Abstract
Purpose
Non-coding RNA activated by DNA damage (NORAD) has been reported to be a cancer-related long non-coding RNA (lncRNA) implicated in the progression of several cancers; however, its role in breast cancer (BC) has not yet been clarified.
Methods
Quantitative real-time polymerase chain reaction was used to examine NORAD, microRNA (miR)-155-5p, and suppressor of cytokine signaling 1 (SOCS1) mRNA expression levels. Western blotting was used to analyze SOCS1 protein expression. The malignancy of BC cells was assessed using the cell counting kit-8 (CCK-8), BrdU, and Transwell assays. Bioinformatics analysis, RNA immunoprecipitation assay, and dual-luciferase reporter gene assays were used to verify the targeted relationship between NORAD and miR-155-5p. Additionally, the regulatory effects of NORAD and miR-155-5p on SOCS1 expression were determined by western blotting.
Results
NORAD expression was significantly reduced in BC cell lines and tissues, and its low expression was associated with poor tumor tissue differentiation. NORAD overexpression repressed BC cell proliferation, migration, and invasion, whereas its knockdown produced the opposite effects. Additionally, miR-155-5p was found to be a target of NORAD, and the biological functions of miR-155-5p and NORAD were counteractive. MiR-155-5p was confirmed to target SOCS1, and SOCS1 was found to be positively regulated by NORAD.
Conclusion
NORAD suppresses miR-155-5p to upregulate SOCS1, thereby repressing the proliferation, migration, and invasion of BC cells.
Keywords: Breast neoplasms, Cell proliferation, Metastasis
INTRODUCTION
Breast cancer (BC) is one of the most common causes of cancer-related deaths among women [1]. Although radical surgery, radiotherapy, chemotherapy, targeted-therapy, and endocrine therapy have reduced the mortality of patients with BC, treatment failures are still common in clinical practice [2,3]. Therefore, there is an urgent need to decipher the mechanism of BC progression and identify novel therapeutic targets.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides in length that do not encode proteins [4]. Over the past few years, several studies have indicated that lncRNAs are vital in regulating tumor growth and metastasis [5]. For instance, lncRNA TUC338 accelerates lung cancer progression by regulating the mitogen-activated protein kinase pathway [6]. Furthermore, lncRNA MALAT1 inhibits BC metastasis, while lncRNA ITGB2-AS1 facilitates BC cell metastasis by upregulating ITGB2 [7,8]. As a member of the lncRNA family, non-coding RNA activated by DNA damage (NORAD) is associated with the progression of multiple cancers, such as gastric, pancreatic, and thyroid cancers [9]. However, the role of NORAD in BC remains controversial, as high NORAD expression was reported to predict poor prognosis in patients with BC and activate the transforming growth factor (TGF)-β pathway to drive the metastasis of BC cells [10]; whereas another study reported that NORAD functioned as a tumor suppressor in BC by repressing S100P-elicited pro-metastatic signaling [11]. Therefore, its role in BC is not fully understood.
In this study, we demonstrated that NORAD was downregulated in BC tissues and cell lines, and functionally, NORAD suppressed the malignancy of BC cells, which indicated that NORAD functions as a tumor-suppressor. Mechanistically, NORAD functions as a competing endogenous RNA (ceRNA) for miR-155-5p, reduces its expression, and positively regulates suppressor of cytokine signaling 1 (SOCS1).
METHODS
Tissue samples and ethics statement
BC and adjacent tissue samples were obtained from 46 patients who underwent surgery. The tissues were immediately stored in liquid nitrogen following resection. The collection and use of human tissue samples were authorized by the Institutional Review Board (IRB) of Yichang Central People's Hospital (IRB approval number: 2017036). All participants provided informed consent.
Cell culture
The normal mammary epithelial cell line MCF-10A and BC cell lines HCC70, MCF-7, SK-BR-3, and T-47D were purchased from the Cell Center of the Chinese Academy of Sciences (Shanghai, China). These cells were cultured in RPMI-1640 medium (Hyclone, South Logan, USA) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin (Invitrogen, Carlsbad, USA), and 10% fetal bovine serum (FBS) (Invitrogen) in a thermostatic incubator perfused with 5% CO2 at 37°C.
Cell transfection
MCF-7 and HCC70 cells in the logarithmic growth phase were selected. After trypsinization, the cell suspension was plated in 6-well plates (1 × 105 cells/well) and cultured for 24 hours in 5% CO2 at 37°C. Subsequently, the cells were transfected with pcDNA-NORAD, NORAD small interfering RNA (siRNA; si-NORAD), miR-155-5p mimics/inhibitors, and their negative controls (Biosettia Inc., San Diego, USA) using Lipofectamine® 3000 transfection reagent (Invitrogen) according to the manufacturer's instructions. At 48 hours after transfection, the transfection efficiency was validated by quantitative real-time polymerase chain reaction (qRT-PCR).
qRT-PCR
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen), and the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) was used to synthesize cDNA. The mRNA expression levels of NORAD, miR-155-5p, and SOCS1 were determined using SYBR® Premix Ex Taq™ (TaKaRa). The relative expression levels of these genes were normalized to U6 or GAPDH using the 2−ΔΔCT method. The primer sequences used in this study are listed in Table 1.
Table 1. Sequences used for qRT-PCR.
| Name | Primer sequences |
|---|---|
| NORAD | Forward: 5′-TGATAGGATACATCTTGGACATGGA-3′ |
| Reverse: 5′-AACCTAATGAACAAGTCCTGACATACA-3′ | |
| miR-155-5p | Forward: 5′-UUAAUGCUAAUUGUGAUAGGGGU-3′ |
| Reverse: 5′-CCCUAUCACGAUUAGCAUUAAUU-3′ | |
| U6 | Forward: 5′-GAGGGTTAATGCTAATCGTGATAGG-3′ |
| Reverse: 5′-GCACAGAATCAACACGACTCACTAT-3′ | |
| SOCS1 | Forward: 5′-GGTCCCCCTGGTTGTTGTA-3′ |
| Reverse: 5′-GTTGGTTGGTGTGTGGGAGC-3′ | |
| GAPDH | Forward: 5′-TAGGAGGTGCGAGTTCAGGT-3′ |
| Reverse: 5′-GGCTGTTGTCATACTTCTCATGG-3′ |
qRT-PCR = quantitative real-time polymerase chain reaction; NORAD = non-coding RNA activated by DNA damage; miR = microRNA; SOCS1 = suppressor of cytokine signaling 1; GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
Cell counting kit-8 (CCK-8) assay
The transfected HCC70 and MCF-7 cells were transferred to 96-well plates (2,000 cells/well) and cultured at 37°C. Cell viability was evaluated every 24 hours using the CCK-8 Kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) according to the manufacturer's instructions. Four days later, the proliferation curves were plotted based on the cell viability for each day.
BrdU assay
In brief, cells were transferred into a culture dish (35 mm; with a cover slip in each well), and after culturing for 1 day, they were synchronized with medium containing 4% FBS for 3 days. Then, BrdU solution (BD Pharmingen, San Diego, USA) was added, and the cells were cultured for 4 hours at 37°C. Next, the culture solution was discarded, the cover slips were rinsed with phosphate-buffered saline (PBS) thrice, and the cells were fixed with methanol and air dried. The cells were blocked with rabbit serum. Subsequently, the slides were washed with PBS, and anti-BrdU antibody (Abcam, Shanghai, China) was added to the cells and incubated at 37°C for 2 hours. DAPI staining solution (BD Pharmingen) was then added to stain the nuclei. Five fields of view (× 200), randomly selected from each slide, were observed under a fluorescence microscope, and the percentage of BrdU-positive cells was calculated.
Transwell assays
For the migration assay, transfected HCC70 and MCF-7 cells were harvested, resuspended in serum-free medium, and the cell concentration was adjusted to 1.0 × 105 cells/mL. Next, 200 μL of the cell suspension was added to the upper chamber of a Transwell® system (Corning, Beijing, China), and the lower chamber was replenished with medium containing 10% FBS. After 24 hours of culture, cotton swabs were used to remove the cells remaining in the upper chamber. The cells on the lower surface of the Transwell membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The membranes were washed with tap water, dried, and the cells were then observed and counted under a microscope. For the invasion assay, the Transwell® membranes were precoated with a layer of Matrigel (BD Biosciences, Franklin Lakes, USA). The remaining steps in the protocol were identical to those used in the migration assay.
Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) assay
Apoptotic cells were determined by TUNEL staining using an In Situ Cell Death Detection Kit (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer's instructions. Images were captured with an Olympus BX41 fluorescence microscope equipped with an OlympusDP72 color digital camera. Five photos (magnification ×250) were randomly captured in each sample. The percentage of TUNEL-positive cells in the total number of cells was used to calculate the apoptotic rate.
Dual-luciferase reporter assay
StarBase (http://starbase.sysu.edu.cn/index.php) was used to predict the binding site between NORAD and miR-155-5p, and PCR was used to amplify the binding fragments between miR-155-5p and NORAD. Subsequently, the amplified products were inserted into pGL3-promoter plasmid vector (Promega, Madison, USA) to construct a wild-type (WT) reporter. Site-directed mutagenesis of the binding fragments was performed to construct a NORAD mutant (MUT) reporter. Next, 293T cells were co-transfected with recombinant reporter plasmids and miR-155-5p mimics or miR-control. At 48 hours after transfection, the cells were collected, and a Dual-luciferase Reporter Kit (Promega) was used to detect luciferase activity. The firefly luciferase activity was normalized to that of the renilla.
RNA immunoprecipitation (RIP) assay
The direct interaction between miR-155-5p and NORAD was examined using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, USA). RIP lysis buffer was used to lyse the BC cells. Next, the lysate was incubated with anti-argonaute 2 (Ago2) or control anti-immunoglobulin G (IgG) antibodies conjugated to magnetic beads for 6 hours at 4°C. After the mixtures were incubated with proteinase K, RNA was extracted from the immunoprecipitates using the TRIzol method. qRT-PCR was then performed to detect the expression of NORAD.
Western blot analysis
Radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) was used to lyse BC cells and tissues, and a BCA Protein Assay Kit (Bio-Rad, Hercules, USA) was used to determine the protein concentration. The protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene fluoride (PVDF) membranes (Beyotime). The membranes were then blocked with 5% non-fat milk followed by incubation with primary anti-SOCS1 (1:1,000, ab62584, Abcam) and anti-γH2AX (1:1,000, ab81299, Abcam) antibodies for 12 h at room temperature. Next, the membranes were washed with Tris-buffered saline containing 0.1% Tween 20 (TBST), and incubated for 1 h with horseradish peroxidase (HRP) conjugated secondary antibodies at room temperature. An Amersham Imager 600 instrument (GE Healthcare, Chicago, USA) was used to visualize the protein bands developed using the ECL Kit (Beyotime).
Statistical analysis
Data are presented as the mean ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism 8.0 Software (GraphPad Inc., San Diego, USA). Student's t-test was used to analyze the differences between two groups, and Pearson's correlation analysis was conducted to examine the correlation between NORAD and miR-155-5p expression. Statistical significance was set at p < 0.05.
RESULTS
NORAD expression is downregulated in BC tissues and cells
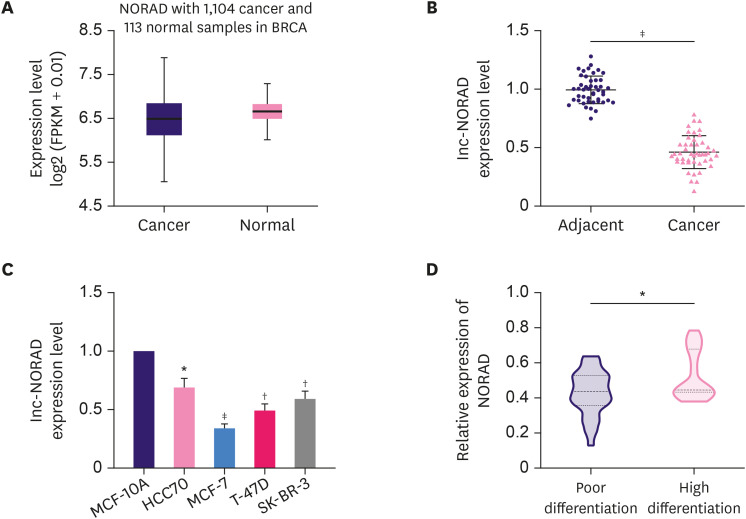
The StarBase database revealed that NORAD expression was markedly reduced in BC samples compared to that in normal samples (Figure 1A). Next, we used qRT-PCR to analyze NORAD expression in 46 paired BC and adjacent tissue samples. Consistent with the StarBase data, qRT-PCR results also showed that NORAD expression was lower in BC tissues compared to that in adjacent tissues (Figure 1B). Subsequently, NORAD expression was analyzed in a normal mammary epithelial cell line (MCF-10A) and four different BC cell lines (HCC70, MCF-7, T-47D, and SK-BR-3). The results showed that NORAD expression was also reduced in BC cell lines (Figure 1C). Additionally, NORAD expression was found to be reduced in patients with poor tumor tissue differentiation (Figure 1D).
Figure 1. NORAD is downregulated in BC and negatively correlates with tumor differentiation in patients with BC.
(A) Analysis of the StarBase database to examine NORAD expression in normal and BC tissues. (B) qRT-PCR analysis of the relative expression of NORAD in 46 pairs of BC and matched normal tissues. (C) qRT-PCR analysis of the relative expression of NORAD in four BC cell lines and the normal breast cell line, MCF-10A. (D) qRT-PCR analysis of NORAD expression in BC tissues with different differentiation status.
NORAD = non-coding RNA activated by DNA damage; BC = breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction.
*p < 0.05, †p < 0.01, and ‡p < 0.001.
NORAD inhibits BC cell proliferation and metastasis
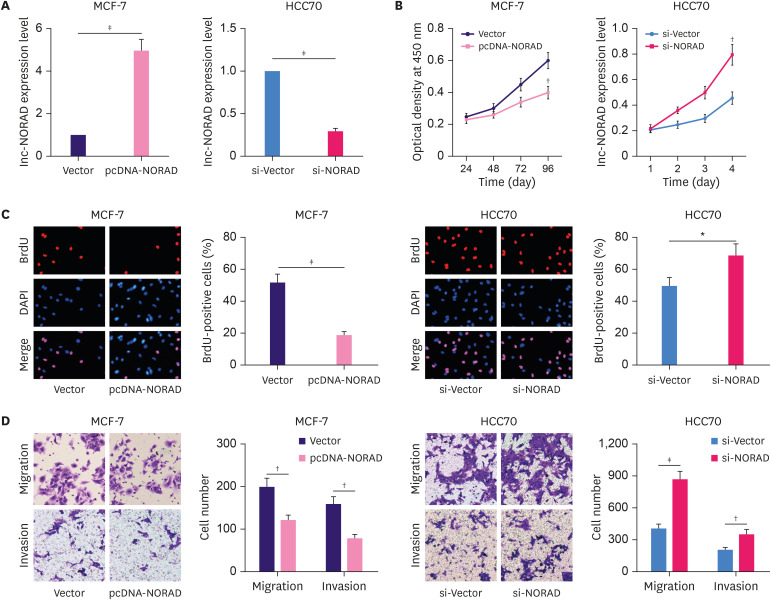
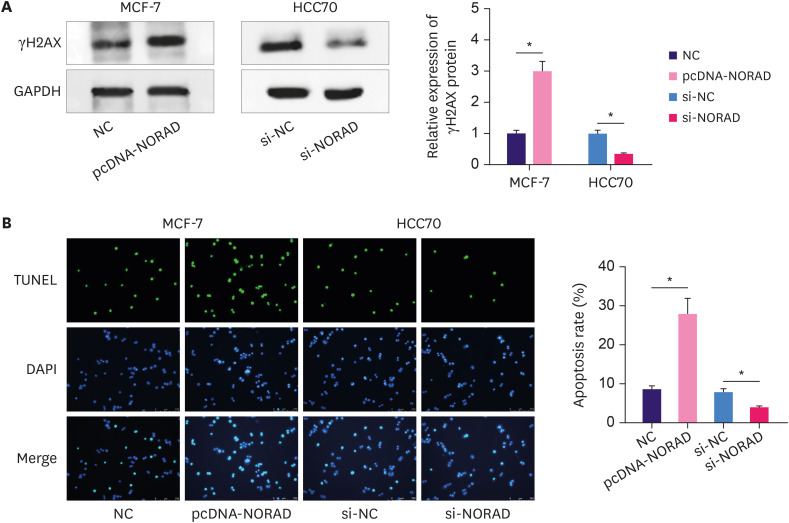
Next, pcDNA-NORAD and si-NORAD were transfected into MCF-7 and HCC70 cells, respectively and qRT-PCR confirmed that NORAD overexpression and knockdown were effective (Figure 2A). Subsequently, CCK-8 and BrdU assays revealed that NORAD overexpression suppressed MCF-7 cell proliferation, whereas NORAD knockdown promoted HCC70 cell proliferation (Figure 2B and C). In addition, the Transwell assay indicated that NORAD knockdown markedly promoted BC cell migration and invasion, whereas NORAD overexpression produced the opposite effect (Figure 2D). Furthermore, western blotting showed that overexpression of NORAD increased the expression of γH2AX, a marker of DNA damage, whereas NORAD knockdown inhibited γH2AX expression (Figure 3A). TUNEL assay revealed that NORAD overexpression significantly promoted the apoptosis of BC cells, whereas its knockdown inhibited apoptosis (Figure 3B). These findings confirm that NORAD suppresses malignancy of BC cells.
Figure 2. NORAD suppresses BC cell proliferation, migration, and invasion.
(A) qRT-PCR was used to verify the relative expression of NORAD in MCF-7 and HCC70 cells transfected with pcDNA-NORAD or siRNA targeting NORAD. (B and C) BrdU and CCK-8 assays were conducted to analyze the proliferation of BC cells. (D) Transwell assays were used to analyze migration and invasion of BC cells.
NORAD = non-coding RNA activated by DNA damage; BC = breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction; siRNA = small interfering RNA; CCK-8 = cell counting kit-8; DAPI = 4′,6-diamidino-2-phenylindole.
*p < 0.05, †p < 0.01, and ‡p < 0.001.
Figure 3. NORAD promotes DNA damage and cell apoptosis.
(A) Western blotting was used to detect the expression of the DNA damage marker γH2AX, following overexpression or knockdown of NORAD. (B) TUNEL assay was used to detect apoptotic cells.
NORAD = non-coding RNA activated by DNA damage; TUNEL = terminal deoxynucleotidyl transferase dUTP Nick-End Labeling; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; DAPI = 4′,6-diamidino-2-phenylindole; NC = negative control.
*p < 0.001.
NORAD serves as a molecular sponge of miR-155-5p
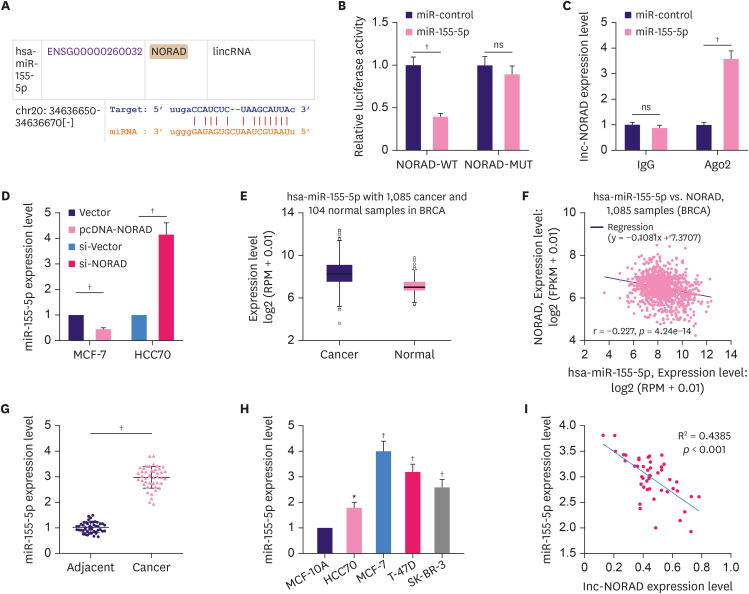
The “ceRNA network” is a well-known classic regulatory mechanism by which lncRNAs regulate various biological processes. LncRNAs can sponge miRNAs to repress their expression and regulate their effect on target mRNAs. The StarBase database suggested a potential binding site between miR-155-5p and NORAD (Figure 4A). To confirm this, we performed a dual-luciferase reporter assay. The reporter assay indicated that miR-155-5p inhibited luciferase activity of NORAD WT reporter but not that of the MUT reporter (Figure 4B). Furthermore, RIP assay indicated that in contrast with control IgG, NORAD and miR-155-5p were preferentially enriched in Ago2-containing microribonucleoproteins (Figure 4C). Moreover, NORAD overexpression reduced miR-155-5p expression in MCF-7 cells, whereas NORAD knockdown increased miR-155-5p expression in HCC70 cells (Figure 4D). Notably, the data from the StarBase database suggested that compared to normal tissues, miR-155-5p expression was higher in BC tissues (Figure 4E), and the expression of NORAD and miR-155-5p were negatively correlated with each other in these samples (Figure 4F). Consistently, qRT-PCR showed that compared to normal controls, miR-155-5p expression was markedly enhanced in both BC tissues and cells (Figure 4G and H). Furthermore, miR-155-5p expression was negatively correlated with NORAD expression in BC tissues (Figure 4I). These data suggest that miR-155-5p is a downstream target of NORAD in BC.
Figure 4. NORAD targets miR-155-5p and negatively regulates its expression.
(A) Bioinformatics analysis was used to predict the binding site between NORAD and miR-155-5p. Dual-luciferase reporter assay (B), and RIP assay (C) were conducted to validate the interaction between miR-155-5p and NORAD in BC cells. (D) Relative miR-155-5p expression in BC cells transfected with NORAD-overexpression/control plasmids and si-NORAD/control plasmids. Analysis of the StarBase database to examine miR-155-5p expression (E), and the correlation between miR-155-5p and NORAD in BC (F). qRT-PCR was conducted to analyze miR-155-5p expression in BC tissues (G), and cell lines (H). (I) Analysis of the correlation between NORAD and miR-155-5p in BC tissues.
NORAD = non-coding RNA activated by DNA damage; miR = microRNA; RIP = RNA immunoprecipitation; BC = breast cancer; WT = wild-type; MUT = mutant; IgG = immunoglobulin G; Ago2 = argonaute 2; ns, no statistical significance.
*p < 0.01 and †p < 0.001.
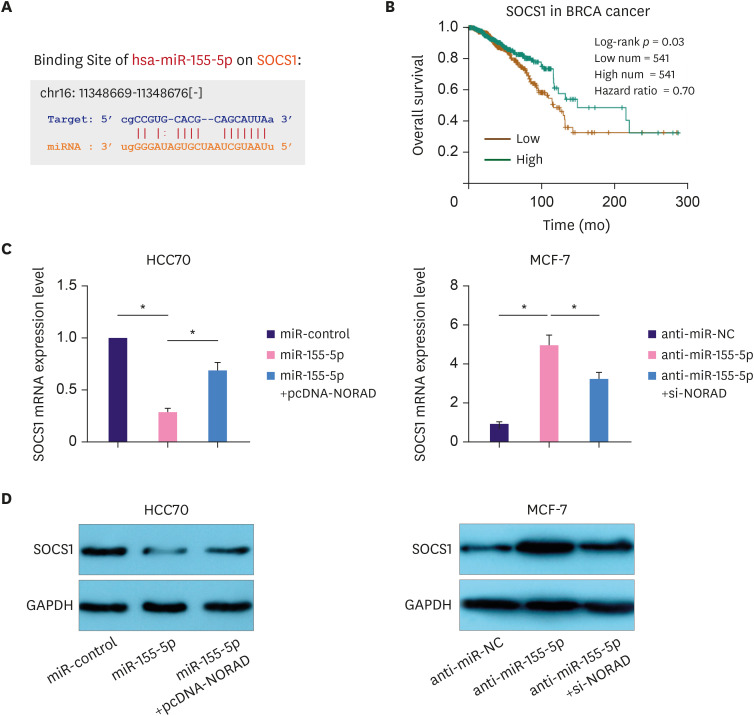
Tumor suppressor SOCS1 is regulated by the NORAD/miR-155-5p axis
SOCS1 is a target gene of miR-155-5p (Figure 5A) [12]. Importantly, survival analysis in the StarBase database indicated that the overall survival rate of patients with BC with high SOCS1 expression was higher than that of the low expression group (Figure 5B). Moreover, western blotting and qRT-PCR showed that NORAD enhanced SOCS1 expression and reversed the miR-155-5p-induced SOCS1 downregulation in HCC70 cells. Conversely, NORAD knockdown reduced SOCS1 expression and attenuated miR-155-5p antagonist-induced SOCS1 upregulation in MCF-7 cells (Figure 5C and D). These data suggest that the NORAD/miR-155-5p axis regulates BC progression by modulating the expression of SOCS1.
Figure 5. NORAD sponges miR-155-5p to elevate SOCS1 expression.
(A) StarBase database was used to examine binding site for miR-155-5p in the SOCS1 3′-UTR. (B) StarBase database was used to analyze the relationship between the overall survival of patients with BC and SOCS1 expression. qRT-PCR (C) and western blotting (D) were performed to analyze SOCS1 expression in BC following NORAD and miR-155-5p regulation.
NORAD = non-coding RNA activated by DNA damage; miR = microRNA; SOCS1 = suppressor of cytokine signaling 1; UTR = untranslated region; BC = breast cancer; qRT-PCR = quantitative real-time polymerase chain reaction; GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
*p < 0.001.
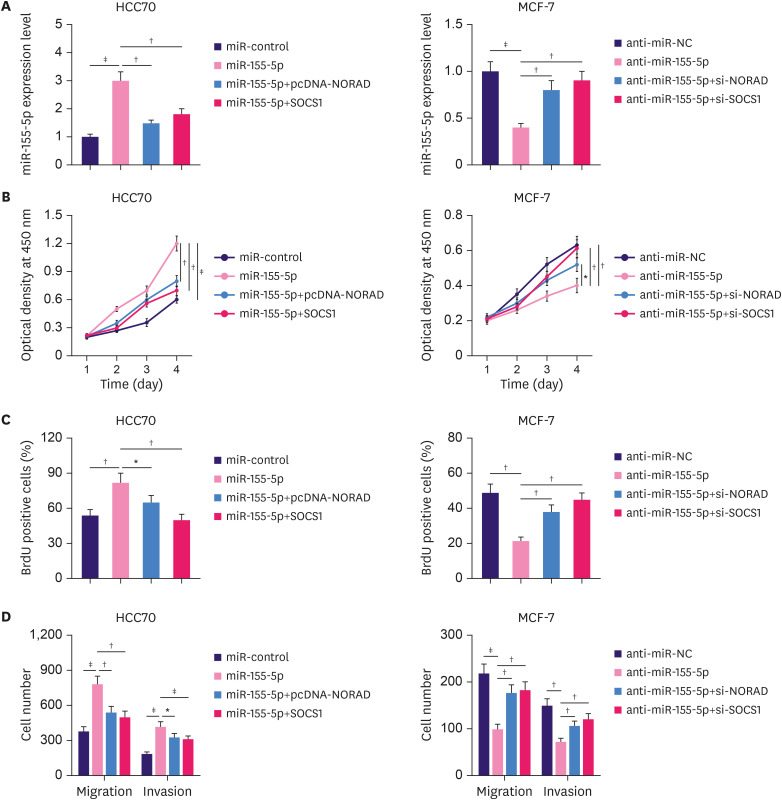
Overexpression of NORAD/SOCS1 reverses the promotional effects of miR-155-5p on BC cells
To further investigate the significance of the NORAD/miR-155-5p/SOCS1 axis in regulating BC progression, NORAD or SOCS1 overexpression plasmids and miR-155-5p mimics were co-transfected into HCC70 cells. Similarly, NORAD or SOCS1 siRNA and miR-155-5p inhibitors were co-transfected into MCF-7 cells (Figure 6A). Transfection with miR-155-5p mimics promoted the proliferation, migration, and invasion of BC cells, whereas inhibition of miR-155-5p reduced the malignancy of BC cells (Figure 6B-D). However, NORAD/SOCS1 overexpression and knockdown counteracted the effects of miR-155-5p mimics and inhibitors, respectively (Figure 6B-D). These findings imply that NORAD inhibits the malignant behaviors of BC cells partly by repressing miR-155-5p/SOCS1.
Figure 6. NORAD counteracts the effects of miR-155-5p in BC cells.
(A) HCC70 cells were transfected with control miRNA, miR-155-5p mimics, miR-155-5p mimics + NORAD overexpression plasmid, or miR-155-5p mimics + SOCS1 overexpression plasmid. MCF-7 cells were transfected with miRNA inhibitor control, miR-155-5p inhibitors, miR-155-5p inhibitors + NORAD siRNA, or miR-155-5p inhibitors + SOCS1 siRNA. Then, qRT-PCR was used to analyze the expression of miR-155-5p in each group of cells. (B) CCK-8, (C) BrdU, and (D) Transwell assays were conducted to analyze proliferation, migration, and invasion of BC cells.
NORAD = non-coding RNA activated by DNA damage; miR = microRNA; BC = breast cancer; siRNA = small interfering RNA; SOCS1 = suppressor of cytokine signaling 1; qRT-PCR = quantitative real-time polymerase chain reaction; CCK-8 = cell counting kit-8.
*p < 0.05, †p < 0.01, and ‡p < 0.001.
DISCUSSION
LncRNAs are crucial regulators of tumorigenesis and progression. Small nucleolar RNA host gene 16 (SNHG16) is upregulated in cervical cancer tissues and cells, and knockdown of SNHG16 suppresses cancer cell proliferation and epithelial-mesenchymal transition (EMT) by modulating the cell cycle and apoptosis [13]. In BC, the upregulation of basal-like breast cancer-associated transcript 1, resulting from hypomethylation at CpG islands in the promoter, reduces apoptosis and promotes the invasion of cancer cells [14]. Nuclear transcription factor nuclear factor (NF)-κB-interacting lncRNA represses TGF-β-induced EMT by blocking NF-κB signaling in BC [15]. LncRNA cancer susceptibility candidate 9 functions as an oncogene in BC by sponging the miR-195/497 cluster and promoting BC cell proliferation and survival [16]. NORAD is dysregulated in various tumors. However, its role in BC remains controversial. We demonstrated that NORAD is downregulated in BC tissues and cells, and that the downregulation of NORAD is associated with unfavorable pathological characteristics of the patients. Loss- and gain-of-function experiments revealed that NORAD repressed the malignancy of BC cells. Our data support that NORAD functions as a tumor suppressor, and not an oncogene, in BC.
MiRNAs are non-coding RNAs 21–25 nucleotides in length that are involved in the pathogenesis of many human diseases [17]. They are also key regulators of BC tumorigenesis and progression. MiR-330-3p promotes BC metastasis by targeting collagen and calcium-binding epidermal growth factor domain-1 [18]. Another study showed that miR-193b inhibits metastasis and angiogenesis by suppressing dimethylarginine dimethylaminohydrolase 1 in BC [19]. MiR-155-5p has also drawn a lot of attention in cancer research. MiR-155-5p promotes cervical cancer cell metastasis by targeting tumor protein p53-inducible nuclear protein 1 [20]. MiR-155-5p is pivotal in cancer-associated fibroblast-induced tumor angiogenesis via inhibition of SOCS1 expression and activation of Janus kinase (JAK2)/signal transducers and activators of transcription (STAT3) signaling pathway in melanoma [12]. Importantly, miR-155-5p is markedly upregulated in the plasma of patients with BC [21]. Functionally, miR-155-5p promotes BC cell proliferation and migration by targeting matrix metallopeptidase 16 and SOCS1 [22]. In the present work, we also validated that miR-155-5p had the properties of an oncomiR, was upregulated in BC, and promoted the malignant behaviors of BC cells.
LncRNAs serve as ceRNAs to regulate the translation of mRNAs by competing for shared miRNAs [23]. This mechanism has been shown to be a crucial process during cancer progression. For example, lncRNA small nucleolar RNA host gene 5 facilitates BC cell proliferation by sponging miR-154-5p and upregulating proliferating cell nuclear antigen [24]. LINC01116 upregulates estrogen receptor 1 expression by competing with miR-145, thus facilitating BC progression [25]. MiR-155-5p is also involved in ceRNA networks. The lncRNA, myocardial infarction-associated transcript, facilitates BC progression and functions as a ceRNA by sponging miR-155-5p [26]. In this study, NORAD was identified as a ceRNA for miR-155-5p, and NORAD positively regulated SOCS1 by sponging miR-155. Importantly, NORAD or SOCS1 overexpression counteracted the tumor-promoting effects of miR-155-5p in BC. Therefore, our results support the view that NORAD/miR-155-5p/SOCS1 is a novel ceRNA network in BC progression.
As a member of the SOCS family, SOCS1 participates in multiple biological processes, including tumorigenesis [27]. SOCS1 contains an SH2 domain, a C-terminal SOCS box motif, and an N-terminal variable sequence. Based on its protein structure, SOCS has two main functions: first, SOCS1 regulates cell proliferation and apoptosis as a signal inhibitor; second, SOCS1 regulates cell growth by participating in ubiquitination and mediating the degradation of ubiquitination substrate [28]. With the C-terminal SOCS box motif, SOCS1 can function as a tumor suppressor by reducing oncoproteins via ubiquitination mechanism [29]. SOCS1 directly or indirectly regulates many cancer-related molecules or pathways, including cyclin D1, cyclin-dependent kinase 4 (CDK4), mitogen-activated protein kinase (MAPK)/p38, p53, programmed cell death receptor ligand 1 (PD-L1), STAT3, STAT1, STAT6, etc. [27]. In previous studies, SOCS1 was shown to be regulated by miRNAs. For example, miR-221 represses SOCS1 expression, thereby facilitating prostate cancer cell proliferation and metastasis [30]. SOCS1 is targeted and negatively regulated by miR-155-5p [12]. The present work demonstrated that low expression of SOCS1 was associated with poor prognosis of patients with BC, suggesting a tumor-suppressor role of SOCS1 in BC. Moreover, we validated that the NORAD/miR-155-5p axis regulates the expression of SOCS1. Our findings provide a novel explanation for the dysregulation of SOCS1 in cancers.
In summary, this study revealed that low expression of NORAD indicates poor prognosis for patients with BC. NORAD contributes to BC cell proliferation and high potential of metastasis by modulating the miR-155-5p/SOCS1 axis. However, further studies, including experiments based on animal models, are needed to validate the tumor-suppressor functions of NORAD in BC.
ACKNOWLEDGMENTS
We thank Hubei Yican Health Industry Co., Ltd. (Luminescience) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Conflict of Interest: The authors declare that they have no competing interests.
- Conceptualization: Liu W, Chen A.
- Data curation: Liu W, Chen A.
- Formal analysis: Zhou X.
- Investigation: Zhou X, Li Y, Jiang H.
- Methodology: Zhou X, Li Y, Jiang H.
- Validation: Li Y.
- Visualization: Jiang H.
- Writing - original draft: Liu W, Zhou X, Chen A.
- Writing - review & editing: Liu W, Chen A.
References
- 1.Fahad Ullah M. Breast cancer: current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51–64. doi: 10.1007/978-3-030-20301-6_4. [DOI] [PubMed] [Google Scholar]
- 2.Song JL, Chen C, Yuan JP, Sun SR. Progress in the clinical detection of heterogeneity in breast cancer. Cancer Med. 2016;5:3475–3488. doi: 10.1002/cam4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J, Cheng L, Zhang J, Chen L, Wang D, Guo X, et al. Predictive value of circulating cell-free DNA in the survival of breast cancer patients: a systemic review and meta-analysis. Medicine (Baltimore) 2018;97:e11417. doi: 10.1097/MD.0000000000011417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850–3856. [PubMed] [Google Scholar]
- 5.Lei T, Lv ZY, Fu JF, Wang Z, Fan Z, Wang Y. LncRNA NBAT-1 is down-regulated in lung cancer and influences cell proliferation, apoptosis and cell cycle. Eur Rev Med Pharmacol Sci. 2018;22:1958–1962. doi: 10.26355/eurrev_201804_14721. [DOI] [PubMed] [Google Scholar]
- 6.Zhang YX, Yuan J, Gao ZM, Zhang ZG. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci. 2018;22:443–449. doi: 10.26355/eurrev_201801_14193. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M, Gou L, Xia J, Wan Q, Jiang Y, Sun S, et al. LncRNA ITGB2-AS1 could promote the migration and invasion of breast cancer cells through up-regulating ITGB2. Int J Mol Sci. 2018;19:1866. doi: 10.3390/ijms19071866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Zhao Y, Lin G, Zhou X, Jiang X, Zhao H. Noncoding RNA activated by DNA damage (NORAD): Biologic function and mechanisms in human cancers. Clin Chim Acta. 2019;489:5–9. doi: 10.1016/j.cca.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Zhou K, Ou Q, Wang G, Zhang W, Hao Y, Li W. High long non-coding RNA NORAD expression predicts poor prognosis and promotes breast cancer progression by regulating TGF-β pathway. Cancer Cell Int. 2019;19:63. doi: 10.1186/s12935-019-0781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan BS, Yang MC, Singh S, Chou YC, Chen HY, Wang MY, et al. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene. 2019;38:5612–5626. doi: 10.1038/s41388-019-0812-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Yan T, Huang C, Xu Z, Wang L, Jiang E, et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J Exp Clin Cancer Res. 2018;37:242. doi: 10.1186/s13046-018-0911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W, Guo L, Liang Z, Liu Y, Yao Z. Lnc-SNHG16/miR-128 axis modulates malignant phenotype through WNT/β-catenin pathway in cervical cancer cells. J Cancer. 2020;11:2201–2212. doi: 10.7150/jca.40319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han YJ, Boatman SM, Zhang J, Du XC, Yeh AC, Zheng Y, et al. LncRNA BLAT1 is upregulated in basal-like breast cancer through epigenetic modifications. Sci Rep. 2018;8:15572. doi: 10.1038/s41598-018-33629-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J, et al. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. Int J Cancer. 2018;143:2213–2224. doi: 10.1002/ijc.31605. [DOI] [PubMed] [Google Scholar]
- 16.Shao G, Wang M, Fan X, Zhong L, Wang Z, Zhang P, et al. lncRNA CASC9 positively regulates CHK1 to promote breast cancer cell proliferation and survival through sponging the miR‑195/497 cluster. Int J Oncol. 2019;54:1665–1675. doi: 10.3892/ijo.2019.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuire A, Brown J A L, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34:145–155. doi: 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesci A, Huang X, Taeb S, Jahangiri S, Kim Y, Fokas E, et al. Targeting of CCBE1 by miR-330-3p in human breast cancer promotes metastasis. Br J Cancer. 2017;116:1350–1357. doi: 10.1038/bjc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulin JA, Tommasi S, Elliot D, Hu DG, Lewis BC, Mangoni AA. MiR-193b regulates breast cancer cell migration and vasculogenic mimicry by targeting dimethylarginine dimethylaminohydrolase 1. Sci Rep. 2017;7:13996. doi: 10.1038/s41598-017-14454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N, Cui T, Guo W, Wang D, Mao L. MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther. 2019;12:3181–3196. doi: 10.2147/OTT.S193097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalighfard S, Alizadeh AM, Irani S, Omranipour R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci Rep. 2018;8:17981. doi: 10.1038/s41598-018-36321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Chen CJ, Guo GL. MiR-155 promotes the proliferation and migration of breast cancer cells via targeting SOCS1 and MMP16. Eur Rev Med Pharmacol Sci. 2018;22:7323–7332. doi: 10.26355/eurrev_201811_16269. [DOI] [PubMed] [Google Scholar]
- 23.Gao C, Li H, Zhuang J, Zhang H, Wang K, Yang J, et al. The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. Cancer Manag Res. 2018;11:1–11. doi: 10.2147/CMAR.S182521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y, et al. SNHG5 promotes breast cancer proliferation by sponging the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. 2019;17:138–149. doi: 10.1016/j.omtn.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu HB, Chen Q, Ding SQ. LncRNA LINC01116 competes with miR-145 for the regulation of ESR1 expression in breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1987–1993. doi: 10.26355/eurrev_201804_14726. [DOI] [PubMed] [Google Scholar]
- 26.Luan T, Zhang X, Wang S, Song Y, Zhou S, Lin J, et al. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget. 2017;8:76153–76164. doi: 10.18632/oncotarget.19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma J, Larkin J., 3rd Therapeutic implication of SOCS1 modulation in the treatment of autoimmunity and cancer. Front Pharmacol. 2019;10:324. doi: 10.3389/fphar.2019.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ying J, Qiu X, Lu Y, Zhang M. SOCS1 and its potential clinical role in tumor. Pathol Oncol Res. 2019;25:1295–1301. doi: 10.1007/s12253-019-00612-5. [DOI] [PubMed] [Google Scholar]
- 29.Lessard F, Saint-Germain E, Mignacca L, Ferbeyre G. SOCS1: phosphorylation, dimerization and tumor suppression. Oncoscience. 2019;6:386–389. doi: 10.18632/oncoscience.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao N, Ma G, Zhang J, Zhu W. miR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018;18:14. doi: 10.1186/s12894-018-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]