Abstract
Background
Immunothrombosis and coagulopathy in the lung microvasculature may lead to lung injury and disease progression in coronavirus disease 2019 (COVID‐19). We aim to identify biomarkers of coagulation, endothelial function, and fibrinolysis that are associated with disease severity and may have prognostic potential.
Methods
We performed a single‐center prospective study of 14 adult COVID‐19(+) intensive care unit patients who were age‐ and sex‐matched to 14 COVID‐19(−) intensive care unit patients, and healthy controls. Daily blood draws, clinical data, and patient characteristics were collected. Baseline values for 10 biomarkers of interest were compared between the three groups, and visualized using Fisher's linear discriminant function. Linear repeated‐measures mixed models were used to screen biomarkers for associations with mortality. Selected biomarkers were further explored and entered into an unsupervised longitudinal clustering machine learning algorithm to identify trends and targets that may be used for future predictive modelling efforts.
Results
Elevated D‐dimer was the strongest contributor in distinguishing COVID‐19 status; however, D‐dimer was not associated with survival. Variable selection identified clot lysis time, and antigen levels of soluble thrombomodulin (sTM), plasminogen activator inhibitor‐1 (PAI‐1), and plasminogen as biomarkers associated with death. Longitudinal multivariate k‐means clustering on these biomarkers alone identified two clusters of COVID‐19(+) patients: low (30%) and high (100%) mortality groups. Biomarker trajectories that characterized the high mortality cluster were higher clot lysis times (inhibited fibrinolysis), higher sTM and PAI‐1 levels, and lower plasminogen levels.
Conclusions
Longitudinal trajectories of clot lysis time, sTM, PAI‐1, and plasminogen may have predictive ability for mortality in COVID‐19.
Keywords: biomarkers, coronavirus, fibrinolysis, observational study, thrombosis
Essentials
-
•
COVID‐19 is an immunothrombotic disease that leads to respiratory failure.
-
•
It is unknown why some critically ill patients recover while some decompensate.
-
•
We identified four potential biomarkers of fibrinolysis and endotheliopathy associated with mortality.
-
•
If validated, these biomarkers may have predictive ability for worse outcomes.
Alt-text: Unlabelled Box
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), which is responsible for the novel coronavirus disease 2019 (COVID‐19) pandemic, is a newly emergent zoonotic coronavirus that appears to have originated in Wuhan, China.1 First reported in December 2019, COVID‐19 has spread to more than 200 countries and territories, infecting more than 94 million people worldwide and killing over 2 million.2
SARS‐CoV‐2 is thought to target the angiotensin converting enzyme 2 receptor and heparan sulfate on the surface of alveolar endothelial cells, binding through spike S proteins on the viral envelope.3., 4., 5., 6. Subsequent internalization of the virus diminishes remaining membrane‐bound angiotensin converting enzyme 2 receptors, potentially leading to increased angiotensin II levels, further promoting hypoxemia and resulting lung injury.7 Consequently, COVID‐19 presents as a lower respiratory tract infection, with severe cases progressing to acute respiratory distress syndrome with intravascular and extravascular fibrin deposition.8., 9., 10., 11.
Increased rates of systemic thrombotic complications are a prominent feature in COVID‐19 patients. Emerging observations suggest that the immunothrombotic responses to SARS‐CoV‐2 in the lung microvasculature may contribute to disease progression. Fibrin deposition in the lungs, elevated levels of D‐dimer, and rates of thrombosis ranging from 5% to 30% despite thromboprophylaxis12., 13., 14., 15., 16. are hallmarks of COVID‐19 pneumonia.17., 18., 19. Therefore, modulation of the immunothrombosis and coagulopathy may prevent lung injury in COVID‐19.
Current evidence suggest dysregulation of the coagulation and fibrinolytic systems in COVID‐19 patients.20., 21., 22., 23., 24., 25., 26., 27., 28., 29. Although the coagulation system has been studied extensively,30., 31. the fibrinolytic system has not been longitudinally evaluated in intensive care unit (ICU) patients with or without COVID‐19. To address this gap, we explored the time course of markers of coagulation (fibrinogen, D‐dimer, thrombin‐antithrombin [TAT] complex), endothelial function (soluble thrombomodulin [sTM]), and fibrinolytic activity (plasminogen, plasminogen activator inhibitor‐1 [PAI‐1], plasmin‐antiplasmin [PAP] complex, thrombin‐activatable fibrinolysis inhibitor [TAFI], activated TAFI [TAFIa], clot lysis time) in patients with COVID‐19 admitted to the ICU and associations with mortality that may inform future predictive modelling efforts.
2. EXPERIMENTAL PROCEDURE
2.1. Study design and setting
This was a prospective cohort study conducted at a single ICU at an academic tertiary care hospital in London, Canada.32., 33., 34., 35., 36. The study was approved by the Western University Human Research Ethics Board and written informed consent obtained. We followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement for cohort studies in the preparation of this manuscript.37
2.2. Participants
Consecutive patients ≥18 years of age admitted to the ICU meeting clinical criteria for suspected COVID‐1938 were prospectively enrolled and analyzed. Patients were included from March 16, 2020, to April 24, 2020, corresponding to the initial COVID‐19 outbreak in our health region. If not known at clinical presentation, COVID‐19 status was confirmed by two positive polymerase chain reaction tests for the SARS‐CoV‐2 virus. Patients that subsequently tested negative for SARS‐CoV‐2 were maintained to form a COVID‐19(−) critically ill control group. Thus, both groups initially met the same inclusion criteria at the time of ICU admission. Daily blood draws were initiated at the time of ICU admission and were continued up to day 3 for COVID‐19(−) patients, and up to day 10 for COVID‐19(+) patients. All COVID‐19(+) patients were age‐ and sex‐matched with a COVID‐19(−) patient from the prospectively collected pool. Previously collected blood samples from a healthy control group were assembled from age‐ and sex‐matched participants held at the Translational Research Centre in London, Ontario.39., 40.
2.3. Clinical data
Baseline patient characteristics included age, sex, comorbidities, and presenting chest x‐ray findings. Disease severity was classified using the Sequential Organ Failure Assessment (SOFA) and Multiple Organ Dysfunction scores (MODS). Both patient groups were characterized as having confirmed or suspected sepsis diagnosis using Sepsis 3.0 criteria.41 Clinical data were prospectively collected including the lowest or worst perfusion‐ventilation (P/F) ratio, mean arterial pressure, and standard laboratory values. Recorded interventions included the use of antibiotics or antivirals, systemic corticosteroids, vasopressors, respiratory support, renal replacement, antiplatelet agents, and anticoagulants. Patients that survived to hospital discharge were considered survivors for the purposes of these analyses.
2.4. Materials
Human tissue‐type plasminogen activator (tPA; Alteplase) was purchased from Kingston General Hospital pharmacy. Human α‐thrombin was from Enzyme Research Laboratories. A recombinant plasminogen derivative labeled with fluorescein and C9‐maleimide‐QSY‐labeled fibrin degradation products were prepared as described previously.42 The human PAP complex ELISA kit was purchased from Biomatik. Human plasminogen, VisuLize TAFI, TAT complex, and fibrinogen antigen ELISA kits were from Affinity Biologicals Inc. The human PAI‐1 total antigen ELISA kit was from Molecular Innovations. D‐dimer was quantified using an ELISA kit from RayBiotech. sTM was quantified using the thrombomodulin/BDCA‐3 Quantikine ELISA kit from R&D Systems. All of the assays were performed according to the manufacturers’ protocols. The intra‐ and interassay variability of the commercial ELISAs are respectively as follows: PAP complex: 8% and 10%; TAFI: 5.8% and 8%; PAI‐1: 5.9% and 6.5%; D‐dimer: <12% and <10%; sTM: 2.9% and 6.9%; and fibrinogen: 9.4% and 13.4%. These values for plasminogen and TATs are unknown.
2.5. Functional assays of fibrinolysis
Clot lysis assays were performed as described previously.43 Briefly, 96‐well clear flat‐bottom microtiter plates were pretreated for at least 1 h with 0.02 M HEPES, 0.15 M NaCl, pH 7.4 (HBS) containing 1% Tween 80 (Sigma‐ Aldrich), and washed thoroughly with water. Plasma diluted 1:3 in HBS was added to the wells and clotting and lysis were initiated with 5 nM thrombin and 1 nM tPA, respectively, in the presence of 10 mM CaCl2, and absorbance was monitored at 405 nm for 2 h at 37°C at 15‐s intervals using a SpectraMax M2 plate reader (Molecular Devices). Clotting time and lysis time were determined as the time to reach half‐maximal increase and decrease in absorbance as determined by the instrument software.
Functional levels of TAFIa were quantified as described previously.42 Briefly, 50 µl of a solution containing 50 nM recombinant plasminogen derivative labeled with fluorescein and 100 nM C9‐maleimide‐QSY‐labeled fibrin degradation products in HBS was placed in wells of a 96‐well white U‐bottom microtiter plate. Baseline fluorescence readings were measured at 1‐min intervals at 25°C, with excitation and emission wavelengths of 495 nm and 535 nm, respectively, with a cutoff at 515 nm. After equilibration, a plasma solution (50 µl) with a final 1:5 dilution was added. Reactions were monitored for 2 h and TAFIa levels were measured by quantifying rates of fluorescence increase.
2.6. Statistical analysis
Baseline clinical characteristics were compared between COVID‐19(+) patients and age‐ and sex‐matched COVID‐19(−) controls. Paired Wilcoxon rank‐sum tests were used for continuous variables and Fisher's exact tests for categorical variables. Biomarkers determined on ICU admission day 1 (baseline) were compared in COVID‐19(+) patients, COVID‐19(−) patients, and healthy controls in a similar manner. Baseline biomarkers between the three groups were further visualized using Fisher's linear discriminant function, a nonparametric dimensionality reduction technique.44 Before dimensionality reduction, biomarkers were normalized, and the results were displayed on a biplot.
Individual biomarker trajectories were plotted in COVID‐19(+) patients, stratified by survivors and nonsurvivors. Variable selection for modelling was performed using individual repeated measures linear mixed models, comparing log‐transformed biomarker values between survivors and nonsurvivors. Mean differences in biomarker values that reached our prespecified p‐value cutoff of .25 were included, a typical threshold when performing p‐value screening on a small dataset.45 These biomarkers were further analyzed using a more complex mixed model by additionally including the day of measurement in the model and an interaction term between day of measurement and vital status, beginning at the first day of ICU admission. Where the interaction term was nonsignificant, and there was no apparent trajectory difference between groups on visual inspection, the interaction term was removed from the model. First‐order autoregressive covariance structures were used for all mixed modelling.
Selected biomarkers were then included in a longitudinal, multivariate k‐means clustering algorithm (kml3d package in R),46., 47. with the number of known clusters set at two. It was our a priori hypothesis that clustering may be able to divide the cohort into higher and lower disease severity clusters. Cluster sizes up to six were also tested and resulted in single membership in clusters, and lower performance scores (Calinski‐Harabatz Index).48 The machine learning algorithm was naïve to death status or any patient characteristic aside from the selected biomarkers. A sensitivity analysis that excluded one patient who died before day 10 did not change the results. Mean and individual biomarker values, and patient characteristics were described for the two clusters.
Missing biomarker data were imputed with linear interpolation, which is a more prudent method than mean imputation when the values before and after the missing value are known. All analyses were performed using SAS version 9.4 (SAS Institute Inc.) and RStudio (RStudio Inc.). All statistical tests were two‐sided, and a p‐value ≤.05 was considered statistically significant. Because this study was primarily exploratory and hypothesis‐generating, adjustments for multiple comparisons were not made.49
3. RESULTS
3.1. Patient characteristics
Over the study period, 14 COVID‐19(+) patients were identified in our ICU, as were age‐ and sex‐matched with 14 COVID‐19(−) critically ill patients and 14 healthy controls. Patient characteristics are presented in Table 1 . Thirteen of the 14 COVID‐19(+) patients received anticoagulation treatment with prophylactic dose low‐molecular weight heparin (dalteparin; 5000 or 7500 units/day), whereas one patient received acetylsalicylic acid alone (81 mg/d). Four of the 13 patients treated with dalteparin also received acetylsalicylic acid (81 mg/d). Compared with COVID‐19(−) patients, those with COVID‐19 were significantly more likely to have bilateral pneumonia, lymphopenia, and require high‐flow nasal oxygen. The COVID‐19(+) cohort also had small but significant increases in international normalized ratio (INR) and activated partial thromboplastin time values. Although not statistically significant, mortality among COVID‐19(+) patients was 50% (7/14) compared with 14.3% (2/14) in the critically ill controls.
TABLE 1.
Patient and clinical characteristics between COVID‐19(+) and age‐ and sex‐matched COVID‐19(−) critically ill patients
| Patient Characteristics | COVID‐19(+) (n = 14) | COVID‐19(−) (n = 14) | p Value |
|---|---|---|---|
| Age, y | 61 (54–67) | 58.5 (52.5–63) | .4616 |
| Male | 6 (43%) | 6 (42.9%) | 1 |
| Comorbidities | |||
| Diabetes | 5 (35.7%) | 5 (35.7%) | 1 |
| Hypertension | 7 (50.0%) | 9 (64.3%) | .7036 |
| Coronary artery disease | 2 (14.3%) | 2 (14.3%) | 1 |
| Congestive heart failure | 0 | 2 (14.3%) | .4815 |
| Chronic kidney disease | 2 (14.3%) | 1 (7.1%) | 1 |
| Cancer | 2 (14.3%) | 1 (7.1%) | 1 |
| COPD | 1 (7.1%) | 3 (21.4%) | .5956 |
| Chest x‐ray findings | |||
| Normal | 0 | 3 (21.4%) | <.001 |
| Unilateral pneumonia | 1 (7.1%) | 8 (57.1%) | |
| Bilateral pneumonia | 13 (92.9%) | 2 (14.3%) | |
| Interstitial/atypical findings | 0 | 1 (7.1%) | |
| MODS | 4 (3–5.5) | 6 (3–8) | .3262 |
| SOFA | 4.5 (2–9.25) | 6 (4.25–10.5) | .1555 |
| Mean arterial pressure | 84 (72.75–97.5) | 75.5 (59.25–107.25) | .7695 |
| P/F ratio | 107 (65.5–161.675) | 172 (137.75–312) | .1514 |
| WBC | 8.45 (6.9–16.075) | 15.25 (11.05–20.45) | .104 |
| Lymphocytes | 0.7 (0.55–1) | 1.3 (0.5–1.75) | .03581 |
| Neutrophils | 7.3 (5.6–12.55) | 12.2 (8.1–15.725) | .2734 |
| Lactate | 1.5 (1–2) | 1.2 (0.9–1.6) | 1 |
| Platelets | 206 (133.5–293.75) | 201.5 (163.75–259.5) | 1 |
| Hemoglobin | 121.5 (101.5–134.5) | 123.5 (101.75–137.75) | .8752 |
| Creatinine | 81.5 (57.5–187) | 75 (54.25–113) | .9442 |
| INR | 1.2 (1.1–1.3) | 1.05 (1–1.125) | .04022 |
| aPTT | 28 (25–31) | 23 (19.5–25.25) | .006323 |
| Treatments | |||
| Antibiotics | 14 (100%) | 14 (100%) | 1 |
| Antivirals | 3 (21.4%) | 2 (14.3%) | 1 |
| Steroids | 3 (21.4%) | 5 (35.7%) | .6776 |
| Vasoactive medications | 11 (78.6%) | 8 (57.1%) | .4197 |
| Renal replacement therapy | 2 (14.3%) | 1 (7.1%) | 1 |
| Antiplatelet agent | 5 (35.7%) | 7 (50.0%) | .7036 |
| Anticoagulation | 13 (92.9%) | 14 (100%) | 1 |
| Respiratory support | |||
| High‐flow nasal oxygen | 8 (57.1%) | 1 (7.1%) | .01275 |
| NIMV | 6 (42.9%) | 8 (57.1%) | .7064 |
| Invasive ventilation | 10 (71.4%) | 11 (78.6%) | 1 |
| Died | 7 (50.0%) | 2 (14.3%) | .1032 |
Note
Values shown are median (interquartile range) and N (%). Statistical tests used were paired Wilcoxon rank‐sum tests and Fisher's exact tests.
Abbreviations: COPD, chronic obstructive pulmonary disease; COVID‐19, coronavirus disease 19; INR, international normalized ratio; MODS, Multi‐organ Dysfunction Score; NIMV, noninvasive mechanical ventilation; P/F ratio, perfusion/ventilation ratio; aPTT, activated partial thromboplastin time; SOFA, sequential organ failure assessment score; WBC, white blood cell
3.2. Biomarkers and COVID‐19 status
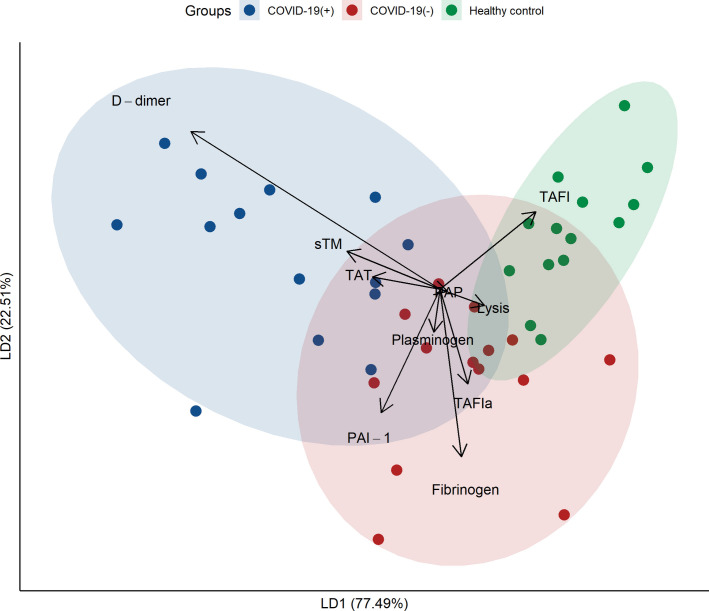
The 10 biomarkers of interest at baseline are presented for each group in Table 2 . Univariate comparisons between COVID‐19(+) and (−) patients demonstrated no significant differences, except for D‐dimer, which was higher in COVID‐19(+) patients. Both groups had impaired clot lysis, with approximately one‐half of the patients having lysis times above 100 mins. Compared with healthy controls, COVID‐19(+) patients had significantly higher levels of PAI‐1, sTM, D‐dimer, fibrinogen, and TAT; lower levels of plasminogen and TAFI; and longer clot lysis times. Dimensionality reduction was performed using Fisher's linear discriminant function. Figure 1 shows a biplot with the patient groups plotted according to the first two linear discriminants, which explains 77.49% of the variance of the centroids. D‐dimer was the strongest contributor toward identifying COVID‐19 status, followed by sTM.
TABLE 2.
Biomarkers between COVID‐19(+) patients, age‐ and sex‐matched COVID‐19(−) patients, and age‐ and sex‐matched healthy controls
| Biomarker at Baseline | COVID‐19(+) (n = 14) | COVID‐19(−) (n = 14) | Healthy Controls (n = 14) | COVID‐19(+) to COVID‐19(−) p Values | COVID‐19(+) to Healthy Controls p Values |
|---|---|---|---|---|---|
| PAI‐1 (ng/ml) |
40.9 (30.7–56.5) | 52.3 (21.3–90.2) | 9.8 (0.7–14.4) | .391 | <.001 |
| Plasminogen (µM) |
1.2 (1.1–1.4) | 1.3 (1.1–1.5) | 1.8 (1.4–2.0) | .2958 | .01074 |
| PAP (µg/ml) |
0.7 (0.5–1.6) | 0.6 (0.2–1.5) | 0.8 (0.2–1.2) | .6257 | .7609 |
| TAFI (nM) |
106.9 (77.1–115.0) | 121.3 (100.2–132.0) | 129.8 (118.8–175.2) | .1353 | .008545 |
| TAFIa (pM) |
75.9 (23.4–124.3) | 162.3 (67.0–217.4) | 26.6 (0.0–87.3) | .1353 | .2166 |
| sTM (ng/ml) |
5.1 (4.2–8.1) | 5.7 (3.8–7.4) | 3.3 (3.0–3.4) | 1 | .003052 |
| D‐dimer (µg/ml) |
3.5 (2.5–5.4) | 1.3 (0.9–1.7) | 0.7 (0.6–0.8) | <.001 | <.001 |
| Fibrinogen (mg/ml) |
10.7 (8.9–11.3) | 10.3 (8.4–11.2) | 7.1 (5.7–7.6) | .9032 | .005249 |
| Lysis time (min) | |||||
| <20 | 3 (21.4%) | 2 (14.3%) | 12 (85.7%) | .7844 | .00226 |
| 20–59.9 | 4 (28.6%) | 6 (42.9%) | 2 (14.3%) | ||
| 60–99.9 | 0 | 0 | 0 | ||
| 100+ | 7 (50.0%) | 6 (42.9%) | 0 | ||
| TAT (µg/L) |
54.2 (38.8–143.5) | 30.9 (19.8–98.6) | 9.3 (6.3–30.5) | .5016 | <.001 |
Note
Values shown are median (interquartile range) and N (%). Statistical tests used were paired Wilcoxon rank‐sum tests and Fisher's exact tests.
Abbreviations: COVID‐19, coronavirus disease 19; PAI‐1, plasminogen activator inhibitor 1; PAP, plasmin‐antiplasmin complex; sTM, soluble thrombomodulin; TAFI, thrombin‐activatable fibrinolysis inhibitor; TAFIa, activated thrombin‐activatable fibrinolysis inhibitor; TAT, thrombin‐antithrombin complex
FIGURE 1.
Biplot from Fisher's linear discriminant function, showing the three patient groups in two dimensions based on the baseline coagulopathy biomarkers. The first linear discriminant axis explains 77.49% of the variance of the centroids, on which D‐dimer is the highest contributor. PAI‐1, plasminogen activator inhibitor 1; PAP, plasmin‐antiplasmin complex; TAFI, thrombin‐activatable fibrinolysis inhibitor; TAFIa, activated thrombin‐activatable fibrinolysis inhibitor; sTM, soluble thrombomodulin; TAT, thrombin‐antithrombin complex
3.3. Biomarkers and survival
Mortality among COVID‐19(+) patients in our cohort was 50%, with follow‐up extending until hospital discharge. Patient characteristics in survivors and nonsurvivors are presented in Table S1. As expected, nonsurvivors had trends toward more comorbidities and more abnormal laboratory values and were more likely to require invasive ventilation and vasoactive medications.
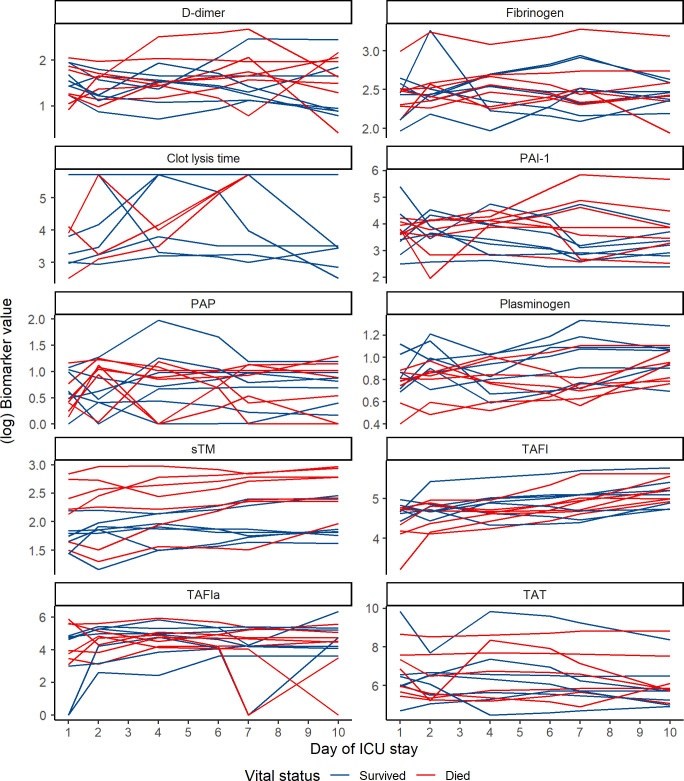
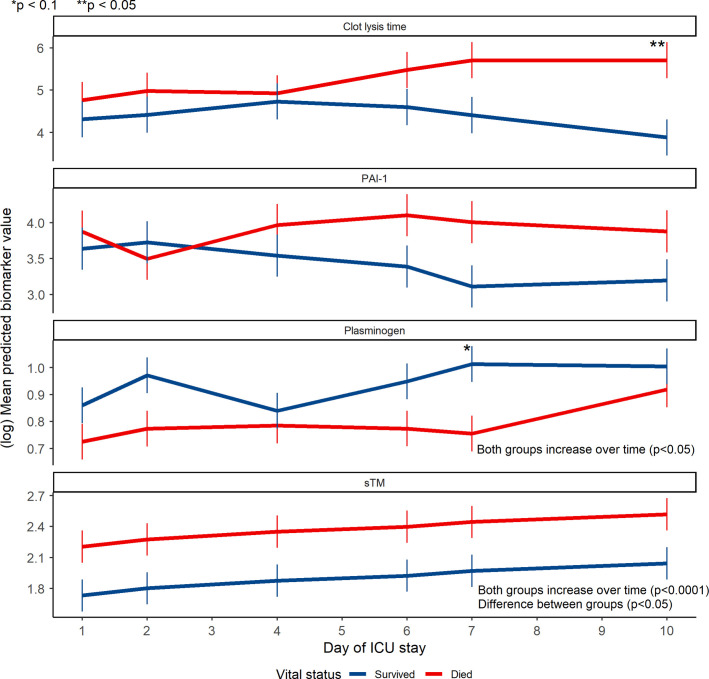
We explored the relationships between our 10 biomarkers of interest and survival. Individual trajectories for all biomarkers stratified by survivors and nonsurvivors are presented in Figure 2 . After variable screening, four biomarkers that had associations with death that met the p value cutoff of <.25: clot lysis time, sTM, PAI‐1, and plasminogen (Table S2). More complex repeated‐measures mixed models were specified for these biomarkers (Figure 3 ). At baseline, there was a significant difference between survivors and nonsurvivors in the mean sTM values (p = .0423). There was a significant temporal trend among all COVID‐19(+) patients for mean plasminogen (p = .0003) and sTM (p < .0001) values, with both biomarkers increasing over time. Individual interaction term parameters indicated there was significantly higher mean clot lysis time among nonsurvivors compared with survivors at day 10 (p = .0041). Although individual interactions were nonsignificant for day‐to‐day comparisons in plasminogen, the overall inclusion of an interaction was significant (p = .0495), suggesting the two groups had different trajectories.
FIGURE 2.
Individual biomarker trajectories of COVID‐19(+) patients, stratified by survivors and nonsurvivors. COVID‐19, coronavirus disease 19; PAI‐1, plasminogen activator inhibitor 1; PAP, plasmin‐antiplasmin complex; TAFI, thrombin‐activatable fibrinolysis inhibitor; TAFIa, activated thrombin‐activatable fibrinolysis inhibitor; sTM, soluble thrombomodulin; TAT, thrombin‐antithrombin complex
FIGURE 3.
Mean predicted trajectories of select biomarker in COVID‐19(+) survivors and nonsurvivors as determined by repeated measures linear mixed models. Error bars indicate standard errors. Differences between groups at individual time points were determined by interaction terms between day of measurement and vital status (*p < .1; **p < .05). Overall changes over time were determined by tests of fixed effects (ANOVA). The difference between groups for sTM was the main effect for vital status, with no interaction term included. COVID‐19, coronavirus disease 19; PAI‐1, plasminogen activator inhibitor 1; sTM, soluble thrombomodulin
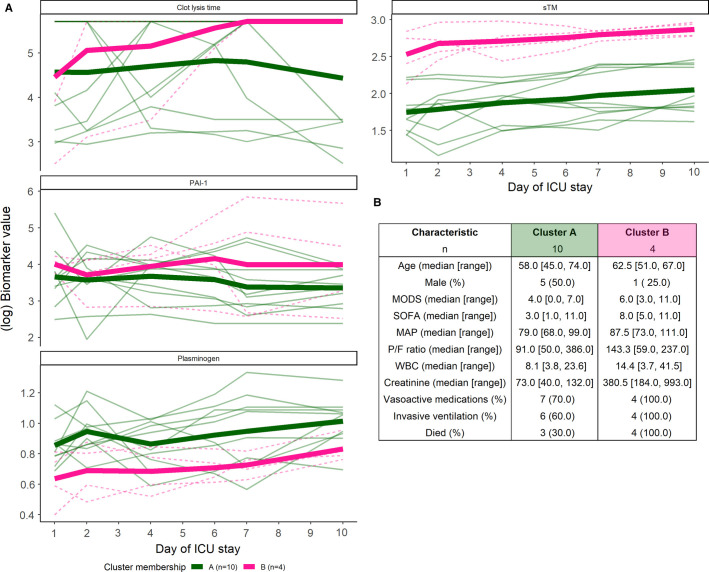
These four biomarkers were then entered into an unsupervised longitudinal k‐means clustering algorithm. This algorithm assigned the 14 COVID‐19(+) patients into two clusters based only on the joint trajectories of the biomarkers (Figure 4 ). One cluster represented low mortality (cluster A; 30% [3/10]), whereas the other cluster represented high mortality (cluster B; 100% [4/4]). Patients in cluster B also had trends toward higher SOFA scores, MODS white blood cell count, and creatinine. This is consistent with creatinine being identified as a mortality predictor using metabolomics analysis.34 More patients in cluster B required vasoactive medications and invasive ventilation. The biomarker trends that defined the severe disease/high mortality cluster were longer clot lysis times, higher levels of sTM and PAI‐1, and lower levels of plasminogen.
FIGURE 4.
(A) COVID‐19(+) patients clustered into two groups: cluster A (green, solid line) and cluster B (pink, dashed line) based on the selected biomarkers using a longitudinal k‐means clustering algorithm. Mean (bold lines) and individual trajectories are displayed. (B) Select patient characteristics in each cluster. COVID‐19, coronavirus disease 19; PAI‐1, plasminogen activator inhibitor 1; sTM, soluble thrombomodulin
4. DISCUSSION
This initial, hypothesis‐generating study is part of a larger effort to investigate the potential of markers of the coagulation and fibrinolytic systems to predict clinical courses and outcomes in critically ill COVID‐19(+) patients (COVID‐BEACONS [COVID‐19: Comprehensive biomarker analysis for prediction of clinical course and patient treatment outcomes]). To the best of our knowledge, our study using repeated measure modelling and unsupervised machine learning is the first of its kind to attempt to provide trajectory profiles of important biomarkers of coagulation and fibrinolysis in critically ill COVID‐19(+) patients that are associated with death. Similar analyses on endotheliopathy were reported by Fraser et al.35 using the same cohort. We report evidence of longitudinally increased coagulation, impaired fibrinolysis, and endothelial activation. Independent analyses of the various biomarkers suggest that there are differences in the baseline and time‐dependent trajectories of plasminogen, PAI‐1, sTM, and clot lysis times between survivors and nonsurvivors. These results are supported by our unsupervised longitudinal clustering algorithm, which similarly identified longer clot lysis times (i.e., inhibition of fibrinolysis), higher levels of PAI‐1 and sTM, and lower levels of plasminogen in clusters of COVID‐19(+) patients with higher mortality and signs of more severe disease.
sTM had the strongest longitudinal association with death in the ICU. Compared with those that survived, patients that died had elevated baseline and longitudinal sTM values. sTM levels increased for all COVID‐19 patients during their ICU stay. These data corroborate our current understanding of COVID‐19 disease pathophysiology that seemingly involves endothelial dysfunction, which is reflected by increased levels of (1) solubilized forms of membrane proteins such as thrombomodulin and syndecan‐1 and (2) von Willebrand factor, which is stored in Weibel‐Palade bodies of endothelial cells.35
Based on other reported studies that suggest inhibition of fibrinolysis,20., 25. we also hypothesize that inhibitors of fibrinolysis (such as PAI‐1 and α2‐antiplasmin) are involved. The implication is that although coagulation (as demonstrated by elevated TATs) and fibrinolysis are both increased, inhibition of fibrinolysis tips the balance toward overall thrombosis. In our cohort of COVID‐19(+) patients, we demonstrate that PAI‐1 levels are elevated, and the values are higher in those with worse outcomes, although this difference did not reach statistical significance. This is in agreement with the genomic analyses performed by Gill et al.,36 suggesting that transcription of SERPINE1, the gene for PAI‐1, is up‐regulated in this COVID‐19(+) patient cohort compared with COVID‐19(−) critically ill patients. Increased PAI‐1 levels are consistent with our functional findings whereby overall fibrinolysis measured by clot lysis times is impaired, particularly in the nonsurvivors. The impairment of clot breakdown in COVID‐19 could potentially reflect alterations in the clot structure because of components such as neutrophil extracellular traps or cell‐free DNA,50 which has been reported to possess antifibrinolytic properties.51 Taken together with other factors that may promote coagulation such as endothelial dysfunction and inflammation,35., 52., 53., 54. platelet hyperreactivity,14 and tissue factor expression in monocytes upon infection,14., 55. may all be involved in the overall thrombotic phenotype.
Other factors of fibrinolysis were investigated. The plasminogen level in the nonsurvivor group is 50% lower than that in the survivor group, suggesting consumption and/or activation of plasminogen may be involved in COVID‐19, which likely generates elevated D‐dimer levels. However, it is unclear why enhanced plasminogen activation (i.e., plasmin generation) would not be accompanied by elevated levels of the PAP complex. One explanation is that the PAP complex has a short half‐life (~0.5 days), which fails to capture acute plasminogen consumption. Similar disparities were observed between TAFI and TAFIa. Levels of TAFIa not mirroring early consumption of TAFI may also be due to short functional half‐life of TAFIa, which is ~7 min in vivo.56 Another potential mechanism is that neutrophil‐derived elastase is elevated in COVID‐19 patients,57., 58. which was also observed in this same cohort.34., 36. Elastase modifies plasminogen to generate mini‐plasminogen, a truncated form of plasminogen. Although activation of full‐length or mini‐plasminogen by tPA is comparable,58 inhibition of activated mini‐plasmin by α2‐antiplasmin is 100‐fold slower than the full‐length plasmin.59., 60. It is also possible that the PAP ELISA kit is not sensitive toward mini‐plasmin‐antiplasmin complex; however, this was not confirmed in this study.
Several studies to date have reported elevated levels of D‐dimer in COVID‐19(+) patients, particularly those with moderate to severe disease.19., 61., 62. We confirmed elevated D‐dimer levels in our cohort of COVID‐19(+) patients, which were significantly higher than the levels in both COVID‐19(−) critically ill patients and healthy controls. Although D‐dimer was the single largest identifier of COVID‐19(+) status, D‐dimer was not associated with death. Our findings suggest that D‐dimer lacks prognostic power to characterize the clinical course of patients with COVID‐19, which is consistent with the apparent discrepancy between elevated levels and the thrombotic phenotype of COVID‐19. These analyses also support earlier reports that suggest D‐dimer testing upon hospital admission may not be a reliable predictor of thrombotic complications or treatment outcomes, demonstrated by a modest sensitivity and specificity of ~85%.12., 19. D‐dimer level, however, was reported to decrease upon intensive prophylactic and therapeutic anticoagulation in COVID‐19 patients, which correlated with reduced need for mechanical ventilation, increased gas exchange, and improved 30‐day mortality outcome.63., 64. It is possible that with more rigorous and standardized reporting,65 along with longitudinal monitoring, D‐dimers may have more prognostic value.
Fibrinolytic factors such as plasminogen66., 67., 68. and uPA69 have been implicated in wound healing, whereas fibrinogen,70., 71. TAFI,72., 73., 74. plasminogen,75., 76., 77. and its receptors78., 79. have been implicated in inflammation. It is unclear how the altered fibrinolytic factor levels found in the critically ill COVID‐19 patients would affect the clinical course and outcome by impacting systems beyond coagulation and fibrinolysis. Increasing the sample size will provide additional mechanistic details, whereas further defining the potential prognostic value of other fibrinolytic factors such as TAFI, which was excluded in our exploratory analyses but was approaching the p value cutoff (p = .29). Analyses of additional cohorts will also be needed to validate our initial findings.
There are limitations to this study. The patient population consisted of a small cohort recruited from a single ICU relatively early in the COVID‐19 pandemic. This phase was characterized by concentrated outbreaks among vulnerable populations such as long‐term care residents, and ICU care was not yet standardized among COVID‐19 patients. This may be reflected in the unusually high mortality rate in this cohort. Therefore, it is unclear how our results will generalize to other patient populations. Furthermore, ELISAs may not be widely available because of cost, whereas carrying out clot lysis assays may require experienced technical staff, making quantification of these biomarkers challenging in a hospital setting. We attempted to address the small sample size by refraining from multivariable regression, and using a conservative p value cutoff for variable selection. Finally, although the results of this work have implications for the understanding of coagulopathy of COVID‐19, we did not directly measure thrombotic outcomes. This will be the focus of future work.
Taken together, our findings improve the mechanistic understanding of COVID‐19‐associated coagulopathy. If validated, our findings have the potential to make direct impact in COVID‐19 prognostication by identifying patients that are at greater risk of decompensation based on the joint trajectories of key biomarkers of fibrinolysis and endothelial dysfunction.
CONFLICT OF INTEREST
COVID‐BEACONS investigators include Paul Y. Kim, Calvin H. Yeh, Matthew Castelo, Bettina Hansen, Bernardo Trigatti, Jeffrey I. Weitz, Patricia C. Liaw, Alison Fox‐Robichaud, Peter L. Gross, Geoff Werstuck, Colin A. Kretz, Keyvan Karkouti, Stuart McCluskey, and Claudia dos Santos. Dr. Weitz has received personal fees from Bayer, Boehringer Ingelheim, Bristol‐Myers Squibb, Daiichi‐Sankyo, Ionis Pharmaceuticals, Janssen, Merck, Novartis, Pfizer, and Portola, outside the submitted work. All other authors declare that they have no conflicts of interest with the contents of this article.
AUTHOR CONTRIBUTIONS
Douglas D. Fraser, Marat Slessarev, and Claudio Martin collected the patient samples and associated deidentified clinical data. Calvin H. Yeh, Scott McGilvray, Peter L. Gross, Patricia C. Liaw, Jeffrey I. Weitz, and Paul Y. Kim designed the experiments. Ganeem K. Juneja, Samantha E. Cerroni, James E. Chessum, Joel Abraham, Erblin Cani, and Douglas D. Fraser generated the data. Matthew Castelo, Bettina E. Hansen, and Paul Y. Kim analyzed the data.
ACKNOWLEDGMENTS
The authors thank the patients and their families who have been affected in the pandemic. The tireless efforts of frontline medical and support workers must be commended; in particular, the daily lifesaving services provided by those in food and environmental services, personal support workers, clinical support services, paramedics, nurses, and allied health. P. Y. K. is supported by the Department of Medicine Career Award (McMaster University). J. I. W. holds the Canada Research Chair (Tier I) in Thrombosis and the Heart and Stroke Foundation J. Fraser Mustard Chair in Cardiovascular Research at McMaster University and is supported by a Canadian Institutes of Health Research Foundation Grant. This work is partly supported by the Canadian Institutes of Health Research COVID‐19 Rapid Response (VR2‐172768). D. D. F. received funding from Western University (Research), the Department of Medicine and Department of Pediatrics at Western University, the Lawson Health Research Institute (https://www.lawsonresearch.ca/), the London Health Sciences Foundation (https://lhsf.ca/), and the AMOSO Innovation Fund.
Canadian Institutes of Health Research FoundationVR2‐172768
Western University
Lawson Health Research Institute
London Health Sciences Foundation
AMOSO Innovation Fund
Footnotes
Ganeem K. Juneja and Matthew Castelo contributed equally to this study.
Manuscript handled by: Jean Connors.
Supporting Information
Table S1‐S2
REFERENCES
- 1.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID‐19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2.Organization WH. Coronavirus disease (COVID‐19). 2020.
- 3.Clausen T.M., Sandoval D.R., Spliid C.B., et al. SARS‐CoV‐2 infection depends on cellular heparan sulfate and ACE2. bioRxiv Prepr Serv Biol. 2020 doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connors J.M., Levy J.H. Thromboinflammation and the hypercoagulability of COVID‐19. J Thromb Haemost. 2020;18(7):1559–1561. doi: 10.1111/jth.14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine‐Weber H., Schroeder S., et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malha L., Mueller F.B., Pecker M.S., et al. COVID‐19 and the renin‐angiotensin system. Kidney Int Rep. 2020;5(5):563–565. doi: 10.1016/j.ekir.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gattinoni L., Coppola S., Cressoni M., et al. COVID‐19 does not lead to a “typical” acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;201(10):1299–1300. doi: 10.1164/rccm.202003-0817LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Idell S. Coagulation, fibrinolysis, and fibrin deposition in acute lung injury. Crit Care Med. 2003;31(4 Suppl):S213–S220. doi: 10.1097/01.CCM.0000057846.21303.AB. [DOI] [PubMed] [Google Scholar]
- 10.Yuki K., Fujiogi M., Koutsogiannaki S. COVID‐19 pathophysiology: a review. Clin Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Zhou P., Wei Y., et al. Histopathologic changes and SARS‐CoV‐2 immunostaining in the lung of a patient with COVID‐19. Ann Intern Med. 2020;172(9):629–632. doi: 10.7326/M20-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui S., Chen S., Li X., Liu S., Wang F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J Thromb Haemost. 2020;18(6):1421–1424. doi: 10.1111/jth.14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klok F.A., Kruip M.J.H.A., van der Meer N.J.M., et al. Incidence of thrombotic complications in critically ill ICU patients with COVID‐19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manne B.K., Denorme F., Middleton E.A., et al. Platelet gene expression and function in patients with COVID‐19. Blood. 2020;136(11):1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miesbach W., Makris M. COVID‐19: Coagulopathy, risk of thrombosis, and the rationale for anticoagulation. Clin Appl Thromb. 2020;26 doi: 10.1177/1076029620938149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rali P., O'Corragain O., Oresanya L., et al. Incidence of venous thromboembolism in coronavirus disease 2019: an experience from a single large academic center. J Vasc Surg Venous Lymphat Disord. 2020;9(3):585–591. doi: 10.1016/j.jvsv.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Xiao M., Zhang S., et al. Coagulopathy and antiphospholipid antibodies in patients with Covid‐19. N Engl J Med. 2020;382(17) doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh C.H., de Wit K., Levy J.H., et al. Hypercoagulability and COVID‐19 associated hypoxemic respiratory failure: mechanisms and emerging management paradigms. J Trauma Acute Care Surg. 2020;89(6) doi: 10.1097/TA.0000000000002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu B., Li X., Chen J., et al. Evaluation of variation in D‐dimer levels among COVID‐19 and bacterial pneumonia: a retrospective analysis. J Thromb Thrombolysis. 2020;50(3):548–557. doi: 10.1007/s11239-020-02171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nougier C., Benoit R., Simon M., et al. Hypofibrinolytic state and high thrombin generation may play a major role in SARS‐COV2 associated thrombosis. J Thromb Haemost. 2020;18(9):2215–2219. doi: 10.1111/jth.15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seheult J.N., Seshadri A., Neal M.D. Fibrinolysis shutdown and thrombosis in severe COVID‐19. J Am Coll Surg. 2020;231(2):203–204. doi: 10.1016/j.jamcollsurg.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright F.L., Vogler T.O., Moore E.E., et al. Fibrinolysis shutdown correlation with thromboembolic events in severe COVID‐19 infection. J Am Coll Surg. 2020;231(2):193–203. doi: 10.1016/j.jamcollsurg.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadd C., Rowe T., Nazeef M., et al. Thromboelastography to detect hypercoagulability and reduced fibrinolysis in coronavirus disease 2019 acute respiratory distress syndrome patients. Crit Care Explor. 2020;2(9) doi: 10.1097/CCE.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z., Shi L., Wang Y., et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo Y., Warnock M., Harbaugh A., et al. Plasma tissue plasminogen activator and plasminogen activator inhibitor‐1 in hospitalized COVID‐19 patients. medRxiv. 2020 doi: 10.1038/s41598-020-80010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creel‐Bulos C, Auld SC, Caridi‐Scheible M, et al. Fibrinolysis shutdown and thrombosis in a COVID‐19 ICU. Shock. 55(3):316‐320. [DOI] [PMC free article] [PubMed]
- 27.Bakchoul T., Hammer S., Lang P., Rosenberger P. Fibrinolysis shut down in COVID‐19 patients: report on two severe cases with potential diagnostic and clinical relevance. Thromb Updat. 2020;1 doi: 10.1016/j.tru.2020.100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goshua G., Pine A.B., Meizlish M.L., et al. Endotheliopathy in COVID‐19‐associated coagulopathy: evidence from a single‐centre, cross‐sectional study. Lancet Haematol. 2020;7(8) doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouck E.G., Denorme F., Holle L.A., et al. COVID‐19 and sepsis are associated with different abnormalities in plasma procoagulant and fibrinolytic activity. Arterioscler Thromb Vasc Biol. 2020;41(1):401–414. doi: 10.1161/ATVBAHA.120.315338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao D., Zhou F., Luo L., et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID‐19: a retrospective cohort study. Lancet Haematol. 2020;7(9) doi: 10.1016/S2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser D.D., Cepinskas G., Patterson E.K., et al. Novel outcome biomarkers identified with targeted proteomic analyses of plasma from critically Ill coronavirus disease 2019 patients. Crit Care Explor. 2020;2(9) doi: 10.1097/CCE.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser D.D., Cepinskas G., Slessarev M., et al. Inflammation profiling of critically Ill coronavirus disease 2019 patients. Crit Care Explor. 2020;2(6) doi: 10.1097/CCE.0000000000000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser D.D., Slessarev M., Martin C.M., et al. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit Care Explor. 2020;2(10) doi: 10.1097/CCE.0000000000000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser D.D., Patterson E.K., Slessarev M., et al. Endothelial injury and glycocalyx degradation in critically Ill coronavirus disease 2019 patients: implications for microvascular platelet aggregation. Crit Care Explor. 2020;2(9) doi: 10.1097/CCE.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gill S.E., Dos Santos C.C., O’Gorman D.B., et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive care Med Exp. 2020;8(1):75. doi: 10.1186/s40635-020-00361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Elm E., Altman D.G., Egger M., et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370(9596):1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 38.Centers for Disease Control and Prevention. Overview of testing for SARS‐CoV‐2 (COVID‐19). 2020.
- 39.Brisson A.R., Matsui D., Rieder M.J., Fraser D.D. Translational research in pediatrics: tissue sampling and biobanking. Pediatrics. 2012;129(1):153–162. doi: 10.1542/peds.2011-0134. [DOI] [PubMed] [Google Scholar]
- 40.Gillio‐Meina C., Cepinskas G., Cecchini E.L., Fraser D.D. Translational research in pediatrics II: blood collection, processing, shipping, and storage. Pediatrics. 2013;131(4):754–766. doi: 10.1542/peds.2012-1181. [DOI] [PubMed] [Google Scholar]
- 41.Singer M., Deutschman C.S., Seymour C.W., et al. The Third International Consensus definitions for sepsis and septic shock (Sepsis‐3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim P.Y., Foley J., Hsu G., Kim P.Y., Nesheim M.E. An assay for measuring functional activated thrombin‐activatable fibrinolysis inhibitor in plasma. Anal Biochem. 2008;372(1):32–40. doi: 10.1016/j.ab.2007.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Kim P.Y., Stewart R.J., Lipson S.M., Nesheim M.E. The relative kinetics of clotting and lysis provide a biochemical rationale for the correlation between elevated fibrinogen and cardiovascular disease. J Thromb Haemost. 2007;5(6):1250–1256. doi: 10.1111/j.1538-7836.2007.02426.x. [DOI] [PubMed] [Google Scholar]
- 44.Krzanowski W.J. The performance of fisher’s linear discriminant function under non‐optimal conditions. Technometrics. 1977;19(2):191–200. [Google Scholar]
- 45.Chowdhury M.Z.I., Turin T.C. Variable selection strategies and its importance in clinical prediction modelling. Fam Med Community Heal. 2020;8(1) doi: 10.1136/fmch-2019-000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genolini C., Falissard B. KmL: k‐means for longitudinal data. Comput Stat. 2010;25(2):317–328. [Google Scholar]
- 47.Genolini C, Alacoque X, Sentenac M, Arnaud C. kml and kml3d: R packages to cluster longitudinal data. J. Stat. Software; Vol 1, Issue 4 . 2015;
- 48.Caliński T., Harabasz J. A dendrite method for cluster analysis. Commun Stat. 1974;3(1):1–27. [Google Scholar]
- 49.Bender R., Lange S. Adjusting for multiple testing–when and how? J Clin Epidemiol. 2001;54(4):343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 50.Leppkes M., Knopf J., Naschberger E., et al. Vascular occlusion by neutrophil extracellular traps in COVID‐19. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gould T.J., Vu T.T., Stafford A.R., et al. Cell‐free DNA modulates clot structure and impairs fibrinolysis in sepsis. Arterioscler Thromb Vasc Biol. 2015;35(12):2544–2553. doi: 10.1161/ATVBAHA.115.306035. [DOI] [PubMed] [Google Scholar]
- 52.Levi M., van der Poll T. Two‐way interactions between inflammation and coagulation. Trends Cardiovasc Med. 2005;15(7):254–259. doi: 10.1016/j.tcm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Nicolai L., Leunig A., Brambs S., et al. Immunothrombotic dysregulation in COVID‐19 pneumonia is associated with respiratory failure and coagulopathy. Circulation. 2020;142(12):1176–1189. doi: 10.1161/CIRCULATIONAHA.120.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bikdeli B., Madhavan M.V., Jimenez D., et al. COVID‐19 and thrombotic or thromboembolic disease: implications for prevention, antithrombotic therapy, and follow‐up: JACC state‐of‐the‐art review. J Am Coll Cardiol. 2020;75(23):2950–2973. doi: 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaid Y., Puhm F., Allaeys I., et al. Platelets can associate with SARS‐Cov‐2 RNA and are hyperactivated in COVID‐19. Circ Res. 2020;127(11):1404–1418. doi: 10.1161/CIRCRESAHA.120.317703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Foley J.H., Kim P.Y., Mutch N.J., Gils A. Insights into thrombin activatable fibrinolysis inhibitor function and regulation. J Thromb Haemost. 2013;11(Suppl 1):306–315. doi: 10.1111/jth.12216. [DOI] [PubMed] [Google Scholar]
- 57.Wu H.L., Chang B.I., Wu D.H., et al. Interaction of plasminogen and fibrin in plasminogen activation. J Biol Chem. 1990;265:19658–19664. [PubMed] [Google Scholar]
- 58.Kim P.Y., Tieu L.D., Stafford A.R., Fredenburgh J.C., Weitz J.I. A high affinity interaction of plasminogen with fibrin is not essential for efficient activation by tissue‐type plasminogen activator. J Biol Chem. 2012;287(7):4652–4661. doi: 10.1074/jbc.M111.317719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiman B., Collen D. On the kinetics of the reaction between human antiplasmin and plasmin. Eur J Biochem. 1978;84:573–578. doi: 10.1111/j.1432-1033.1978.tb12200.x. [DOI] [PubMed] [Google Scholar]
- 60.Schaller J., Gerber S.S. The plasmin‐antiplasmin system: structural and functional aspects. Cell Mol Life Sci. 2011;68(5):785–801. doi: 10.1007/s00018-010-0566-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in china: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323(13):1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 63.Hsu A., Liu Y., Zayac A.S., Olszewski A.J., Reagan J.L. Intensity of anticoagulation and survival in patients hospitalized with COVID‐19 pneumonia. Thromb Res. 2020;196:375–378. doi: 10.1016/j.thromres.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemos A.C.B., do Espírito Santo D.A., Salvetti M.C., et al. Therapeutic versus prophylactic anticoagulation for severe COVID‐19: a randomized phase II clinical trial (HESACOVID) Thromb Res. 2020;196:359–366. doi: 10.1016/j.thromres.2020.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thachil J., Longstaff C., Favaloro E.J., et al. The need for accurate D‐dimer reporting in COVID‐19: communication from the ISTH SSC on fibrinolysis. J Thromb Haemost. 2020;18(9):2408–2411. doi: 10.1111/jth.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Romer J., Bugge T.H., Pyke C., et al. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2(3):287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 67.Rømer J., Bugge T.H., Pyke C., et al. Plasminogen and wound healing. Nat Med. 1996;2(7):725. doi: 10.1038/nm0796-725a. [DOI] [PubMed] [Google Scholar]
- 68.Sulniute R., Shen Y., Guo Y.Z., et al. Plasminogen is a critical regulator of cutaneous wound healing. Thromb Haemost. 2016;115(05):1001–1009. doi: 10.1160/TH15-08-0653. [DOI] [PubMed] [Google Scholar]
- 69.Madhyastha H.K., Radha K.S., Nakajima Y., Omura S., Maruyama M. uPA dependent and independent mechanisms of wound healing by C‐phycocyanin. J Cell Mol Med. 2008;12(6B):2691–2703. doi: 10.1111/j.1582-4934.2008.00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flick M.J., Du X., Witte D.P., et al. Leukocyte engagement of fibrin(ogen) via the integrin receptor αMβ2/Mac‐1 is critical for host inflammatory response in vivo. J Clin Invest. 2004;113(11):1596–1606. doi: 10.1172/JCI20741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luyendyk J.P., Schoenecker J.G., Flick M.J. The multifaceted role of fibrinogen in tissue injury and inflammation. Blood. 2019;133(6):511–520. doi: 10.1182/blood-2018-07-818211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell W.D., Lazoura E., Okada N., Okada H. Inactivation of C3a and C5a octapeptides by carboxypeptidase R and carboxypeptidase N. Microbiol Immunol. 2002;46(2):131–134. doi: 10.1111/j.1348-0421.2002.tb02669.x. [DOI] [PubMed] [Google Scholar]
- 73.Myles T., Nishimura T., Yun T.H., et al. Thrombin activatable fibrinolysis inhibitor, a potential regulator of vascular inflammation. J Biol Chem. 2003;278(51):51059–51067. doi: 10.1074/jbc.M306977200. [DOI] [PubMed] [Google Scholar]
- 74.Shinohara T., Sakurada C., Suzuki T., et al. Pro‐carboxypeptidase R cleaves bradykinin following activation. Int Arch Allergy Immunol. 2004;103(4):400–404. doi: 10.1159/000236661. [DOI] [PubMed] [Google Scholar]
- 75.Sugimoto M.A., Ribeiro A.L.C., Costa B.R.C., et al. Plasmin and plasminogen induce macrophage reprogramming and regulate key steps of inflammation resolution via annexin A1. Blood. 2017;129(21):2896–2907. doi: 10.1182/blood-2016-09-742825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baker S.K., Chen Z.‐.L., Norris E.H., et al. Blood‐derived plasminogen drives brain inflammation and plaque deposition in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2018;115(41) doi: 10.1073/pnas.1811172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silva L.M., Lum A.G., Tran C., et al. Plasmin‐mediated fibrinolysis enables macrophage migration in a murine model of inflammation. Blood. 2019;134(3):291–303. doi: 10.1182/blood.2018874859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Godier A., Hunt B.J. Plasminogen receptors and their role in the pathogenesis of inflammatory, autoimmune and malignant disease. J Thromb Haemost. 2013;11(1):26–34. doi: 10.1111/jth.12064. [DOI] [PubMed] [Google Scholar]
- 79.Miles L.A., Baik N., Lighvani S., et al. Deficiency of plasminogen receptor, Plg‐R(KT), causes defects in plasminogen binding and inflammatory macrophage recruitment in vivo. J Thromb Haemost. 2017;15(1):155–162. doi: 10.1111/jth.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S2