Summary
Modifications in HLA‐I expression are found in many viral diseases. They represent one of the immune evasion strategies most widely used by viruses to block antigen presentation and NK cell response, and SARS‐CoV‐2 is no exception. These alterations result from a combination of virus‐specific factors, genetically encoded mechanisms, and the status of host defences and range from loss or upregulation of HLA‐I molecules to selective increases of HLA‐I alleles. In this review, I will first analyse characteristic features of altered HLA‐I expression found in SARS‐CoV‐2. I will then discuss the potential factors underlying these defects, focussing on HLA‐E and class‐I‐related (like) molecules and their receptors, the most documented HLA‐I alterations. I will also draw attention to potential differences between cells transfected to express viral proteins and those presented as part of authentic infection. Consideration of these factors and others affecting HLA‐I expression may provide us with improved possibilities for research into cellular immunity against viral variants.
Keywords: HLA class‐I, HLA‐E, MICA, NK cells, SARS‐CoV‐2
Abbreviations
- aa
amino acid
- ACE2
angiotensin‐converting enzyme 2
- ADAM17
a disintegrin and metalloprotease 17
- ADCC
antibody‐dependent cell‐mediated cytotoxicity
- ARDS
acute respiratory distress syndrome
- Covid‐19
coronavirus disease 19
- CTLs
CD8+ cytotoxic T lymphocytes
- HIF‐1α
hypoxia‐inducible factor 1α
- HLA
human leucocyte antigen
- IFN
interferon
- IL
interleukin
- ILC 2
innate lymphoid cell type 2
- KIR
Killer immunoglobulin‐like receptor
- KLRC2
killer cell lectin‐like receptor C 2
- MICA
class‐I‐related (like) molecules
- MULT1
murine UL16 binding protein‐like transcript
- NK
natural killer
- ORF
open reading frame
- Rae‐1
retinoic acid early inducible gene 1
- S
surface spike
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2 virus
- SP1
spike 1 protein
- TMPRSS2
type 2 TM serine protease
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus2 (SARS‐CoV‐2) targets the respiratory tract and causes coronavirus disease 19 (Covid‐19) characterised by varying the severity of lung lesions and reducing gas exchange preceded by alveolar and interstitial oedema. 1 Individuals responses to SARS‐CoV‐2 range in complexity from self‐limited inflammatory responses to excessive inflammation that accompanies acute respiratory distress syndrome (ARDS). 2 Differences in the behaviour of the virus observed between patients result from complex interactions between the virus 3 and components of the human immune system. 4 Among these factors, natural killer (NK) subsets and conventional cytotoxic T CD8+ cells (CTLs), as well as genetic determinants, have been proposed to at least partly participate in the resolution or severity of Covid‐19. 5 , 6 , 7 Expression of human leucocyte antigen (HLA)‐I molecules might potentially affect the susceptibility or resistance to this illness 8 since they are essential for immune control. 9
HLA‐I molecules can be dividing into the classic HLA‐A, ‐B and ‐C molecules, the class‐I‐related (like) molecules (MICA, MICB), and the nonclassic HLA‐E, ‐F and ‐G molecules. HLA‐I are glycoproteins that present antigen derived from the endogenous route of antigen processing to CTLs. They also function as ligands for activating or inhibitory receptors whose expression and function in NK cells and CTLs are just beginning to be understood. The significance of the interaction between HLA‐I molecules and their receptors depends on the HLA‐I cell surface expression state or HLA genotype of the individuals. Thus, aberrant or reduced expression of these molecules might be clinically relevant since, in exposed patients, including older 10 , 11 and adults with comorbidities (cancer, male sex), 12 , 13 they could escape from CTL and NK cell killing, 14 eventually leading to viral persistence 15 , 16 and subsequent severity of Covid‐19.
This review will analyse characteristic features of altered HLA‐I expression found in SARS‐CoV‐2, focussing on the most documented HLA‐I alterations.
2. HLA‐I INTERACTIONS WITH NK LYMPHOCYTES AND CTLs TO PRODUCE SPECIFIC CYTOTOXICITY
SARS‐CoV‐2 uses angiotensin‐converting enzyme‐2 (ACE2) and type 2 transmembrane serine protease (TMPRSS2) to bind and enter into target cells such as alveolar epithelial cells. 17 , 18 During the time preceding stimulation of T‐ and B‐cell responses, innate immunity is mediated by interferon (IFN) α/β, which prevents infection, and NK cells, which eliminate infected cells. NK cells control viral replication and spreading by spontaneous cytolysis or by secreting cytokines such as IFN‐γ and chemokines. 19 HLA‐E can present self‐signal peptides derived from other HLA molecules (HLA‐A, ‐ B, ‐C and ‐G) 20 to NK cells through direct interaction with NKG2/CD94 complex (NKG2A or NKG2C), 21 a C‐type lectin receptor expressed mainly on NK lymphocytes. It interacts with inhibitory NKG2A with higher affinities than activating NKG2C. 22 Once SARS‐CoV‐2 has evaded innate immunity, 23 viral control relies on cellular and humoral adaptive immunity. Naïve CD8+ T cells are activated to proliferate extensively after recognition of HLA‐I/viral epitope complex on dendritic cells in the draining lymph nodes and subsequently differentiate into functional effector CTLs. During the convalescent phases following Covid‐19, CTLs migrate to extralymphoid tissues, such as lung, to recognise the classical HLA‐ I/peptide complex on target cells, which in turn exert a potent inhibitory action on viral replication. 24 , 25 This step is crucial for clearance and to avoid the virus spreading in tissues. 14 , 26 Since HLA‐I proteins are essential for T cell stimulation and NK cell modulation, I will address immune evasion mechanisms that target antigen‐presentation and NK cell responses.
3. POTENTIAL SARS‐CoV‐2 PROTEINS THAT DISRUPT HLA‐I ANTIGEN PRESENTATION PATHWAYS
The classical HLA‐I proteins and their associated endogenous route of antigen processing are involved in antiviral CD8 T cell responses during SARS‐CoV infection. 27 Given the abilities of SARS‐CoV‐2 to mutate and increase virion spike density and infectivity, 28 the simplest ways for SARS‐CoV‐2 to avoid TCD8+ recognition are to mutate their immunoevasin proteins to inhibit HLA‐I antigen presentation pathways. Viral proteins produced by this virus such as surface spike (S), nucleocapsid, open reading frame‐1ab (ORF1ab), and ORF8 show frequent mutations. 29 The S protein contains neutralisation and CTL epitopes, which are essential to induce adaptive immune responses. It carries spike 1 (SP1) and SP2 domains, 30 which mediate receptor binding and downstream membrane fusion, respectively. 31 In vivo, there is no proof of reduced expression of HLA‐A/B/C following infection with SARS‐CoV‐2. However, in vitro data show that S protein may modulate the expression of HLA‐I. 32 When transferred at a higher level (1 μg) to lung epithelial cells, intracellular S protein reduces classical HLA‐I (A, B, and C) expression. Indeed, the aberrant expression of HLA‐A/B/C was more pronounced when using SP1 than SP2 and S proteins. Whether SP1 affects the expression of other genes involved in the endogenous route of antigen presentation and at which step inhibition occurs in the antigen processing is unknown. Like other human respiratory RNA viruses, 33 variation in the S protein indirectly could potentially manipulate HLA‐I antigen presentation pathways by mutating the S protein‐specific CTL epitopes binding to HLA‐I molecules. 34 However, further experiments will be required to completely understand the effect of three mutations (K417N, E484K and N501Y) 35 in spike on HLA‐I antigen presentation pathways. While the immunological effects of the observed decrease are not yet known, I speculate that the loss of HLA‐I often leads to a reduction in CTL responses. A similar function was likely attributed to the ORF8 protein, causing degradation of newly synthesised classical HLA‐I molecules in vitro, 36 , 37 but these preliminary data warrant confirmation. Deletion in ORF8 (Δ382 variant) was associated with more effective T‐cell responses and mild stage of Covid‐19, 38 suggesting that the binding of ORF8 protein to HLA‐I reduced CTL‐mediated antiviral activity. In a preprint study, Weingarten‐Gabbay et al. 39 assume that SARS‐CoV‐2 may interfere with HLA‐I antigen presentation pathways through both proteasome maturation protein depletion and by altering ubiquitination enzymes, thereby preventing presentation of highly expressed SARS‐CoV‐2 proteins by infected cells in vitro. 40 This potential strategy for evasion of production of HLA‐I peptides through ubiquitin‐proteasome pathway warrants further investigation. In the same way, Middle East respiratory syndrome‐coronavirus alters antigen presentation pathways and downregulates classical HLA‐I molecules in vitro. 41
4. IMPACT OF KIRs AND LILRB1 ON NK CELLS Covid‐19 RESPONSE
Multiple NK cell receptors (NKRs) govern the NK cell responsiveness capacity through direct or indirect recognition of HLA‐I molecules. 41 , 42 Such receptors, based on their function, could be subdivided mainly into three groups. NKRs, which utilise classical HLA‐I as their ligands, include the killer immunoglobulin‐like (KIRs) and human leucocyte immunoglobulin‐like (LILRB1 or LIR‐1) receptors. The second subgroup of activating NKp30, 44 and 46 receptors recognise virus‐derived peptides. 43 The last subgroup includes the NKG2/CD94 complex (NKG2A and NKG2C) and NKG2D. The ligand for NKG2A/C‐CD94 is HLA‐E. NKG2D does not dimerise with CD94 and recognises at least six ligands (UL16‐binding protein, MICA/B, Rae‐1, MULT1, H60). 44 The ‘missing‐self axis’ is controlled by inhibitory receptors (NKG2A and KIRs). Whereas KIRs directly scan surface HLA‐I molecules, the HLA‐E‐NKG2A interaction is an indirect immune‐surveillance mechanism for the HLA‐I expression. 45 Maucourant et al. showed that cytotoxic function of CD56dim NK cells expressing different combinations of inhibitory KIRs and NKG2A in acute Covid‐19 occurred independently of inhibitory KIRs expression. Furthermore, another study focussing on circulating NK cells found that only a smaller NK cell subset displayed upregulation of the inhibitory (KIR2DL1 and NKG2A) and activating (KIR2DS1) receptors in patients with ARDS. 46 These data suggest that KIRs do not participate in Covid‐19 disease progression. Of note, the presence of other NK receptors such as NKG2A has been correlated with disease severity. 47 , 48 The impact of LIR‐1 on NK cells' Covid‐19 response is more conflicting. There is some evidence that an increase in messenger RNA (mRNA) levels of LIR‐1 on monocytes may lead to a mild stage of Covid‐19, 49 though it can or cannot reflect increased levels in protein expression. Contrary to this, some studies have failed to replicate these findings and refuted any correlation between LIR‐1 and critical Covid‐19. 50 It is too early to deduce from a patient that LIR‐1 may not affect the severity of Covid‐19.
5. MODULATION OF HLA‐E AND MICA EXPRESSION BY SARS‐CoV‐2: HOW EXHAUSTED NK EFFECTOR CELLS FUNCTION
Several mechanisms can lead to NK cell exhaustion, including dysregulation of NKG2A/D receptors, detrimental modulations of the expression of non‐classical HLA‐E and MICA molecules, and suppressive effects by excessive levels of cytokines (Figure 1).
FIGURE 1.
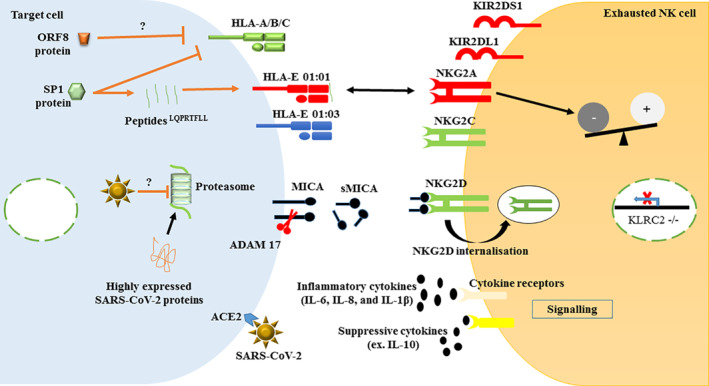
Major modifications in the expression of HLA‐I molecules (HLA‐A/B/C, HLA‐E and MICA) and their receptors on NK cells. The right panel represents the most documented modifications in NK cell receptors expression during SARS‐CoV‐2 infection. The left panel shows how viral proteins or SARS‐CoV‐2 alter the expression of HLA‐I molecules on target cells in vitro, therefore lead to NK cells exhaustion. Red: increased expression; Green: decreased expression; Dark blue: stable expression; Black: induced soluble factors; Brown lines and arrows: virus antagonistic tactics. SP1, spike 1 protein; ORF8, open reading frame 8; HLA, human leucocyte antigen; MICA, class‐I‐related (like) molecules; ADAM17, metalloprotease and disintegrin 17; NK, natural killer cells; NKG2A, inhibitory NK cell; NKG2C, activator NK cell; NKG2D, activator NK cell; KIRs, Killer Ig‐like receptors; KLRC2, Killer‐cell lectin‐like receptor C 2; IL, interleukin; ACE2, angiotensin‐converting enzyme 2
5.1. Factors affecting HLA‐E
The overexpression of nonclassical HLA‐E is a common observation through a wide range of viral infectious diseases. These molecules have been extensively studied 51 and expressed at low levels in most tissues under physiological conditions. 52 To counteract NK cell activation via the ‘missing self’‐axis, SARS‐CoV‐2 encodes immune evasion proteins that allow it to elude the NK cell activation. Bortolotti et al. 33 showed that in vitro, higher levels of SP1 (1 μg) on lung epithelial cells increase GATA3 transcription factor and alter cell surface HLA‐E expression. Of most relevance to HLA‐E is the SP1 subunit, a 671 amino acid glycoprotein (14–685 residues) containing a 291 amino acid N‐terminal domain (14–305 residues) 53 with an HLA‐E binding 8mer leader peptide (270–277; LQPRTFLL). Such peptides efficiently stabilise HLA‐E*01:01 on the transfected cell surface in comparison with HLA‐E*01:03. I hypothesised at least two potential implications of this. Perhaps the HLA‐E*01:01 allele, due to its lower binding affinity to standard HLA‐E specific peptides, binds SARS‐CoV‐2 SP1 derived peptides more efficiently. Therefore, HLA‐E may be differentially expressed according to cell types, and thus the effect of SP1 might be cell‐type specific. Consequently, HLA‐E molecules might exhibit differing affinities with SP1 peptides in a way that correlates with the differing stability and cell surface expression. In agreement, another study suggested that differential S protein stability (and binding to ACE2) may contribute to the varying disease courses. 54 In this study, aa variation in S protein (D614G mutation) correlated with increased viral loads and high transmission but low mortality. Thus, SARS‐CoV‐2 control appears to have more to do with the status of host defences than the virulence of the virus. Among genetic factors, polymorphic HLA‐E may also be responsible for differential HLA‐E expression, and therefore Covid‐19 severity. HLA‐E*01:01 allele and heterozygous HLA‐E*01:01/03 genotype are associated with severe Covid‐19. 55 Thus, the genetic difference at HLA‐E alleles may account for individual variations in the NK cell response against SARS‐CoV‐2 infection.
5.2. Evasion of NK cells through HLA‐E‐CD94/NKG2A axis
HLA‐E preferentially presents signal peptides derived from other HLA molecules (HLA‐A, ‐B, ‐C and ‐G) 56 or from viruses. 57 They also function as ligands for activating or inhibitory receptors, whose expression and function in NK cells, NKT cells, and CTLs are just beginning to be understood. The immunological consequences of the HLA‐Epeptide‐NKG2A interaction were revealed in a series of studies by several laboratories. Zheng et al. 47 showed an increased expression of inhibitory NKG2A in circulating lymphocytes (NK and T cells) in parallel with decreased IFN‐γ secretion and cytotoxic function in Covid‐19 patients. NKG2A upregulation may be caused by excessive levels of interleukin‐6 (IL‐6) and IL‐10, 58 the most important mediators of cytokine storm in fatal Covid‐19. 59 , 60 , 61 However, the lack of strong linkages between these soluble factors and NKG2A+NK cell increase suggests that their phenotype is driven mainly by receptor‐ligand engagement. Whereas there is some knowledge about NKG2A, our understanding of the NKG2‐HLA‐E interaction in SARS‐CoV‐2 infection—including those of HLA‐E‐NKG2A/C—is incomplete.
The binding affinity between CD94/NKG2A receptor and HLA‐E is affected by noncanonical peptides derived from viruses. Bortolotti et al. 32 wanted to determine in vitro whether SP1 binds NKG2 receptors. They focused their efforts on identifying a conserved target of NKG2A receptors. As part of immune evasion, SP1 peptide LQPRTFLL can mimic classical HLA‐I leader sequence peptides and bind to HLA‐E, ensuring it is continually increased, whereby it inhibits cytolysis of NK cells through interaction with CD94/NKG2A receptor. 32 Also, this interaction depends on HLA‐E allelic variations. Despite this relevant study, it is unclear how these observations relate to exhausted NK cells exposed to SARS‐CoV‐2 infection in vivo. These findings have been supported by the study of the conserved target of NKG2A receptors in HIV‐1 infected T cells, showing that the HLA‐E/capsid peptide complex prevents NK cell stimulation through its binding to CD94/NKG2A receptor. 62 , 63
Genetic variations of the NKG2C‐HLA‐E axis may also have an impact on the NK cell Covid‐19 response. NK‐cell lectin‐like receptor C 2 (KLRC2−/−) haplotype deletion and, to a lesser extent, HLA‐E allelic variant (HLA‐E*01:01) are correlated with the development of severe Covid‐19. 55 In this context, it can promote the possibility of progress in Covid‐19 as HLA‐E‐bearing virally infected cells are not being recognised by NK cells, while the probability of inhibition by the NKG2A receptor may increase.
Thus the HLA‐E/SP1LQPRTFLL complex interacts with NKG2A and, together with genetic factors and excessive levels of inflammatory cytokines, contributes to increased inhibitory signalling, leading to NK cell exhaustion at the early stages of Covid‐19 (Figure 1).
5.3. SARS‐CoV‐2 escape from NKG2D recognition
NK cell stimulation via NKG2D‐MICA interaction serves as a viral subversion target. The MICA and MICB proteins are different from other HLA class I antigens, which neither bind β2 microglobulin nor present peptides. 64 They are expressed as ‘danger signals’ on infected cells. However, many virus‐infected cells release a soluble MICA (sMICA) to escape from NKG2D recognition. 65 A similar mechanism might be used by SARS‐CoV‐2 to escape from NKG2D recognition as elevated plasma levels of sMICA 66 and lower expression of NKG2D on NK cells were both correlated with severe Covid‐19. 67 Among the proteinase family, a disintegrin and metalloproteinase17 (ADAM17) might be responsible for the shedding of MICA as transcription of these proteolytic enzymes is upregulated by several factors, including hypoxia‐inducible factor 1α (HIF‐1α), 68 SP1‐ACE2 interaction 69 and IL‐1β 70 during SARS‐CoV‐2 infection. Besides, Varchetta et al. 67 speculate that downregulated NKG2D expression in severe Covid‐19, probably driven by an uncontrolled secretion of inflammatory cytokines (IL‐6, IL‐8 and IL‐1β) by monocytes, could be an additional way to reduce immature NK CD56bright cells ability to secrete IFN‐ γ. Moreover, this cell population was decreased, particularly in patients who later died.
6. ACTIVATION OF INNATE LYMPHOCYTES (NK CELLS AND ILC2) DURING SEVERE Covid‐19
Overstimulation of NK cells can both increase function and cause exhaustion, often simultaneously. It occurs when activating signals are higher than the inhibiting signals. Many clinical observations suggest that mature NK cells are highly activated after infection with SARS‐CoV‐2 and participate to worsen lung injury observed in some patients with severe or fatal outcomes. In this context, Maucourant et al. 45 found an increased frequency of peripheral blood and lung adaptive NKG2C+CD57+ NK cells that display signs of proliferation and activation in patients with severe Covid‐19 in Sweden. Besides, they found an increase in HLA‐E mRNA contents in the immune and epithelial cells of bronchoalveolar lavage fluid in Covid‐19 patients, suggesting an NKG2C/HLA‐E driven proliferation of adaptive NK cells. However, confirmatory experiments are required to support these results. The adaptive NK cells activated in this context could also be induced directly by excessive levels of inflammatory cytokines. Further analysis of NK cells was performed by Varchetta et al. 67 using peripheral blood lymphocytes from Covid‐19 patients. The authors found an increased proportion of CD57+ FcεRIγneg adaptive NK cells in patients with fatal disease compared with survivors. These data suggest that adaptive NK lymphocytes do not appear to use NKG2C, NKp46, NKp30 and CD16 receptors as an efficient mechanism to cause lung injury. Circulating innate lymphoid cells 2 (ILC2) expressing NKG2D, on the other hand, increased significantly in severe compared to mild patients, indicating that this particular cell can control infection and reduce the number of Covid‐19 subjects requiring mechanical ventilation. 71 IL‐18 might be responsible for NKG2D upregulation on ILC2.
7. MORE QUESTIONS THAN ANSWERS
Much remains to be explored regarding the role of HLA‐I and NK cells during SARS‐CoV‐2.
Can the results obtained by transfecting cells to express S protein reach valid conclusions about the role of this protein for the pathogenesis of infection with SARS‐CoV‐2? Altogether, experimental approaches using SARS‐CoV‐2‐infected cells are needed to answer question about the linkage between classical and nonclassical HLA‐I alteration and viral immunoevasins. Although an increased number of more mature NK cells expressing activation markers is consistently observed in convalescent Covid‐19 patients, 47 it is unclear whether this increase confers protection from SARS‐CoV‐2 reinfection? There are two potential mechanisms responsible for this varying protection. Perhaps adaptive FcεRIγ negative CD56+/CD57+ NK cells may potentially play a significant role in protection from Covid‐19 through antibody‐dependent cell‐mediated cytotoxicity (ADCC) when IgG antibodies against S protein are present. These antibodies were correlated with a reduced risk of reinfection by SARS‐CoV‐2. 72 While eliciting neutralising IgG antibodies is a goal for an effective Covid‐19 vaccine, it is becoming clear that IgG that mediates ADCC can provide protective immunity against influenza. 73 Furthermore, important information on immune mechanisms capable of controlling Covid‐19 may come from children undergoing an intense immunisation program. 74 The trained immunity elicited by human Bacillus Calmette and Guerin vaccination cannot be excluded to improve this process 75 , 76 , 77 through a more potent cross immune defence in these individuals. Equally important is the question of how the modulation of HLA‐I expression by immunoevasins exhausted CTL cytotoxicity, especially in severe cases. As discussed above, there is some evidence NKG2A overexpression on TCD8+ is involved in functional exhaustion of CTLs and disease progression in the early stage of Covid‐19. However, the lack of stability of the HLA‐E‐SP1 complex identified to date limits our ability to study their CTLs responses in detail.
As mentioned previously, one mechanism that SARS‐CoV‐2 uses to evade adaptive immunity is antigenic variation, in which structural variants emerge that potentially alter HLA‐I antigen presentation pathways and enable this virus to persist or infect previously immunised subjects. As a result, it may be challenging to control new strains of SARS‐CoV‐2 using existing Covid‐19 vaccines. Moreover, the complexity of the molecular mechanisms underlying HLA‐I alteration expression and other related molecules involved in NK cell recognition makes selecting a therapeutic target to potentiate antiviral immune responses very difficult.
CONFLICT OF INTEREST
The author declares no conflict of interest.
AUTHOR CONTRIBUTIONS
The author confirms sole responsibility for the following: design and writing the manuscript, analysis and discussion of results. The author approved submitted version.
ACKNOWLEDGEMENT
The author thanks Faiza BOUAYAD and Ahmed Amine EL OUMRI for their enormous technical assistance.
Bouayad A. Features of HLA class I expression and its clinical relevance in SARS‐CoV‐2: what do we know so far? Rev Med Virol. 2021;31(6):e2236. 10.1002/rmv.2236
REFERENCES
- 1. Schenck EJ, Hoffman K, Goyal P, et al. Respiratory mechanics and gas exchange in COVID‐19‐associated respiratory failure. Ann Am Thorac Soc. 2020;17(9):1158‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2): an update. Cureus. 2020;12(3):e7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdullahi IN, Emeribe AU, Ajayi OA, Oderinde BS, Amadu DO, Osuji AI. Implications of SARS‐CoV‐2 genetic diversity and mutations on pathogenicity of the COVID‐19 and biomedical interventions. J Taibah Univ Med Sci. 2020;15(4):258‐264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS‐CoV‐2 infection and COVID‐19 disease severity. Hum Genom. 2020;14(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuri‐Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID‐19. Sci Immunol. 2020;5(49):eabd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang Y, Wei X, Guan J, et al. COVID‐19 pneumonia: CD8+ T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin Immunol. 2020;218:108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS‐CoV‐2 genomic variations associated with mortality rate of COVID‐19. J Hum Genet. 2020;65(12):1075‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shankarkumar U, Ghosh K, Mohanty D. The human leucocyte antigen (HLA) system. J Assoc Physicians India. 2002;50:916‐926. [PubMed] [Google Scholar]
- 10. Vrillon A, Hourregue C, Azuar J, et al. COVID ‐19 in older adults: a series of 76 patients aged 85 years and older with COVID‐19. J Am Geriatr Soc. 2020;68(12):2735‐2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kadambari S, Klenerman P, Pollard AJ. Why the elderly appear to be more severely affected by COVID‐19: the potential role of immunosenescence and CMV. Rev Med Virol. 2020;30(5):e2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison SL, Fazio‐Eynullayeva E, Lane DA, Underhill P, Lip GYH. Comorbidities associated with mortality in 31,461 adults with COVID‐19 in the United States: a federated electronic medical record analysis. PLoS Med. 2020;17(9):e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta S, Hayek SS, Wang W, et al. Factors associated with death in critically ill patients with coronavirus disease 2019 in the US. JAMA Intern Med. 2020;180(11):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lucas M, Karrer U, Lucas A, Klenerman P. Viral escape mechanisms—escapology taught by viruses. Int J Exp Pathol. 2001;82(5):269‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110(2):163‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Westrich JA, Warren CJ, Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Res. 2017;231:21‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan HW, Xu YM, Lau AT. Angiotensin‐converting enzyme 2: the old door for new severe acute respiratory syndrome coronavirus 2 infection. Rev Med Virol. 2020;30(5):e2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin‐converting enzyme 2 (ACE2) as a SARS‐CoV‐2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586‐590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raulet D. Natural Killer Cells. Fundamental Immunology. Lippincott Williams & Wilkins Publishers; 2003:671‐721. [Google Scholar]
- 20. Fang M, Orr MT, Spee P, Egebjerg T, Lanier LL, Sigal LJ. CD94 is essential for NK cell‐mediated resistance to a lethal viral disease. Immunity. 2011;34(4):579‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braud VM, Allan DSJ, O'Callaghan CA, et al. HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795‐799. [DOI] [PubMed] [Google Scholar]
- 22. Kaiser BK, Barahmand‐Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA‐E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174(5):2878‐2884. [DOI] [PubMed] [Google Scholar]
- 23. Bouayad A. Innate immune evasion by SARS‐CoV‐2: comparison with SARS‐CoV. Rev Med Virol. 2020;30(6):1‐9. [DOI] [PubMed] [Google Scholar]
- 24. Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181(7):1489‐1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neidleman J, Luo X, Frouard J, et al. SARS‐CoV‐2‐specific T cells exhibit phenotypic features of helper function, lack of terminal differentiation, and high proliferation potential. Cell Rep Med. 2020;1(6):100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sekine T, Perez‐Potti A, Rivera‐Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID‐19. Cell. 2020;183(1):158‐168.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J, Wu P, Gao F, et al. Novel immunodominant peptide presentation strategy: a featured HLA‐A*2402‐restricted cytotoxic T‐lymphocyte epitope stabilized by intrachain hydrogen bonds from severe acute respiratory syndrome coronavirus nucleocapsid protein. J Virol. 2010;84(22):11849‐11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang L, Jackson CB, Mou H, et al. SARS‐CoV‐2 spike‐protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11(1):6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS‐CoV‐2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. 2020;85:104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23(6):101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bortolotti D, Gentili V, Rizzo S, Rotola A, Rizzo R. SARS‐CoV‐2 spike 1 protein controls natural killer cell activation via the HLA‐E/NKG2A pathway. Cells. 2020;9(9):1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price GE, Ou R, Jiang H, Huang L, Moskophidis D. Viral escape by selection of cytotoxic T cell‐resistant variants in influenza A virus pneumonia. J Exp Med. 2000;191(11):1853‐1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo E, Guo H. CD8 T cell epitope generation toward the continually mutating SARS‐CoV‐2 spike protein in genetically diverse human population: implications for disease control and prevention. PLoS One. 2020;15(12):e0239566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang JW, Toovey OTR, Harvey KN, Hui DDS. Introduction of the South African SARS‐CoV‐2 variant 501Y.V2 into the UK. J Infect. 2021;S0163‐4453(21)00030‐X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park MD. Immune evasion via SARS‐CoV‐2 ORF8 protein? Nat Rev Immunol. 2020;20(7):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zinzula L. Lost in deletion: the enigmatic ORF8 protein of SARS‐CoV‐2. Biochem Biophys Res Commun. 2021;538:116‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Young BE, Fong S‐W, Chan Y‐H, et al. Effects of a major deletion in the SARS‐CoV‐2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396(10251):603‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weingarten‐Gabbay S, Klaeger S, Sarkizova S, et al. SARS‐CoV‐2 infected cells present HLA‐I peptides from canonical and out‐of‐frame ORFs. bioRxiv. 2020;2020.10.02.324145. [Google Scholar]
- 40. Menachery VD, Schäfer A, Burnum‐Johnson KE, et al. MERS‐CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc Natl Acad Sci U S A. 2018;115(5):E1012‐E1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kumar S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology. 2018;154(3):383‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saunders PM, Vivian JP, O'Connor GM, et al. A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol Rev. 2015;267(1):148‐166. [DOI] [PubMed] [Google Scholar]
- 43. Koch J, Steinle A, Watzl C, Mandelboim O. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 2013;34(4):182‐191. [DOI] [PubMed] [Google Scholar]
- 44. Lanier LL. NKG2D receptor and its ligands in host defense. Cancer Immunol Res. 2015;3(6):575‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maucourant C, Filipovic I, Ponzetta A, et al. Natural killer cell immunotypes related to COVID‐19 disease severity. Sci Immunol. 2020;5(50):eabd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Demaria O, Carvelli J, Batista L, et al. Identification of druggable inhibitory immune checkpoints on natural killer cells in COVID‐19. Cell Mol Immunol. 2020;17(9):995‐997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17(5):533‐535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bouadma L, Wiedemann A, Patrier J, et al. Immune alterations in a patient with SARS‐CoV‐2‐related acute respiratory distress syndrome. J Clin Immunol. 2020;40(8):1082‐1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saheb Sharif‐Askari N, Saheb Sharif‐Askari F, Mdkhana B, et al. Enhanced expression of immune checkpoint receptors during SARS‐CoV‐2 viral infection. Mol Ther Methods Clin Dev. 2021;20:109‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang S, Gan J, Chen BG, et al. Dynamics of peripheral immune cells and their HLA‐G and receptor expressions in a patient suffering from critical COVID‐19 pneumonia to convalescence. Clin Transl Immunology. 2020;9(5):e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pietra G, Romagnani C, Moretta L, Mingari M. HLA‐E and HLA‐E‐bound peptides: recognition by subsets of NK and T cells. Curr Pharm Des. 2009;15(28):3336‐3344. [DOI] [PubMed] [Google Scholar]
- 52. Djaoud Z, Riou R, Gavlovsky P‐J, et al. Cytomegalovirus‐infected primary endothelial cells trigger NKG2C+ natural killer cells. J Innate Immun. 2016;8(4):374‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang Y, Yang C, Xu X‐F, Xu W, Liu S‐W. Structural and functional properties of SARS‐CoV‐2 spike protein: potential antivirus drug development for COVID‐19. Acta Pharmacol Sin. 2020;41(9):1141‐1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Groves DC, Rowland‐Jones SL, Angyal A. The D614G mutations in the SARS‐CoV‐2 spike protein: implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104‐107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vietzen H, Zoufaly A, Traugott M, Aberle J, Aberle SW, Puchhammer‐Stöckl E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA‐E variants are risk factors for severe COVID‐19 [published online ahead of print, 2021 Jan 26]. Genet Med. 2021;1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petersdorf E, Socié G. The HLA system in hematopoietic stem cell transplantation Immune Biology of Allogeneic Hematopoietic Stem Cell Transplantation. Elsevier; 2019:15‐32. [Google Scholar]
- 57. Allard M, Tonnerre P, Nedellec S, et al. HLA‐E‐restricted cross‐recognition of allogeneic endothelial cells by CMV‐associated CD8 T cells: a potential risk factor following transplantation. PLoS One. 2012;7(11):e50951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Antonioli L, Fornai M, Pellegrini C, Blandizzi C. NKG2A and COVID‐19: another brick in the wall. Cell Mol Immunol. 2020;17(6):672‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID‐19 patients reveals IL‐6 and IL‐10 are disease severity predictors. Emerg Microbes Infect. 2020;9(1):1123‐1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cai L, Zhou X, Wang M, et al. Predictive nomogram for severe COVID‐19 and identification of mortality‐related immune features. J Allergy Clin Immunol Pract. 2021;9(1):177‐184.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Y, Tan W, Chen H, et al. Dynamic changes in lymphocyte subsets and parallel cytokine levels in patients with severe and critical COVID‐19. BMC Infect Dis. 2021;21(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nattermann J, Nischalke HD, Hofmeister V, et al. HIV‐1 infection leads to increased HLA‐E expression resulting in impaired function of natural killer cells. Antivir Ther. 2005;10(1):95‐107. [DOI] [PubMed] [Google Scholar]
- 63. Davis ZB, Cogswell A, Scott H, et al. A conserved HIV‐1‐derived peptide presented by HLA‐E renders infected T‐cells highly susceptible to attack by NKG2A/CD94‐bearing natural killer cells. PLoS Pathog. 2016;12(2):e1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Beck S, Trowsdale J. The human major histocompatibility complex: lessons from the DNA sequence. Annu Rev Genom Hum Genet. 2000;1(1):117‐137. [DOI] [PubMed] [Google Scholar]
- 65. Kumar V, Yi Lo PH, Sawai H, et al. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV‐induced hepatocellular carcinoma. PLoS One. 2012;7(9):e44743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. De Biasi S, Meschiari M, Gibellini L, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID‐19 pneumonia. Nat Commun. 2020;11(1):3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Varchetta S, Mele D, Oliviero B, et al. Unique immunological profile in patients with COVID‐19. Cell Mol Immunol. 2021;18(3):604‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baginska J, Viry E, Paggetti J, et al. The critical role of the tumor microenvironment in shaping natural killer cell‐mediated anti‐tumor immunity. Front Immunol. 2013;4:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gheblawi M, Wang K, Viveiros A, et al. Angiotensin‐converting enzyme 2: SARS‐CoV‐2 receptor and regulator of the renin‐angiotensin system. Circ Res. 2020;126(10):1456‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kohga K, Tatsumi T, Tsunematsu H, et al. Interleukin‐1β enhances the production of soluble MICA in human hepatocellular carcinoma. Cancer Immunol Immunother. 2012;61(9):1425‐1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gomez‐Cadena A, Spehner L, Kroemer M, et al. Severe COVID‐19 patients exhibit an ILC2 NKG2D+ population in their impaired ILC compartment. Cell Mol Immunol. 2021;18(2):484‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lumley SF, O’Donnell D, Stoesser NE, et al. Antibody status and incidence of SARS‐CoV‐2 infection in health care workers. N Engl J Med. 2021;384(6):533‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vanderven HA, Jegaskanda S, Wheatley AK, Kent SJ. Antibody‐dependent cellular cytotoxicity and influenza virus. Curr Opin Virol. 2017;22:89‐96. [DOI] [PubMed] [Google Scholar]
- 74. Castagnoli R, Votto M, Licari A, et al. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection in children and adolescents. JAMA Pediatr. 2020;174(9):882‐889. [DOI] [PubMed] [Google Scholar]
- 75. Cao Q, Chen Y‐C, Chen C‐L, Chiu C‐H. SARS‐CoV‐2 infection in children: transmission dynamics and clinical characteristics. J Formos Med Assoc. 2020;119(3):670‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kleinnijenhuis J, Quintin J, Preijers F, et al. BCG‐induced trained immunity in NK cells: role for non‐specific protection to infection. Clin Immunol. 2014;155(2):213‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Netea MG, Giamarellos‐Bourboulis EJ, Domínguez‐Andrés J, et al. Trained immunity: a tool for reducing susceptibility to and the severity of SARS‐CoV‐2 infection. Cell. 2020;181(5):969‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]


