Abstract
The ability of the new coronavirus SARS‐CoV‐2 to spread and contaminate is one of the determinants of the COVID‐19 pandemic status. SARS‐CoV‐2 has been detected in saliva consistently, with similar sensitivity to that observed in nasopharyngeal swabs. We conducted ultrasound‐guided postmortem biopsies in COVID‐19 fatal cases. Samples of salivary glands (SGs; parotid, submandibular, and minor) were obtained. We analyzed samples using RT‐qPCR, immunohistochemistry, electron microscopy, and histopathological analysis to identify SARS‐CoV‐2 and elucidate qualitative and quantitative viral profiles in salivary glands. The study included 13 female and 11 male patients, with a mean age of 53.12 years (range 8–83 years). RT‐qPCR for SARS‐CoV‐2 was positive in 30 SG samples from 18 patients (60% of total SG samples and 75% of all cases). Ultrastructural analyses showed spherical 70–100 nm viral particles, consistent in size and shape with the Coronaviridae family, in the ductal lining cell cytoplasm, acinar cells, and ductal lumen of SGs. There was also degeneration of organelles in infected cells and the presence of a cluster of nucleocapsids, which suggests viral replication in SG cells. Qualitative histopathological analysis showed morphologic alterations in the duct lining epithelium characterized by cytoplasmic and nuclear vacuolization, as well as nuclear pleomorphism. Acinar cells showed degenerative changes of the zymogen granules and enlarged nuclei. Ductal epithelium and serous acinar cells showed intense expression of ACE2 and TMPRSS receptors. An anti‐SARS‐CoV‐2 antibody was positive in 8 (53%) of the 15 tested cases in duct lining epithelial cells and acinar cells of major SGs. Only two minor salivary glands were positive for SARS‐CoV‐2 by immunohistochemistry. Salivary glands are a reservoir for SARS‐CoV‐2 and provide a pathophysiological background for studies that indicate the use of saliva as a diagnostic method for COVID‐19 and highlight this biological fluid's role in spreading the disease. © 2021 The Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Keywords: COVID‐19; autopsy; infection control; salivary gland; RT‐PCR, SARS‐CoV‐2; saliva
Introduction
Since the WHO declared a pandemic status for COVID‐19, governments and health care organizations have created a series of strategies to mitigate the spread of its etiological agent, SARS‐CoV‐2. The contagion occurs through infected droplets that are disseminated directly by coughing and sneezing [1]. Salivary secretions are the main components of small speech droplets and thus play an essential role in the contamination pattern of COVID‐19. The presence of SARS‐CoV‐2 RNA in saliva droplets has been demonstrated consistently for different stages of the disease and has been used as a reliable COVID‐19 diagnostic tool [2, 3]. In a series of 70 COVID‐19 patients, Iwasaki et al detected more copies of SARS‐CoV‐2 RNA in saliva samples than in the gold standard diagnostic method, nasopharyngeal smear samples [4].
Saliva is a complex biological fluid composed of salivary gland secretion, crevicular fluid, respiratory secretion, and exfoliated epithelial cells. The presence of SARS‐CoV‐2 in saliva may be related to viral proliferation and RNA secretion in any cells and tissues involved in production of salivary components, such as periodontal tissue, salivary glands, and cells of the upper respiratory tract [5]. For instance, we have previously demonstrated the presence of SARS‐CoV‐2 RNA in periodontal tissue [6]. Determining how each tissue contributes as a reservoir for SARS‐CoV‐2 may be a path towards better understanding of the SARS‐CoV‐2 profile in saliva and for developing strategies for improving diagnosis, as well as for mitigating contamination through salivary droplets.
SARS‐CoV‐2 infection of the host's cells depends on the cleavage of one of its spike subunits by furin [7], thus allowing the cleaved spike protein to interact with angiotensin‐converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS) receptors. These interactions initiate cell endocytosis and begin the viral replication cycle [8]. Animal studies have shown the interaction of these receptors and SARS‐CoV‐2, suggesting that ACE2, TMPRSS, and furin present in salivary gland tissues are early targets of coronavirus infection [9].
Therefore, to better understand the basis of transmission patterns, it is crucial to verify whether SARS‐CoV‐2 RNA in the saliva is related to viral infection and replication within glandular epithelial cells or is related, instead, only to the respiratory secretion and periodontal component of saliva.
Materials and methods
We conducted postmortem biopsies of the major (parotid and submandibular) and minor salivary glands (lower lip) during ultrasound‐guided minimally invasive autopsy (US‐MIA) of patients who died of COVID‐19. Institutional and federal review boards approved this study under protocol number 30364720.0.0000.0068. We performed the US‐MIA after obtaining informed consent of the next‐of‐kin. The procedure consisted of a verbal autopsy questionnaire to gather clinical and medical information, followed by ultrasound‐guided postmortem biopsies to obtain samples, following established safety protocols described previously [10].
In each autopsy, we identified the parotid and submandibular glands using a portable SonoSite M‐Turbo R (Fujifilm, Bothell, WA, USA) ultrasound system with an HFL38X (13–6 MHz linear) transducer. We performed postmortem biopsies using Tru‐Cut® semi‐automatic percutaneous 14G coaxial needles (20 cm) (Supplementary materials and methods and supplementary material, Figure S1). Punctures were made by percutaneous access to avoid the risk of salivary contamination. In order to access the minor salivary glands, initially we wiped the inner lip mucosa area using gauze soaked in an enzymatic detergent (Riozyme; Rioquímica, São José do Rio Preto, São Paulo, Brazil) to clean all superficial contamination, and we performed the biopsy using a 0.3‐mm punch.
Samples were frozen and stored at −80 °C. Tissue samples were macerated, and nucleic acid extracted using the TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). Molecular detection of SARS‐CoV‐2 was performed using the SuperScriptTM III PlatinumTM One‐Step RT‐qPCR Kit (Invitrogen) and primers/probes sets for E and N (N1) gene amplification [11].
The human RNase P gene was also amplified as a nucleic acid extraction control [3]. RT‐qPCR reactions were performed using the 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA). They consisted of a step of reverse transcription at 55 °C for 10 min, then 95 °C for 3 min, followed by 45 cycles of 95 °C for 15 s and 58 °C (E gene)/55 °C (N and RNAse P genes) for 30 s.
Additional samples were fixed in buffered 10% formalin solution and embedded in paraffin wax. We prepared 3‐μm‐thick sections and mounted them on glass slides for H&E staining and immunohistochemistry for identification of SARS‐CoV‐2, ACE2, and TMPRSS (protocol details are available in supplementary material, Supplementary materials and methods).
To perform ultrastructural analyses, we reprocessed the formalin‐fixed, paraffin‐embedded biological tissue. This procedure is especially suitable whenever it is desirable to select a specific area from the sample for transmission electron microscopy.
We identified ductal and acinar areas with pathological tissue derangement on H&E‐stained slides of the parotid and submandibular salivary glands and marked them using a fiber tip marker on the glass. By macroscopic comparison, we identified corresponding regions on paraffin block surfaces. The target areas were cut out using a razor blade. The resulting small fragments were deparaffinized in xylene, rehydrated through a graded alcohol series, and re‐fixed using 2% glutaraldehyde in 0.15 m phosphate buffer, pH 7.2, followed by post‐fixation in 1% OsO4 and staining in 1% aqueous uranyl acetate overnight. The specimens were then embedded in an epoxy resin. Ultrathin sections were cut using a Leica‐Reichert ultratome (Leica, Wetzlar, Giessen, Germany) and double‐stained using uranyl acetate and lead citrate. Micrographs were obtained using a Gatan 792 BioScan 1K by 1K wide angle CCD camera (Gatan, Pleasanton, CA, USA) coupled to a JEOL JEM 1010, 80 kV electron microscope (JEOL, Tokyo, Japan).
Results and discussion
For all patients, the diagnosis was confirmed by RT‐qPCR of nasopharyngeal swab specimens. We evaluated 45 salivary gland (SG) samples (20 parotid, 15 submandibular, and 10 minor SGs) from 24 deceased patients. In selected cases, we also performed immunohistochemistry for SARS‐CoV‐2, ACE2, and TMPRSS receptors (15 cases), and electron microscopy (two cases) (see Supplementary materials and methods for details).
The study included 13 female and 11 male patients, with a mean age of 53.12 years (range 8–83 years). The mean timespan between symptoms' onset and death was 21.12 days (range 4–47 days). RT‐qPCR for SARS‐CoV‐2 was positive in 30 SG samples from 18 patients (60% of total SG samples and 75% of all COVID‐19 cases).
Ultrastructural analyses showed spherical 70–100 nm viral particles, consistent in size and shape with the Coronaviridae family, in the ductal lining cell cytoplasm, acinar cells, and ductal lumen of SG (Figure 1A,B). There was also degeneration of organelles in what appeared to be virally infected cells (Figure 1C). Although cellular degeneration may occur due to the fact that we are using postmortem samples from paraffin blocks, the high abundance of particles indicates that the damage was probably at least partially due to viral infection.
Figure 1.
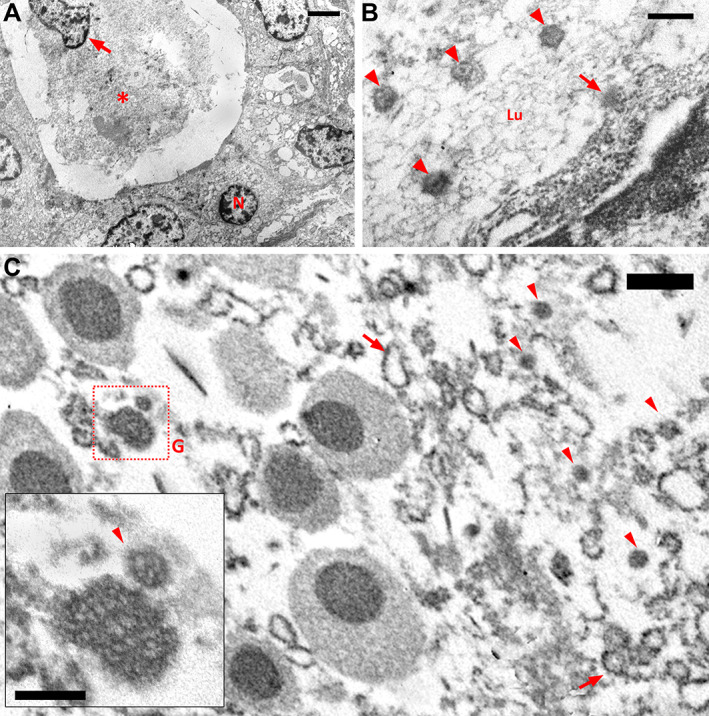
Postmortem biopsy histological findings: (A) Low‐magnification electron micrograph of an intralobular duct of the submandibular gland. The ductal epithelium consists of a single layer of cuboidal cells that have a centrally located nucleus (N). The ductal lumen was almost completely obliterated by accumulation of debris (asterisk), including an isolated cell nucleus (arrow). Bar = 2 μm. (B) Electron micrograph showing the apical zone of a ductal epithelial cell of the parotid gland. In addition to the viral particles (arrowheads) inside the ductal lumen (Lu), there is a viral particle leaving the cell by budding through the membrane (arrow). Bar = 200 nm. (C) Electron micrograph showing part of an acinar cell of the submandibular gland. On the left side of the image, the cytoplasm contains seromucous secretory granules (G) typically formed by strongly stained spherules surrounded by an unstained component. On the right side, the cytoplasm shows degeneration, with viral particles (arrowhead) and microsomal vesicles (arrows). The inset shows a mature viral particle (arrowhead). Bar = 500 nm; inset bar = 200 nm.
Qualitative histopathological analysis showed morphologic alterations in the duct lining epithelium characterized by cytoplasmic and nuclear vacuolization as well as nuclear pleomorphism. Acinar cells showed degenerative changes of the zymogen granules and enlarged nuclei (Figure 2A,B) when qualitatively compared with controls (Figure 2C,D). By immunohistochemistry, both ductal epithelium and serous acinar cells showed intense expression of ACE2 and TMPRSS receptors (Figure 2E,F). The anti‐SARS‐CoV‐2 antibody was positive in eight (53%) of the 15 tested cases in duct lining epithelial cells and acinar cells of major SGs (Figure 2G,H). Only two (13%) of the minor salivary glands were positive for SARS‐CoV‐2 by immunohistochemistry.
Figure 2.
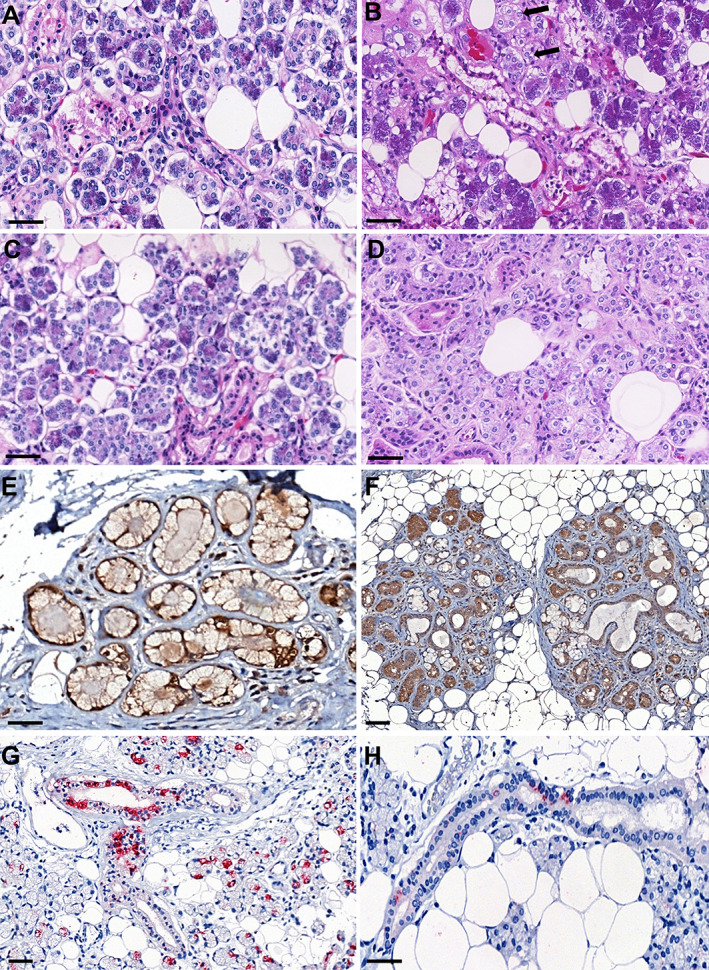
Postmortem biopsy histological findings: (A) Parotid COVID‐19 patient – H&E. (B) Submandibular COVID‐19 patient – H&E; duct lining epithelium characterized by nuclear pleomorphism. Acinar cells showing enlarged nuclei (arrows); condensation of zymogen granules. (C) Parotid control patient – H&E. (D) Submandibular control patient – H&E. (E) ACE2 receptor – parotid immunohistochemistry targeting the human ACE2 protein (brown) showed staining in acinar cells. (F) TMPRSS receptor – submandibular immunohistochemistry targeting the human ACE2 protein (brown) showed staining in acinar and ductal cells. Immunohistochemistry targeting SARS‐CoV‐2. (G) Parotid showing positive staining for SARS‐CoV‐2 in intercalated duct and striated duct. Acinar cell staining characterized by an apical localization. (H) Submandibular SG showing diffuse positive staining for SARS‐CoV‐2 in a striated duct. Scale bars: 50 μm.
The study of SARS‐CoV‐2 organotropism is important for understanding the disease's pathogenesis and infection patterns [12]. Salivary glands were reported as a virus reservoir for prevalent diseases such as herpes simplex, EBV, HHV‐7, and cytomegalovirus [5, 13]. Viral replication within the SGs seems to be an efficient dissemination strategy as the contaminated droplets expelled during coughs, sneezes, and speech are mainly composed of saliva excreta [14]. Even patients from our study who died from non‐respiratory causes – including tumors, neurologic events, and vascular causes, presented SARS‐CoV‐2 infections in salivary gland cells.
For the first time and using different methods, we demonstrate the presence of SARS‐CoV‐2 infection and its replication in the major and minor salivary glands. We also present the expression of the cellular viral targets, ACE2 and TMPRSS receptors, in patients with severe COVID‐19. Our findings demonstrate that salivary glands are a reservoir for SARS‐CoV‐2 and provide a pathophysiology background to the recent studies that indicate the use of saliva as a diagnostic method for COVID‐19 and highlight this biological fluid's role in spreading the disease.
Author contributions statement
BM contributed to conception, design, acquisition, interpretation, and data analysis, and drafted and critically revised the manuscript. MD contributed to conception, design, acquisition, interpretation, and data analysis, and critically revised the manuscript. GM and SCG contributed to design and acquisition, and drafted the manuscript. DS and NA contributed to analysis and interpretation, and drafted and critically revised the manuscript. AZ contributed to acquisition, analysis, and interpretation, and drafted and critically revised the manuscript. AN‐D, RM and CK contributed to conception, acquisition, and analysis, and critically revised the manuscript. JRP and MG‐G contributed to design, analysis, and interpretation, and critically revised the manuscript. SM contributed to design, analysis, and interpretation, and drafted and critically revised the manuscript. TM contributed to conception, design, and interpretation, and critically revised the manuscript. PS contributed to conception and design, and critically revised the manuscript. PB‐S and EC contributed to conception, analysis, and interpretation, and critically revised the manuscript. LFS contributed to conception, design, acquisition, analysis, and interpretation, and critically revised the manuscript. All the authors gave final approval and agree to be accountable for all aspects of the work.
Supporting information
Supplementary materials and methods
Supplementary figure legends
Figure S1. Postmortem salivary gland ultrasound and biopsy procedure showing the Tru‐Cut® needle
Figure S2. Salivary gland morphology and immunohistochemistry for SARS‐CoV‐2 (referred to in Supplementary materials and methods)
Table S1. Clinical information of patients included (referred to in Supplementary materials and methods)
Table S2. Summary of RT‐qPCR from all samples (cycle threshold values) (referred to in Supplementary materials and methods)
Table S3. Summary of immunohistochemistry results for SARS‐CoV‐2 (referred to in Supplementary materials and methods)
Acknowledgements
We wish to thank Kely Cristina Soares Bispo, Jair Theodoro Filho, Gustavo Linari Rodrigues, Angela BG dos Santos, Sandra de Moraes Fernezlian, Reginaldo Silva do Nascimento, Glaucia Aparecida dos Santos Bento, Thábata Larissa, Luciano Ferreira Leite, and Catia Sales de Moura for their technical support. We would also like to acknowledge all health care providers involved in the care of patients with COVID‐19 and all Hospital (HC‐FMUSP) and São Paulo Autopsy Service staff who have taken part in the Coronavirus Crisis Task Force during the epidemic season. We acknowledge and are deeply thankful to all relatives and legal representatives who gave their consent to the postmortem examinations of their beloved relatives lost to COVID‐19. This study was funded by the following organizations: Fundação de Amparo à Pesquisa do Estado de São Paulo, 2013/17159‐2 (Funder DOI: 10.13039/501100001807); ekBill and Melinda Gates Foundation, INV‐002396 (Funder DOI: 10.13039/100000865); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), 401825/2020‐5.
No conflicts of interest were declared.
References
- 1. Huff HV, Singh A. Asymptomatic transmission during the COVID‐19 pandemic and implications for public health strategies. Clin Infect Dis 2020; 71: 2752–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wyllie AL, Fournier J, Casanovas‐Massana A, et al. Saliva or nasopharyngeal swab specimens for detection of SARS‐CoV‐2. N Engl J Med 2020; 383: 1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. To KK, Tsang OT, Yip CC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 2020; 71: 841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iwasaki S, Fujisawa S, Nakakubo S, et al. Comparison of SARS‐CoV‐2 detection in nasopharyngeal swab and saliva. J Infect 2020; 81: e145–e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen T, Hudnall SD. Anatomical mapping of human herpesvirus reservoirs of infection. Mod Pathol 2006; 19: 726–737. [DOI] [PubMed] [Google Scholar]
- 6. Fernandes Matuck B, Dolhnikoff M, Maia GVA, et al. Periodontal tissues are targets for Sars‐Cov‐2: a post‐mortem study. J Oral Microbiol 2020; 13: 1848135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jose RJ, Manuel A. COVID‐19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med 2020; 8: e46–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus‐induced lung injury. Nat Med 2005; 11: 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu L, Wei Q, Alvarez X, et al. Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol 2011; 85: 4025–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Duarte‐Neto AN, Monteiro RAA, da Silva LFF, et al. Pulmonary and systemic involvement in COVID‐19 patients assessed with ultrasound‐guided minimally invasive autopsy. Histopathology 2020; 77: 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill 2020; 25: 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med 2020; 383: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laane CJ, Murr AH, Mhatre AN, et al. Role of Epstein–Barr virus and cytomegalovirus in the etiology of benign parotid tumors. Head Neck 2002; 24: 443–450. [DOI] [PubMed] [Google Scholar]
- 14. Ferreiro MC, Dios PD, Scully C. Transmission of hepatitis C virus by saliva? Oral Dis 2005; 11: 230–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods
Supplementary figure legends
Figure S1. Postmortem salivary gland ultrasound and biopsy procedure showing the Tru‐Cut® needle
Figure S2. Salivary gland morphology and immunohistochemistry for SARS‐CoV‐2 (referred to in Supplementary materials and methods)
Table S1. Clinical information of patients included (referred to in Supplementary materials and methods)
Table S2. Summary of RT‐qPCR from all samples (cycle threshold values) (referred to in Supplementary materials and methods)
Table S3. Summary of immunohistochemistry results for SARS‐CoV‐2 (referred to in Supplementary materials and methods)


