Summary
Severe acute respiratory syndrome related coronavirus‐2 (SARS‐CoV‐2) is the cause of Covid‐19 which was classified as a global pandemic in March 2020. The increasing global health and economic burden of SARS‐CoV‐2 has necessitated urgent investigations into the pathogenesis of disease and development of therapeutic and vaccination regimens. Human trials of vaccine and antiviral candidates have been undertaken, but basic pathogenetic studies are still required to inform these trials. Gaps in understanding of cellular infection by, and immunity to, SARS‐CoV‐2 mean additional models are required to assist in improved design of these therapeutics. Human organoids are three‐dimensional models that contain multiple cell types and mimic human organs in ex vivo culture conditions. The SARS‐CoV‐2 virus has been implicated in causing not only respiratory injury but also injury to other organs such as the brain, liver and kidneys. Consequently, a variety of different organoid models have been employed to investigate the pathogenic mechanisms of disease due to SARS‐CoV‐2. Data on these models have not been systematically assembled. In this review, we highlight key findings from studies that have utilised different human organoid types to investigate the expression of SARS‐CoV‐2 receptors, permissiveness, immune response, dysregulation of cellular functions, and potential antiviral therapeutics.
Keywords: antivirals, Covid‐19, immune response, organoids, pathogenesis, SARS‐CoV‐2
Abbreviations
- ACE2
angiotensin I converting enzyme 2
- AD
Alzheimer's disease
- AKI
acute kidney injury
- ARDS
acute respiratory distress syndrome
- AT1
alveolar type I
- AT2
alveolar type II
- Aβ
β‐amyloid
- CFTR
cystic fibrosis transmembrane conductance regulator
- CLDN1
Claudin 1
- Covid‐19
coronavirus disease 2019
- Dpi,
days post‐infection
- GBS
Guillain–Barre syndrome
- Hpi
hours post‐infection
- hPSC
human pluripotent stem cells
- hrsACE2
human recombinant soluble ACE2
- HSV‐1
herpes simplex virus type 1
- IFN
interferon
- iPSC
induced pluripotent stem cells
- ISGs
interferon‐stimulated genes
- MERS‐CoV
Middle Eastern respiratory syndrome related coronavirus
- MPA
mycophenolic acid
- NPCs
neural progenitor cells
- QNHC
quinacrine dihydrochloride
- SARS‐CoV
severe acute respiratory syndrome related coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome related coronavirus‐2
- SEAM
self‐formed ectodermal autonomous multi‐zone
- SLC10A2
solute carrier family 10 member 2
- TMPRSS2
Transmembrane serine protease 2
- WHO
World Health Organisation
- ZIKV
Zika virus
1. INTRODUCTION
Severe acute respiratory syndrome related coronavirus‐2 (SARS‐CoV‐2) is a member of the Coronaviridae family and is a single‐stranded positive‐sense enveloped RNA virus. 1 , 2 Human coronaviruses are one of the leading causes of the common cold and tend to result in mild respiratory tract symptoms, although recent data suggest they can cause more severe disease. 3 Novel human coronaviruses have emerged over the last two decades including severe acute respiratory syndrome related coronavirus (SARS‐CoV), the Middle Eastern respiratory syndrome related coronavirus (MERS‐CoV), and recently SARS‐CoV‐2. 2 , 4 , 5 Covid‐19 is caused by SARS‐CoV‐2 infection and was classified as a pandemic by the World Health Organisation (WHO) in March 2020. 6 The symptoms of Covid‐19 range widely, presenting as asymptomatic in many individuals or fever, myalgia, cough, chest tightness, and in severe cases pneumonia, occasionally resulting in death. 7
The current pandemic has developed globally rapidly, meaning there is urgency around studies of the pathogenetic mechanisms of SARS‐CoV‐2 infection to inform development of antiviral drugs and vaccines. A key method of investigating pathogenesis of infection, assisting design and development of therapeutics, is the employment of three‐dimensional, multi‐cell‐type culture models known as organoids. These allow (i) investigation in a human, biologically relevant model, (ii) utilisation of multicellular models to demonstrate the effects of infection and treatment on different cell lines, and (iii) dissection of the molecular response of individual cells within a human organ.
1.1. Overview of traditional models for disease pathogenesis research
Two‐dimensional cell monocultures and animal models are frequently used to model human viral infections. Monocultures are the primary model used in the isolation and propagation of viruses, including the first culture of SARS‐CoV‐2 in the Vero/hSLAM cell line. 8 However, monocultures do not accurately represent the complexity of in vivo tissues. Different cell types in organs, such as the brain, have specific gene regulation patterns which would otherwise not be taken into account in single‐cell culture models. 9 , 10 This has been further confirmed by reported alterations in cell signalling networks between two‐ and three‐dimensional culture systems. 11 , 12 In order to overcome these limitations, researchers have begun utilising alternative three‐dimensional disease modelling systems, such as organoids. These more closely mimic the complex multi‐cell‐type composition and structure of human organs. 13 , 14
Animal models are multi‐cell‐type, multi‐organ models that facilitate the translation from basic research to clinical trials. Animal models are essential in informing the development of antiviral treatments and vaccines and have recently been used to evaluate Covid‐19 vaccine candidates in models such as Rhesus macaques. 15 , 16 , 17 However, the differing developmental patterns between non‐human animals and humans 18 have necessitated the development of alternative human organ models such as organoids. Furthermore, the ethical considerations involved in animal research are monitored through the 3Rs principle (replacement, reduction and refinement of animal research) in an effort to maximise high‐quality data while minimising harm to animals during research. 19 This principle dictates the replacement of animal models where possible through the use of alternative culture systems such as human organoids. 20
1.2. Organoid generation and use in modelling viral infection
Organoids are organ‐like tissue models from pluripotent stem cells or progenitor cells through differentiation. 13 During the differentiation process, multiple cell types arise and self‐organize to form cellular organisation and tissue morphology resembling in vivo human organs, which can be used to model infection. 13 , 14
Organoids have successfully been used to model neurological infections such as with Zika virus (ZIKV). 21 Congenital ZIKV infection has been associated with the development of foetal neural malformation such as microcephaly. 22 Researchers have utilised cerebral organoids to study pathogenesis of ZIKV infection, including brain‐region‐specific forebrain organoids. 21 In this model, ZIKV induced increased cell death and reduced cellular proliferation, subsequently decreasing neuronal volume, inducing microcephaly‐like pathophysiology, 21 suggesting a mechanism for ZIKV‐induced neural malformation.
Different organoids have been utilised in SARS‐CoV‐2 mechanistic investigations. The respiratory tract is the major organ system affected by Covid‐19 disease, although injury to other organs has been observed including acute kidney injury, cardiovascular disease and neurological disease including encephalopathy, encephalitis, Guillain–Barre syndrome and acute stroke. 23 , 24 The wide array of organs affected in Covid‐19 disease necessitates the use of a variety of organoid models to extrapolate the mechanisms of SARS‐CoV‐2 infection in different systems. Specific growth and differentiation factors that facilitate the development and generation of a variety of organoids have been used to produce organoids simulating different human organs, allowing a biologically relevant, three‐dimensional model for disease. 13 , 14 In this review, we detail the conclusions drawn from 16 studies investigating SARS‐CoV‐2 infection of different human organoid models.
2. SARS‐CoV‐2 AND ORGANOIDS
2.1. Expression of SARS‐CoV‐2 receptors in organoids
The SARS‐CoV‐2 spike (S) protein, which is structurally similar to that of SARS‐CoV, mediates viral attachment and cellular entry through the binding of the N‐terminal S1 subunit to the metallopeptidase, angiotensin I converting enzyme 2 (ACE2) receptor (Figure 1). 25 , 26 , 27 Interestingly, SARS‐CoV‐2 binds to the ACE2 receptor with a higher affinity than does SARS‐CoV. 25 Hoffmann et al. suggest the SARS‐CoV‐2 S protein is primed by the transmembrane serine protease 2 (TMPRSS2), inducing virus‐host cell fusion, similar to SARS‐CoV (Figure 1). 26 Using single‐cell RNA sequencing datasets, several studies have determined the ACE2 and TMPRSS2 receptors are expressed in the human lung, eye, heart, oesophagus, ileum, kidney, colon, liver, gallbladder and testis (pre‐print). 28 , 29 , 30 Investigating the presence of these host cell receptors in various organoid models is an important first step in determining their ability to support viral infection. The detection of these receptors in various organoids is summarised in Figure 1.
FIGURE 1.
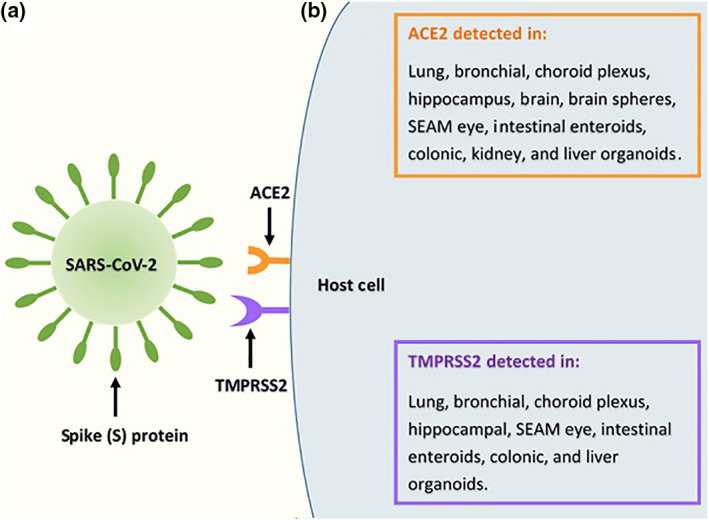
Severe acute respiratory syndrome related coronavirus‐2 (SARS‐CoV‐2) and host cell receptors. (a) Viral attachment and cellular entry is mediated by the SARS‐CoV‐2 spike (S) protein. The N‐terminal subunit, S1 binds to the receptor, angiotensin I converting enzyme 2 (ACE2). 25 , 26 , 27 Hoffmann et al. suggest the S protein is primed by the transmembrane serine protease 2 (TMPRSS2), inducing host cell fusion. 26 (b) Using single cell RNA sequencing datasets, various human organs have been identified as expressing the target receptors for SARS‐CoV‐2 infection, ACE2 and TMPRSS2. 28 , 29 , 30 Subsequently, numerous organoid models have been utilised to detect ACE2 and TMPRSS2. 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43
In organoids, the ACE2 receptor was detected in lung (pre‐print), 31 , 32 bronchial (pre‐print), 33 the choroid plexus, 34 hippocampus, 34 brain, 35 self‐formed ectodermal autonomous multi‐zone (SEAM) eye (pre‐print), 36 intestinal enteroids, 37 , 38 colonic, 32 kidney 39 , 40 and liver 41 , 42 organoids, as well as brain spheres. 43 Interestingly, ACE2 was also expressed in multipotent neural progenitor cells (NPCs), from which brain spheres are generated through differentiation. 43 During foetal cerebral organogenesis, NPCs are a crucial population of cells, with the capability of differentiating into oligodendrocytes, astrocytes and neurons that populate the developed adult brain. 44 , 45 The presence of the ACE2 receptor in this population of cells suggests SARS‐CoV‐2 tropism, and further research is required to determine the effects of infection on foetal brain development. However, most guidelines suggest effects on the foetus are unlikely (UK guidelines). 24 , 46
The TMPRSS2 receptor was expressed in lung (pre‐print), 31 , 32 bronchial (pre‐print), 33 choroid plexus, 34 hippocampal, 34 SEAM eye (pre‐print), 36 intestinal enteroid, 37 colonic 32 and liver 41 organoids. However, TMPRSS2 receptor expression was reportedly below the limit of detection in brain spheres. 43
Expression of these target cellular receptors localised in certain cell populations in organoids. In lung organoids, ACE2 was detected predominantly in club 31 and alveolar type II (AT2)‐like cells (pre‐print). 31 , 32 Interestingly, in alveolar type I (AT1) and AT2‐like cultures, SARS‐CoV‐2 preferentially infected AT2 cells over AT1 cells, 47 which may be explained by the reported predominant detection of ACE2 31 , 32 in the AT2 population of cells. Enrichment of TMPRSS2 was reported in AT2‐like 31 , 32 and club 31 cells in lung organoids (pre‐print). Furthermore, results from Suzuki et al. suggested both ACE2 and TMPRSS2 are expressed in the basal cells of bronchial organoids, but only TMPRSS2 is expressed in ciliated cells (pre‐print). 33
Single‐cell RNA sequencing of SEAM eye organoids revealed ACE2 may be predominantly expressed in the limbus, conjunctiva and a subset of ocular surface ectoderm cells (pre‐print). 36 The TMPRSS2 receptor was expressed in corneal cells in SEAM eye organoids (pre‐print). 36 Single‐cell profiling of kidney organoids revealed the proximal tubule and podocyte II cell clusters express ACE2. 39
Organoids have allowed researchers to identify specific cell populations that express SARS‐CoV‐2 receptors in the lung, 31 , 32 , 33 eye 36 and kidney. 39 Understanding the localisation and enrichment of host cell receptors that are utilised by SARS‐CoV‐2 during infection not only indicates the cell types that are more permissive to infection in specific organs, but also can potentially inform development of targeted therapeutics to these vulnerable cell populations.
Although organoids have facilitated the detection of target cell receptors in different organ types, this alone is not sufficient to determine the organ's permissiveness to SARS‐CoV‐2 infection, nor the organs' ability to support viral replication. Yang et al. reported that despite the expression of ACE2 in human pluripotent stem‐cell‐derived macrophages, cortical neurons and endothelial cells, they had little to no permissiveness to SARS‐CoV‐2 or SARS‐CoV‐2 pseudo‐entry virus, suggesting other factors are involved in determining viral tropism. 42 Therefore, supplementing viral receptor studies with those specifically investigating the tropism of different organoids are necessary to ascertain their effectiveness as models for SARS‐CoV‐2 infection.
2.2. Permissiveness of organoids to SARS‐CoV‐2 infection
In a study using autopsy tissue samples from patients who had died from Covid‐19, SARS‐CoV‐2 was detected in the lungs, pharynx, brain, heart, liver and kidneys, suggesting viral tropism in these organs. 48 Additionally, monocultures have been utilised to model and establish SARS‐CoV‐2 tropism in cell lines representative of different organ types including Caco2 (intestinal epithelial carcinoma), Calu3 (lung epithelial adenocarcinoma) and U251 (glioblastoma). 49
Studies investigating the permissiveness of different types of organoids to SARS‐CoV‐2 infection are detailed in Table 1. 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 41 , 42 , 43 , 50 , 51 , 52
TABLE 1.
Permissiveness to SARS‐CoV‐2 infection in various organoid models
| Organoid type | Source | SARS‐CoV‐2 strain | Permissiveness |
|---|---|---|---|
| Lung | Human pluripotent stem cells (hPSC) |
|
Permissive and supported robust viral replication. 32 |
| |||
| Distal lung epithelial cells and MRC5 human lung fibroblast cells |
|
Intact alveolar organoids were refractory to viral infection. Gentle physical and enzymatic disruption to organoids made them permissive to infection and viral replication (pre‐print). 50 | |
| Distal airway cells from patient lung tissue |
|
Permissive to infection. Organoids required everting so cells would be relocated and the ACE2 receptor would face outward (pre‐print). 31 | |
| Normal human bronchial epithelial cells |
|
Permissive to infection and viral replication (pre‐print). 33 | |
| Brain spheres and cerebral organoids | Induced pluripotent stem cells (iPSC) |
|
Permissive to infection. Small fraction of neural cells contained viral particles. Increased viral RNA indicative of replication. 43 |
| iPSC |
|
Choroid plexus organoids are permissive to productive infection. 34 | |
| iPSC |
|
Brain organoids are permissive to infection but do not support active viral replication. 35 | |
| SEAM eye organoid | Human embryonic stem cells (hESC) |
|
Permissive to infection (pre‐print). 36 |
| Liver | iPSC |
|
Liver hepatocyte organoids. Permissive in ALB + hepatocytes and supported robust replication. 42 |
| |||
| Liver bile duct‐derived progenitor cells |
|
Liver ductal organoids. Permissive and supported robust replication. Infected cholangiocytes formed syncytia. 41 | |
| hPSC |
|
Cholangiocyte organoids. Permissive in CK19+ cholangiocytes and supported robust replication. 42 | |
| |||
| Enteroids/Intestinal organoids | Intestinal samples from patients |
|
Enteroids are permissive to infection. Most infected cells were Villin+, indicating enterocytes are the predominant target cells for infection. 37 |
| Patient tissue |
|
Duodenal organoids are permissive to infection. 38 | |
| |||
| Patient tissue |
|
Ileum‐derived organoids are permissive to infection and support robust viral replication. 38 | |
| |||
| Patient tissue |
|
Colon‐derived organoids are permissive and support viral replication. 38 | |
| Patient tissue |
|
Small intestinal organoids are permissive and support productive infection. Viral particles detected in the lumen of the organoid. 51 | |
| Stem cells isolated from human tissue |
|
Permissive to infection, supports viral replication, and de novo infectious virus production. 52 | |
| Colonoids/Colonic organoids | Colonic sample from patient |
|
Permissive to infection and robust viral replication. 30 |
| hPSC |
|
Permissive. 32 | |
| Capillary | iPSC |
|
Permissive to active viral replication. 39 |
| Kidney | hESC |
|
Permissive to viral replication. 39 |
Note: Table summarising the tropism of different strains of SARS‐CoV‐2 in various organoid types as well as their ability to support viral replication when specified.
Abbreviations: ACE2, angiotensin I converting enzyme 2; hESC, human embryonic stem cells; hPSC, human pluripotent stem cells; iPSC, induced pluripotent stem cells; SARS‐CoV‐2, severe acute respiratory syndrome related coronavirus‐2; SEAM, self‐formed ectodermal autonomous multi‐zone.
2.3. SARS‐CoV‐2 and the immune response
Increased production of cytokines has been implicated in the immunopathology of disease in patients with Covid‐19, resulting in what is commonly referred to a ‘cytokine storm'. 53 , 54 Various types of organoids have been utilised to model this immunological phenomenon. A Th1 cytokine response was elicited following SARS‐CoV‐2 infection in alveolar (pre‐print), 50 choroid plexus 34 and intestinal, 37 , 51 organoids (Table 2). In a study comparing post‐mortem lung samples from Covid‐19 patients to biopsies from healthy lung tissue from uninfected individuals, transcriptional analysis revealed high chemokine signatures in Covid‐19 patients including CCL2 (MCP‐1), CCL8 (MCP‐2) and CCL11. 53 Similarly, induction of chemokines or chemokine transcripts were detected in SARS‐CoV‐2 infected lung, 32 hepatocyte, 42 cholangiocyte, 42 intestinal 37 , 51 and colonic 32 organoids (Table 2).
TABLE 2.
Expression profiles of cytokines and chemokines in organoid models following SARS‐CoV‐2 infection
| Organoid type | Expression of cytokines and chemokines following SARS‐CoV‐2 infection | Downregulation of cytokines and chemokines following SARS‐CoV‐2 infection |
|---|---|---|
| Alveolar (pre‐print) | IFNB1 50 | .. |
| Lung a | CXCL2, CXCL3, CXCL5, CCL2, and CCL20 32 | .. |
| Choroid plexus | CCL7, IL‐32, CCL2 (MCP1), IL‐18, and IL‐8 34 | .. |
| Intestinal/intestinal enteroids | CCR1, CCR8, IL16, IL3 37 CXCL10 (IP10) 37 , 51 | CCR2, CCR5 and IL5 37 |
| Hepatocyte | CXCL1, CXCL3, CXCL5, CXCL6 (GCP‐2), and CCL20 (MIP3α) 42 | .. |
| Cholangiocyte | CXCL1, CXCL2 (MIP‐2α), CXCL3, and CCL2 (MCP‐1) 42 | .. |
| Colonic a | CXCL6, CXCL8, CXCL11, IL‐1α, IL‐1β 32 | .. |
Note: Table summarising the expression profiles of different cytokines and chemokines in a variety of SARS‐CoV‐2 infected organoid models. These data are indicative of the induction of cytokine and chemokines during infection in a majority of organoid types.
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome related coronavirus‐2.
Data extrapolated from volcano plots of gene expression profiles from mock and SARS‐CoV2‐infected organoids in Han et al. 32 study.
Among the consequences of viral infection induced cytokine storms in the lung are cellular apoptosis, vascular leakage, insufficient T‐cell response and the development of acute respiratory distress syndrome (ARDS). 55 , 56 A recent cohort study reported up to 85% of Covid‐19 patients admitted to the ICU had developed ARDS, 57 and this was associated with increased mortality rates. 58 , 59 The SARS‐CoV‐2‐mediated increase in cytokine and chemokine signatures observed in both organoid models and clinical settings may reflect the immunopathological mechanism for the development of further Covid‐19‐related sequelae.
Interferons (IFNs) are innate immune response proteins secreted by host cells and are responsible for inducing and regulating antiviral mechanisms following viral infection. 60 , 61 The effects of SARS‐CoV‐2 infection on IFN signalling have been summarised in Table 3. Transcriptomics analysis of infected alveolar organoids 2 days post‐infection (dpi) revealed IFN signalling was the most upregulated canonical pathway (pre‐print). 50 Similarly, SARS‐CoV‐2 infection of bronchial organoids induced a moderate increase in type I IFN and IFN‐stimulated genes (ISGs) at 5 dpi (pre‐print). 33 Upregulation of type III (IFN‐ λ) 52 as well as modest expression of ISGs, such as ISG15 51 were also observed in infected colon and intestinal organoids at 24 and 60 hours post‐infection (hpi), respectively. Similarly, SARS‐CoV‐2 infection of intestinal enteroids induced expression of IFNL2 and IFNL3 at 48 hpi. 37
TABLE 3.
IFN response in different organoid types following SARS‐CoV‐2 infection
| Organoid type | Induction of the IFN response following SARS‐CoV‐2 infection | Minimal induction or downregulation of the IFN response following SARS‐CoV‐2 infection | Hours/days post infection |
|---|---|---|---|
| Alveolar (pre‐print) | Upregulated 50 | .. | 2 dpi 50 |
| Bronchial (pre‐print) | Moderate increase in type I IFN and IFN‐stimulated genes (ISGs) 33 | .. | 5 dpi 33 |
| Intestinal/intestinal enteroids | Modest expression of ISGs 51 | .. | 60 hpi 51 |
| Induction of IFNL2 and IFNL3 37 | IFN‐α, IFN‐β and IFN‐γ were “barely” induced 37 | 48 hpi 37 | |
| Colonic | Upregulation of type III IFN (IFN‐λ) 52 | No upregulation of type I IFN (IFN‐β1) 52 | 24 hpi 52 |
| SEAM eye (pre‐print) | .. | No upregulation of type I or type III detected 36 | .. a |
Note: Induction of IFN response in a variety of SARS‐CoV‐2 infected organoid types. The hours/days post infection appear to influence the induction of an IFN response, where organoids analysed after shorter time periods after SARS‐CoV‐2 infection exhibited minimal induction or no upregulation of some IFNs (i.e., 24–48 hpi).
Abbreviations: dpi, days post‐infection; hpi, hours post‐infection; IFN, interferon; SARS‐CoV‐2, severe acute respiratory syndrome related coronavirus‐2; SEAM, self‐formed ectodermal autonomous multi‐zone.
Hpi unclear in study.
However, contrary to these findings, no upregulation of type I or type III IFNs was reported in SARS‐CoV‐2‐infected SEAM eye organoids, although the hpi was unclear from the published studies (pre‐print). 36 Similarly, no upregulation of type I IFN (IFN‐β1) was observed in colon organoids at 24 hpi 52 and expression of IFN‐α, IFN‐β and IFN‐γ was barely induced in infected intestinal enteroids at 48 hpi. 37 These data are reflective of SARS‐CoV‐2‐infected human ex vivo lung tissues which exhibited no significant induction of IFN I, II or III. 62
It is possible that the difference in results between the aforementioned studies is due to differential IFN responses in different organ types in response to SARS‐CoV‐2 infection. Variances in experimental design may also impact the induction of IFNs in different tissues. Longer infection times in future investigations would allow researchers to fully elucidate whether the observed lack of IFN induction in certain organoids is a result of SARS‐CoV‐2‐mediated suppression, or an infection‐induced delay in the response. Lei et al. reported a delayed IFN response in infected Calu‐3 cells (airway epithelial cell line) where SARS‐CoV‐2 induced a substantial but delayed type I IFN response in which IFN‐β and ISG56 only modestly increased at 12 hpi, but were significantly increased by 24 hpi. 63 In mice, SARS‐CoV‐2 infection similarly induced a prolonged yet delayed type I IFN signal which triggered innate inflammatory monocyte‐macrophages, resulting in increased cytokine and chemokines, and impaired T‐cell response. 64 Dysregulation of the T‐cell response is also reported in Covid‐19 patients, 65 suggesting a mechanistic link between SARS‐CoV‐2‐mediated IFN dysregulation and immune sequelae. The potential delay in IFN production and its downstream mechanistic effects during SARS‐CoV‐2 infection are yet to be completely explained, and further research combining clinical findings and organoids is necessary before any conclusions are drawn.
Dysregulation of immune pathways was also observed in SARS‐CoV‐2 infected organoids. Infection induced expression of various immune pathways in lung and liver organoids including cytokine–cytokine receptor interaction, 32 , 42 IL‐17, 32 , 42 chemokine, 42 TNF, 32 , 42 NF‐ κB, 42 , 50 TLR, 50 and IL1. 50 In bronchial organoids, SARS‐CoV‐2 infection increased expression of genes involved in the regulation of immune effectors, inflammatory response, and interferon‐gamma production (pre‐print). 33
Investigations involving organoids have thus far allowed researchers to model the effects of SARS‐CoV‐2 infection on the immune response in representative organ types, revealing the induction of cytokine, chemokine, and other immune pathways. Further research examining naturally infected lung tissue using immunohistochemistry compared to ex vivo infection of lung organoids could be used to evaluate the immunopathology of SARS‐CoV‐2 infection. This approach has been successfully utilised in other tissues, for example, analysis of the effects of cytomegalovirus infection on cytokine expression in naturally infected placentae compared to an ex vivo placental villous explant histoculture model. 66
2.4. SARS‐CoV‐2 dysregulation of cellular function
Dysregulation of essential cellular proteins and functions during SARS‐CoV‐2 infection has been reported in a number of models including brain, lung and liver organoids. Cell death was observed in SARS‐CoV‐2‐infected brain 35 and choroid plexus organoids, 34 and an increase in pyknotic cells and cell bursting was observed in infected bronchial organoids (pre‐print) 33 and brain spheres, 43 respectively. Furthermore, markers of apoptosis such as caspase‐3 and CD40 were increased in SARS‐CoV‐2‐infected alveolar (pre‐print), 50 brain 35 and liver ductal 41 organoids. Interestingly, from 25 to 72 hpi, SARS‐CoV‐2‐infected choroid plexus organoids exhibited increased cell death in both infected (1·4%–5·9%) and uninfected (1·9% 5·4%) transthyretin positive cells. 34 Similarly, a proportion of apoptotic cells in infected alveolar organoids was uninfected (pre‐print). 50 This suggests SARS‐CoV‐2 induces cytopathic effects on both infected and neighbouring uninfected cells. These findings are reflective of clinical observations of cellular apoptosis and necrosis in a number of tissues of Covid‐19 patients including the lung, thyroid and liver. 67 , 68 This has also been observed in influenza A infection, which causes cell death in the respiratory tract and lung parenchyma, which can, in severe cases, result in increased inflammation, a compromised epithelial cell barrier and lung failure. 69 The published studies to date suggest SARS‐CoV‐2‐mediated cell death may contribute to the damage observed clinically in the organs of Covid‐19 patients. 70 , 71
In brain organoids, SARS‐CoV‐2‐induced phosphorylation and re‐localisation of tau (neuronal marker) from the axon of infected neurons to the soma. 35 The tau protein is responsible for stabilising neuronal microtubules and promoting axonal growth. 72 Hyper‐phosphorylation and abnormal tau aggregation are characteristics of neurodegenerative diseases known as tauopathies such as Alzheimer's disease (AD). 72 , 73 , 74 In fact, in a three‐dimensional brain‐like model, herpes simplex virus type 1 (HSV‐1)‐induced hyper‐phosphorylation of tau and resulted in an AD‐like pathology including the development of β‐amyloid (Aβ) plaque‐like formations, a marker of AD. 75 Tau phosphorylation and re‐localisation, as well as cell death in SARS‐CoV‐2‐infected brain organoids, indicate potential neuronal cell damage 35 and should be further investigated as a potential mechanism for SARS‐CoV‐2‐induced cerebral sequelae.
In choroid plexus organoids, SARS‐CoV‐2 infection resulted in the downregulation of a number of important genes, the dysregulation of which is implicated in neurological malformations. Gene ontology analysis of infected choroid plexus organoids at 72 hpi revealed cell junction genes were downregulated, which Jacob et al. suggested could cause disruption to blood–cerebrospinal fluid barrier function. 34 This disruption to the barrier has been implicated in increased permeability, allowing entry of pathogens through the barrier. 76 Furthermore, SARS‐CoV‐2‐downregulated genes involved in ion channels and transmembrane transport, 34 indicating possible mechanisms of pathogenesis and the development of neurological symptoms observed in Covid‐19 patients. 77 , 78 , 79
Zhao et al. reported expression of claudin 1 (CLDN1), a constituent of tight junctions, was decreased in SARS‐CoV‐2‐infected liver ductal organoids. 41 Infection also decreased enrichment of cell junction organisation genes. 41 The blood–bile barrier function is typically enabled by tight junctions, and chronic liver injury is associated with a loss of function of these tight junctions in murine models. 80 Furthermore, SARS‐CoV‐2 infection of liver organoids significantly reduced mRNA expression of the major bile acid transporter genes; solute carrier family 10 member 2 (SLC10A2) and cystic fibrosis transmembrane conductance regulator (CFTR). 41 This suggests the liver and hepatic injury observed in Covid‐19 patients 81 is potentially mediated through SARS‐CoV‐2‐induced dysregulation of tight junctions and subsequent accumulation or leakage of bile acid into the liver parenchyma. 41
2.5. Therapeutics
The rapid progression and burden of the Covid‐19 pandemic have driven a prompt development of therapeutics. Thus far, the efficacy of a number of treatment options has been evaluated including controlled trials that have shown promising results of remdesivir and 82 dexamethasone, 83 both of which are licenced therapeutics. 84 , 85 However, recent findings from the WHO Solidarity trial have indicated remdesivir treatment had little or no effect on hospitalised Covid‐19 patients (pre‐print). 86 A randomised control trial involving combined therapy of remdesivir and baricitinib showed this improved patient recovery time and clinical status of Covid‐19 patients in comparison to treatment with remdesivir alone. 87 Inhaled nebulised interferon beta‐1a treatment report clinical benefit in improving the outcome of Covid‐19 disease in infected patients. 88 Additionally, several promising vaccine candidates have undergone phase 3 randomised clinical trials including Pfizer (95%), 89 Moderna (94.1% efficacy) 90 and Gamaleya (92% efficacy). 91
In tandem with these randomised controlled trials, research using organoid models would provide excellent assistance in the timely evaluation of the safety and efficacy of various treatment options. Lung organoids pre‐treated with imatinib, mycophenolic acid (MPA), quinacrine dihydrochloride (QNHC) and chloroquine blocked luciferase activity following SARS‐CoV‐2‐entry virus infection, suggesting potential efficacy in decreasing SARS‐CoV‐2 infection. 32 In SARS‐CoV‐2‐infected alveolar organoids, viral nucleoprotein (N) RNA was reduced following the treatment with IFN‐β1 (3·2 log), hydroxychloroquine (2·4 log) and remdesivir (9 log) compared to infected, untreated organoids. 50 However, hydroxychloroquine exhibited variable effects on viral replication and gene expression that were dependant on the donor epithelium (pre‐print). 50 Pre‐treatment of colon organoids with both IFN‐β1 and IFN‐λ significantly impaired infection and this was associated with a decrease in viral genome copies. 52
Clinical‐grade human recombinant soluble ACE2 (hrsACE2) has been tested in phase 1 92 and phase 2 93 clinical trials, and is reportedly well tolerated in patients with ARDS, a clinical feature of Covid‐19. In capillary and kidney organoids, ACE2 decreased SARS‐CoV‐2 infection. 39
Camostat, a TMPRSS2 inhibitor, is a promising therapeutic candidate for SARS‐CoV‐2 in Caco‐2 and Vero‐TMPRSS2 cell culture models. 26 Suzuki et al. reported a reduction of SARS‐CoV‐2 genome in infected bronchial organoids to 2% of that of infected, untreated organoids (pre‐print). 33 Furthermore, camostat treatment suppressed the SARS‐CoV‐2 mediated increase in genes involved in the immune response (pre‐print). 33
3. ORGANOID LIMITATIONS AND FUTURE CONSIDERATIONS
Despite the advantages of organoids as an alternative to traditional culture systems, limitations exist in the organoid model. One such limitation is the variability between individual organoids. Little et al. reported transcriptional variability between kidney organoids from different experimental batches, particularly in genes related to temporal maturation. 94 However, other studies have reported little variability in cortical and dorsal forebrain organoids, comparing the variation as being similar to that of endogenous brains. 95 , 96
It is important to take into consideration the degree of accuracy to which organoids mimic the cell composition and structure of human organ development. Bhaduri et al. reported smaller numbers of cell subtypes in cortical organoids compared to primary brain tissue, and a portion of organoid cells co‐expressed markers for both radial glial and neuronal cells, indicating broader cell type characterisation in organoids. 97 Structurally, the human cortex consists of stratified laminae with distinct neuronal populations; however, cerebral organoids reportedly contain neurons of different layers interspersed throughout the organoid, indicating a lack of stratified organisation. 98 Other studies have, however, reported the development of different brain regions and stratified layers within cerebral organoids. 21 , 99
Unlike monocultures which are excellent for large‐scale experiments, the time‐consuming and technically challenging nature of organoid generation has brought into question the scalability of this alternative model. 14 Equipment such as the mini‐bioreactor can improve organoid scalability as it allows for the consistent culture of multiple organoids and larger‐scale organoid production that is necessary for applications such as compound testing and drug efficacy investigations. 21
The original source of the cells used to differentiate organoids is an important consideration for future studies. The reviewed SARS‐CoV‐2 infection studies (Table 1) utilised organoids derived from a variety of cell types, including induced pluripotent stem cells (iPSC), human embryonic stem cells (hESC), and cells obtained from patient‐derived tissue samples. Organoids that are derived from patient‐derived tissue biopsies without the cells first being reprogramed into iPSCs reportedly develop simpler structures, comprising mostly of epithelial cell types. 100 In comparison, organoids that are derived from stem cells undergo a multi‐step differentiation process resulting in more complex structures with multiple germ layers. 20 , 99 Further research is required to examine differences in structural complexity and cellular composition between organoid types, and this should be taken into account in future studies of organoid infection by SARS‐CoV‐2.
Organoids have proven to be an amenable culture system with many new developments in place to improve upon the limitations discussed above. Organoids are a multi‐cell‐type, three‐dimensional human organ model that can be used in tandem with traditional animal and two‐dimensional monocultures. The comparable effects of SARS‐CoV‐2 infection in organoids and human and post‐mortem studies (particularly cellular tropism and immunopathology) that were highlighted in this review, emphasises the benefits of using this model for the study of other viral infections. Of note, the availability of single‐cell sequencing datasets have been useful in assessing the presence of SARS‐CoV‐2 viral receptors in human organs, 28 , 29 , 30 and will continue to assist the transition to organoid research and inform researchers of key target genes in the investigation of other infectious diseases.
4. CONCLUDING STATEMENT
The onset of the Covid‐19 pandemic has resulted in a global health crisis, resulting in increasing numbers of people infected worldwide. Three‐dimensional, biologically relevant organoids are an excellent tool for Covid‐19 disease modelling and can be used to supplement other forms of research into the pathogenetic mechanisms of SARS‐CoV‐2 infection. Organoids have been shown to express SARS‐CoV‐2 receptors and studies have demonstrated viral pathogenesis and tropism occurs, in many ways similar to that seen in human and post‐mortem studies. The immunopathological consequences of SARS‐CoV‐2 infection of organoids have closely mirrored the inflammatory responses observed in patients with Covid‐19, demonstrating the biological relevance of organoid models. Several studies have shown virus‐induced pathogenetic effects on cells such as apoptosis, and dysregulation of significant pathways and genes such as immune and cell junctions.
Further research is needed to demonstrate the mechanisms of SARS‐CoV‐2 causing clinical sequelae during Covid‐19 disease. The use of organoids in tandem with traditional two‐dimensional cell culture, animal models, clinical studies and human clinical trials can be utilised to identify important host and viral targets. These targets can then be exploited in the development of safe and effective antiviral interventions and vaccination programmes.
CONFLICT OF INTERESTS
We declare no competing interests.
AUTHOR CONTRIBUTIONS
William D Rawlinson designed the review, editing and writing throughout the process. Ece Egilmezer reviewed the results of the search strategy, wrote and edited the manuscript. All authors reviewed the manuscript for the final draft.
ACKNOWLEDGEMENTS
No funding sources to declare. All authors had full access to the manuscript and accept responsibility to submit for publication.
Egilmezer E, Rawlinson WD. Review of studies of severe acute respiratory syndrome related coronavirus–2 pathogenesis in human organoid models. Rev Med Virol. 2021;31(6):e2227. 10.1002/rmv.2227
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
References
- 1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cui J, Li F, Shi Z‐L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li D, Wolk DM, Cantor MN. Comparing clinical characteristics of influenza and common coronavirus infections using electronic health records. J Infect Dis. 2020. 10.1093/infdis/jiaa626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moriyama M, Hugentobler WJ, Iwasaki A. Seasonality of respiratory viral infections. Annu Rev Virol. 2020;7(1):83–101. [DOI] [PubMed] [Google Scholar]
- 5. Paules CI, Marston HD, Fauci AS. Coronavirus infections—more than just the common cold. JAMA. 2020;323(8):707–708. [DOI] [PubMed] [Google Scholar]
- 6. Whitworth J. COVID‐19: a fast evolving pandemic. Trans R Soc Trop Med Hyg. 2020;114(4):241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharma R, Agarwal M, Gupta M, Somendra S, Saxena SK. Clinical Characteristics and Differential Clinical Diagnosis of Novel Coronavirus Disease 2019 (COVID‐19). Coronavirus Disease 2019 (COVID‐19). Singapore: Springer; 2020:55–70. [Google Scholar]
- 8. Caly L, Druce J, Roberts J, et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS‐CoV‐2) from the first patient diagnosed with COVID‐19 in Australia. Med J Aust. 2020;212(10):459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Darmanis S, Sloan SA, Zhang Y, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U. S. A. 2020;112(23):7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oldham MC, Konopka G, Iwamoto K, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simian M, Bissell MJ. Organoids: a historical perspective of thinking in three dimensions. J Cell Biol. 2017;216(1):31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F, Weaver VM, Petersen OW, et al. Reciprocal interactions between β1‐integrin and epidermal growth factor receptor in three‐dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci U. S. A. 1998;95(25):14821–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125‐1–1247125‐9. [DOI] [PubMed] [Google Scholar]
- 14. Lancaster MA, Huch M. Disease modelling in human organoids. Dis Model Mech. 2019;12(7):dmm039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiros TG, Levast B, Auray G, Strom S, van Kessel J, Gerdts V. The importance of animal models in the development of vaccines. In Innovation in Vaccinology. Dordrecht: Springer D; 2012:251–264. [Google Scholar]
- 16. van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV‐19 vaccine prevents SARS‐CoV‐2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. N Engl J Med. 2020;383(16):1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson EL, Lu H. Three‐dimensional models for studying development and disease: moving on from organisms to organs‐on‐a‐chip and organoids. Integr Biol (Camb). 2016;8(6):672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tannenbaum J, BT B. Russell and Burch's 3Rs then and now: the need for clarity in definition and purpose. J Am Assoc Lab Anim Sci. 2015;54(2):120–132. [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21(10):571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qian X, Nguyen HN, Song MM, et al. Brain‐region‐specific organoids using mini‐bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vissoci JRN, Rocha TAH, da Silva NC, et al. Zika virus infection and microcephaly: evidence regarding geospatial associations. PLoS Negl Trop Dis. 2018;12(4):e0006392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zaim S, Chong JH, Sankaranarayanan V, Harky A. COVID‐19 and multi‐organ response. Curr Probl Cardiol. 2020;45(8):100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ellul, M , Benjamin, L , Singh, B , et al. Neurological associations of COVID‐19. Lancet Neurol. 2020;19(9):767–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoffmann, M , Kleine‐Weber, H , Schroeder, S , et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2): 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walls AC, Park Y‐J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu H, Gai S, Wang X, et al. Single‐cell analysis of SARS‐CoV‐2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc Res. 2020;116(10):1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qi J, Zhou Y, Hua J, et al. The scRNA‐seq expression profiling of the receptor ACE2 and the cellular protease TMPRSS2 reveals human organs susceptible to COVID‐19 infection. bioRxiv. 2020. 10.1101/2020.04.16.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sungnak W, Huang N, Bécavin C, et al. SARS‐CoV‐2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26(5):681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salahudeen AA, Choi SS, Rustagi A, et al. Progenitor identification and SARS‐CoV‐2 infection in long‐term human distal lung organoid cultures. bioRxiv. 2020. 10.1101/2020.07.27.212076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han Y, Duan X, Yang L, et al. Identification of SARS‐CoV‐2 inhibitors using lung and colonic organoids. Nature. 2020;589:270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki T, Itoh Y, Sakai Y, et al. Generation of human bronchial organoids for SARS‐CoV‐2 research. bioRxiv. 2020. 10.1101/2020.05.25.115600. [DOI] [Google Scholar]
- 34. Jacob F, Pather S, Huang W‐K, et al. Human pluripotent stem cell‐derived neural cells and brain organoids reveal SARS‐CoV‐2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27(6):937–950.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramani A, Müller L, Ostermann PN, et al. SARS‐CoV‐2 targets neurons of 3D human brain organoids. EMBO J. 2020; 39:e106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makovoz B, Moeller R, Zebitz Eriksen A, tenOever BR, Blenkinsop TA. SARS‐CoV‐2 infection of ocular cells from human adult donor eyes and hESC‐derived eye organoids. SSRN. 2020. 10.2139/ssrn.3650574. [DOI] [Google Scholar]
- 37. Zhou, J , Li, C , Liu, X , et al. Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med, 26(7):1077–1083. [DOI] [PubMed] [Google Scholar]
- 38. Zang R, Castro MFG, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS‐CoV‐2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5(47):eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4). 905–913.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xia S, Wu M, Chen S, et al. Long term culture of human kidney proximal tubule epithelial cells maintains lineage functions and serves as an ex vivo model for coronavirus associated kidney injury. Virol Sin. 2020;35(3):311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao B, Ni C, Gao R, et al. Recapitulation of SARS‐CoV‐2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11(10):771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang L, Han Y, Nilsson‐Payant BE, et al. A human pluripotent stem cell‐based platform to study SARS‐CoV‐2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27(1):125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bullen CK, Hogberg HT, Bahadirli‐Talbott A, et al. Infectability of human BrainSphere neurons suggests neurotropism of SARS‐CoV‐2. ALTEX. 2020;37(4):665–671. [DOI] [PubMed] [Google Scholar]
- 44. Temple S. The development of neural stem cells. Nature. 2001;414(6859). 112–117. [DOI] [PubMed] [Google Scholar]
- 45. Schwartz PH, Brick DJ, Stover AE, Loring JF, Müller F‐J. Differentiation of neural lineage cells from human pluripotent stem cells. Methods. 2008;45(2):142–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gynaecologists RCoOa . Coronavirus (COVID‐19) infection in pregnancy‐ information for healthcare professionals. 2020. https://www.rcog.org.uk/globalassets/documents/guidelines/2020‐07‐24‐coronavirus‐covid‐19‐infection‐in‐pregnancy.pdf. Accessed November 14 2020. [Google Scholar]
- 47. Hou YJ, Okuda K, Edwards CE, et al. SARS‐CoV‐2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med. 2020;383(6):590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chu H, Chan JF‐W, Yuen TT‐T, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS‐CoV‐2 and SARS‐CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID‐19: an observational study. Lancet Microb. 2020;1(1):E14–E23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mulay A, Konda B, Garcia G, et al. SARS‐CoV‐2 infection of primary human lung epithelium for COVID‐19 modeling and drug discovery. bioRxiv. 2020. 10.1101/2020.06.29.174623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lamers MM, Beumer J, van der Vaart J, et al. SARS‐CoV‐2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stanifer ML, Kee C, Cortese M, et al. Critical role of type III interferon in controlling SARS‐CoV‐2 infection in human intestinal epithelial cells. Cell Rep. 2020;32(1):107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blanco‐Melo D, Nilsson‐Payant BE, Liu W‐C, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costela‐Ruiz VJ, Illescas‐Montes R, Puerta‐Puerta JM, Ruiz C, Melguizo‐Rodríguez L. SARS‐CoV‐2 infection: the role of cytokines in COVID‐19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de Lucena TMC, da Silva Santos AF, de Lima Fabrício BR, de Albuquerque Borborema ME, de Azevêdo Silva J. Mechanism of inflammatory response in associated comorbidities in COVID‐19. Diabetes Metab Syndr. 2020;14(4):597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ziehr DR, Alladina J, Petri CR, et al. Respiratory pathophysiology of mechanically ventilated patients with COVID‐19: a cohort study. Am J Respir Crit Care Med. 2020;201(12):1560–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grasselli G, Tonetti T, Protti A, et al. Pathophysiology of COVID‐19‐associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med. 2020;8(12):1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou J‐H, Wang Y‐N, Chang Q‐Y, Ma P, Hu Y, Cao X. Type III interferons in viral infection and antiviral immunity. Cell Physiol Biochem. 2018;51(1):173–185. [DOI] [PubMed] [Google Scholar]
- 61. García‐Sastre A, Biron CA. Type 1 interferons and the virus‐host relationship: a lesson in detente. Science. 2006;312(5775):879–882. [DOI] [PubMed] [Google Scholar]
- 62. Chu H, Cha, JF‐W , Wang Y, et al. Comparative replication and immune activation profiles of SARS‐CoV‐2 and SARS‐CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID‐19. Clin Infect Dis. 2020;71(6):1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lei X, Dong X, Ma R, et al. Activation and evasion of type I interferon responses by SARS‐CoV‐2. Nat Commun. 2020;11(1):3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte‐macrophage responses cause lethal pneumonia in SARS‐CoV‐infected mice. Cell Host Microbe. 2016;19(2):181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID‐19 patients. Cell Mol Immunol. 2020;17(5):533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hamilton ST, Scott G, Naing Z, et al. Human cytomegalovirus‐induces cytokine changes in the placenta with implications for adverse pregnancy outcomes. PLoS One. 2012;7(12):e52899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ding Y, Wang H, Shen H, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200(3):282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chau, TN , Lee, KC , Yao, H , et al. SARS‐associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology. 2004;39(2):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Atkin‐Smith GK, Duan M, Chen W, Poon IK. The induction and consequences of Influenza A virus‐induced cell death. Cell Death Dis. 2020;9:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li S, Jiang L, Li XLF, et al. Clinical and pathological investigation of patients with severe COVID‐19. JCI Insight. 2020;5(12):138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID‐19. Nat Med. 2020;26:1017–1032. [DOI] [PubMed] [Google Scholar]
- 72. Wang, Y , Mandelkow, E . Tau in physiology and pathology. Nat Rev Neurosci. 2016;17(1):22–35. [DOI] [PubMed] [Google Scholar]
- 73. Grundke‐Iqbal I, Iqbal K, Tung Y‐C, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule‐associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U. S. A. 1986;83(13):4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iqbal K, Liu F, Gong C‐X, Grundke‐Iqbal I. Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res. 2010;7(8):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cairns DM, Rouleau N, Parker RN, Walsh KG, Gehrke, L , Kaplan, DL . A 3D human brain–like tissue model of herpes‐induced Alzheimer's disease. Sci Adv. 2020;6(19):eaay8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lauer AN, Tenenbaum T, Schroten H, Schwerk C. The diverse cellular responses of the choroid plexus during infection of the central nervous system. Am J Physiol Cell Physiol. 2018;314(2):C152–C165. [DOI] [PubMed] [Google Scholar]
- 77. Najjar S, Najjar A, Chong DJ, et al. Central nervous system complications associated with SARS‐CoV‐2 infection: integrative concepts of pathophysiology and case reports. J Neuroinflamm. 2020;17(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Filatov A, Sharma P, Hindi F, Espinosa PS. Neurological complications of coronavirus disease (COVID‐19): encephalopathy. Cureus. 2020;12(3):e7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou, L , Zhang, M , Wang, J , Gao, J . SARS‐CoV‐2: underestimated damage to nervous system. Trav Med Infect Dis. 2020;36:101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pradhan‐Sundd, T , Vats, R , Russell, JO , et al. Dysregulated bile transporters and impaired tight junctions during chronic liver injury in mice. Gastroenterology. 2018;155(4):1218–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang Y, Liu S, Liu H, et al. SARS‐CoV‐2 infection of the liver directly contributes to hepatic impairment in patients with COVID‐19. J Hepatol. 2020;73(4):807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 — final report. N Engl J Med. 2020;383(19):1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lim WS, Emberson, JR , Mafham, M , et al. Dexamethasone in hospitalized patients with Covid‐19 – preliminary report. N Engl J Med. 2020;384(8):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lamb YN. Remdesivir: first approval. Drugs. 2020;80(13):1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Theoharides TC,Conti P. Dexamethasone for COVID‐19? Not so fast. J Biol Regul Homeost Agents. 2020;34(3):1241–1243. [DOI] [PubMed] [Google Scholar]
- 86. WHO Solidarity Trial Consortium. Repurposed antiviral drugs for COVID‐19 –interim WHO SOLIDARITY trial results. medRxiv. 2020. 10.1101/2020.10.15.20209817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kalil AC, Patterson TF, Mehta AK, et al. Baricitinib plus remdesivir for hospitalized adults with Covid‐19. N Engl J Med. 2020. 10.1056/nejmoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Monk PD, Marsden RJ, Tear VJ, et al. Safety and efficacy of inhaled nebulised interferon beta‐1a (SNG001) for treatment of SARS‐CoV‐2 infection: a randomised, double‐blind, placebo‐controlled, phase 2 trial. Lancet Respir Med. 2020;9(2):196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;83(27):2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Baden LR, Sahly HMEL, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2020;384:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. TGN Centre . The first interim data analysis of the Sputnik V vaccine against COVID‐19 phase III clinical trials in the Russian Federation demonstrated 92% efficacy. 2020. https://sputnikvaccine.com/newsroom/pressreleases/the‐first‐interim‐data‐analysis‐of‐the‐sputnik‐v‐vaccine‐against‐covid‐19‐phase‐iii‐clinical‐trials‐/. Accessed January 25 2021. [Google Scholar]
- 92. Haschke M, Schuster M, Poglitsch M, et al. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin‐converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52(9):783–792. [DOI] [PubMed] [Google Scholar]
- 93. Khan A, Benthin C, Zeno B, et al. A pilot clinical trial of recombinant human angiotensin‐converting enzyme 2 in acute respiratory distress syndrome. Crit Care. 2017;21(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Phipson, B , Er, PX , Combes, AN , et al. Evaluation of variability in human kidney organoids. Nat Methods. 2019;16(1):79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Velasco S, Kedaigle AJ, Simmons SK, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570(7762). 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yoon SJ, Elahi LS, Pașca AM, et al. Reliability of human 3D cortical organoid generation. Nat Methods. 2019;16(1):75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bhaduri A, Andrews MG, Mancia Leon W, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578(7793):142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Andrews MG, Nowakowski TJ Human brain development through the lens of cerebral organoid models. Brain Res. 2019;1725(146470). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lancaster MA, Knoblich JA . Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sato T, Stange DE, Ferrante M, et al. Long‐term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


