Summary
The coronavirus disease (Covid‐19) pandemic is the most serious event of the year 2020, causing considerable global morbidity and mortality. The goal of this review is to provide a comprehensive summary of reported associations between inter‐individual immunogenic variants and disease susceptibility or symptoms caused by the coronavirus strains severe acute respiratory syndrome‐associated coronavirus, severe acute respiratory syndrome‐associated coronavirus‐2, and two of the main respiratory viruses, respiratory syncytial virus and influenza virus. The results suggest that the genetic background of the host could affect the levels of proinflammatory and anti‐inflammatory cytokines and might modulate the progression of Covid‐19 in affected patients. Notably, genetic variations in innate immune components such as toll‐like receptors and mannose‐binding lectin 2 play critical roles in the ability of the immune system to recognize coronavirus and initiate an early immune response to clear the virus and prevent the development of severe symptoms. This review provides promising clues related to the potential benefits of using immunotherapy and immune modulation for respiratory infectious disease treatment in a personalized manner.
Keywords: Covid‐19, cytokine storm, genetic susceptibility, influenza virus, immune‐related variants, RSV
Abbreviations
- AHSG
alpha 2‐HS glycoprotein
- C3
complement molecule C3
- C5
complement molecule C5
- CCL2
C‐C motif chemokine ligand 2
- CCL5
C‐C motif chemokine ligand 5
- CCR5
C‐C motif chemokine receptor 5
- CD14
cluster of differentiation 14
- CD209
cluster of differentiation 209
- CD55
complement decay‐accelerating factor
- CLEC4M
C‐type lectin domain family 4 member
- Covid‐19:
coronavirus 2019 disease
- CX3CR1
C‐X3‐C motif chemokine receptor 1
- CXCL9
C‐X‐C motif chemokine receptor 6
- CXCR6
C‐X‐C motif chemokine receptor 6
- DC
dendritic cell
- FCGR2A
Fc fragment of IgG receptor 2A
- FcR
Fc (fragment crystallizable) receptor
- H1N1
influenza A virus subtype H1N1 (A/H1N1)
- H3N2
influenza A virus subtype H3N2 (A/H3N2)
- HIV
human immunodeficiency virus
- HLA
human leukocyte antigen
- ICU
intensive care unit
- IFITM3
interferon induced transmembrane protein 3
- IFN
interferon
- IFNA5
IFN alpha 5
- IFNAR1
IFN alpha and beta receptor subunit 1
- IFNAR2
IFN alpha and beta receptor subunit 2
- IFNG
IFN gamma
- IFNL3
IFN lambda 3
- IL
interleukin
- IL10
interleukin 10
- IL12A
interleukin 12A
- IL12RB
interleukin 12 receptor beta 1 subunit
- IL13
interleukin 13
- IL17A
interleukin 17A
- IL18
interleukin 18
- IL19
interleukin 19
- IL1A
interleukin 1 alpha
- IL1B
interleukin 1 beta
- IL1RL1
interleukin 1 receptor like 1
- IL20
interleukin 20
- IL27
interleukin 27
- IL4
interleukin 4
- IL4R
interleukin 4 receptor
- IL5
interleukin 5
- IL6
interleukin 6
- IL7
interleukin 7
- IL9
interleukin 9
- IRF
IFN regulatory factor
- LD
linkage disequilibrium
- LOF
loss of function
- LRTI
lower respiratory tract infection
- MBL2
mannose‐binding lectin 2
- MX1
MX dynamin like GTPase 1
- NSP1
nonstructural protein 1
- OAS1
2′‐5′‐oligoadenylate synthetase 1
- RSV
respiratory syncytial virus
- SARS
severe acute respiratory syndrome
- SARS‐CoV
severe acute respiratory syndrome‐associated coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome‐associated coronavirus‐2
- SNP
single nucleotide polymorphism
- TLR
toll‐like receptor
- TNF
tumor necrosis factor
- TNFRSF1B
tumor necrosis factor receptor superfamily member 1B
1. INTRODUCTION
The recent global coronavirus disease 2019 (Covid‐19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is the defining global health crisis. Covid‐19 shows a wide range of disease severity among affected individuals from asymptomatic carriers to patients who have severe respiratory failure. While the details of the clinical picture of the Covid‐19 pandemic are being increasingly clarified, there remain considerable unanswered questions regarding the role of host genetic background in the susceptibility and vulnerability to SARS‐CoV‐2 infection. 1
Valuable genetic susceptibility insight into immunity‐related genes has been previously gained from the widespread respiratory viruses such as the respiratory syncytial virus (RSV) and influenza virus. On the other hand, association studies concerning the earlier strain of coronavirus, namely SARS‐CoV, could provide some clues since SARS‐CoV‐2 has 78% genetic identity to SARS‐CoV. Regarding the dysfunction of the immune response to Covid‐19, two scenarios have been proposed: (1) the immune response is insufficient to clear the virus and prevent the development of severe disease resulting from various factors such as deficiencies in virus recognition and misdirection of antigen presentation; (2) an overactive immune system due to genetic predisposition to immune dysregulation may induce an immunopathological condition named ‘cytokine storm’ resulting in collateral damage to neighboring tissue, organ failure, and even death. 2 , 3 , 4 Notably, the immune‐related genetic susceptibility plays considerable roles in both scenarios. However, the dynamic and complex nature of the immune response during acute infection is still not fully understood. In this review, we focus on the host genetic variations involved in both scenarios in the context of respiratory viruses, with particular emphasis on coronavirus.
2. PATHOGEN RECOGNITION, PRESENTATION AND ELIMINATION
The innate immune system establishes the first line of host defense during infection through early recognition of pathogens and subsequently induction of proinflammatory responses. On the other hand, the elimination of pathogens in late infection and generation of immunological memory is arranged by the adaptive immune system.
Several genetic variants in the immune system compartments, which influence either the protein structure or expression, have been uncovered and shown to be implicated in human respiratory tract infections (Table 1).
TABLE 1.
Summary of studies which reported the pathogen recognition‐, presentation‐ and elimination‐related genetic variants behind the susceptibility and vulnerability to coronavirus, respiratory syncytial virus and influenza
| Gene | Variant | Virus | Country | Allele/genotype effect |
|---|---|---|---|---|
| Pathogen recognition, presentation and elimination | ||||
| CD14 | rs2569190 | SARS‐CoV‐1 | China | CC genotype/risk |
| CD209 | rs4804803 | SARS‐CoV‐1 | China | G allele/protective |
| CD55 | rs2564978 | Influenza | China | TT genotype/risk |
| CLEC4M | rs71179137 | SARS‐CoV‐1 | China | Homozygous for indel/protective |
| FCGR2A | rs1801274 | SARS‐CoV‐1 | China | GG genotype/risk |
| HLA | HLA‐B*0703 | SARS‐CoV‐1 | China | Risk |
| HLA‐DRB1*0301 | SARS‐CoV‐1 | China | Risk | |
| HLA‐DRB1*12 | SARS‐CoV‐1 | Vietnam and China | Risk | |
| HLA‐DRB4*01010101 | SARS‐CoV‐1 | China | Risk | |
| HLA‐B*4601 | SARS‐CoV‐1 | Taiwan | Risk | |
| HLA‐Cw*0801 | SARS‐CoV‐1 | Taiwan | Risk | |
| HLA‐B*46:01 | SARS‐CoV‐2 | In silico analysis | Risk | |
| HLA‐B*15:03 | SARS‐CoV‐2 | In silico analysis | Protective | |
| Haplotype HLA‐A*:01:01 ‐B*08:01 ‐C*07:01 ‐DRB1*03:01 | SARS‐CoV‐2 | Italy | Risk | |
| Haplotype HLA‐A*02.01 ‐B*18.01 ‐C*07.01 ‐DRB1*11.04 | SARS‐CoV‐2 | Italy | Protective | |
| IRF3 | p.Glu49del | SARS‐CoV‐2 | Bolivian/Spain | Heterozygote for TCC deletion/risk |
| p.Asn146Lys | SARS‐CoV‐2 | Italy | C allele/risk | |
| IRF7 | p.Arg7fs | SARS‐CoV‐2 | Italy | C allele/risk |
| p.Phe95Ser | SARS‐CoV‐2 | Turkey | G allele/risk | |
| p.Pro246fs | SARS‐CoV‐2 | Spain | Heterozygote for 13‐bp deletion/risk | |
| p.Pro364fs | SARS‐CoV‐2 | Italy | Homozygote for insertion C/risk | |
| p.Arg369Gln | SARS‐CoV‐2 | Italy | T allele/risk | |
| p.Gln185* | SARS‐CoV‐2 | France | A allele/risk | |
| p.Met371Val | SARS‐CoV‐2 | Turkey | CC genotype/risk | |
| MBL2 | rs7096206 | SARS‐CoV‐1 | China | C allele/risk |
| rs1800450 | SARS‐CoV‐1 | China | A allele/risk | |
| TLR3 | p.Ser339fs | SARS‐CoV‐2 | Spain | Heterozygote for deletion T/risk |
| p.Pro554Ser | SARS‐CoV‐2 | Italy | T allele/risk | |
| p.Met870Val | SARS‐CoV‐2 | Colombian/Spain | G allele/risk | |
| p.Trp769* | SARS‐CoV‐2 | Italy | A allele/risk | |
| TLR4 | rs4986791 | Influenza | Greece | C allele; CC genotype/risk |
| TLR7 | c.2129_2132del | SARS‐CoV‐2 | A family in Netherlands | Frameshift deletion/risk |
| c.2383G>T | SARS‐CoV‐2 | A family in Netherlands | T allele/risk | |
2.1. Mannose‐binding lectin 2
Mannose‐binding lection 2 (MBL2) is a soluble, Ca2+‐dependent serum protein that is expressed primarily in the liver and secreted into the blood. MBL2 is a vital component of the innate immune response that provides a front line of host defense. 5 Genetic deficiency of MBL2 gene boosts susceptibility to viral and bacterial agents such as human immunodeficiency virus (HIV), influenza A and Neisseria meningitides. 6 , 7 , 8
2.1.1. Relevance to coronavirus
The differences in the vulnerability of individuals to the coronavirus infections have been proposed to be linked to the key role of MBL2 in early host defense mechanisms, especially before producing specific antibodies. 9 Polymorphisms in the promoter (rs7096206, C/G) and exon 1 (rs1800450, G/A) have a significant association with SARS‐CoV susceptibility. 10 , 11 Moreover, the alleles associated with low expression of MBL2 are increasingly present in patients with other respiratory infectious diseases, such as invasive pneumococcal disease 12 and chronic obstructive pulmonary disease. 13 , 14
2.2. Type C lectin
Dendritic cells (DCs) express pattern recognition receptors, such as CD209 and CLEC4M that directly recognize a wide range of microorganisms, for example, hepatitis C, HIV, SARS‐CoV‐1 coronavirus, and Ebola. Human CD209 and CLEC4M proteins are a C‐type lectin, which comprises a Ca2+‐dependent complementarity‐determining region followed by a flexible neck region at the C‐terminal domain, a transmembrane domain and a cytoplasmic N‐terminal region.
The extended neck region in the extracellular domain is encoded by tandem repeats, which is involved in the recognition of pathogens. 15
2.2.1. Relevance to coronavirus
A genetic‐risk association study revealed that homozygosity of the rs71179137 variant in CLEC4M tandem repeat plays a protective role for SARS‐CoV infection. 16 However, this finding has not been approved by a case‐control study in northern China. 17 The G allele of the rs4804803 polymorphism located on the CD209 promoter is associated with low promoter activity. 18 It has been suggested that the G allele could protect the lung from injury during the progression of SARS‐CoV infection 19 and patients with the AA genotype had a 60% chance for developing severe symptoms. 18 In contrast, another study among 181 SARS patients and 172 controls reported that no genetic predisposition allele in the lectin genes cluster at 19p13.3 is involved in SARS‐CoV‐1 infection. 20
2.3. Toll‐like receptor signaling compartments
Toll‐like receptors (TLRs) are type I transmembrane glycoproteins expressed by various immune cells such as DCs, macrophages, natural killer cells, and neutrophils as well as epithelial and endothelial cells. 21 Moreover, B and T cells also express the restricted types of TLRs (TLR2, TLR7 and TLR9). These receptors play a pivotal role in the innate immune response by recognizing structurally conserved molecules derived from pathogens. 22
2.3.1. Relevance to coronavirus
Early in the process of SARS‐CoV‐2 infection, the ssRNA and dsRNA structure of the virus is, respectively, recognized by TLR7 and TLR3 in the respiratory tract, which results in inducing proinflammatory cytokines and interferons (IFNs) 23 (Figure 1).
FIGURE 1.
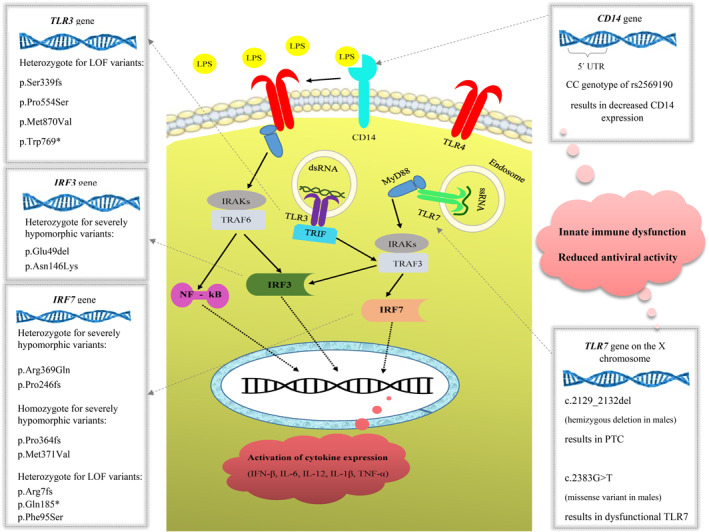
Toll‐like receptor (TLR) signaling and coronavirus susceptibility. dsRNA, double‐stranded RNA; IL, interleukin; IRF, interferon regulatory factor 3; LPS, lipopolysaccharides; NF‐κB, nuclear factor kappa B; PTC, premature termination codon; TNF, tumor necrosis factor; ssRNA, single‐stranded RNA
Sequencing methods applied to young males without any underlying diseases from two independent families who showed the severe form of Covid‐19 have revealed that rare mutations in TLR7 could be associated with severe symptoms and even death. 23 The four‐nucleotide hemizygous deletion (c.2129_2132del) in the TLR7 gene, which is likely to generate a premature termination codon, was detected in two severely affected brothers (29 and 32 years old) and their uninfected mother with the heterozygous state, a condition consistent with X‐linked inheritance. In a second family, a missense variant (c.2383G>T; p.Val795Phe), which was predicted as deleterious in all in silico prediction tools, was identified in two brothers, age 21 and 23 years, with severe Covid‐19 complicated by pulmonary embolisms requiring mechanical ventilation in the intensive care unit (ICU).
Four heterozygote loss of function (LOF) variants in TLR3 (p.Ser339fs, p.Pro554Ser, p.Trp769* and p.Met870Val) in patients with life‐threatening Covid‐19 pneumonia was found 24 and some rare variants in IRF3 and IRF7 genes have been reported (Table 1).
CD14 is a glycosylphosphatidylinositol‐anchored receptor known to serve as a coreceptor for several TLRs both at the cell surface and in the endosomal compartment, which plays a unique role in initiating innate immunity through TLRs‐mediated cytokine response. 25
A possible functional role of the CD14‐c.159T/C polymorphism (rs2569190) in determining the course of SARS development has been reported in Chinese patients. 26 It is suggested that CD14‐c.159CC carriers may have reduced antiviral activity that results in enhanced viral toxicity.
2.3.2. Relevance to RSV
Investigation of TLR4 single nucleotide polymorphisms (SNPs) including rs4986790 and rs4986791 in a group of hospitalized infants with severe RSV infection (case group) and two control groups demonstrated an overrepresentation of both TLR4 SNPs in the case group compared to controls. Therefore, these SNPs were associated with a severe RSV bronchiolitis. 27 Another study on high‐risk infants and young children showed that the same SNPs were strongly associated with symptomatic RSV infection 28 while in other populations, either one or both of these SNPs demonstrated marginal 29 or no association with severe RSV disease. 30 , 31 The rs5743836 SNP in the promoter of TLR9 was weakly associated with severe RSV, 32 and TLR9 rs352162 SNP was associated with the requirement for mechanical ventilation. Furthermore, rs352162 and rs187084 SNPs were associated with ICU admission. 33 As for TLR10, haplotype analysis demonstrated a weak yet significant association with RSV infection even though no single SNP was identified as the major polymorphism contributing to this demonstration. 32
2.3.3. Relevance to influenza virus
Investigation of TLR4 polymorphisms in individuals infected with the influenza virus revealed that these polymorphisms confer protection in the tonsils against Haemophilus influenzae. In addition, there was an association between the rs4986791 SNP and reduced risk of infection with H. influenzae. 34
2.4. Human leukocyte antigen
The human leukocyte antigens (HLA), which are critical components of the viral antigen presentation pathway, consist of the most polymorphic genes (Figures 2 and 3). They have been shown in various reports to confer differential viral susceptibility and severity of disease. The associations between HLA genotypes and severity of infections extend broadly to several unrelated viruses.
FIGURE 2.
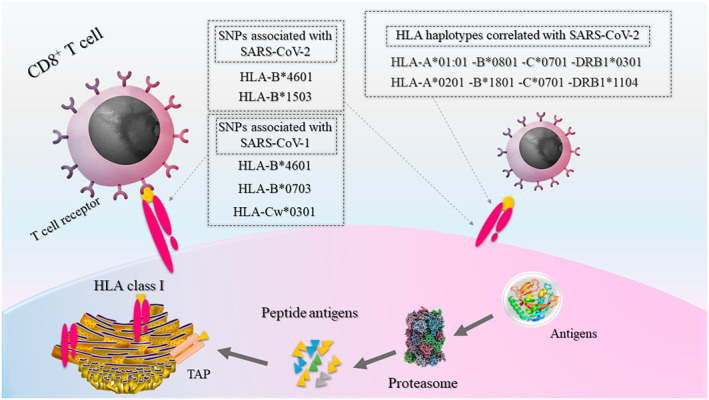
Antigen presentation by HLA class I proteins to CD8+ T cells (cytotoxic T cells). Proteins inside the cytosol of cells are degraded by the proteasome into peptide antigens. These peptides further pass through TAP into the ER, and will then bind to HLA proteins. Finally, antigen‐bound receptors will translocate through the Golgi apparatus (not shown) to the plasma membrane and present the antigens to cytotoxic T cells. Associations between genetic variants in HLA class I genes with SARS‐CoV‐1 and ‐2 are shown in gray boxes. ER, endoplasmic reticulum; HLA, human leukocyte antigen; SARS‐CoV, severe acute respiratory syndrome coronavirus; TAP, transporter associated with antigen processing
FIGURE 3.
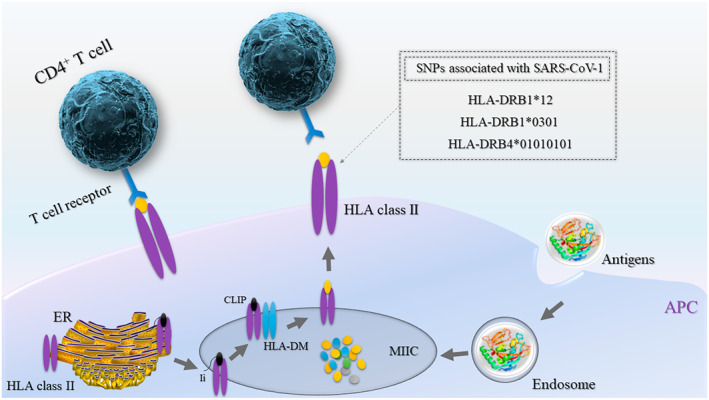
Antigen presentation by HLA class II proteins to CD4+ T cells (helper T cells). Extracellular proteins that enter APCs through phagocytosis are then transported into and degraded in MIIC. HLA class II and Ii proteins produced on ribosomes are translocated into the ER to form heterotrimer units. Upon further maturation of trimers in the Golgi apparatus (not shown) and transportation to MIIC, li is degraded into CLIP, and HLA‐DM induces the replacement of CLIP with peptide antigens within MIIC. Finally, antigen‐bound HLAs translocate to the plasma membrane and present the antigens to CD4+ T cells. Associations between genetic variants in HLA class II genes with SARS‐CoV‐1 are shown in the gray box. APC, antigen‐presenting cell; CLIP, class II‐associated invariant chain peptide; ER, endoplasmic reticulum; HLA, human leukocyte antigen; li, invariant chain; MIIC, MHC class II compartment
2.4.1. Relevance to coronavirus
In studies of the HLA‐A, HLA‐B, HLA‐DRA and HLA‐DQA1 alleles among Chinese patients, two HLA‐B*0703 and HLA‐DRB1*0301 alleles showed a significant association with SARS‐CoV infection. 35 In the Vietnamese population, a case‐control study comprising SARS‐CoV infected patients, healthy individuals without contact history and staff members who had contact with SARS‐CoV infected patients, but had not developed the infection, revealed that HLA‐DRB1*12 showed a significant positive association with developing SARS‐CoV. 36 The tendency of patients with the HLA‐DRB1*12 allele to develop infection has been supported in a large study from southern China. 37 The HLA‐DRB4*01010101 allele is another variant in the Chinese population that was found to be significantly associated with genetic susceptibility to infection by SARS‐CoV. 38 Furthermore, in Taiwanese association studies, the HLA‐B*4601 and HLA‐Cw*0801 alleles were also found to associate with susceptibility to SARS‐CoV infection. 39 , 40 In studies on SARS‐CoV‐2, a comprehensive in silico analysis of viral peptide‐MHC class I binding affinity across 145 HLA‐A, HLA‐B and HLA‐C genotypes for all SARS‐CoV‐2 peptides has revealed that the HLA‐B*46:01 allele has the fewest predicted binding peptides for SARS‐CoV‐2. Conversely, the HLA‐B*15:03 allele shows the greatest capacity to present highly conserved SARS‐CoV‐2 peptides, suggesting that it could play a protective role through activating T‐cell‐based immunity. 41
Another study has uncovered that the two most frequent HLA haplotypes in the Italian population, HLA‐A*:01:01, HLA‐B*08:01, HLA‐C*07:01, HLA‐DRB1*03:01 and HLA‐A*02.01, HLA‐B*18.01, HLA‐C*07.01, HLA‐DRB1*11.04, show a positive (suggestive of susceptibility factor) and negative (suggestive of protective factor), respectively, significant correlation with both Covid‐19 incidence and severity. 42
2.5. Fc fragment of immunoglobulin G receptors
2.5.1. Relevance to coronavirus
Receptors for the constant region of antibodies (FcR) play a crucial role in the regulation of immunity and initiation of local inflammation. The human FCGR2A is composes a crucial link between the humoral branch and the effector cells of the immune system. A functional polymorphism (rs1801274, c.500A>G) in the FCGR2A gene, a subclass of immunoglobulin G receptors, shows clinical implications in infectious diseases, either at the level of disease susceptibility or at the level of disease severity. 43
Genotyping of this SNP in a Chinese study in patients with a moderate course of SARS infection, patients with severe symptoms (ICU admission), patients who died from SARS‐CoV infection, and healthy controls demonstrated that ICU patients and deceased subgroups had a significant increase of the GG genotype compared to controls (23% vs. 9%). 44 It is suggested that the GG genotype could be a risk factor for developing a more severe course of SARS‐CoV‐1 infection, while the AA genotype might show a protective role in the patient outcomes.
2.6. Complement decay‐accelerating factor
2.6.1. Relevance to influenza virus
Complement decay‐accelerating factor (CD55) is a prominent complement‐regulatory protein that impedes C5 and C3 convertase activation and thus prevents the activation of C5 and C3 proteins. The rs2564978 SNP in the promoter of CD55 is associated with severe influenza A(H1N1)pdm09; the rs2564978 TT genotype is associated with remarkably reduced transcriptional activity of the promoter compared to the CC genotype. 45
3. INFLAMMATION: A DOUBLE‐EDGED SWORD
Inflammation has vital roles in orchestrating innate and adaptive immune responses to pathogens and tissue injury. The production of a variety of proinflammatory cytokines and chemokines induces a counter regulatory anti‐inflammatory response to avoid excessive injury. However, sometimes such counter‐regulation fails and results in excessive infiltration of inflammatory cells into infected tissues. 2 , 46
3.1. Cytokines
Cytokines are a group of small proteins, including IFNs, interleukins (ILs), chemokines, colony‐stimulating factors and tumor necrosis factor‐alpha (TNF‐α) that are released by specific cells to regulate intercellular signaling (Figure 4). In this section, the elements of inflammation in coronavirus, RSV and influenza infection will be discussed (Table 2).
FIGURE 4.
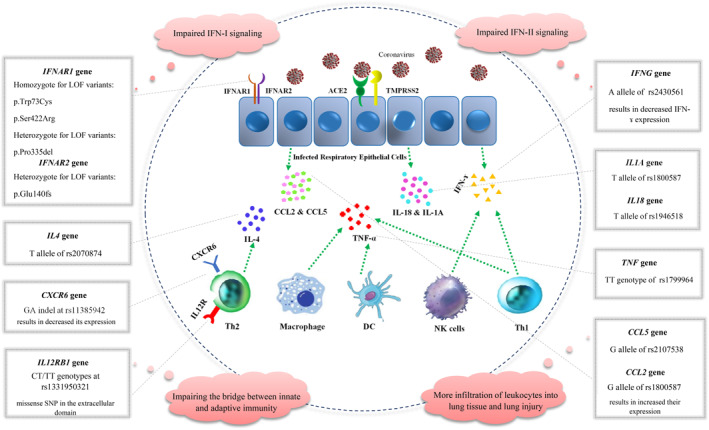
Variants in inflammation‐related genes underlying the susceptibility to coronavirus. CCL, C‐C motif chemokine ligand; DC, dendritic cell; IFN, interferon; IL, interleukin; NK, natural killer cell; Th, T‐helper cell
TABLE 2.
Summary of studies which reported the inflammation‐related genetic variants behind the susceptibility and vulnerability to coronavirus, respiratory syncytial virus and influenza
| Gene | Variant | Virus | Country | Allele/genotype effect |
|---|---|---|---|---|
| Inflammation | ||||
| AHSG | rs2248690 | SARS‐CoV‐1 | China | AA genotype/protective |
| CCL2 | rs1024611 | SARS‐CoV‐1 | China | G allele/risk |
| CCL5 | rs2107538 | SARS‐CoV‐1 | China | G allele/risk |
| rs2107538 | RSV | Brazil | CT genotype/risk | |
| CCR5 | rs1799987 | RSV | UK | G allele/risk |
| rs2734648 | RSV | UK | T allele/risk | |
| CX3CR1 | rs3732378 | RSV | Greece | TT and TC genotypes/risk |
| CXCR6 | rs11385942 | SARS‐CoV‐2 | Spain and Italy | Indel of GA/risk |
| IFITM3 | rs12252 | Influenza | China | CC genotype/risk |
| rs34481144 | Influenza | African American | A allele/risk | |
| IFNA5 | rs10757212 | RSV | Netherlands | T allele/risk |
| IFNAR1 | p.Trp73Cys | SARS‐CoV‐2 | Turkey | CC genotype/risk |
| p.Ser422Arg | SARS‐CoV‐2 | Pakistan | CC genotype/risk | |
| p.Pro335del | SARS‐CoV‐2 | China | Heterozygote for TTC deletion/risk | |
| IFNAR2 | p.Glu140fs | SARS‐CoV‐2 | Belgium | Heterozygote for 13‐bp deletion/risk |
| IFNG | rs2430561 | SARS‐CoV‐1 | China | A allele/risk |
| rs3138557 | RSV | China | CA12‐/CA12‐ genotype/risk | |
| IFNL3 | rs8099917 | Influenza | Iran | TT genotype/risk |
| IL10 | rs1800872 | Influenza | Iran | GG and GT genotypes/risk |
| rs1800872 | Influenza | Mexico | C allele/risk | |
| rs1800870 | Influenza | Mexico | A allele/risk | |
| IL12RB1 | rs1331950321 | SARS‐CoV‐1 | China | T allele/risk |
| IL17A | rs2275913 | Influenza | Iran | GG and AG genotypes/risk |
| IL18 | rs1946518 | SARS‐CoV‐1 | Taiwan | T allele/risk |
| rs360721 | RSV | Germany | G allele/risk | |
| IL1A | rs17561 | Influenza | China | T allele/risk |
| rs1800587 | SARS‐CoV‐1 | Taiwan | T allele/risk | |
| IL1B | rs16944 | Influenza | Iran | AA genotype/protection |
| rs1143627 | Influenza | China | C allele/risk | |
| IL1RL1 | rs1921622 | RSV | Netherlands | G allele/risk |
| IL4 | rs2070874 | Respiratory infections (RSV, influenza, SARS‐CoV‐1, and pneumonia) | Meta‐analysis of different populations | T allele/Risk |
| rs2243250 | RSV | Korea | T allele/risk | |
| rs2243289 | RSV | Korea | G allele/risk | |
| c.8412A | RSV | Korea | A allele/risk | |
| IL4R | rs1801275 | RSV | Netherlands | G allele/risk |
| IL9 | rs1799962 & rs2069885 | RSV | Netherlands | TT haplotype/risk in girls |
| MX1 | rs17000900 | SARS‐CoV‐1 | China | A allele/protective |
| rs2071430 | SARS‐CoV‐1 | China | G allele/risk | |
| OAS1 | rs3741981 | SARS‐CoV‐1 | Vietnam | G allele/risk |
| rs2660 | SARS‐CoV‐1 | China | G allele/protective | |
| TNF | rs1799964 | SARS‐CoV‐1 | China | CT genotype/protective |
| rs1800629 | Influenza | Mexico | G allele/risk | |
3.1.1. IFNs
IFNs are a subgroup of cytokines that have a fundamental role in the immune response against both viral and microbial pathogens. 47 Three major types of IFNs (IFN‐I, IFN ‐II and IFN ‐III) are characterized based on their receptor specificity.
3.1.1.1. Relevance to coronavirus
IFN gamma (IFNG) is the only member of IFN‐II, which is located on chromosome 12. The ability of IFNG to interfere with viral infections through inducing the production of free radicals and other proinflammatory cytokines have made it an intriguing factor to study in determining genetic variants involving both susceptibility and vulnerability to viral infections.
The immunopathological events related to the IFN‐mediated pathway have a critical role in the immune response against SARS‐CoV infection. 48 In China, a case‐control study genotyped the IFNG +874 A/T polymorphism (rs2430561) in 476 SARS patients and 449 healthy controls; the IFNG c.874A allele was significantly overrepresented in patients compared to the controls. Particularly, the IFNG c.874A allele shows a significant association with susceptibility to SARS‐CoV infection. 49 This polymorphism has been previously reported to be associated with other infectious diseases such as parvovirus, tuberculosis, and hepatitis B virus infection. 50 , 51 The T allele of IFNG c.874A>T variant prepares a site for binding transcription factor nuclear factor kappa B (NF‐κB). 52 It is suggested that the c.874A allele results in the downexpression of IFNG, which could impair antiviral responses.
In studies related to Covid‐19, a recent investigation uncovered two LOF homozygote variants (p.Trp73Cys, p.Ser422Arg) and one LOF heterozygote variant (p.Pro335del) in the IFNAR1 gene, as well as one LOF heterozygote variant (p.Glu140fs) in the IFNAR2 gene in patients with life‐threatening Covid‐19. 24 IFNAR1 and IFNAR2 subunits comprise the IFN receptor that binds to the type I IFNs including IFN‐α and IFN‐β.
3.1.1.2. Relevance to RSV
Members of the IFN type I family are involved in RSV‐related infection, as the rs10757212 SNP in IFNA5 is associated with RSV‐related bronchiolitis. 53 Moreover, IFNA13 rs643070 and IFNAR2 rs7279064 were associated with RSV‐related bronchiolitis in term‐born children and IFNG rs1861493 was associated with RSV‐related bronchiolitis in premature birth. 54
Another type of genetic variation associated with RSV‐related bronchiolitis severity and susceptibility is attributed to the polymorphism in the CA microsatellite (rs3138557) in the first intron of IFNG. The number of repeats varies from 11 to 17, and the highest level of IFNG is produced when the number of repeats equals 12 (CA12). Children with the CA12+/CA12+ or CA12+/CA12− genotypes demonstrated a significantly lower RSV bronchiolitis respiratory score compared to those carrying the CA12−/CA12− genotype. Furthermore, the CA12 repeat frequency was lower in RSV‐infected children compared to controls; patients in whom CA repeats did not reach 12 had high respiratory scores and severe disease. 55
3.1.1.3. Relevance to influenza virus
Several lines of evidence indicate that IFN‐III contributes greatly in defense against pathogens that infect respiratory tracts such as influenza A and B virus, which predominantly affect the elderly population. Association of IFNL3 SNP rs8099917 with influenza virus is controversial as this SNP was associated with influenza virus infection in one Iranian study 56 while no association was observed in another report. 57
3.1.2. ILs
ILs are regulatory cytokines involved in the activation and differentiation of immune cells. They could show both pro‐ and anti‐inflammatory functions.
3.1.2.1. Relevance to coronavirus
IL18 and IL1A are constitutively expressed in the lungs. 58 Investigation of 281 SNPs in the Taiwanese patients with SARS‐CoV‐1 infection revealed that detectable nasopharyngeal shedding of the virus is associated with the T allele of rs1946518 in the promoter of the IL18 gene and the T allele of rs1800587 in the IL1A gene. 59 It is postulated that these polymorphisms could affect IL gene expression, thereby participating in the clearance of viral infection.
IL12A is another inflammatory cytokine that is naturally produced by macrophages, neutrophils and DCs. A study on cases with SARS‐CoV infection and two control groups including healthy individuals without contact history with SARS‐CoV‐infected patients and 141 healthy individuals with close contacts discovered a significant association between susceptibility to SARS‐CoV infection and c.1664 CT/TT genotypes (rs1331950321) in the IL12RB1 gene. 60 This variant is a missense SNP (p.P534L) in the extracellular coding sequence of IL12RB1 protein.
IL6 is transiently produced in response to infectious agents. Two variants—c.‐572C/G (rs1800797) and c.‐174G/C (rs1800795)—have been confirmed to affect both the transcription and secretion levels of IL6. 61 Interestingly, the c.‐174 G/C variant of the IL6 gene is significantly associated with the severity of pneumonia. 62 It was demonstrated that carriers of the C allele show a higher IL6 expression and 2.42‐fold higher risk for septic shock. 63 , 64 Moreover, susceptibility to acute lung injury is associated with an IL6 gene haplotype that spans −1363 to +4835 from the transcription start site. 65 , 66 It is suggested that IL6 polymorphisms are involved in the progression of coronavirus infection, and suppression of the IL6 signaling cascade is suggested as a promising therapeutic strategy against severe SARS‐CoV‐2 infection. 62
IL4 plays a fundamental role in shaping the immune reaction to pathogens through inducing both T‐cell and B‐cell differentiation and its dysregulation could interfere with the balance between Th1 and Th2 responses. 67 A meta‐analysis of studies on respiratory infectious diseases including RSV, influenza virus, SARS‐CoV and pneumonia has revealed that the T allele of rs2070874 polymorphism from the IL4 gene is significantly associated with the risk of respiratory infections. 68
3.1.2.2. Relevance to RSV
Some SNPs in IL genes are believed to be involved in bronchiolitis caused by RSV. In this regard, the IL1RN SNPs rs315952 and IL27 rs181206 are associated with RSV‐related bronchiolitis in premature birth, 54 and the IL1RL1 SNP rs1921622 is associated with RSV bronchiolitis severity. 69
Investigation of IL4, IL5 and IL13 genes in Korean children revealed various SNPs in various regions of these genes, among which the SNPs c.589T (rs2243250), c.‐33T (rs2070874), c.8375G (rs2243289) and c.8412A form a haplotype and each was significantly associated with severe RSV infection. However, after statistical correction, the results related to each SNP were marginal. 70
It is noteworthy that association studies demonstrated different results in different populations; that is, a specific SNP might be significantly associated with a specific disease in one population, but not in another. This is the case for the IL4 promoter variant rs2243250, which was reported as an RSV‐associated variant in Korea 70 while there was no such association observed in Canadian 71 or German cohorts. 72 A similar story holds true for the IL4R SNP rs1801275 that was associated with severe RSV in the study of Hoebee et al., 73 while there was no significant association reported by Marr et al. 71 Similarly, IL13 SNP 1112C/T (rs1800925) was associated with severe RSV in the study of Puthothu et al. 72 in contrary to the report of Choi et al. 70 The two IL13 SNPs rs20541 and rs1881457 were investigated for possible association with severe RSV. However, there was no significant association observed. 72
Other SNPs in ILs that were associated with RSV infection include the IL18 SNPs 133G/C (rs360721 74 ), and IL6 SNP ‐174G/C rs1800795, for which the GG and GC genotypes were associated with shorter hospital stay. 75 The IL10 SNP rs1800872 C allele was also associated with a higher risk of RSV‐related wheeze reported by parents at the corrected age of 1. 76 Association of genetic diversity in other IL including IL19 rs2243191 and rs2243188, IL20 rs2981573, IL4R rs1805015 and IL7 rs2583762 SNPs, with post‐lower respiratory tract infection (LRTI) recurrent wheeze caused by RSV has also been reported. 77
It is noteworthy that RSV has a gender‐specific pattern of infection, as it is more predominant in boys. Investigation of possible genetic factors underlying gender specificity revealed that IL9 SNP rs2069885 confers disparate effects in boys and girls. The major allele of this SNP in boys was associated with higher susceptibility to severe forms of the disease, but reduced susceptibility in girls. On the other hand, haplotype analysis demonstrated that the TT haplotype was associated with the highest risk of bronchiolitis requiring hospitalization in girls. This haplotype includes the IL9 SNPs rs1799962 and rs2069885. 78
3.1.2.3. Relevance to influenza virus
Variations in IL1A and IL1B genes are associated with susceptibility to pandemic A/H1N1 influenza virus. Allele T in IL1A rs17561 and allele C in IL1B rs1143627 are possibly associated with a higher risk of A(H1N1)pdm09 infection. 79
In the Iranian population, the AA genotype of IL1B rs16944 is associated with higher protection against influenza B infection. 57 However, another study stated that the decreased risk of infection is associated with the GG genotype of IL1B rs16944. 56 For IL17A SNP rs2275913, the lack of allele A is associated with a higher predisposition to Influenza virus. 57
The rs1554286 SNP in IL10 is significantly associated with the epiglottitis caused by invasive H. influenzae serotype B infection in immunized children. This SNP is in strong LD with two other polymorphisms within the promoter and the recessive genotype of this SNP is associated with epiglottitis. 80
The IL10 −1082A (rs1800870) and −592C (rs1800872) alleles were overrepresented in influenza patients and associated with a higher risk of being affected with severe infection. In the case of inflammatory conditions, the IL10 rs1800870 A allele is associated with lower levels of cytokine production, whereas the IL10 rs1800870 G allele leads to higher levels of cytokine production. Individuals with the IL10 rs1800870 AA genotype are more susceptible to severe influenza virus infection. 81 Moreover, the GG and TG genotypes of the IL10 rs1800872 SNP were associated with severe influenza A/H3N2 disease in the Iranian population 56 while in another study, no association was observed between this IL10 SNP and severe influenza disease. 57
3.1.3. Chemokines
Chemokines are small proteins that belong to the category of cell signaling molecules and induce chemotaxis in nearby cells. 82 Concerning their central roles in inflammatory responses, many chemokines and chemokine receptors have been introduced as potential therapeutic targets for several inflammatory diseases. 83
3.1.3.1. Relevance to coronavirus
C‐C motif chemokine ligand 5 (CCL5), also known as RANTES, has a fundamental role in the recruitment and migration of T cells toward inflammation sites during acute infections.
Law et al. 84 reported that nonstructural protein 1 (NSP1) of SARS coronavirus can strongly induce CCL5 expression in human lung epithelial cells. Regarding the SARS‐CoV epidemic in China, it was found that the G allele of CCL5 c.‐28C/G polymorphism (rs2280788) was significantly associated with more severe symptoms and admission to ICU or deaths. 85 This SNP could be involved in regulating CCL5 expression by modifying the binding site of the NF‐κB transcription factor. 86
CCL2 is involved in the chemoattraction of monocytes and polarization of T‐helper cells. CCL2 is one of the upregulated chemokines in monocytes and lung epithelial cells during the early stage of SARS‐CoV infection. 87 It has been revealed that the higher plasma levels of CCL2 in patients with SARS‐CoV‐1 infection is significantly correlated with severe symptoms. 88 A functional SNP (rs1024611G/A) is located in CCL2 promoter that affects the expression level. The G allele triggers higher expression of CCL2 and is associated with more infiltration of leukocytes into tissues compared to the A allele. 89 It has been suggested that the rs1024611 variant could be involved in interindividual differences of host vulnerability to SARS‐CoV infection. 90
A genome‐wide association study on patients with severe Covid‐19 (defined as respiratory failure) and control participants from Italy and Spain has been recently conducted. 91 This investigation has uncovered two association signals on chromosome 3p21.31 and 9q34.2 which cover a cluster of six genes (SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6 and XCR1) and the ABO blood group, respectively. Accordingly, the rs11385942 insertion–deletion GA or G variant at 3p21.31 locus is associated with Covid‐19‐induced respiratory failure with genome‐wide significance. Interestingly, the locus contains genes encoding chemokine receptors, including CCR9 and CXCR6. The risk allele GA is associated with lower expression of CXCR6 and higher expression of SLC6A20, and LZTFL1 in human lung cells. CXCR6 regulates the location of lung‐resident memory CD8+ T cells during the immune reaction to airway pathogens. It should be noted that further studies will be needed to reveal the functional consequences of detected associations.
3.1.3.2. Relevance to RSV
The CCL5 SNP rs2107538 is associated with RSV and RSV subtype A‐associated bronchiolitis. 33 Polymorphisms located within the promoter (−408G/A rs2107538; −28G/C rs2280788) and the first intron (In1.1T/C rs2280789) of CCL5 are in strong LD and it was demonstrated that a common combined genotype of these polymorphisms is associated with severe RSV infection. 92
In the category of chemokines, the CX3CR1 SNP rs3732378 (p.T280M; c.935C/T) variant was associated with RSV‐related bronchiolitis, which is due to an interruption in the affinity of CX3CR1 to its ligand fractalkine. Carriers of the 280M allele were more prevalent among patients and individuals with genotypes containing this allele were more likely to be hospitalized due to RSV‐related bronchiolitis. 93
CCR5 is the receptor of CCL5 and CCL3 that recruit various immune cells such as monocytes, T cells and basophils, and so on. Regarding RSV infection, the −2459G (rs1799987) and −2554T (rs2734648) variants of CCR5 are associated with severe RSV‐related bronchiolitis. 94 The effect of rs2734648 is not functionally clear, and the impact of rs1799987 is debating as it leads to both increased and decreased expression of CCR5. 75
Chemokine (C‐X‐C motif) ligand 9 (CXCL9)/induced by gamma IFN is a small cytokine of CXC chemokine and participates in several cellular and immune‐related processes. 95 There is a significant association between CXCL9 rs2276886 and post‐RSV LRTI recurrent wheeze. 77
3.1.4. TNF
TNF is vital for the innate immune response against pathogens and is a key mediator of inflammatory response. Dysregulation of this cytokine could lead to tissue injuries through the inflammation cascade and therefore, its gene regulation is a considerable factor in the pathobiology of inflammation. 96
3.1.4.1. Relevance to coronavirus
A case‐control study in China including 75 SARS patients, 41 healthcare workers, and 92 healthy controls revealed that the CT genotype compared to the TT genotype at the c.‐204 locus of TNF gene (rs1799964) could be a protective factor for SARS‐CoV infection. 97
3.1.4.2. Relevance to RSV
TNF receptor superfamily member 1B (TNFRSF1B) is a TNF‐binding receptor that mediates the effects of TNF in autoimmunity, inflammation and tumorigenesis. 98 In patients affected with RSV, TNFRSF1B rs1061622 was associated with post‐RSV LRTI recurrent wheeze. 77
3.1.4.3. Relevance to influenza virus
The TNF SNP c.‐308G rs1800629 was overrepresented in patients affected with influenza and was associated with disease severity. 81
3.2. Other inflammation‐related genes
3.2.1. IFN‐inducible genes: Inhibitors of viral replication or viral entry
3.2.1.1. Relevance to coronavirus
IFN‐induced GTP‐binding protein Mx1 (MX1) is an antiviral protein that prevents viral replication. A large case‐control study in China, the A allele of the MX1 c.‐123C/A variant (rs17000900) is significantly associated with a lower risk of SARS coronavirus infection. 99 Moreover, the G‐allele of c.‐88G/T polymorphism (rs2071430) was found more frequently in the hypoxemic group compared to the nonhypoxemic group of SARS‐CoV infected patients. 100 Interestingly, the G allele induces lower promoter activity rather than the T allele. 101
Another IFN‐inducible gene is the OAS1, which is a member of the 2′‐5′ oligoadenylate synthetase family. A study on Vietnamese SARS‐CoV‐1 patients and controls revealed that the G allele of nonsynonymous A/G SNP in exon 3 of the OAS1 gene (rs3741981) affects disease susceptibility and progression of SARS. 100 In China, genotyping of SARS‐CoV cases and close‐contact uninfected controls for the OAS1 gene indicated that the G allele of rs2660 polymorphism in the 3′‐untranslated region is associated with a protective effect on SARS infection. 102
3.2.1.2. Relevance to influenza virus
IFN induced transmembrane proteins (IFITMs) are located within the endolysosomal and plasma membranes to disrupt membrane fusion between the host cell and viruses, and confront viral entry. SNP rs12252 and rs34481144 in the promoter of IFITM3 are associated with severe influenza virus including. 103 Although some studies declared an association between rs12252C as a risk allele with severe influenza A virus, other studies suggest that homozygosity for this allele might predispose individuals to a mild form of influenza but not the severe form, 104 or suggest that this allele was not associated with pediatric influenza virus infection. 105 This SNP was not associated with seasonal influenza‐related hospitalization. 106
On the other hand, allele C of the IFITM3 SNP rs12252 leads to a change of splice site sequence and was significantly overrepresented in hospitalized patients. Moreover, the minor IFITM3 CC genotype was linked to decreased restriction of influenza virus in vitro. In conclusion, IFITM3 is an important factor in morbidity and mortality of influenza virus infection. 107
4. PRIMARY IMMUNODEFICIENCY AND COVID‐19
Primary immunodeficiency (PID) patients are characterized by an impaired immune function that could result in susceptibility to severe infections. It is suggested that the frequency of severe Covid‐19 is significantly higher in these patients. However, an opposite possibility is that PID patients are protected from Covid‐19 due to suppression of cytokine storm, which is caused by a deficient inflammatory response in these patients. 108 Therefore, regarding the two mentioned scenarios in the introduction, PID could affect individuals' susceptibility to Covid‐19. Similar to general populations, PID patients with Covid‐19 demonstrate variable disease severity and outcome depending on the type of immune deficiency. 109 , 110 , 111 , 112 Patients with a mutation in antiviral immunity‐related genes are severely affected with Covid‐19. Studies have reported that mutations in STK4, RAB27A, IL1RN, IFNAR2, IRF7 and TLRs genes were associated with Covid‐19 severity in PID patients. 24 , 113 , 114 , 115 , 116
In contrast, patients with inflammatory pathway dysfunction show milder Covid‐19 manifestations. 108 A recent investigation on two agammaglobulinemia and five common variable immune deficiency (CVID) patients revealed that the agammaglobulinemia patients showed mild Covid‐19 symptoms. In contrast, CVID patients manifested a severe form of Covid‐19 and required ICU admission. 117 It is suggested that the absence of B cells in agammaglobulinemia patients results in suppressed development of inflammatory cascade and cytokine storm. Similarly, two agammaglobulinemia patients with Bruton's tyrosine kinase (BTK) germline mutation (S578Y) recovered from Covid‐19 without intensive care. 118
Collectively, a recent systematic review and meta‐analysis reported that PID patients have a 1.55‐fold higher risk of manifesting severe Covid‐19, but this observation is statistically nonsignificant. 119
5. CONCLUSION
The attention given to the roles of interindividual immunogenic variants in infectious diseases has become more remarkable over the past decade. The results of host immunogenetic‐focused studies suggest that the genetic background of the host could affect the levels of proinflammatory and anti‐inflammatory cytokines and might modulate the progression of Covid‐19 in affected patients. This review provides promising clues related to the potential benefits of using immunotherapy and immune modulation for respiratory infectious disease treatment in a personalized manner. Moreover, these results provide insights for risk assessment and subsequent preventive interventions. Employment of high throughput sequencing and powerful bioinformatics methods, now broadly available, could further help verify the involved genetic variants in different ethnic groups and are expected to continuously contribute to the genetic basis of Covid‐19 susceptibility.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
AUTHOR CONTRIBUTIONS
Farzaneh Darbeheshti and Mojdeh Mahdiannasser equally carried out the literature review, visualization, and writing‐original draft. Bruce D. Uhal, Shuji Ogino and Sudhir Gupta critically revised the work. Nima Rezaei contributed to the conceptualization, project administration, investigation and validation. All authors contributed to the article and approved the submitted version.
ACKNOWLEDGEMENT
The authors gratefully acknowledge Paul Griffiths, Professor of Virology, University College London, for reviewing, suggestions and editing.
Darbeheshti F, Mahdiannasser M, Uhal BD, Ogino S, Gupta S, Rezaei N. Interindividual immunogenic variants: susceptibility to coronavirus, respiratory syncytial virus and influenza virus. Rev Med Virol. 2021;31(6):e2234. 10.1002/rmv.2234
Farzaneh Darbeheshti and Mojdeh Mahdiannasser contributed equally to this study as first co‐authors.
REFERENCES
- 1. Darbeheshti F, Rezaei N. Genetic predisposition models to COVID‐19 infection. Med Hypotheses. 2020;142:109818. 10.1016/j.mehy.2020.109818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buonaguro FM, Ascierto PA, Morse GD, et al. Covid‐19: time for a paradigm change. Rev Med Virol. 2020;30:e2134. 10.1002/rmv.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng Y, Ling Y, Bai T, et al. COVID‐19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380‐1388. 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ezekowitz RA. Role of the mannose‐binding lectin in innate immunity. J Infect Dis. 2003;187(suppl 2):S335‐S339. 10.1086/374746. [DOI] [PubMed] [Google Scholar]
- 6. Garred P, Madsen HO, Balslev U, et al. Susceptibility to HIV infection and progression of AIDS in relation to variant alleles of mannose‐binding lectin. Lancet. 1997;349:236‐240. 10.1016/S0140-6736(96)08440-1. [DOI] [PubMed] [Google Scholar]
- 7. Hartshorn KL, Sastry K, White MR, et al. Human mannose‐binding protein functions as an opsonin for influenza A viruses. J Clin Invest. 1993;91:1414‐1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hibberd ML, Sumiya M, Summerfield JA, Booy R, Levin M. Association of variants of the gene for mannose‐binding lectin with susceptibility to meningococcal disease. Meningococcal Research Group. Lancet. 1999;353:1049‐1053. 10.1016/s0140-6736(98)08350-0. [DOI] [PubMed] [Google Scholar]
- 9. Peiris J, Lai S, Poon L, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319‐1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ip WK, Chan KH, Law HK, et al. Mannose‐binding lectin in severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2005;191:1697‐1704. 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Zhou G, Zhi L, et al. Association between mannose‐binding lectin gene polymorphisms and susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2005;192:1355‐1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy S, Knox K, Segal S, et al. MBL genotype and risk of invasive pneumococcal disease: a case‐control study. Lancet. 2002;359:1569‐1573. 10.1016/S0140-6736(02)08516-1. [DOI] [PubMed] [Google Scholar]
- 13. Yang I, Seeney S, Wolter J, et al. Mannose‐binding lectin gene polymorphism predicts hospital admissions for COPD infections. Gene Immun. 2003;4:269‐274. [DOI] [PubMed] [Google Scholar]
- 14. Engler R, Klote M, Spooner C, Reardon G, Halsey J, Nelson M. Mannan‐binding lectin (MBL) serum levels in a healthy adult population: gender differences in prevalence of MBL deficiency. J Allergy Clin Immunol. 2010;125:AB10. [Google Scholar]
- 15. Barreiro LB, Patin E, Neyrolles O, Cann HM, Gicquel B, Quintana‐Murci L. The heritage of pathogen pressures and ancient demography in the human innate‐immunity CD209/CD209L region. Am J Hum Genet. 2005;77:869‐886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan VS, Chan KY, Chen Y, et al. Homozygous L‐SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat Genet. 2006;38:38‐46. 10.1038/ng1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhi L, Zhou G, Zhang H, et al. Lack of support for an association between CLEC4M homozygosity and protection against SARS coronavirus infection. Nat Genet. 2007;39:692‐693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan KYK, Xu M‐S, Ching JCY, et al. CD209 (DC‐SIGN) −336A>G promoter polymorphism and severe acute respiratory syndrome in Hong Kong Chinese. Hum Immunol. 2010;71:702‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan K, Xu M, Ching J, et al. Association of a single nucleotide polymorphism in the CD209 (DC‐SIGN) promoter with SARS severity. Hong Kong Med J. 2010;16:37. [PubMed] [Google Scholar]
- 20. Li H, Tang NL, Chan PK, et al. Polymorphisms in the C‐type lectin genes cluster in chromosome 19 and predisposition to severe acute respiratory syndrome coronavirus (SARS‐CoV) infection. J Med Genet. 2008;45:752‐758. 10.1136/jmg.2008.058966. [DOI] [PubMed] [Google Scholar]
- 21. Kawai T, Akira S. Toll‐like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637‐650. [DOI] [PubMed] [Google Scholar]
- 22. Heil F, Hemmi H, Hochrein H, et al. Species‐specific recognition of single‐stranded RNA via toll‐like receptor 7 and 8. Science. 2004;303:1526‐1529. 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 23. Park A, Iwasaki A. Type I and type III interferons–induction, signaling, evasion, and application to combat COVID‐19. Cell Host Microbe. 2020;27(6):870‐878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science. 2020;370:eabd4570. 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Z, Zhang Z, Lei Z, Lei P. CD14: biology and role in the pathogenesis of disease. Cytokine Growth Factor Rev. 2019;48:24‐31. [DOI] [PubMed] [Google Scholar]
- 26. Yuan FF, Boehm I, Chan PK, et al. High prevalence of the CD14‐159CC genotype in patients infected with severe acute respiratory syndrome‐associated coronavirus. Clin Vaccine Immunol. 2007;14:1644‐1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tal G, Mandelberg A, Dalal I, et al. Association between common toll‐like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis. 2004;189:2057‐2063. 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- 28. Awomoyi AA, Rallabhandi P, Pollin TI, et al. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high‐risk infants and young children. J Immunol. 2007;179:3171‐3177. 10.4049/jimmunol.179.5.3171. [DOI] [PubMed] [Google Scholar]
- 29. Puthothu B, Forster J, Heinzmann A, Krueger M. TLR‐4 and CD14 polymorphisms in respiratory syncytial virus associated disease. Dis Markers. 2006;22:303‐308. 10.1155/2006/865890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kresfelder TL, Janssen R, Bont L, Pretorius M, Venter M. Confirmation of an association between single nucleotide polymorphisms in the VDR gene with respiratory syncytial virus related disease in South African children. J Med Virol. 2011;83:1834‐1840. 10.1002/jmv.22179. [DOI] [PubMed] [Google Scholar]
- 31. Paulus SC, Hirschfeld AF, Victor RE, Brunstein J, Thomas E, Turvey SE. Common human toll‐like receptor 4 polymorphisms—role in susceptibility to respiratory syncytial virus infection and functional immunological relevance. Clin Immunol. 2007;123:252‐257. 10.1016/j.clim.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 32. Mailaparambil B, Krueger M, Heinze J, Forster J, Heinzmann A. Polymorphisms of toll like receptors in the genetics of severe RSV associated diseases. Dis Markers. 2008;25:59‐65. 10.1155/2008/619595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alvarez AE, Marson FAL, Bertuzzo CS, et al. Association between single nucleotide polymorphisms in TLR4, TLR2, TLR9, VDR, NOS2 and CCL5 genes with acute viral bronchiolitis. Gene. 2018;645:7‐17. 10.1016/j.gene.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liadaki K, Petinaki E, Skoulakis C, et al. Toll‐like receptor 4 gene (TLR4), but not TLR2, polymorphisms modify the risk of tonsillar disease due to Streptococcus pyogenes and Haemophilus influenzae . Clin Vaccine Immunol. 2011;18:217‐222. 10.1128/CVI.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ng MH, Lau KM, Li L, et al. Association of human‐leukocyte‐antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515‐518. 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keicho N, Itoyama S, Kashiwase K, et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum Immunol. 2009;70:527‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiong P, Zeng X, Song M, et al. Lack of association between HLA‐A,‐B and‐DRB1 alleles and the development of SARS: a cohort of 95 SARS‐recovered individuals in a population of Guangdong, southern China. Int J Immunogenet. 2008;35:69‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ng M, Cheng S, Lau K, et al. Immunogenetics in SARS: a case‐control study. Hong Kong Med J. 2010;16:29. [PubMed] [Google Scholar]
- 39. Lin M, Tseng H‐K, Trejaut JA, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen YM, Liang SY, Shih YP, et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol. 2006;44:359‐365. 10.1128/JCM.44.2.359-365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen A, David JK, Maden SK, et al. Human leukocyte antigen susceptibility map for SARS‐CoV‐2. J Virol. 2020;94:e00510–e00520. 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pisanti S, Deelen J, Gallina AM, et al. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid‐19. J Transl Med. 2020;18(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brouwer KC, Lal RB, Mirel LB, et al. Polymorphism of Fc receptor IIa for IgG in infants is associated with susceptibility to perinatal HIV‐1 infection. AIDS. 2004;18:1187‐1194. [DOI] [PubMed] [Google Scholar]
- 44. Yuan FF, Tanner J, Chan PK, et al. Influence of FcgammaRIIA and MBL polymorphisms on severe acute respiratory syndrome. Tissue Antigens. 2005;66:291‐296. 10.1111/j.1399-0039.2005.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou J, To KK, Dong H, et al. A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J Infect Dis. 2012;206:495‐503. 10.1093/infdis/jis378. [DOI] [PubMed] [Google Scholar]
- 46. Zhou Y, Fu B, Zheng X, et al. Aberrant pathogenic GM‐CSF+ T cells and inflammatory CD14+CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2020. 10.1101/2020.02.12.945576. [DOI] [Google Scholar]
- 47. Fensterl V, Sen GC. Interferons and viral infections. Biofactors. 2009;35:14‐20. [DOI] [PubMed] [Google Scholar]
- 48. Cameron MJ, Ran L, Xu L, et al. Interferon‐mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81:8692‐8706. 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chong, WP , Ip, WK , Tso, GH , et al. The interferon gamma gene polymorphism ?874A/T is associated with severe acute respiratory syndrome. BMC Infect Dis. 2006;6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ben‐Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144‐150. [DOI] [PubMed] [Google Scholar]
- 51. Tso H, Ip W, Chong W, Tam C, Chiang A, Lau YL. Association of interferon gamma and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Gene Immun. 2005;6:358‐363. [DOI] [PubMed] [Google Scholar]
- 52. Pravica V, Perrey C, Stevens A, Lee J‐H, Hutchinson IV. A single nucleotide polymorphism in the first intron of the human IFN‐γ gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN‐γ production. Hum Immunol. 2000;61:863‐866. [DOI] [PubMed] [Google Scholar]
- 53. Janssen R, Bont L, Siezen CL, et al. Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis. 2007;196:826‐834. 10.1086/520886. [DOI] [PubMed] [Google Scholar]
- 54. Siezen CL, Bont L, Hodemaekers HM, et al. Genetic susceptibility to respiratory syncytial virus bronchiolitis in preterm children is associated with airway remodeling genes and innate immune genes. Pediatr Infect Dis J. 2009;28:333‐335. 10.1097/INF.0b013e31818e2aa9. [DOI] [PubMed] [Google Scholar]
- 55. Huang J, Zhang M, Zhang X, Lu A, Wang L, Chen C. IFN‐gamma CA microsatellite polymorphism is associated with susceptibility to respiratory syncytial virus Infection and severity. Acta Paediatr. 2014;103:e544‐e547. 10.1111/apa.12767. [DOI] [PubMed] [Google Scholar]
- 56. Rogo LD, Rezaei F, Marashi SM, et al. Seasonal influenza A/H3N2 virus infection and IL‐1beta, IL‐10, IL‐17, and IL‐28 polymorphisms in Iranian population. J Med Virol. 2016;88:2078‐2084. 10.1002/jmv.24572. [DOI] [PubMed] [Google Scholar]
- 57. Keshavarz M, Namdari H, Farahmand M, Mehrbod P, Mokhtari‐Azad T, Rezaei F. Association of polymorphisms in inflammatory cytokines encoding genes with severe cases of influenza A/H1N1 and B in an Iranian population. Virol J. 2019;16:79. 10.1186/s12985-019-1187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Allan SM, Tyrrell PJ, Rothwell NJ. Interleukin‐1 and neuronal injury. Nat Rev Immunol. 2005;5:629‐640. 10.1038/nri1664. [DOI] [PubMed] [Google Scholar]
- 59. Chen W‐J, Yang J‐Y, Lin J‐H, et al. Nasopharyngeal shedding of severe acute respiratory syndrome—associated coronavirus is associated with genetic polymorphisms. Clin Infect Dis. 2006;42:1561‐1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tang F, Liu W, Zhang F, et al. IL‐12 RB1 genetic variants contribute to human susceptibility to severe acute respiratory syndrome infection among Chinese. PloS One. 2008;3:e2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tong Y, Wang Z, Geng Y, et al. The association of functional polymorphisms of IL‐6 gene promoter with ischemic stroke: analysis in two Chinese populations. Biochem Biophys Res Commun. 2010;391:481‐485. [DOI] [PubMed] [Google Scholar]
- 62. Ulhaq ZS, Soraya GV. Anti‐IL‐6 receptor antibody treatment for severe COVID‐19 and the potential implication of IL‐6 gene polymorphisms in novel coronavirus pneumonia. Med Clin. 2020;155(12):548‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Feng B, Mao ZR, Pang K, Zhang SL, Li L. Association of tumor necrosis factor alpha ‐308G/A and interleukin‐6 ‐174G/C gene polymorphism with pneumonia‐induced sepsis. J Crit Care. 2015;30:920‐923. 10.1016/j.jcrc.2015.04.123. [DOI] [PubMed] [Google Scholar]
- 64. Jerrard‐Dunne P, Sitzer M, Risley P, et al. Interleukin‐6 promoter polymorphism modulates the effects of heavy alcohol consumption on early carotid artery atherosclerosis: the Carotid Atherosclerosis Progression Study (CAPS). Stroke. 2003;34:402‐407. 10.1161/01.str.0000053849.09308.b2. [DOI] [PubMed] [Google Scholar]
- 65. Flores C, Ma S‐F, Maresso K, Wade MS, Villar J, Garcia JG. IL6 gene‐wide haplotype is associated with susceptibility to acute lung injury. Transl Res. 2008;152:11‐17. [DOI] [PubMed] [Google Scholar]
- 66. Mao Z‐R, Zhang S‐L, Feng B. Association of IL‐10 (‐819T/C,‐592A/C and‐1082A/G) and IL‐6‐174G/C gene polymorphism and the risk of pneumonia‐induced sepsis. Biomarkers. 2017;22:106‐112. [DOI] [PubMed] [Google Scholar]
- 67. Choi P, Reiser H. IL‐4: role in disease and regulation of production. Clin Exp Immunol. 1998;113:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Patarcic I, Gelemanovic A, Kirin M, et al. The role of host genetic factors in respiratory tract infectious diseases: systematic review, meta‐analyses and field synopsis. Sci Rep. 2015;5:16119. 10.1038/srep16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Faber TE, Schuurhof A, Vonk A, et al. IL1RL1 gene variants and nasopharyngeal IL1RL‐a levels are associated with severe RSV bronchiolitis: a multicenter cohort study. PLoS One. 2012;7:e34364. 10.1371/journal.pone.0034364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Choi EH, Lee HJ, Yoo T, Chanock SJ. A common haplotype of interleukin‐4 gene IL4 is associated with severe respiratory syncytial virus disease in Korean children. J Infect Dis. 2002;186:1207‐1211. 10.1086/344310. [DOI] [PubMed] [Google Scholar]
- 71. Marr N, Hirschfeld AF, Lam A, Wang S, Lavoie PM, Turvey SE. Assessment of genetic associations between common single nucleotide polymorphisms in RIG‐I‐like receptor and IL‐4 signaling genes and severe respiratory syncytial virus infection in children: a candidate gene case‐control study. PLoS One. 2014;9:e100269. 10.1371/journal.pone.0100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Puthothu B, Krueger M, Forster J, Heinzmann A. Association between severe respiratory syncytial virus infection and IL13/IL4 haplotypes. J Infect Dis. 2006;193:438‐441. 10.1086/499316. [DOI] [PubMed] [Google Scholar]
- 73. Hoebee B, Rietveld E, Bont L, et al. Association of severe respiratory syncytial virus bronchiolitis with interleukin‐4 and interleukin‐4 receptor alpha polymorphisms. J Infect Dis. 2003;187:2‐11. 10.1086/345859. [DOI] [PubMed] [Google Scholar]
- 74. Puthothu B, Krueger M, Forster J, Heinze J, Weckmann M, Heinzmann A. Interleukin (IL)‐18 polymorphism 133C/G is associated with severe respiratory syncytial virus infection. Pediatr Infect Dis J. 2007;26:1094‐1098. 10.1097/INF.0b013e3181453579. [DOI] [PubMed] [Google Scholar]
- 75. Miyairi I, DeVincenzo JP. Human genetic factors and respiratory syncytial virus disease severity. Clin Microbiol Rev. 2008;21:686‐703. 10.1128/CMR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Drysdale SB, Prendergast M, Alcazar M, et al. Genetic predisposition of RSV infection‐related respiratory morbidity in preterm infants. Eur J Pediatr. 2014;173:905‐912. 10.1007/s00431-014-2263-0. [DOI] [PubMed] [Google Scholar]
- 77. Ermers MJ, Janssen R, Onland‐Moret NC, et al. IL10 family member genes IL19 and IL20 are associated with recurrent wheeze after respiratory syncytial virus bronchiolitis. Pediatr Res. 2011;70:518‐523. 10.1203/PDR.0b013e31822f5863. [DOI] [PubMed] [Google Scholar]
- 78. Schuurhof A, Bont L, Siezen CL, et al. Interleukin‐9 polymorphism in infants with respiratory syncytial virus infection: an opposite effect in boys and girls. Pediatr Pulmonol. 2010;45:608‐613. 10.1002/ppul.21229. [DOI] [PubMed] [Google Scholar]
- 79. Liu Y, Li S, Zhang G, et al. Genetic variants in IL1A and IL1B contribute to the susceptibility to 2009 pandemic H1N1 influenza A virus. BMC Immunol. 2013;14:37. 10.1186/1471-2172-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ladhani SN, Davila S, Hibberd Martin L, et al. Association between single‐nucleotide polymorphisms inMal/TIRAP and interleukin‐10 genes and susceptibility to invasive Haemophilus influenzae serotype b infection in immunized children. Clin Infect Dis. 2010;51:761‐767. 10.1086/656236. [DOI] [PubMed] [Google Scholar]
- 81. Martinez‐Ocana J, Olivo‐Diaz A, Salazar‐Dominguez T, et al. Plasma cytokine levels and cytokine gene polymorphisms in Mexican patients during the influenza pandemic A(H1N1)pdm09. J Clin Virol. 2013;58:108‐113. 10.1016/j.jcv.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 82. Wang J, Knaut H. Chemokine signaling in development and disease. Development. 2014;141:4199‐4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Proudfoot AE. Chemokine receptors: multifaceted therapeutic targets. Nat Rev Immunol. 2002;2:106‐115. 10.1038/nri722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Law AH, Lee DC, Cheung BK, Yim HC, Lau AS. Role for nonstructural protein 1 of severe acute respiratory syndrome coronavirus in chemokine dysregulation. J Virol. 2007;81:416‐422. 10.1128/JVI.02336-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ng MW, Zhou G, Chong WP, et al. The association of RANTES polymorphism with severe acute respiratory syndrome in Hong Kong and Beijing Chinese. BMC Infect Dis. 2007;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Moriuchi H, Moriuchi M, Fauci AS. Nuclear factor‐kappa B potently up‐regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J Immunol. 1997;158:3483‐3491. [PubMed] [Google Scholar]
- 87. Law HK, Cheung CY, Ng HY, et al. Chemokine up‐regulation in sars‐coronavirus–infected, monocyte‐derived human dendritic cells. Blood. 2005;106:2366‐2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wong C, Lam C, Wu A, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. McDermott DH, Yang Q, Kathiresan S, et al. CCL2 polymorphisms are associated with serum monocyte chemoattractant protein‐1 levels and myocardial infarction in the Framingham Heart Study. Circulation. 2005;112:1113‐1120. [DOI] [PubMed] [Google Scholar]
- 90. Tu X, Chong WP, Zhai Y, et al. Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. J Infect. 2015;71:101‐109. 10.1016/j.jinf.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Severe Covid GG, Ellinghaus D, Degenhardt F, et al. Genomewide association study of severe covid‐19 with respiratory failure. N Engl J Med. 2020;383:1522‐1534. 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Amanatidou V, Sourvinos G, Apostolakis S, et al. RANTES promoter gene polymorphisms and susceptibility to severe respiratory syncytial virus‐induced bronchiolitis. Pediatr Infect Dis J. 2008;27:38‐42. 10.1097/INF.0b013e31814d4e42. [DOI] [PubMed] [Google Scholar]
- 93. Amanatidou V, Sourvinos G, Apostolakis S, Tsilimigaki A, Spandidos DA. T280M variation of the CX3C receptor gene is associated with increased risk for severe respiratory syncytial virus bronchiolitis. Pediatr Infect Dis J. 2006;25:410‐414. 10.1097/01.inf.0000214998.16248.b7. [DOI] [PubMed] [Google Scholar]
- 94. Hull J, Rowlands K, Lockhart E, Moore C, Sharland M, Kwiatkowski D. Variants of the chemokine receptor CCR5 are associated with severe bronchiolitis caused by respiratory syncytial virus. J Infect Dis. 2003;188:904‐907. 10.1086/377587. [DOI] [PubMed] [Google Scholar]
- 95. Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40‐47. 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sariban E, Imamura K, Luebbers R, Kufe D. Transcriptional and posttranscriptional regulation of tumor necrosis factor gene expression in human monocytes. J Clin Invest. 1988;81:1506‐1510. 10.1172/JCI113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang S, Wei M, Han Y, et al. Roles of TNF‐α gene polymorphisms in the occurrence and progress of SARS‐Cov infection: a case‐control study. BMC Infect Dis. 2008;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li H, Anderson SK. Association of TNFRSF1B promoter polymorphisms with human disease: further studies examining T‐regulatory cells are required. Front Immunol. 2018;9:443. 10.3389/fimmu.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ching JC‐Y, Chan KYK, Lee EHL, et al. Significance of the Myxovirus resistance A (MxA) gene—123C> a single‐nucleotide polymorphism in suppressed interferon β induction of severe acute respiratory syndrome coronavirus infection. J Infect Dis. 2010;201:1899‐1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hamano E, Hijikata M, Itoyama S, et al. Polymorphisms of interferon‐inducible genes OAS‐1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005;329:1234‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fernandez‐Arcas N, Blanco A, Gaitan MJ, Nyqvist M, Alonso A, Reyes‐Engel A. Differential transcriptional expression of the polymorphic myxovirus resistance protein A in response to interferon‐alpha treatment. Pharmacogenetics. 2004;14:189‐193. 10.1097/00008571-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 102. He J, Feng D, de Vlas SJ, et al. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case‐control study. BMC Infect Dis. 2006;6:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zani A, Yount JS. Antiviral protection by IFITM3 in vivo. Curr Clin Microbiol Rep. 2018;5:229‐237. 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lopez‐Rodriguez M, Herrera‐Ramos E, Sole‐Violan J, et al. IFITM3 and severe influenza virus infection. No evidence of genetic association. Eur J Clin Microbiol Infect Dis. 2016;35:1811‐1817. 10.1007/s10096-016-2732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Randolph AG, Yip WK, Allen EK, et al. Evaluation of IFITM3 rs12252 association with severe pediatric influenza infection. J Infect Dis. 2017;216:14‐21. 10.1093/infdis/jix242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Carter TC, Hebbring SJ, Liu J, et al. Pilot screening study of targeted genetic polymorphisms for association with seasonal influenza hospital admission. J Med Virol. 2018;90:436‐446. 10.1002/jmv.24975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Everitt AR, Clare S, Pertel T, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519‐523. 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Babaha F, Rezaei N. Primary immunodeficiency diseases in COVID‐19 pandemic: a predisposing or protective factor? Am J Med Sci. 2020;360:740‐741. 10.1016/j.amjms.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shields AM, Burns SO, Savic S, Richter AG. COVID‐19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J Allergy Clin Immunol. 2021;147(3):870‐875.e1. 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life‐threatening COVID‐19. Science. 2020:370:eabd4585. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pairo‐Castineira E, Clohisey S, Klaric L, et al. Genetic mechanisms of critical illness in COVID‐19. Nature. 2021;591(7848):92‐98. 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 112. Marcus N, Frizinsky S, Hagin D, et al. Minor clinical impact of COVID‐19 pandemic on patients with primary immunodeficiency in Israel. Front Immunol. 2021;11:614086. 10.3389/fimmu.2020.614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Delavari S, Abolhassani H, Abolnezhadian F, et al. Impact of SARS‐CoV‐2 pandemic on patients with primary immunodeficiency. J Clin Immunol. 2021;41:345‐355. 10.1007/s10875-020-00928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Meyts I, Bucciol G, Quinti I, et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520‐531. 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Klocperk A, Parackova Z, Dissou J, et al. Case report: systemic inflammatory response and fast recovery in a pediatric patient with COVID‐19. Front Immunol. 2020;11:1665. 10.3389/fimmu.2020.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. van der Made CI, Simons A, Schuurs‐Hoeijmakers J, et al. Presence of genetic variants among young men with severe COVID‐19. J Am Med Assoc. 2020;324:663‐673. 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Quinti I, Lougaris V, Milito C, et al. A possible role for B cells in COVID‐19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146:211‐213.e4. 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Soresina A, Moratto D, Chiarini M, et al. Two X‐linked agammaglobulinemia patients develop pneumonia as COVID‐19 manifestation but recover. Pediatr Allergy Immunol. 2020;31:565‐569. 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gao Y, Chen Y, Liu M, Shi S, Tian J. Impacts of immunosuppression and immunodeficiency on COVID‐19: a systematic review and meta‐analysis. J Infect. 2020;81:e93‐e95. 10.1016/j.jinf.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


