Abstract
Background.
Although recent work has described the spatiotemporal diffusion of influenza viruses worldwide, comprehensive data on spatiotemporal patterns of influenza from the African continent and Madagascar are still lacking.
Methods.
National Influenza Centers from 5 countries–Cameroon, Côte d’Ivoire, Madagascar, Niger, and Senegal–collected specimens from patients presenting with influenza-like illness who visited sentinel surveillance clinics during a 2-year period (2008–2009). Isolates were genetically and antigenically characterized.
Results.
Overall, 8312 specimens were tested. Seasonal influenza A virus subtypes H1N1 and H3N2 and influenza B viruses were detected in 329, 689, and 148 specimens, respectively. In 2009, pandemic influenza A virus subtype H1N1 was detected in Madagascar most commonly (98.5% of cases). Influenza activity was either significant year-round or occurred during a specific period of the year in the African countries we evaluated.
Conclusions.
Our results demonstrate that, from Madagascar to Senegal, the epidemiologic and virologic characteristics of influenza viruses are diverse in terms of spatiotemporal circulation of the different virus types, subtypes, and strains. Our data highlight the importance of country-specific surveillance and of data and virus sharing, and they provide a rational basis to aid policy makers to develop strategies, such as vaccination at the right moment and with the right formulation, aimed at reducing the disease burden in Africa and Madagascar.
Acute respiratory infections, including those due to influenza virus, are one of the most important causes of morbidity and mortality worldwide, especially in children. It is estimated that 1 million deaths each year are due to influenza virus infection [1]. According to the World Health Organization (WHO), 20% of all deaths among children aged <5 years are attributable to influenza-like illnesses (ILI) [2], and 70% of those deaths occur in Africa and Southeast Asia [3]. Next to infants, elderly individuals and immunocompromised persons are also at risk to develop serious complications due to ILI [4–6]. Despite an increased interest in influenza surveillance, little is known about the burden and public health importance of influenza on the African continent. This could be partially explained by the fact that health concerns on the continent are focused on other widespread and high-impact infectious diseases, like AIDS and malaria. As such the impact of seasonal influenza remains unknown. Studies over the last 10 years in tropical areas have shown that the impact of influenza was an important contributor to respiratory illness [7–10], but data from Africa have been rare.
Although some countries, such as Madagascar and Senegal, have conducted national influenza surveillance for many years, several countries on the continent only began conducting surveillance for influenza recently. From the few data reported from sub-Saharan countries, it is difficult to comprehensively map and define the seasonality of influenza in the region. Some countries, like South Africa, Zambia [11], and Senegal [12], seem to have well-defined patterns of seasonality, with a period of influenza circulation lasting from June through October that corresponds to the drier and cooler months in South Africa and Zambia but to the hot and humid period in Senegal. Madagascar described a yearly circulation of influenza with 2 distinct periods of increased influenza circulation, one that corresponds to the northern hemisphere winter (ie, January–March) and a second that corresponds to the southern hemisphere winter (ie, June–October) [7, 13]. In Asia, these 2 active periods of circulation were also described in Hong Kong and Singapore [14, 15]. The aim of this collaborative study was to describe the spatiotemporal circulation and genetic characteristics of influenza viruses detected between 2008 and 2009 in 5 countries located in different latitudes and longitudes on the African continent.
MATERIALS AND METHODS
Ethical Considerations
Specimens and data were collected as part of national public health sentinel surveillance systems, which is considered a nonresearch activity. Influenza surveillance protocols were approved by the respective ministries of health. Before collecting each specimen, physicians explained the purpose of the surveillance system. After that, patients could refuse to participate. Oral consent was documented in the patient’s study records. Specimens and data were processed anonymously.
Study Design and Specimen Collection
This study was conducted in the framework of influenza sentinel surveillance implemented from January 2008 through December 2009 in 5 countries (Cameroon, Côte d’Ivoire, Madagascar, Niger, and Senegal) hosting laboratories that were part of the Institut Pasteur International Network. Each national sentinel system except for the one in Seychelles encompasses private and/or public clinics located in urban or semiurban areas and covers all ages. For Seychelles, specimens were collected during an outbreak. All these sites send daily epidemiological data about ILI cases. In each of the 5 countries, throat and/or nasopharyngeal swab specimens were collected from outpatients visiting sentinel sites and presenting with ILI, which was defined according to a WHO standard case definition [16]. Specimens were shipped to the National Influenza Center (NIC) in each country, except in Niger, where specimens were collected at the virology laboratory from the Centre de Recherche Médicale et Sanitaire de Niamey. Since Seychelles did not have the laboratory capacity, specimens collected were shipped to the NIC in Madagascar for characterization.
Laboratory Procedures
Specimens were tested for influenza by a real-time reverse-transcription polymerase chain reaction (rRT-PCR) and/or viral isolation by inoculation on Madin-Darby canine kidney (MDCK) cells. rRT-PCR was carried out according to protocols from the Centers for Disease Control and Prevention (CDC, Atlanta GA) or the Institut Pasteur (Paris, France). Protocols and are available upon request. After virus isolation on MDCK cells, identification of influenza virus isolates was performed using a hemagglutination inhibition (HI) assay and the WHO Influenza Reagent Kit (CDC) according to WHO protocol [17]. For some of the influenza A virus subtype H3N2 (A[H3N2]) isolates, a more detailed antigenic characterization was performed by HI analysis, using postinfection ferret sera against A/Brisbane/10/2007 and A/Perth/16/2009 (kindly provided by J. McCauley, WHO Collaborating Center, National Institute for Medical Research, London, United Kingdom). A representative subset of isolates was selected for virological analysis. We attempted to collect 30 seasonal influenza type A virus isolates (ie, subtype H1N1 [A{H1N1}] and A[H3N2]) from all 5 countries both during and between the epidemic periods.
The hemagglutinin (HA) and neuraminidase (NA) genes of A(H1N1) and A(H3N2) were amplified with specific primers by RT-PCR, using a protocol developed by the National Influenza Center for Northern France at the Institut Pasteur in Paris, which is available upon request. Sequencing was performed using the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems). Sequence chromatograms from both strands were obtained on the automated sequence analyzer ABI3730XL (Applied Biosystems) with the PCR primers. Contigs assembly was performed independently by distinct software, using either BioNumerics version 6.5 (Applied-Maths, Sint-Martens-Latem, Belgium) or CLC main workbench 6.0 (Aarhus, Denmark). Bayesian inference of phylogeny was used, and phylogenetic trees were inferred using the Mr Bayes 3.1 software package [18, 19]. All sequences newly reported from this study were deposited in the GenBank database (accession numbers pending).
Testing for Resistance to NA Inhibitors
Selected isolates were tested for resistance to NA inhibitors both genetically, by NA sequencing, and phenotypically, by a previously described fluorescence-based 2’-(4-methylumbelli-feryl)-alpha-D-N-acetylneuraminic acid (MUNANA) NA inhibition assay [20, 21].
RESULTS
Influenza Activities in Senegal, Niger, Côte d’Ivoire, Cameroon, and Madagascar
Four of the 5 countries had 2 full years of surveillance. Niger started influenza surveillance in May 2009. During the study period (January 2008 through December 2009), 8312 specimens were collected and tested by rRT-PCR, with 2480 and 5832 specimens collected in 2008 and 2009, respectively (Table 1). In 2008, the number of specimens tested ranged from 255 (Cameroon) to 890 (Côte d’Ivoire), and percentage of specimens positive for influenza virus ranged from 8.2% (Côte d’Ivoire) to 25.9% (In Senegal). During this year, seasonal A(H1N1) was the main virus detected, followed by influenza B virus in Cameroon, Madagascar, and Senegal, while in Côte d’Ivoire, the number of specimens that tested positive for A(H1N1) and influenza B virus was similar (29 and 31, respectively).
Table 1.
Results of Testing for and Subtyping of Influenza Virus in 5 African Countries, 2008 and 2009
| No. of Specimens Tested | Percentage Positive for Influenza | Influenza AVirus Subtype |
Influenza B Virus | |||||
|---|---|---|---|---|---|---|---|---|
| Country | Year | H1N1 | H3N2 | H1N1pdm09 | Unsubtyped | |||
| Senegal | 2008 | 522 | 25.9 | 74 | 1 | ... | ... | 60 |
| 2009 | 936 | 29.3 | 3 | 271 | ... | ... | ... | |
| Niger | 2009a | 113 | 18.6 | ... | 19 | ... | 1 | 1 |
| Côte d’Ivoire | 2008 | 890 | 8.2 | 29 | ... | ... | 13 | 31 |
| 2009 | 1583 | 29.9 | 29 | 171 | 5 | 134 | 135 | |
| Cameroon | 2008 | 255 | 26.7 | 52 | ... | ... | ... | 16 |
| 2009 | 556 | 25.9 | 4 | 129 | 9 | ... | 2 | |
| Madagascar | 2008 | 813 | 17.7 | 119 | 20 | ... | ... | 5 |
| 2009 | 2644 | 48.1 | 19 | 88 | 935 | ... | 231 | |
| Total | 2008 | 2480 | 16.9 | 274 | 21 | ... | 13 | 112 |
| 2009 | 5832 | 37.5 | 55 | 678 | 949 | 135 | 368 | |
Abbreviation: H1N1pdm09, 2009 pandemic influenza A virus subtype H1N1.
Surveillance started in May 2009.
Overall, A(H1N1)pdm09 was the main virus detected in 2009. Nevertheless, most of the specimens that tested positive for A(H1N1)pdm09 (935) were isolated in Madagascar. For the remaining 4 countries, only Cameroon and Côte d’Ivoire detected sporadic cases of infection due to A(H1N1)pdm09 (9 and 5 cases, respectively. Beside A(H1N1)pdm09, 2009 was dominated by A(H3N2) and influenza B virus, with 678 and 368 specimens, respectively, testing positive.
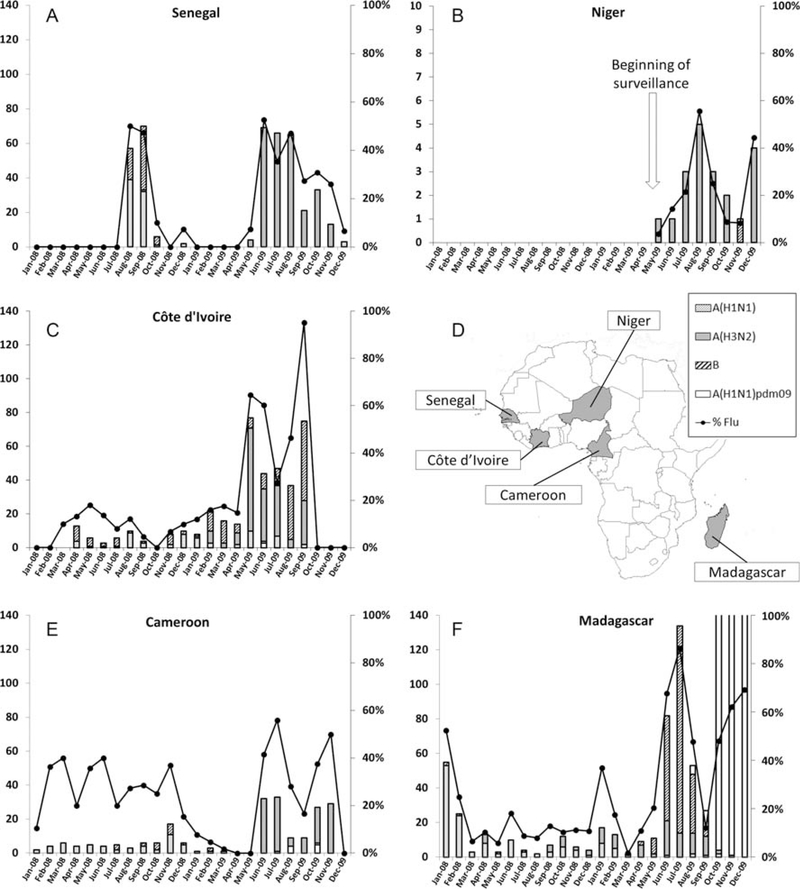
In 2008, influenza virus was detected year-round in Côte d’Ivoire, Cameroon, and Madagascar, with an increase in activity from January through February in Madagascar. In Senegal, circulation was detected from August through October (Figure 1). In 2009, the period in which activity was highest was from May through November for all countries except Madagascar, in which 2 periods of high influenza virus activity (during January–March and May–September) were detected. A third period of influenza virus activity was due to the circulation of A(H1N1)pdm09.
Figure 1.
Incidence of the influenza virus detection from January 2008 through December 2009. Specimens from outpatients that consult for influenza-like illness were analyzed using real-time reverse-transcription polymerase chain reaction for influenza A virus subtypes H1N1 (A[H1N1]) and H3N2 (A[H3N2]), 2009 pandemic influenza A virus subtype H1N1 (A[H1N1]pdm09), and influenza B virus. Each panel shows the monthly incidence of influenza viruses in Senegal (A), Niger (B), Côte d’Ivoire (C), Cameroon (E), and Madagascar (F). Location of countries in the African sub-continent where specimens were collected is also represented (D). Bars represent the cumulative number of specimens that were positive for influenza (corresponding to the left y-axis). The curve shows the monthly proportion of influenza-positive specimens among tested specimens (corresponding to the right y-axis).
Genetic Characterization of Seasonal A(H1N1) Isolates
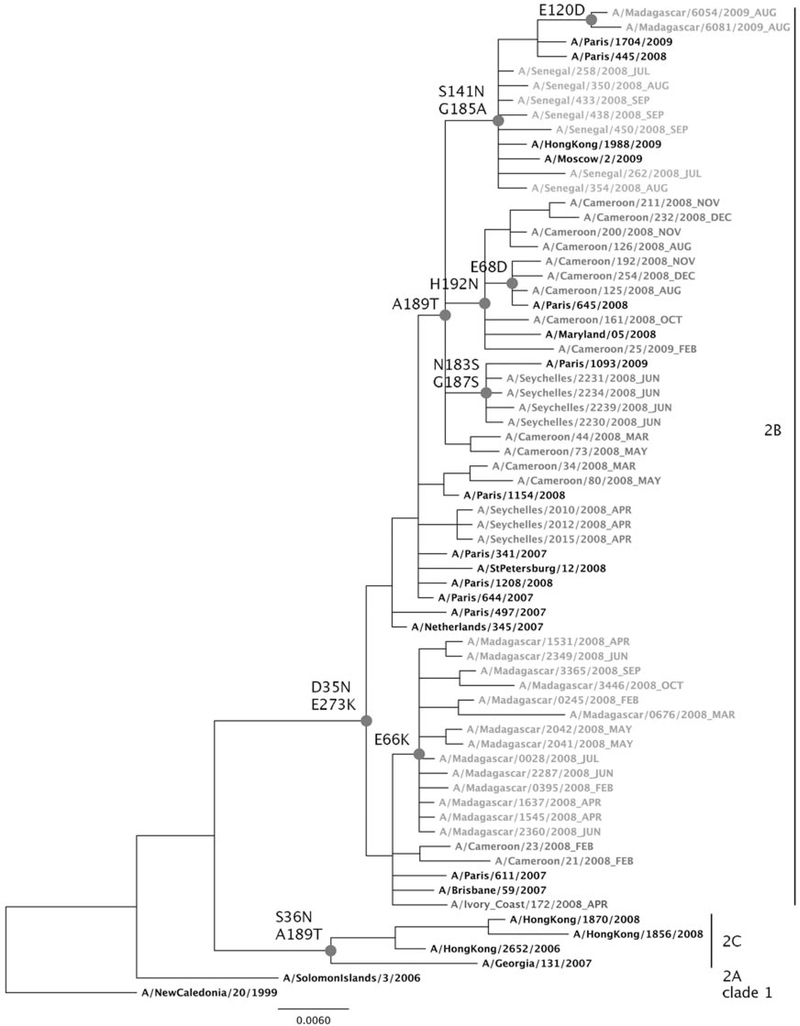
For the analysis of seasonal influenza A(H1N1) isolates, 97 sequences were generated by the Institut Pasteur laboratories, and 16 sequences were generated by the WHO Collaborative Centre in London. All were made available in the Global Initiative on Sharing Avian Influenza Database (available at: http://platform.gisaid.org/). Overall, phylogenetic analyses were performed on 28 sequences from Cameroon, 24 from Côte d’Ivoire, 21 from Madagascar, 33 from Senegal, and 7 from Seychelles. Sequences from reference strains, vaccine strains (from 1999 through 2007), and French isolates generated by NIC Northern France were added to the analysis. For clarity, one set of representative sequences by country was selected to construct the phylogenetic tree. Phylogenetic analysis of the HA1 genes showed that sequences of all 113 A(H1N1) isolates clustered in clade 2B together with the A/Brisbane/59/2007 strain, the vaccine strain recommended for the 2008–2009 influenza season in the northern hemisphere (Figure 2). At the amino acid level, in contrast with the A/NewCaledonia/20/1999 vaccine strain, these 113 isolates harbored the D35N and E273K substitutions observed in the A/Brisbane/59/2007 (H1N1) vaccine strain. However, some genetic drifts were observed in some countries. In Cameroon, where seasonal A (H1N1) was detected throughout the year, 2 of the 3 viruses of February 2008 were more closely related to the A/Brisbane/59/2007 virus than those isolated later, indicating a gradual evolution of the viruses. Most of the latter isolates were characterized by the A189T substitution, and all 21 viruses isolated during August 2008 had the additional H192N substitution. In Senegal, all 33 sequences belonged to a same cluster characterized by A189T, with additional S141N and G185A substitutions. In Côte d’Ivoire, except for 2 isolates recovered during April 2008 that were closely related to A/Brisbane/59/2007, all 22 sequenced isolates harbored the A189T substitution in association with either H192N (in 8) or S141N and G185A (in 14) changes. In Seychelles, the 7 sequences recovered formed 2 distinct clusters, with one harboring the A189T, N183S, and G187S substitutions. By contrast, all 19 of the 2008 A(H1N1) isolates from Madagascar characterized by the amino acid change E66K were closely related to A/Brisbane/59/2007. Thus, all isolates collected after May 2008, except the Malagasy isolates, fell within a cluster characterized by the A189T substitution.
Figure 2.
Phylogenetic analysis of nucleotide sequences of the hemagglutinin 1 gene (nucleotide 886) of influenza A virus subtype H1N1 isolates from Cameroon, Côte d’Ivoire, Madagascar, Senegal, and Seychelles, as well as references viruses. The tree was constructed using an ML method. The scale bar shows the genetic distance expressed as nucleotide substitutions per site. Online, a specific color is used for isolates from each country (red for Cameroon, green for Madagascar, orange for Senegal, light blue for Seychelles, and dark blue for Côte d’Ivoire). The strains name includes the isolation date (year_month). Major genetic clades are indicated on the right.
Phylogenetic analysis of the NA genes showed that all isolates harboring the A189T HA substitution possessed the H275Y NA mutation conferring oseltamivir resistance and the D354G substitution. All isolates collected in Madagascar during 2008, 2 isolates from Cameroon collected during February 2008, 3 isolates collected in Seychelles during April 2008, and 1 isolate collected in Côte d’Ivoire during April 2008 were the only ones without the H275Y NA substitution (Supplementary Figure 1).
Genetic Characterization of the A(H3N2) Isolates
For characterization of A(H3N2) isolates, 151 sequences from 2008 through 2010 were generated, including 36 from Cameroon, 25 from Côte d’Ivoire, 42 from Madagascar, 12 from Niger, and 36 from Senegal. As seen in Figure 4, the HA1 from A(H3N2) formed 4 different clades evolving from the A/Brisbane/10/2007 vaccine strain clade to the A/Victoria/208/2009 clade. For clarity, only 1 set of representative sequences by country is shown in the phylogenetic tree (Supplementary Figure 2). Overall, 38 of 151 viruses (25%) from the 5 countries belonged to the Brisbane/10/2007 clade. These viruses were mostly from Madagascar (27 of 38 [71%]) and were sampled before July 2009 (28 of 38 [74%]). The remaining 10 isolates (10 of 38 [26%]) were collected from July through September 2009. A total of 38 of 151 other viruses (25%) from 3 countries (26 collected in Cameroon, 9 in Côte d’Ivoire, and 3 in Senegal) fell within a new genetic clade characterized by the V112I substitution. All 38 viruses from this new genetic clade were obtained between May and August 2009. Thirty-five of 151 viruses (23%) from 3 countries (32 collected in Senegal, 2 in Niger, and 1 in Madagascar) belonged to a third clade, A/Perth/16/2009, characterized by the N144K, K158N, and N189K substitutions. These 35 viruses were collected between May 2009 and January 2010, with most recovered during June–August 2009. Forty of 151 viruses (26%) from 4 countries (14 collected in Madagascar, 9 in Cameroon, 9 in Niger, and 8 in Côte d’Ivoire) belonged to a fourth clade, A/Victoria/208/2009, characterized by the K158N, N189K, and T212A substitutions. Almost half of these viruses were collected from July through November 2009, and half were collected from January through December 2010.
On the basis of results of HI tests performed with postinfection ferret sera against A/Brisbane/10/2007 and A/Perth/16/2009, all viruses from the A/Brisbane/10/2007 clade and from the new clade harboring the V112I substitution were found to be antigenically related to A/Brisbane/10/2007. In contrast, viruses from the A/Perth/16/2009 and A/Victoria/208/2009 clades were found to be antigenically related to A/Perth/16/2009(H3N2) (data not shown).
NA-Inhibitor Resistance
Forty-six A(H1N1) viruses and 69 A(H3N2) viruses selected among the isolates described above were assayed in a fluorescence-based MUNANA NA inhibition assay (Supplementary Table 1). All 69 A(H3N2) viruses were found to be susceptible to both oseltamivir (IC50 0.28 ± 0.17) and zanamivir (IC50 0.88 ± 0.64). For A(H1N1)isolates, resistance to oseltamivir is strictly correlated to the H275Y NA (N1 numbering) mutation. All 21 A(H1N1) isolates with an histidine residue at position 275 in the NA (275H) were found to be sensitive to both oseltamivir (median inhibitory concentration [IC50] ± SD, 0.77 ± 0.23) and zanamivir (IC50 ± SD, 0.85 ± 0.28), whereas all 25 isolates with a tyrosine residue (275Y) were resistant to oseltamivir (IC50 ± SD, 222 ± 63) but not to zanamivir (IC50 ± SD, 0.70 ± 0.17).
DISCUSSION
Our analysis represents the first attempt to describe the circulation of influenza viruses across sub-Saharan Africa. We analyzed viruses from countries located at latitudes and longitudes ranging from 18°54’ S and 47°31’ E (Madagascar) to 14°43’ N and 17°28’ W (Senegal). During the 2-year study period, we found that, in these tropical countries, except for Senegal, seasonality was less pronounced than in temperate countries, a finding that has been previously reported in studies in Africa [11] and other tropical regions [9,22]. Indeed, we found either significant year-round influenza activity or 2 distinct influenza seasons in the African countries we evaluated. In contrast to tropical Brazil, we did not observe a southward traveling wave of influenza in Africa [23]. In 2008, influenza viruses circulated year-round in Cameroon, Côte d’Ivoire, and Madagascar, whereas in 2009, there was more influenza virus activity in all countries during the second part of the year. However, these results could be biased because of the increase in the number of sentinel sites in 2009, combined with the increase in specimen collection that occurred in late 2009 owing to concerns about A(H1N1)pdm09.
In Senegal (Dakar), Côte d’Ivoire (Abidjan), and Niger (Niamey), influenza virus activity was concomitant with the rainy season, probably because of lower temperatures, as described elsewhere for Niger [24]. This same pattern has been observed in the northeastern part of Brazil (Fortaleza) [25]. In contrast, in Cameroon (Yaoundé) and Madagascar (Antananarivo), influenza virus circulation occurred during the cold and dry period, but a second period of circulation was observed from December through March, similar to the northern hemisphere, and coincided with the warm and rainy period. In Madagascar, these 2 periods of influenza activity have been described elsewhere [7], and they have also been observed in some tropical countries, including Hong-Kong and Singapore [14, 15].
In this study, molecular and antigenic analysis of influenza viruses highlighted the circulation of distinct viral lineages, including distinct clades of the same subtype between 2008 and the beginning of 2010. Similar viral circulation and evolution were observed in all countries except Madagascar.
In 2008, seasonal A(H1N1) and influenza B virus cocirculated, but the proportion of influenza viruses that were A (H1N1) varied from West Africa (around 50% in Senegal and Côte d’Ivoire) to Central Africa (76% in Cameroon) and southeast Africa (>96% in Madagascar). The 2008 seasonal A(H1N1) isolates from all African countries were antigenically different from A/Solomon Island/03/2006, the 2008 southern hemisphere vaccine strain, but were closely related to A/Brisbane/59/2007, the 2008–2009 northern hemisphere vaccine strain.
In Europe, the 2007–2008 winter season saw the emergence and spread of A(H1N1) oseltamivir-resistant viruses (ORVs) independently of drug use [21, 26]. A permissive genetic background achieved through mutations that preempted the H275Y substitution and restored viral fitness of H275Y-bearing viruses is likely to account for this spread [27, 28]. The frequency of A(H1N1) ORV detection increased from close to 0% in week 40% to 56% in week 19. At the end of January 2008, ORVs harboring the H275Y mutation in gene encoding NA had been detected in 9 European countries, and the prevalence of resistance was estimated to be around 14% [29]. As early as February 2008, the first ORV was detected in Africa, in Cameroon [30], and from March 2008, all seasonal A(H1N1) Cameroonian isolates were ORVs. Similarly, in Côte d’Ivoire and Senegal, almost all 2008 A(H1N1) isolates (96.5%), collected in May, were ORVs. Thus, resistant viruses rapidly spread from the northern hemisphere to Africa. Interestingly, Madagascar did not have any A(H1N1) ORVs until 2009, even though ORVs were detected in neighboring countries such as Seychelles and South Africa beginning in June 2008 [31, 32]. The cocirculation of minor variants of ORVs, as evidenced by the cluster with the A189T, S141N, and G185A substitutions observed in Senegal and Côte d’Ivoire, the cluster with the A189T and H192N substitutions in Cameroon and Côte d’Ivoire, and even the A/Cameroon/34/2008 and A/Cameroon/80/2008 isolates without the A189T substitution, suggests multiple introductions of ORVs in Africa.
In the African viruses we evaluated, genetic diversity was more pronounced in A(H3N2) than in seasonal A(H1N1). Indeed, for A(H3N2), we identified 4 clades showing broad geographic distribution and evolving over time from the A/Brisbane/10/2007 vaccine strain clade (mostly detected during the first part of 2009) to the A/Victoria/208/2009 clade (detected at the end of 2009). This trend was consistent with the evolution of A(H3N2) observed throughout the world, which led to the change in September 2009 of the A(H3N2) component of the influenza vaccine recommendation for the 2010 season in the southern hemisphere [33, 34]. In Madagascar, the detection of these new Perth-like variants took place later in 2010. Interestingly, in Cameroon, a genetic switch was observed during the peak of A(H3N2) circulation between May and September 2009, with most of the May and August isolates belonging to the clade characterized by the V112I substitution and most of the October and November isolates belonging to the A/Victoria/208/2009 clade. This genetic switch was associated with an antigenic switch; viruses from the clade characterized by the V112I substitution were antigenically related to A/Brisbane/10/2007, while viruses belonging to the A/Victoria/208/2009 clade were antigenically related to A/Perth/16/2009. These new antigenic variants could have been favored initially before eventually becoming dominant, likely because of positive selective pressure owing to low preexisting immunity in the population.
Our interpretation of these virological data has some limitations. Indeed, sentinel sites were different from country to country. In some countries, surveillance sites treated individuals in all age groups, while in other countries, the surveillance system encompassed pediatric clinics and private practitioners. Also, the ILI case definition slightly differed from the standard WHO definition in Madagascar. These differences may have introduced some bias in the total number of ILIs and the rate of influenza positivity. Nevertheless, since we aimed to study the global circulation of influenza viruses in Africa through their genetic and phenotypic characteristics and not the burden of influenza, these biases should not affect our conclusions regarding influenza virus dynamics on the continent. Even if we selected a subset of specimens from during and between the epidemic periods without consideration of the geographical origin of specimens within each country, another limitation is that most of specimens were collected in the capital cities. Thus, our study might not be nationally representative, and we cannot exclude the possibility that different viruses may have circulated in other parts of the countries we studied.
Even if the year-round influenza activity observed in this study could suggest local persistence of influenza viruses, the large diversity of circulating influenza viruses without clear clustering by geographical location suggests that multiple importations of viruses from outside of the continent occurred frequently in Africa. The introduction of new viral strains in Africa mirrored changes in virus circulation in the rest of the world. However, in Madagascar, in contrast to the other studied countries, there was a significant delay in the appearance of new variants, with the exception of A(H1N1)pdm09. This could be due at least in part to its insularity, which is attributable to a lower level of international travel.
Our results show that sharing data between countries is important because it can improve understanding of influenza circulation at the regional level. Our results also demonstrate that influenza dynamics in Africa are a complex and that differences in influenza virus activity cannot be explained by the simple dichotomy between northern and southern hemispheres. These data can be used by policy makers to answer challenging questions regarding the use of the northern and southern hemisphere vaccine, the period of influenza virus circulation on the African continent, and the use of antivirals. More studies are needed to better understand viral circulation within the African continent. For some countries, seasonality remains unresolved because of the diversity of environmental factors and the lack of robust data over time. Future research should combine virological, epidemiological, and environmental data to understand factors that drive influenza virus activity and circulation nationally and regionally.
Supplementary Material
Acknowledgments.
We recognize the hard work of all individuals who provided care to patients, collected specimens, provided data and information, and kept the public informed, including general practitioners, nurses, and other healthcare workers; personnel at participating virology laboratories; and personnel at participating ministries of health. We also specifically acknowledge John McCauley and his team (National Institute for Medical Research), for their help and for making sequences of influenza viruses available through GISAID; Laure Diancourt, Anne-Sophie Delannoy-Vieillard, and Jean-Michel Thiberge (Genotyping of Pathogens and Public Health Platform), for sequencing the viruses and for excellent technical assistance; and Mathilde Benassaya, David Briand, Frédérique Cuvelier, Sébastien Le Gal, Vanessa Roca, Girard Marcellin Razafitrimo, Jaovanona Seth, Mohamadou Njankouo Ripa, and Stephane Bouloumegue (participating NICs), for their excellent technical assistance.
Zanamivir was kindly provided by Mark von Itzstein (Institute for Glycomics, Griffith University). Oseltamivir carboxylate (GS4071), the active form of the ethyl ester prodrug oseltamivir phosphate, was kindly provided by Roche.
Financial support. This work and the SURGIRA (SURveillance de la Grippe et des Infections Respiratoires en Afrique) project was supported by the Ministère français de la Santé/EPRUS, de la Jeunesse et des Sports, through the Fonds de Solidarité Vieillesse (Project “Surveillance et étude des souches grippales circulantes”) and the Office of the Assistant Secretary for Preparedness and Response, US Department of Health and Human Services (grant No. 6 IDSEP060001-01-01). The surveillance systems were supported in Madagascar by WHO headquarters (APW/reference OD/AP-08-02451) and Sanofi-Pasteur, in Camroon by Sanofi-Pasteur and in Senegal by WHO headquarters (APW/reference OD/AP-08-02637).
Footnotes
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
References
- 1.Pan American Health Organization (PAHO). Final report of the XVI Meeting on vaccine preventable-disease of the pan American health organization. Washington, District of Columbia: PAHO, 2004. http://www.paho.org/English/AD/FCH/IM/TAG16_FinalReport_2004.pdf. Accessed 25 April 2011. [Google Scholar]
- 2.World Health Organization. Acute respiratory infections in children. Geneva: WHO, 2010. http://www.who.int/fch/depts/cah/resp_infections/en/print.html. Accessed 25 April 2011. [Google Scholar]
- 3.Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2002; 2:25–32. [DOI] [PubMed] [Google Scholar]
- 4.Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: global burden of disease study. Lancet 1997; 349:1269–76. [DOI] [PubMed] [Google Scholar]
- 5.Schnell D, Mayaux J, de Bazelaire C, et al. Risk factors for pneumonia in immunocompromised patients with influenza. Respir Med 2010; 104:1050–6. [DOI] [PubMed] [Google Scholar]
- 6.Sprenger MJ, Mulder PG, Beyer WE, Van Strik R, Masurel N. Impact of influenza on mortality in relation to age and underlying disease, 1967–1989. Int J Epidemiol 1993; 22:334–40. [DOI] [PubMed] [Google Scholar]
- 7.Razanajatovo NH, Richard V, Hoffmann J, et al. Viral etiology of influenza-like illnesses in Antananarivo, Madagascar, July 2008 to June 2009. PLoS One 2011; 6:e17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shek LP, Lee BW. Epidemiology and seasonality of respiratory tract virus infections in the tropics. Paediatr Respir Rev 2003; 4:105–11. [DOI] [PubMed] [Google Scholar]
- 9.Viboud C, Alonso WJ, Simonsen L. Influenza in tropical regions. PLoS Med 2006; 3:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong CM, Yang L, Chan KP, et al. Influenza-associated hospitalization in a subtropical city. PLoS Med 2006; 3:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessner BD, Shindo N, Briand S. Seasonal influenza epidemiology in sub-Saharan Africa: a systematic review. Lancet Infect Dis 2011; 11:223–35. [DOI] [PubMed] [Google Scholar]
- 12.Niang M, Dosseh A, Ndiaye K, et al. Sentinel surveillance for influenza, 1996–2009. J Infect Dis 2012; 206(Suppl 1):S129–35. [DOI] [PubMed] [Google Scholar]
- 13.Rabarijaona LP, Rakotondrarija NT, Rousset D, Soares JL, Mauclere P. Influenza epidemiologic and virologic surveillance in Antananarivo from 1995 to 2002. Arch Inst Pasteur Madagascar 2003; 69:20–6. [PubMed] [Google Scholar]
- 14.Lee VJ, Yap J, Ong JB, et al. Influenza excess mortality from 1950–2000 in tropical Singapore. PLoS One 2009; 4:e8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, Wong CM, Lau EH, Chan KP, Ou CQ, Peiris JS. Synchrony of clinical and laboratory surveillance for influenza in Hong Kong. PLoS One 2008; 3:e1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization. Recommended surveillance standards (WHO/CDS/CSR/ISR/992) 2. Geneva: WHO, 1999. http://www.who.int/csr/resources/publications/surveillance/whocdscsrisr992.pdf. Accessed 10 June 2011. [Google Scholar]
- 17.WorldHealthOrganization.WHO/CDS/CSR/NCS/2002.5–WHOmanual on animal influenza diagnosis and surveillance. Geneva: WHO, 2002. http://www.who.int/vaccine_research/diseases/influenza/WHO_manual_on_animal-diagnosis_and_surveillance_2002_5.pdf. Accessed 10 June 2011. [Google Scholar]
- 18.Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL. Bayesian phylogenetic analysis of combined data. Syst Biol 2004; 53:47–67. [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572–4. [DOI] [PubMed] [Google Scholar]
- 20.Rameix-Welti MA, Agou F, Buchy P, et al. Natural variation can significantly alter the sensitivity of influenza A (H5N1) viruses to oseltamivir. Antimicrob Agents Chemother 2006; 50:3809–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rameix-Welti MA, Enouf V, Cuvelier F, Jeannin P, van der Werf S. Enzymatic properties of the neuraminidase of seasonal H1N1 influenza viruses provide insights for the emergence of natural resistance to oseltamivir. PLoS Pathog 2008; 4:e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamerius J, Nelson MI, Zhou SZ, Viboud C, Miller MA, Alonso WJ. Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect 2011; 119:439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso WJ, Viboud C, Simonsen L, Hirano EW, Daufenbach LZ, Miller MA. Seasonality of influenza in Brazil: a traveling wave from the Amazon to the subtropics. Am J Epidemiol 2007; 165:1434–42. [DOI] [PubMed] [Google Scholar]
- 24.Jusot JF, Adamou L, Collard JM. Influenza transmission during a one-year period (2009–2010) in a Sahelian city: low temperature plays a major role. Influenza Other Respi Viruses 2012; 6:87–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moura FE, Perdigao AC, Siqueira MM. Seasonality of influenza in the tropics: a distinct pattern in northeastern Brazil. Am J Trop Med Hyg 2009; 81:180–3. [PubMed] [Google Scholar]
- 26.Meijer A, Lackenby A, Hungnes O, et al. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis 2009; 15:552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 2010; 328:1272–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes EC. Virology. Helping the resistance. Science 2010; 328:1243–4. [DOI] [PubMed] [Google Scholar]
- 29.Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Sur-veill 2008; 13:8026. [DOI] [PubMed] [Google Scholar]
- 30.Njouom R, Mba SA, Noah DN, et al. Circulation of human influenza viruses and emergence of Oseltamivir-resistant A(H1N1) viruses in Cameroon, Central Africa. BMC Infect Dis 2010; 10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besselaar TG, Naidoo D, Buys A, et al. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg Infect Dis 2008; 14:1809–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurt AC, Ernest J, Deng YM, et al. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res 2009; 83:90–3. [DOI] [PubMed] [Google Scholar]
- 33.Recommended composition of influenza virus vaccines for use in the 2010 influenza season (southern hemisphere winter). Wkly Epidemiol Rec 2009; 84:421–31. [PubMed] [Google Scholar]
- 34.Recommended viruses for influenza vaccines for use in the 2010–2011 northern hemisphere influenza season. Wkly Epidemiol Rec 2010; 85:81–92. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.