittrich AS, Garratt LW, et al. Interleukin‐1 is associated with inflammation and structural lung disease in young children with cystic fibrosis. J Cyst Fibros. 2018;17:715‐722. [DOI] [PubMed] [Google Scholar]
- 49. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mattoscio D, Evangelista V, Cristofaro R, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) expression in human platelets: impact on mediators and mechanisms of the inflammatory response. FASEB J. 2010;24:3970‐3980. [DOI] [PubMed] [Google Scholar]
- 51. Ringholz FC, Buchanan PJ, Clarke DT, et al. Reduced 15‐lipoxygenase 2 and lipoxin A4/leukotriene B4 ratio in children with cystic fibrosis. Eur Respir J. 2014;44:394‐404. [DOI] [PubMed] [Google Scholar]
- 52. Tam VC, Quehenberger O, Oshansky CM, et al. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154:213‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li C, Hu X, Li L, Li J. Differential microRNA expression in the peripheral blood from human patients with COVID‐19. J Clin Lab Anal. 2020;34:e23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krishnamoorthy S, Recchiuti A, Chiang N, et al. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc Natl Acad Sci USA. 2010;107:1660‐1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Croasdell A, Sime PJ, Phipps RP. Resolvin D2 decreases TLR4 expression to mediate resolution in human monocytes. FASEB J. 2016;30:3181‐3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fredman G, Li Y, Dalli J, Chiang N, Serhan CN. Self‐limited versus delayed resolution of acute inflammation: temporal regulation of pro‐resolving mediators and microRNA. Sci Rep. 2012;2:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kedzierska K, Ellery P, Mak J, Lewin SR, Crowe SM, Jaworowski A. HIV‐1 down‐modulates γ signaling chain of FcγR in human macrophages: a possible mechanism for inhibition of phagocytosis. J Immunol. 2002;168:2895‐2903. [DOI] [PubMed] [Google Scholar]
- 58. Tseng C‐TK, Perrone LA, Zhu H, Makino S, Peters CJ. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J Immunol. 2005;174:7977‐7985. [DOI] [PubMed] [Google Scholar]
- 59. Watanabe Y, Shiratsuchi A, Shimizu K, Takizawa T, Nakanishi Y. Stimulation of phagocytosis of influenza virus‐infected cells through surface desialylation of macrophages by viral neuraminidase. Microbiol Immunol. 2004;48:875‐881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1


