Abstract
Background
Estimates of community spread and infection fatality rate (IFR) of COVID‐19 have varied across studies. Efforts to synthesize the evidence reach seemingly discrepant conclusions.
Methods
Systematic evaluations of seroprevalence studies that had no restrictions based on country and which estimated either total number of people infected and/or aggregate IFRs were identified. Information was extracted and compared on eligibility criteria, searches, amount of evidence included, corrections/adjustments of seroprevalence and death counts, quantitative syntheses and handling of heterogeneity, main estimates and global representativeness.
Results
Six systematic evaluations were eligible. Each combined data from 10 to 338 studies (9‐50 countries), because of different eligibility criteria. Two evaluations had some overt flaws in data, violations of stated eligibility criteria and biased eligibility criteria (eg excluding studies with few deaths) that consistently inflated IFR estimates. Perusal of quantitative synthesis methods also exhibited several challenges and biases. Global representativeness was low with 78%‐100% of the evidence coming from Europe or the Americas; the two most problematic evaluations considered only one study from other continents. Allowing for these caveats, four evaluations largely agreed in their main final estimates for global spread of the pandemic and the other two evaluations would also agree after correcting overt flaws and biases.
Conclusions
All systematic evaluations of seroprevalence data converge that SARS‐CoV‐2 infection is widely spread globally. Acknowledging residual uncertainties, the available evidence suggests average global IFR of ~0.15% and ~1.5‐2.0 billion infections by February 2021 with substantial differences in IFR and in infection spread across continents, countries and locations.
Keywords: bias, COVID‐19, global health, infection fatality rate, meta‐analysis, seroprevalence
Highlights.
Six systematic evaluations have evaluated seroprevalence studies without restrictions based on country and have estimated either total number of people infected or aggregate infection fatality rates for SARS‐CoV‐2.
These systematic evaluations have combined data from 10 to 338 studies (9‐50 countries) each with partly overlapping evidence synthesis approaches.
Some eligibility, design and data synthesis choices are biased, while other differing choices are defendable.
Most of the evidence (78%‐100%) comes from Europe or the Americas.
All systematic evaluations of seroprevalence data converge that SARS‐CoV‐2 infection has been very widely spread globally.
Global infection fatality rate is approximately 0.15% with 1.5‐2.0 billion infections as of February 2021.
1. INTRODUCTION
The extent of community spread of SARS‐CoV‐2 infection and the infection fatality rate (IFR) of COVID‐19 are hotly debated. Many seroprevalence studies have provided relevant estimates. These estimates feed into projections that influence decision‐making. Single studies create confusion, since they leave large uncertainty and unclear generalizability across countries, locations, settings and time points. Some overarching evaluations have systematically integrated data from multiple studies and countries. 1 , 2 , 3 , 4 , 5 , 6 These synthetic efforts probe what are typical estimates of spread and IFR, how heterogeneous they are, and what factors explain heterogeneity. An overview of these systematic evaluations comparing their methods, biases and inferences may help reconcile their findings on these important parameters of the COVID‐19 pandemic.
2. METHODS
2.1. Eligible articles
Articles were eligible if they included a systematic review of studies aiming to assess SARS‐CoV‐2 seroprevalence; there were no restrictions based on country; and an effort was made to estimate either a total number of people infected or aggregate IFRs. Articles were excluded if they considered exclusively studies of particular populations at different risks of infection than the general population (eg only healthcare workers), if they focused on specific countries (by eligibility criteria, not by data availability), and if they made no effort to estimate total numbers of people infected and/or aggregate IFRs.
2.2. Search strategy
Searches were updated until 14 January 2021 in PubMed, medRxiv and bioRxiv with ‘seroprevalence [ti] OR fatality [ti] OR immunity [ti]’ For feasibility, the search in PubMed was made more specific by adding ‘(systematic review OR meta‐analysis OR analysis)’. Communication with experts sought potentially additional eligible analyses (eg unindexed influential reports).
2.3. Extracted information
From each eligible evaluation, the following information was extracted:
Types of information included (seroprevalence, other)
Date of last search, search sources and types of publications included (peer‐reviewed, preprints, reports/other)
Types of seroprevalence designs/studies included
Number of studies, countries, locations included
Seroprevalence calculations: adjustment/correction for test performance, covariates, type of antibodies measured, seroreversion (loss of antibodies over time)
Death count calculations: done or not; adjustments for over‐ or under‐counting, time window for counting COVID‐19 deaths in relationship to seroprevalence measurements
Quantitative synthesis: whether data were first synthesized from seroprevalence studies in the same location/country/other level; whether meta‐analyses were performed across locations/countries and methods used; handling of heterogeneity, stratification and/or regression analyses, including subgroups
Reported estimates of infection spread, under‐ascertainment ratios (total/documented infections) and/or IFR
Global representativeness of the evidence: proportion of the evidence (weight, countries, studies or locations, depending on how data synthesis had been done) from Europe and North America (sensitivity analysis: Europe and America)
2.4. Comparative assessment
Based on the above, the eligible evaluations were compared against each other with focus on features that may lead to bias and trying to decipher the direction of each bias.
3. RESULTS
3.1. Eligible evaluations
Nine potentially eligible articles were retrieved 1 , 2 , 3 , 5 , 6 , 7 , 8 , 9 , 10 And four were rejected (Figure 1). 7 , 8 , 9 , 10 One more eligible report 4 was identified from communication with experts. The six eligible evaluations are named after their first authors or team throughout the manuscript.
FIGURE 1.
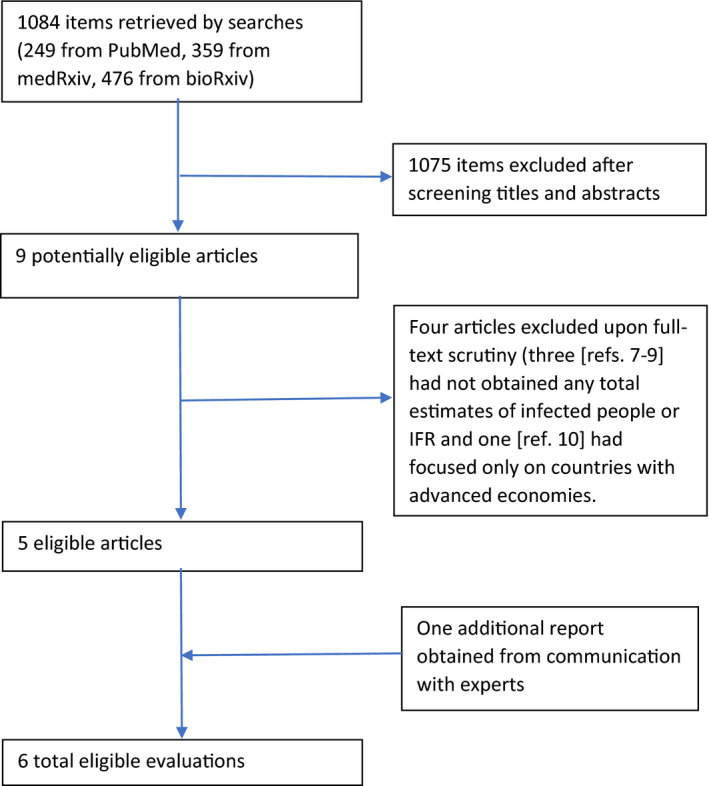
Flow diagram
3.2. Information used
Five evaluations included only seroprevalence studies (Table 1). Meyerowitz‐Katz also included non‐serological and modelling papers; summary IFR was smaller in the seroprevalence studies (0.60% vs 0.84% in others). The six evaluations differed modestly in dates of last search (range, 6/16/2020‐9/9/2020) and in sources searched. Given that few studies outside of Europe and Americas were released early, evaluations with earlier searches have a more prominent dearth of low‐IFR studies from countries with younger populations and fewer nursing home residents.
TABLE 1.
Key features for eligible systematic data syntheses
| Features | Meyerowitz‐Katz | Rostami | Bobrovitz | Imperial college COVID‐19 response team | Ioannidis | O’Driscoll |
|---|---|---|---|---|---|---|
| Types of information included | SP, non‐serological and modelling studies | SP studies | SP studies | SP studies | SP studies | SP studies |
| Last search | 16 June | 14 August | 28 August | Unclear | 9 September | Unclear (1 September?) |
| Search sources | PubMed, preprints (medRxiv, SSRN), Google, Twitter searches, government agency reports eligible | PubMed, Scopus, EMBASE, medRxiv, bioRxiv, research reports eligible | MEDLINE, EMBASE, Web of Science, and Europe PMC, Google, communication with experts | SeroTracker searches (see Bobrovitz) | PubMed (LitCOVID), medRxiv, bioRxiv, Research Square, national reports, communication with experts for additional studies | Unclear |
| Types of SP studies included | Excluded targeted populations with selection bias, also four other studies a | Excluded at‐risk populations (eg HCW), known diseases (eg dialysis, cancer) | All studies included if they reported on sample, date, region and SP estimate | Studies with defined sampling framework, defined geographic area, with availability of test performance, preferentially validation done as part of the study (not just by manufacturers), >100 deaths at SP study mid‐point b ; excluded healthcare workers, symptoms of COVID‐19, self‐referral or self‐selection, narrow age range, confined settings, clinical samples | General population or approximations (including blood donors, excluding high risk, eg HCW, communities), sample size >500, area with population >5000 | Unclear, but eventually it includes some general population studies, some blood donors and some hospital samples |
| Number of studies, countries, locations | 24‐27 studies c , of which 16 serological from 14 countries | 107 data sets from 47 studies from 23 countries | 338 studies (184 from general population) from 50 countries (36 from general population) d | 10 studies (six national, four subnational), nine countries e | 82 estimates, 69 studies, 51 locations, 36 countries (main analysis at the location level) | 25 studies from 20 countries (only 22 national representing 16 countries used in the ensemble model) |
| Studies published in peer‐review journals at the time of the evaluation | 1/16 | 61/107 | 4/40 included in final analysis of under‐ascertainment ratio | 5/10 | 35/82 | 6/20 countries |
Abbreviations: HCW, healthcare workers; IFR, infection fatality rate; SP, seroprevalence.
One study (LA County) 12 with very low IFR was excluded with the justification that it ‘explicitly warned against using its data to obtain an IFR’; as a co‐investigator of the study, both myself and my colleagues are intrigued at the rationale for exclusion; in the publication of the study in JAMA, 12 we did list limitations and caveats, as it is appropriate for any seroprevalence study to do; excluding studies that are honest to discuss limitations would keep only the worst studies that discuss no limitations. Two other studies with low IFR were excluded as well. One was done in Rio Grande do Sul 13 where its authors even report IFR estimates in their paper (0.29%, 0.23%, 0.38% in the three rounds of the serosurvey); the other was done in Boise, 85 where its authors properly discuss limitations but an approximation of IFR is possible; even if not perfectly accurate, it is certainly lower than the IFR estimates included in the Meyerowitz‐Katz meta‐analysis. For the fourth excluded study, 11 the justification offered for its exclusion is that it ‘calculated an IFR, but did not allow for an estimate of confidence bounds’. 1 However, this study presents results of a New York study that Meyerowitz‐Katz did include in their meta‐analysis. Of note, that fourth study 11 also presents a cursory review of seroprevalence studies arriving at a median IFR = 0.31%, half of the summary estimate of Meyerowitz‐Katz.
Clear bias introduced since number of deaths is the numerator itself in the calculation of IFR, and exclusion of studies with low numerator is thus excluding studies likely to have low IFR
Different numbers provided by the authors for total studies in abstract (n = 24), text of the paper (n = 25), tabulated studies (n = 27) and forest plot studies (n = 26)
39 estimates from 17 countries used in main calculation of median under‐ascertainment ratio (N. Bobrovitz, personal communication)
One of the 10 included studies violates the eligibility criterion of the investigators having validated themselves the antibody test used; the ICCRT included this study invoking validation data for the same antibody kit done by a different team in a study in a completely different setting and continent (San Francisco); based on this rationale, perhaps many other studies could have been included, if the same violation of the eligibility criteria was tolerated. The included study was an Italian survey 30 which had only been released in the press with a preliminary report at the time of the ICCRT evaluation and which included crude results on only 64 660 of the intended 150 000 participants (missingness 57%). Its inferred IFR estimate (2.5%) is an extreme outlier, as it is 2‐ to 20‐fold larger than other typical estimates reported from numerous European countries. Moreover, that IFR estimate even matches/exceeds case fatality rates, and thus, it is simply impossible. It is widely accepted that IFR must be several times smaller than case fatality rate, even in locations with substantial testing. Italy had very limited testing in the first wave and modest testing in the second wave. One estimate suggests that the number of infections in Italy at the peak of the first wave was 12 times more than the number of documented cases; that is, the IFR would be more than an order of magnitude lower than the case fatality rate. 31
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Eligibility criteria varied and were sometimes unclear or left room for subjectivity. Consequently, eligible studies varied from 10 to 348 and countries covered with eligible data varied from 9 to 50. Two evaluations 1 , 4 excluded studies in overtly biased ways, leading to inflated IFR estimates.
Specifically, Meyerowitz‐Katz excluded one study with low‐IFR 5 alluding that the study itself ‘explicitly warned against using its data to obtain an IFR’ 1 ; as co‐investigator of the study, both myself and my colleagues are intrigued at this claim. They also excluded two more studies with low‐IFR alluding that it ‘was difficult to determine the numerator (ie number of deaths) associated with the seroprevalence estimate or the denominator (ie population) was not well defined’, 1 while one even presented IFR estimates in its published paper. Another excluded paper 11 tabulated several seroprevalence studies with median IFR = 0.31%, half the Meyerowitz‐Katz estimate.
The Imperial College COVID‐19 Response Team (ICCRT) excluded studies with <100 deaths at the serosurvey mid‐point. 4 This exclusion criterion introduces bias since number of deaths is the numerator in calculating IFR. Exclusion of studies with low numerator excludes studies likely to have low IFR. Indeed, five of six excluded studies with <100 deaths (Kenya, LA County, Rio Grande do Sul, Gangelt, Scotland) 12 , 13 , 14 , 15 , 16 have lower IFR than the 10 ICCRT‐included studies; the sixth (Luxembourg) 17 is in the lower range of the 10 ICCRT‐included studies.
The six evaluations varied on types of populations considered eligible. Table 2 summarizes biases involved in each study population type. General population studies are probably less biased, provided they recruit their intended sample. Conversely, studies of healthcare workers, 18 other high‐risk exposure workers and closed/confined communities may overestimate seroprevalence; these studies were generally excluded, either upfront (5/6 evaluations) or when calculating key estimates (Bobrovitz). Other designs/populations may be biased in either direction, more frequently towards underestimating seroprevalence. 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 Three evaluations (Meyerowitz‐Katz, ICCRT, O’Driscoll) were very aggressive with exclusions.
TABLE 2.
Direction of potential bias in studies with different types of populations
| Type of sampling | Direction of bias |
|---|---|
| General population (entire population or design for representative sample) | Depends on characteristics of individuals who cannot be reached and/or decline participation. If they are more likely to be more disadvantaged (eg have no address/phone/e‐mail) and thus also at higher risk of infection, SP may be underestimated. Potential for bias is more prominent when non‐response/non‐participation is larger. Institutionalized populations and homeless people are typically not included, and these populations often have very high infection rates 19 , 20 ; thus, SP is underestimated |
| Convenience sample (including self‐referral and response to adverts) | Bias could be in either direction. Volunteer bias is common and would tend to recruit more health‐conscious, low‐risk individuals, 21 leading to SP underestimation. Conversely, interest to get tested because of worrying in the presence of symptoms may lead to SP overestimation |
| Blood donors | Bias could be in either direction, but SP underestimation is more likely, since blood donors tend to be more health‐conscious and thus more likely to avoid also risky exposures. An early classic assessment 22 described blood donors as ‘low‐risk takers, very concerned with health, better educated, religious, and quite conservative’—characteristics that would lead to lower infection risk. In countries with large shares of minorities (eg USA and UK), minorities are markedly under‐represented among blood donors. 23 , 24 For example, in the USA, donation rates are 37%‐40% lower in blacks and Hispanics versus whites 23 and in the UK, donation rates range from 1.59 per 1000 among Asian Bangladeshi origin, compared to 22.1 per 1000 among white British origin. 24 These minorities were hit the most by COVID‐19. In European countries, donations are lower in low‐income and low‐education individuals 25 , 26 ; these are also risk factors for COVID‐19 infection. Bobrovitz 3 found median seroprevalence of 3.2% in blood donor studies versus 4.1% in general community/household samples (risk ratio 0.80 in meta‐regression). SP may be overestimated if blood donation is coupled to a free COVID‐19 test in a poor population (as in the case of a study in Manaus, Brazil) |
| Clinical residual samples and patients (eg dialysis, cancer, other) | Bias could be in either direction, but SP underestimation is more likely since patients with known health problems may be more likely to protect themselves in a setting of a pandemic that poses them at high risk. Conversely, repeated exposure to medical facilities may increase risk. Demographic features and socio‐economic status may also affect the size and direction of bias. Bobrovitz 3 found median seroprevalence of 2.9% in studies of residual samples versus 4.1% in general community/household samples (risk ratio 0.63 in meta‐regression). Hospital visitors’ studies had even lower seroprevalence (median 1.4%) |
| Healthcare workers, emergency response, other workers with obvious high risk of exposure | Bias very likely to lead to SP overestimation compared with the general population, because of work‐related contagion hazard; however, this may not always be the case (eg most infections may not happen at work) and any increased risk due to work exposure sometimes may be counterbalanced by favourable socio‐economic profile for some healthcare workers (eg wealthy physicians). Bias may have been more prominent in early days of the pandemic, especially in places lacking protective gear. Across eight studies with data on healthcare workers and other participants, seroprevalence was 1.74‐fold in the former. 3 |
| Other workers | Bias could be in either direction and depends on work experience during the pandemic period and socio‐economic background; for example, SP may be underestimated compared with the general population for workers who are wealthy and work from home during the pandemic and overestimated for essential workers |
| Communities (shelters, religious, other shared‐living) | Likely very strong bias due to high exposure risk leading to SP overestimation compared with the general population. Some of these communities were saturated with very high levels of infection very early. 19 , 20 |
Abbreviations: SP, seroprevalence.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
ICCRT had the most draconian exclusion criteria, excluding 165/175 identified seroprevalence studies. However, ICCRT actually dropped many general population studies (for various reasons), but included two blood donor studies 27 , 28 (out of many such) and one New York study 29 with convenience samples of volunteers recruited while entering grocery stores and through an in‐store flyer. The latter inclusion goes against the stated ICCRT eligibility criteria where self‐selection is reason for exclusion. The New York study 29 had high IFR (from the worst‐hit state in the first wave). The preliminary press‐released report from an Italian general population survey 30 was included in violation of ICCRT eligibility criteria 4 that a study should have performed its own antibody test validation; ICCRT ‘salvaged’ the Italian study by transporting validation data from another study in San Francisco. The Italian study report 30 showed data on only 64 660 of the intended 150 000 participants (missingness 57%). Its inferred IFR estimate (2.5%) is an extreme outlier (2‐ to 20‐fold larger than other reported European estimates) and simply impossible: it matches/exceeds case fatality rates despite probably major under‐ascertainment of infections in Italy. 31
Finally, the six evaluations differed markedly on how many included seroprevalence estimates came from peer‐reviewed publications (journal articles listed in the references) at the time of the evaluation: from only one peer‐reviewed estimate in Meyerowitz‐Katz to 61 in Rostami. Some included seroprevalence estimates that came from preprints/reports published in peer‐reviewed journals by 2/2021; final publications could have minor/modest differences versus preprints/reports. Even journal‐published estimates may get revised; for example, a re‐analysis increased Indiana seroprevalence estimates by a third. 32
3.3. Seroprevalence and death calculations
Three evaluations 3 , 4 , 6 routinely adjusted for test performance, one 5 adjusted for test performance when the authors of the studies had done so, and two were unclear (Table 3). Depending on test sensitivity/specificity, lack of adjustment may inflate or deflate seroprevalence. Ioannidis selected the most fully adjusted seroprevalence estimate, when both adjusted and unadjusted estimates existed; other evaluations were unclear on this issue. Ioannidis corrected the seroprevalence upward when not all three types of antibodies (IgG, IgM, and IgA) were assessed. ICCRT and O’Driscoll considered seroreversion adjustments.
TABLE 3.
Adjustments and corrections for seroprevalence and death counts
| Features | Meyerowitz‐Katz | Rostami | Bobrovitz | Imperial College COVID‐19 response team | Ioannidis | O’Driscoll |
|---|---|---|---|---|---|---|
| Adjustment of SP for test performance | Unclear selection rule | Unclear selection rule | Yes (Bayesian) | Yes | Yes, when done by authors of SP study | Yes (24/25 studies) |
| Adjustment of SP for confounders | Unclear selection rule | Unclear selection rule | Unclear selection rule | Unclear selection rule | Selecting most fully adjusted SP estimated | Unclear selection rule |
| Other SP correction | No | No | No | Seroreversion | Type of antibodies a | Seroreversion, in secondary analysis |
| Death count adjustments | No adjustments | Deaths not assessed | Deaths not assessed | No adjustments | No adjustments | No adjustments |
| Time window for death counts | 10 d after completion of SP study | Deaths not assessed | Deaths not assessed | Distributional (truncated Gaussian and beta), mean 18.3 d from onset to seroconversion, 19.8 d from onset to death | 7 d after mid‐point of SP survey or as chosen by its authors | Distributional (gamma), mean 10 d from onset to seroconversion, 20 d from onset to death |
Abbreviations: d, days; IFR, infection fatality rate; SP, seroprevalence.
one‐tenth adjustment per each not tested antibody (IgG, IgM, IgA).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Rostami and Bobrovitz did not collect death counts to estimate IFR. The other four evaluations did not systematically adjust death counts for under‐ or over‐counting. Finally, ICCRT and O’Driscoll used distributional approaches on the time window for counting deaths (with means between seroconversion and death differing by 1.5 and 10 days, respectively), Ioannidis counted deaths until 7 days after the survey mid‐point (or the date survey authors made a strong case for), and Meyerowitz‐Katz counted deaths up until 10 days after survey end.
3.4. Quantitative synthesis, heterogeneity and main estimates
The six evaluations differed in quantitative synthesis approaches with implications for the main results (Table 4).
TABLE 4.
Quantitative synthesis approaches, stratification and/or regression and main estimates
| Meyerowitz‐Katz | Rostami | Bobrovitz | Imperial College COVID‐19 response team | Ioannidis | O’Driscoll | |
|---|---|---|---|---|---|---|
| Quantitative synthesis | 26 IFR estimates combined at one step with D‐L RE model, I 2 = 99.4% | First step 107 SP estimates combined separately for each country with D‐L RE model, then per region. Also D‐L RE for all 107 estimates, I 2 = 99.7% | Median SP calculated overall and per subgroup of interest. | Log‐linear model for pooling age‐stratified IFR, then age‐stratified estimates extrapolated to the age structure of populations of typical countries | First step, sample size‐weighted summary of SP per location; then median estimated across locations | The ensemble model eventually models age‐stratified IFR in a total of 45 countries with available age‐stratified death counts, but data are used as input from only 16 countries that have IFR data with some age stratification |
| Stratification and/or regression | Subgroup analyses per continent, month of publication, modelling versus serological and risk of bias | Subgroup analyses per age, gender, type of population, serological method, race/ethnicity, income, human development index, latitude/longitude, humidity, temperature, days from onset of pandemic; also RE meta‐regressions | Subgroup analyses per GBD region, scope (national, regional, local, sublocal), risk of bias, days since 100th case (also explored in meta‐regressions); RE inverse variance meta‐analysis of prevalence ratios for demographics (age, sex, race, close contact, HCW status) with I 2 = 85.1%‐99.4% per grouping factor | Focus on age‐strata, also IFR estimates with and without seroreversion, and (for some countries) excluding nursing home deaths | Separate analyses for age <70 years; also subgroup analyses according to level of overall mortality in the location | Focus on age‐strata; also per sex/gender and per country |
| Main estimates | Summary IFR 0.68 (95% CI‐0.53%‐0.82%), 0.60 when limited to serological studies | 263.5 million exposed/infected at the time of the study based on the pooled SP from all 107 data sets; when estimated per region the total is 641 million a | 643 million infected as of 17 November, based on estimated median under‐ascertainment factor of 11.9 (using 9 d before study end date for PCR counts) b | Overall IFR: LIC 0.22 (0.14, 0.39), LMIC) 0.37 (0.25, 0.61), UMIC 0.57 (0.38, 0.92), HIC 1.06 (0.73, 1.64) | Over 500 million infected as of September 12 (vs 29 million documented cases) globally; median IFR 0.23% in the available studies (0.09% in locations with <118 deaths/million), 0.20% in locations with 118‐500 deaths/million, 0.57% in locations with >500 deaths/million | 5.27% of the population of the 45 modelled countries had been infected by 1 September |
Abbreviations: IFR, infection fatality rate; RE, random effects; SP, seroprevalence.
In millions: Europe+North America 47, East+South‐East Asia 47, Latin America 9, South America 6, Sub Saharan Africa 62, Central and South Asia 446, North Africa and West Asia 24
Median under‐ascertainment was 14.5 overall based on 125 study estimates and 11.9 in national estimates, 15.7 in regional estimates and 24.0 in local estimates.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Meyerowitz‐Katz used random effects meta‐analysis of 26 IFRs calculating a summary estimate despite extreme between‐study heterogeneity (I 2 = 99.2%). Such extreme heterogeneity precludes obtaining meaningful summary estimates. Estimates from the same country/location were not combined first, and two multiply‐counted countries (Italy and China) have high IFRs entered in calculations. Meta‐analysis limited to seroprevalence studies yielded slightly lower summary IFR (0.60% vs 0.68%), but extreme between‐study heterogeneity persisted (I 2 = 99.5%); thus, summary estimates remained meaningless. Extreme between‐study heterogeneity persisted also within three risk‐of‐bias categories (I 2 = 99.6%, 98.8% and 94.8%, respectively), within Europe and within America. There was no between‐study heterogeneity for four Asian estimates, but none came from seroprevalence data and their IFR estimate (0.46%) is far higher than many subsequent Asian studies (outside Wuhan) using seroprevalence data 5 instead of modelling.
Rostami also performed random effects meta‐analyses but more appropriately combined at a first step seroprevalence data from studies in the same country, and in the same region, a summary estimate across all 107 estimates in all countries was also obtained. The step‐wise approach avoids the Meyerowitz‐Katz analysis flaw. However, seroprevalence estimates may still vary extremely even within the same location, for example if done at different times. Moreover, the main estimate of the evaluation (‘263.5 million exposed/infected at the time of the study’) extrapolated to the global population the pooled estimate from all 107 data sets. The more appropriate estimate is a sum of the infected per country, or at least per region. Actually, the authors did calculate numbers of people exposed/infected per world region. The sum was 641 million, 2.5‐fold larger. Moreover, these numbers did not reflect ‘the time of the study’: the 107 seroprevalence studies were done 2‐6 months before the Rostami evaluation was written.
Bobrovitz calculated medians (overall and across several subgroups of studies), and Ioannidis calculated sample size‐weighted means per location and then medians across locations. Their approaches avoid multiple counting of locations with many estimates available. Bobrovitz also performed random effects inverse variance meta‐analysis of prevalence ratios for diverse demographics (age, sex, race, close contact, healthcare workers). The approach is defendable, since prevalence ratios were calculated within each study, but still very large between‐study heterogeneity existed (I 2 = 85.1%‐99.4% per grouping factor) making results tenuous. Bobrovitz and Ioannidis reach congruent estimates for total number infected globally (643 million by November 17 and at least 500 million by September 12, respectively) with under‐ascertainment ratios of 11.9 in November and 17.2 in September. Only the latter evaluation calculated IFRs (0.23% overall; 0.05% for those <70 years old).
ICCRT and O’Driscoll focused on age‐stratified estimates. ICCRT extrapolated age‐stratified estimates to the age structure of populations of typical countries, obtaining separate overall IFR estimates for low‐income countries (0.22%), lower‐middle–income countries (0.37%), upper‐middle–income countries (0.57%) and high‐income countries (1.06%). O’Driscoll made extrapolations to 45 countries estimating 5.27% of their population infected by 1 September.
3.5. Global representativeness
Seroprevalence data lacked global representativeness. 72%‐91% of the seroprevalence evidence came from Europe and North America (78%‐100% from Europe or Americas) (Table 5). Lack of representativeness was most prominent in Meyerowitz‐Katz (only one estimate from Asia, none from Africa), ICCRT (no estimates from Asia or Africa) and O’Driscoll (only one estimate from Africa, no estimate from Asia). However, ICCRT extrapolated to all countries globally and O’Driscoll extrapolated to 45 countries including eight in Asia.
TABLE 5.
Global representativeness
| Meyerowitz‐Katz | Rostami | Bobrovitz | Imperial College COVID‐19 response team | Ioannidis | O’Driscoll b | |
|---|---|---|---|---|---|---|
| Estimates (countries) a | ||||||
| Europe | 11 (11) | 52 (13) | 33 (13) | 8 (7) | 22 (21) | 13 (13) |
| North America | 3 (1) | 22 (1) | 1 (1) | 1 (1) | 15 (2) | 1 (1) |
| Latin America | 1 (1) | 17 (2) | 3 (1) | 1 (1) | 3 (3) | 1 (1) |
| Asia | 1 (1) | 14 (5) | 2 (1) | 0 (0) | 10 (9) | 0 (0) |
| Africa | 0 (0) | 2 (2) | 1 (1) | 0 (0) | 1 (1) | 1 (1) |
| Oceania | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Information from Europe and North America | 91% of weight | 72% of data sets | 85% of data sets (82% of countries) | 90% of data sets | 73% of location estimates | 87% of countries |
| Information from Europe and America | 98% of weight | 85% of data sets | 93% of data sets (87% of countries) | 100% of data sets | 78% of location estimates | 94% of countries |
Geographic location of estimates (countries) included in main calculations.
The extrapolated 45 countries on which age‐stratified IFR estimates are obtained also include countries outside the regions that have at least one country represented (Pakistan, Philippines, Bangladesh, Indonesia, China, Thailand, South Korea, Japan) even though not directly measured in any of them.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4. DISCUSSION
This overview of six systematic evaluations of global spread and/or IFR of SARS‐CoV‐2 utilizing seroprevalence data highlights differences in methods, calculations and inferences. Several choices made by some evaluations led to bias. Other choices are defendable and reveal some unavoidable variability on how evidence on these important questions should be handled.
Choices that led to biased inflated IFR estimates are the inclusion of modelling estimates, inappropriate exclusion of low‐IFR studies despite fitting stated inclusion criteria of the evaluators, inappropriate inclusion of high‐IFR studies despite not fitting stated inclusion criteria, and using low death counts as exclusion criterion. Two evaluations (Meyerowitz‐Katz and ICCRT) suffered multiple such problems each. These biases contributed to generate inflated and, sometimes, overtly implausible results. These two evaluations also narrowly selected very scant evidence (16 and 10 studies, including only one and five peer‐reviewed articles, respectively), while hundreds of seroprevalence estimates are available.
Differences in types of study designs and populations considered eligible may be defended with various arguments by each evaluator. Studies of healthcare workers were consistently excluded. No consensus existed on studies of blood donors, clinical samples, workers at no obvious high‐risk occupations and various convenience samples; these designs have variable reliability. Reliability increases with careful adjustment for sampling, demographics and other key factors and when missing data are limited. General population sampling is theoretically best, but general population studies may still suffer large bias from selective missingness. Unreachable individuals, institutionalized people and non‐participating invitees are typically at higher infection risk; if so, some general population studies may substantially underestimate seroprevalence (overestimate IFR). For example, Meyerowitz‐Katz included a Danish government survey press release 33 where only 1071 of 2600 randomly selected invitees participated (missingness 59%); the estimated IFR (0.79%) is probably substantially inflated. 6 , 28
Differences may also ensue from seroprevalence adjustments for test performance and other factors. 34 , 35 Sometimes the change in estimated seroprevalence is substantial. 36 , 37 , 38 Special caution is needed with low seroprevalence. 39 When not all types of antibodies are assessed, a correction may also be useful. Adjustment for test performance may seemingly suffice. However, control samples used to estimate test sensitivity come from PCR‐tested diagnosed patients, while missed diagnoses typically reflect asymptomatic or less symptomatic patients not seeking testing. Sensitivity may be much lower in these people, as many develop no or low‐titre antibodies. 40 , 41 Seroreversion has a similar impact. Preliminary evidence suggests substantial seroreversion. 29 , 42 , 43 , 44 , 45 For example, among healthcare personnel, 28.2% seroreverted in 2 months (64.9% in those with low titres originally). 45 Only ICCRT and O’Driscoll considered corrections for seroreversion, but still did not allow for high seroreversion. All these factors would result in underestimating seroprevalence (overestimating IFR).
Both over‐ and under‐counting of COVID‐19 deaths (the IFR numerator) may exist, 46 , 47 varying across countries with different testing and death coding. Correction of COVID‐19 death counts through excess deaths is problematic. Excess reflects both COVID‐19 deaths and deaths from measures taken. 46 , 47 , 48 , 49 Year‐to‐year variability is substantial, even more so within age‐strata. Comparison against averages of multiple previous years is naïve, worse in countries with substantial demographic changes. For example, in the first wave, an excess of 8071 deaths (SMR 1.03, 95% CI 1.03‐1.04) in Germany became a deficit of 4926 deaths (SMR 0.98, 95% CI 0.98‐0.99) after accounting for demographic changes. 50 The exact timepoint when deaths are counted may affect IFR calculations when surveys happen while many deaths are still accruing. All evaluations that counted deaths allowed for greater time for death to occur than for seroconversion, but Meyerowitz‐Katz used a most extreme delay, considering deaths until 10 days after survey end. Surveys take from one day to over a month; thus, inferred sampling‐to‐death delay may occasionally exceed 6 weeks. Meyerowitz‐Katz defends this choice also in another paper 10 choosing 4 weeks after the serosurvey mid‐point. However, the argument (accounting for death reporting delays) is weak. Several situational reports plot deaths according to date of occurrence rather than date of reporting anyhow. 51 Moreover, infection‐to‐death time varies substantially and may be shorter in developing countries where fewer people are long‐sustained by medical support.
Some quantitative synthesis approaches were problematic, for example calculating summary estimates despite I 2 > 99% or no data combination within the same country/location before synthesis across countries/locations. Another generic problem with meta‐analysis of such data is that it penalizes better studies that allow more appropriately for uncertainty in estimates (eg by accounting for test performance and adjusting for important covariates). Studies with less rigorous or no adjustments may have narrower CIs (smaller variance, thus larger weight). 5 Finally, for IFR meta‐analysis, studies with few deaths may have higher variance (lower weight) and these studies may have the lowest IFR.
Age stratification for IFR estimation and synthesis is a reasonable choice to reduce between‐study heterogeneity driven by steep COVID‐19 death risk age gradient. 52 However, both analyses 4 , 6 that capitalized on granular age stratification made tenuous extrapolations to additional countries from thin or no data. ICCRT lacked seroprevalence data on low‐income and lower‐middle–income countries (~half the global population); upper‐middle–income countries (~35% of global population) were only represented by one estimate from Brazil assuming IFR = 1%, exceeding twofold to fivefold other peer‐reviewed estimates from Brazil. 13 , 53 Estimates used from high‐income countries included an impossible Italian estimate (IFR = 2.5%) 30 and mostly non–peer‐reviewed data. O’Driscoll was more careful, but still some IFR extrapolations appear highly inflated versus data from subsequently accrued seroprevalence studies. Their ensemble model assumed highest IFR in Japan (1.09%) and lowest in Kenya (0.09%) and Pakistan (0.16%). Currently, available seroprevalence studies from these countries show markedly lower IFR estimates: =<0.03%, 54 , 55 , 56 =<0.01% 14 and 0.04%‐0.07%, 57 , 58 respectively. In Japan, infections apparently spread widely without causing detectable excess mortality. 54 In Kenya, under‐ascertainment compared with documented cases was ~1000‐fold. 14 While some COVID‐19 deaths are certainly missed in Africa, containment measures are more deadly. 59
All six evaluations greatly over‐represented Europe and America. Only two (Rostami and Ioannidis) included meaningful amounts of data from Asia and Africa (still less than their global population share) in main estimate calculations. Currently, extensive data suggest high under‐ascertainment ratios in Africa and many Asian countries 5 , 61 and thus much lower IFR in Asia (outside Wuhan) and Africa than elsewhere.
Quality of seroprevalence studies varies. Risk‐of‐bias assessments in prevalence studies are difficult. There are multiple risk‐of‐bias scales/checklists, 62 , 63 , 64 , 65 but bias scores do not translate necessarily to higher or lower IFR estimates, while assessors often disagree in scoring (Appendix S1).
Acknowledging these caveats, four of the six evaluations largely reach congruent estimates of global pandemic spread. O’Driscoll estimated 5.27% of the population of 45 countries had been infected by 1 September 2020, that is 180 million infected among 3.4 billion. Excluding China, the proportion of population infected among the remaining 44 countries would be ~9%, likely >10% after accounting for seroreversion. Countries not included among the 45 include some of the most populous ones with high infection rates (India, Mexico, Brazil, most African countries). Therefore, arguably at least 10% of the non‐China global population (ie at least 630 million) would be infected as of 1 September. This is very similar to the Ioannidis (at least 500 million infected as of 12 September) and Rostami (641 million infected by summer, when numbers are added per region) estimates. The Bobrovitz estimate (643 million infected as of 17 November) should be increased substantially given that only 2 of 17 countries informing the calculated under‐ascertainment ratio were in Asia or Africa, continents with much larger under‐ascertainment ratios. National surveys in India actually estimated 60% seroprevalence in November in urban areas. 66 Therefore, probably infected people globally were ~1 billion (if not more) by 17 November (compared with 54 million documented cases). By extrapolation, one may cautiously estimate ~1.5‐2.0 billion infections as of 21 February 2021 (compared with 112 million documented cases). This corresponds to global IFR ~0.15%—a figure open to adjustment for any over‐ and under‐counting of COVID‐19 deaths (Appendix S2).
Meyerowitz‐Katz and ICCRT reach higher estimates of IFR, but, as discussed above, these are largely due to endorsing selection criteria focusing on high‐IFR countries, violations of chosen selection criteria and obvious flaws that consistently cause IFR overestimation. Similar concerns apply to another publication with implausibly high age‐stratified IFRs by Meyerowitz‐Katz limited to countries with advanced economies, again narrowly selected some of the highest IFR locations and estimates. 12
Even correcting inappropriate exclusions/inclusion of studies, errors and seroreversion, IFR still varies substantially across continents and countries. Overall average IFR may be ~0.3%‐0.4% in Europe and the Americas (~0.2% among community‐dwelling non‐institutionalized people) and ~0.05% in Africa 14 and Asia (excluding Wuhan). Within Europe, IFR estimates were probably substantially higher in the first wave in countries like Spain, 67 UK 68 and Belgium 69 and lower in countries such as Cyprus or Faroe Islands (~0.15%, even case fatality rate is very low), 70 Finland (~0.15%) 71 and Iceland (~0.3%). 72 One European country (Andorra) tested for antibodies 91% of its population. 73 Results 73 suggest an IFR less than half of what sampling surveys with greater missingness have inferred in neighbouring Spain. Moreover, high seroreversion was noted, even a few weeks apart 73 ; thus, IFR may be even lower. Differences exist also within a country; for example within the USA, IFR differs markedly in disadvantaged New Orleans districts versus affluent Silicon Valley areas. Differences are driven by population age structure, nursing home populations, effective sheltering of vulnerable people, 74 medical care, use of effective (eg dexamethasone) 75 or detrimental (eg hydroxychloroquine) 76 treatments, host genetics, 77 viral genetics and other factors.
Infection fatality rate may change over time locally 78 and globally. If new vaccines and treatments pragmatically prevent deaths among the most vulnerable, theoretically global IFR may decrease even below 0.1%. However, there are still uncertainties both about the real‐world effectiveness of new options, as well as the pandemic course and post‐pandemic SARS‐CoV‐2 outbreaks or seasonal re‐occurrence. IFR will depend on settings and populations involved. For example, even ‘common cold’ coronaviruses have IFR~10% in nursing home outbreaks. 79
Admittedly, primary studies, their overviews and the current overview of overviews have limitations. All estimates have uncertainty. Interpretation unavoidably has subjective elements. This challenge is well‐known in the literature of discrepant systematic reviews. 80 , 81 , 82 , 83 , 84 Cross‐linking diverse types of evidence generates even more diverse eligibility/design/analytical options. Nevertheless, one should separate clear errors and directional biases from defendable eligibility/design/analytical diversity.
Allowing for such residual uncertainties, reassuringly the picture from the six evaluations assessed here is relatively congruent: SARS‐CoV‐2 is widely spread and has lower average IFR than originally feared, and substantial global and local heterogeneity. Using more accurate estimates of IFR may yield more appropriate planning, predictions and evaluation of measures.
CONFLICTS OF INTEREST
None.
DISCLOSURES
I am the author of one of the six evaluations assessed in this article.
Supporting information
Appendices
ACKNOWLEDGEMENTS
I am grateful to Niklas Bobrovitz and Rahul Arora for offering clarifications on their important study.
Ioannidis JPA. Reconciling estimates of global spread and infection fatality rates of COVID‐19: An overview of systematic evaluations. Eur J Clin Invest. 2021;51:e13554. 10.1111/eci.13554
Funding information
None; the Meta‐Research Innovation Center at Stanford (METRICS) has been funded with grants from the Laura and John Arnold Foundation.
REFERENCES
- 1. Meyerowitz‐Katz G, Merone L. A systematic review and meta‐analysis of published research data on COVID‐19 infection fatality rates. Int J Infect Dis. 2020;101:138‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rostami A, Sepidarkish M, Leeflang MMG, et al. SARS‐CoV‐2 seroprevalence worldwide: a systematic review and meta‐analysis. Clin Microbiol Infect. 2021;27(3):331‐340. 10.1016/j.cmi.2020.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bobrovitz N, Arora RK, Cao C, et al. Global seroprevalence of SARS‐CoV‐2 antibodies: a systematic review and meta‐analysis. medRxiv. 2020. 10.1101/2020.11.17.20233460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Imperial College COVID‐19 response team . Report 34: COVID‐19 Infection Fatality Ratio: Estimates from Seroprevalence. October 29, 2020. [Google Scholar]
- 5. Ioannidis JPA. The infection fatality rate of COVID‐19 inferred from seroprevalence data. Bull World Health Organ. 2021;99:19‐33F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O'Driscoll M, Ribeiro Dos Santos G, Wang L, et al. Age‐specific mortality and immunity patterns of SARS‐CoV‐2. Nature. 2021;590(7844):140‐145. 10.1038/s41586-020-2918-0 [DOI] [PubMed] [Google Scholar]
- 7. Franceschi VB, Santos AS, Glaeser AB, et al. Population‐based prevalence surveys during the Covid‐19 pandemic: a systematic review. Rev Med Virol. 2020;e2200. 10.1002/rmv.2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen X, Chen Z, Azman AS, et al. Serological evidence of human infection with SARS‐CoV‐2: a systematic review and meta‐analysis. medRxiv. 2020:2020.09.11.20192773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arora RK, Joseph A, Van Wyk J, et al. SeroTracker: A global SARS‐CoV‐2 seroprevalence dashboard. Lancet Infect Dis. 2021;(21):e76‐e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levin AT, Hanage WP, Owusu‐Boaitey N, Cochran KB, Walsh SP, Meyerowitz‐Katz G. Assessing the age specificity of infection fatality rates for COVID‐19: systematic review, meta‐analysis, and public policy implications. Eur J Epidemiol. 2020;35(12):1123‐1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson L. SARS‐CoV2 COVID‐19 infection fatality rate implied by the serology antibody testing in New York City. SSRN 2020; http://refhub.elsevier.com/S1201‐9712(20)32180‐9/sbref0350 [Google Scholar]
- 12. Sood N, Simon P, Ebner P, et al. Seroprevalence of SARS‐CoV‐2‐specific antibodies among adults in Los Angeles County, California, on April 10–11, 2020. JAMA. 2020;323(23):2425‐2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Silveira MF, Barros AJD, Horta BL, et al. Population‐based surveys of antibodies against SARS‐CoV‐2 in Southern Brazil. Nat Med. 2020;26(8):1196‐1199. [DOI] [PubMed] [Google Scholar]
- 14. Uyoga S, Adetifa IMO, Karanja HK, et al. Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Kenyan blood donors. Science. 2021;371(6524):79‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Streeck H, Schulte B, Kümmerer BM, et al. Infection fatality rate of SARS‐CoV2 in a super‐spreading event in Germany. Nat Commun. 2020;11(1):5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson C, Grayson N, Paton RS, Lourenco J, Penman BS, Lee L. Neutralising antibodies to SARS coronavirus 2 in Scottish blood donors – a pilot study of the value of serology to determine population exposure. medRxiv. 2020. 10.1101/2020.04.13.20060467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Snoeck CJ, Vaillant M, Abdelrahman T, et al. Prevalence of SARS‐CoV‐2 infection in the Luxembourgish population: the CON‐VINCE study. medRxiv. 2020. 10.1101/2020.05.11 [DOI] [Google Scholar]
- 18. Galanis P, Vraka I, Fragkou D, Bilali A, Katelidou D. Seroprevalence of SARS‐CoV‐2 antibodies and associated factors in health care workers: a systematic review and meta‐analysis. medRxiv. 2020; 10.1101/2020.10.23.20218289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Imbert E, Kinley PM, Scarborough A, et al. Coronavirus disease 2019 (COVID‐19) outbreak in a San Francisco homeless shelter. Clin Infect Dis. 2020;ciaa1071. 10.1093/cid/ciaa1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. https://covidtracking.com/data/long‐term‐care. Accessed January 18, 2021.
- 21. Burnett JJ. Psychographic and demographic characteristics of blood donors. J Consumer Res. 1981;8:62‐66. [Google Scholar]
- 22. Ganguli M, Lytle ME, Reynolds MD, Dodge HH. Random versus volunteer selection for a community‐based study. J Gerontol A Biol Sci Med Sci. 1998;53(1):M39‐M46. [DOI] [PubMed] [Google Scholar]
- 23. Patel EU, Bloch EM, Grabowski MK. Sociodemographic and behavioral characteristics associated with blood donation in the United States: a population‐based study. Transfusion. 2019;59(9):2899‐2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lattimore S, Wickenden C, Brailsford SR. Blood donors in England and North Wales: demography and patterns of donation. Transfusion. 2015;55:91‐99. [DOI] [PubMed] [Google Scholar]
- 25. Burgdorf KS, Simonsen J, Sundby A, et al. Socio‐demographic characteristics of Danish blood donors. PLoS One. 2017;12:e0169112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Volken T, Weidmann C, Bart T, Fischer Y, Klüter H, Rüesch P. Individual characteristics associated with blood donation: a cross‐national comparison of the German and Swiss population between 1994 and 2010. Transfus Med Hemother. 2013;40(2):133‐138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Erikstrup C, Hother CE, Pedersen OBV, et al. Estimation of SARS‐CoV‐2 infection fatality rate by real‐time antibody screening of blood donors. Clin Infect Dis. 2021;72(2):249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emmenegger M, De Cecco E, Lamparter D, et al. Early plateau of SARS‐CoV‐2 seroprevalence identified by tripartite immunoassay in a large population. medRxiv. 2020. 10.1101/2020.05.31 [DOI] [Google Scholar]
- 29. Rosenberg ES, Tesoriero JM, Rosenthal EM, et al. Cumulative incidence and diagnosis of SARS‐CoV‐2 infection in New York. Ann Epidemiol. 2020;48:23‐29.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Primi risultati dell’ indagine di sieroprevalenza sul SARS‐CoV2. https://www.istat.it/it/archivio/246156. Accessed January 20, 2021. [Google Scholar]
- 31. Russell TW, Golding N, Hellewell J, et al. Reconstructing the early global dynamics of under‐ascertained COVID‐19 cases and infections. BMC Med. 2020;18:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yiannoutsos CT, Halverson PK, Menachemi N. Bayesian estimation of SARS‐CoV‐2 prevalence in Indiana by random testing. Proc Natl Acad Sci USA. 2021;118:e2013906118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Statens Serum Institut . De første foreløbige resultater af undersøgelsen for COVID‐19 i befolkningen er nu klar. https://www.ssi.dk/aktuelt/nyheder/2020/de‐forste‐forelobige‐resultater‐af‐undersogelsen‐for‐covid‐19‐i‐befolkningen‐er‐nu‐klar. Accessed January 18, 2021 [Google Scholar]
- 34. Burgess S, Ponsford MJ, Gill D. Are we underestimating seroprevalence of SARS‐CoV‐2? BMJ. 2020;370:m3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deeks JJ, Dinnes J, Takwoingi Y, et al. Cochrane COVID‐19 diagnostic test accuracy group. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database Syst Rev. 2020;6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bendavid E, Mulaney B, Sood N, et al. COVID‐19 antibody seroprevalence in Santa Clara County, California. Int J Epidemiol. 2021;dyab010. 10.1093/ije/dyab010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shakiba M, Nazemipour M, Salari A, et al. Seroprevalence of SARS‐CoV‐2 in Guilan Province, Iran, April 2020. Emerg Infect Dis. 2021;27(2):636‐638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nawa N, Kuramochi J, Sonoda S, et al. Seroprevalence of SARS‐CoV‐2 IgG antibodies in Utsunomiya City, Greater Tokyo, after first pandemic in 2020 (U‐CORONA): a household‐ and population‐based study. medRxiv. 2020. 10.1101/2020.07.20.20155945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cai B, Ioannidis JP, Bendavid E, Tian L. Exact inference for disease prevalence based on a test with unknown specificity and sensitivity. arXiv. 2020; https://arxiv.org/abs/2011.14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Long Q‐X, Tang X‐J, Shi Q‐L, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26:1200‐1204. [DOI] [PubMed] [Google Scholar]
- 41. Solbach W, Schiffner J, Backhaus I, et al. Antibody profiling of COVID‐19 patients in an urban low‐incidence region in Northern Germany. Front Public Health. 2020;8:570543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stringhini S, Wisniak A, Piumatti G, et al. Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Geneva, Switzerland (SEROCoV‐POP): a population‐based study. Lancet. 2020;396(10247):313‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ibarrondo FJ, Fulcher JA, Goodman‐Meza D, et al. Rapid decay of anti–SARS‐CoV‐2 antibodies in persons with mild covid‐19. N Engl J Med. 383(11):1085‐1087. 10.1056/NEJMc2025179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shioda K, Lau MS, Kraay AN, et al. Estimating the cumulative incidence of SARS‐CoV‐2 infection and the infection fatality ratio in light of waning antibodies. medRxiv. 2020; 2020.11.13.20231266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Self WH, Tenforde MW, Stubblefield WB, et al. Decline in SARS‐CoV‐2 antibodies after mild infection among frontline health care personnel in a multistate hospital network — 12 States, April–August 2020. MMWR. 2020;69:1762‐1766. 10.15585/mmwr.mm6947a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ioannidis JPA. Global perspective of COVID‐19 epidemiology for a full‐cycle pandemic. Eur J Clin Invest. 2020;7:e13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. VanderWeele TJ. Challenges estimating total lives lost in COVID‐19 decisions: consideration of mortality related to unemployment, social isolation, and depression. JAMA. 2020;324:445‐446. [DOI] [PubMed] [Google Scholar]
- 48. Bauchner H, Fontanarosa PB. Excess deaths and the Great Pandemic of 2020. JAMA. 2020;324:1504‐1505. [DOI] [PubMed] [Google Scholar]
- 49. Woolf SH, Chapman DA, Sabo RT, Weinberger DM, Hill L. Excess deaths from COVID‐19 and other causes, March‐April 2020. JAMA. 2020;324(5):510‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stang A, Standl F, Kowall B, et al. Excess mortality due to COVID‐19 in Germany. J Infect. 2020;81(5):797‐801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ioannidis JPA, Axfors C, Contopoulos‐Ioannidis DG. Population‐level COVID‐19 mortality risk for non‐elderly individuals overall and for non‐elderly individuals without underlying diseases in pandemic epicenters. Environ Res. 2020;188:109890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williamson EJ, Walker AJ, Bhaskaran K, et al. Factors associated with COVID‐19‐related death using OpenSAFELY. Nature. 2020;584(7821):430‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buss LF, Prete CA Jr, Abrahim CMM, et al. Three‐quarters attack rate of SARS‐CoV‐2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371(6526):288‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hibino S, Hayashida K, Ahn AC, Hayashida Y. Dynamic change of COVID‐19 seroprevalence among asymptomatic population in Tokyo during the second wave. medRxiv. 2020. 10.1101/2020.09.21.20198796 [DOI] [Google Scholar]
- 55. Takita M, Matsumura T, Yamamoto K, et al. Geographical Profiles of COVID‐19 Outbreak in Tokyo: An analysis of the primary care clinic‐based point‐of‐care antibody testing. J Prim Care Community Health. 2020;11:2150132720942695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Doi A, Iwata K, Kuroda H, et al. Estimation of seroprevalence of novel coronavirus disease (COVID‐19) using preserved serum at an outpatient setting in Kobe, Japan: a cross‐sectional study. medRxiv. 2020. 10.1101/2020.04.26.20079822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nisar I, Ansari N, Amin M, et al. Serial population based serosurvey of antibodies to SARS‐CoV‐2 in a low and high transmission area of Karachi, Pakistan. medRxiv. 2020. 10.1101/2020.07.28.20163451 [DOI] [Google Scholar]
- 58. Javed W, Baqar J, Abidi SHB, Farooq W. Sero‐prevalence findings from metropoles in Pakistan: implications for assessing COVID‐19 prevalence and case‐fatality within a dense, urban working population. medRxiv. 2020; 10.1101/2020.08.13.20173914 [Google Scholar]
- 59. Roberton T, Carter ED, Chou VB, et al. Early estimates of the indirect effects of the COVID‐19 pandemic on maternal and child mortality in low‐income and middle‐income countries: a modelling study. Lancet Glob Health. 2020;8(7):e901‐e908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Poustchi H, Darvishian M, Mohammadi Z, et al. SARS‐CoV‐2 antibody seroprevalence in the general population and high‐risk occupational groups across 18 cities in Iran: a population‐based cross‐sectional study. Lancet Infect Dis. 2021;21(4):473‐481. 10.1016/S1473-3099(20)30858-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Abu Raddad LJ, Chemaitelly H, Ayoub HH, et al. Characterizing the Qatar advanced‐phase SARS‐CoV‐2 epidemic. medRxiv. 2020; 10.1101/2020.07.16.20155317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shamliyan T, Kane RL, Dickinson S. A systematic review of tools used to assess the quality of observational studies that examine incidence or prevalence and risk factors for diseases. J Clin Epidemiol. 2010;63:1061‐1070. [DOI] [PubMed] [Google Scholar]
- 63. Munn Z, Moola S, Lisy K, Riitano D, Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and incidence data. Int J Evid Based Health. 2015;13:147‐153. [DOI] [PubMed] [Google Scholar]
- 64. Munn Z, Moola S, Riitano D, Lisy K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int J Health Policy Manag. 2014;3:123‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hoy D, Brooks P, Woolf A, et al. Assessing risk of bias in prevalence studies: modification of an existing tool and evidence of interrater agreement. J Clin Epidemiol. 2012;65:934‐939. [DOI] [PubMed] [Google Scholar]
- 66. Ghosh A. India is missing about 90 infections for every COVID case, latest government analysis shows. https://theprint.in/health/india‐is‐missing‐about‐90‐infections‐for‐every‐covid‐case‐latest‐govt‐analysis‐shows/567898/. Accessed January 18, 2021 [Google Scholar]
- 67. Pollán M, Pérez‐Gómez B, Pastor‐Barriuso R, et al. Prevalence of SARS‐CoV‐2 in Spain (ENE‐COVID): a nationwide, population‐based seroepidemiological study. Lancet. 2020;396(10250):535‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ward H, Atchinson C, Whitaker M, et al. Antibody prevalence for SARS‐CoV‐2 following the peak of the pandemic in England: REACT2 study in 100,000 adults. medRxiv. 2020; 10.1101/2020.08.12 [DOI] [Google Scholar]
- 69. Herzog S, De Bie J, Abrams S, et al. Seroprevalence of IgG antibodies against SARS coronavirus 2 in Belgium – a prospective cross sectional study of residual samples. medRxiv. 2020; 10.1101/2020.06.08.20125179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Worldometer . Cyprus and Faroe Islands, www.worldometer.com, Accessed February 21, 2021. [Google Scholar]
- 71. Finland Department of Health and Welfare . Weekly report of the population serology survey of the corona epidemic. Helsinki: Finland Department of Health and Welfare. 2020. [Finnish]. https://www.thl.fi/roko/cov‐vaestoserologia/seroreport_weekly.html Accessed January 18, 2021 [Google Scholar]
- 72. Gudbjartsson DF, Norddahl GL, Melsted P, et al. Humoral immune response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020. NEJMoa2026116 [online ahead of print]. 10.1056/NEJMoa2026116 PMID: 32871063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Royo‐Cebrecos C, Vilanova D, Lopez J, et al. Mass SARS‐CoV‐2 serological screening for the Principality of Andorra. Research Square. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ioannidis JPA. Precision shielding for COVID‐19: Metrics of assessment and feasibility of deployment. BMJ Glob Health. 2021;6:e004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Horby P, Lim WS, Emberson JR, et al. Dexamethasone in hospitalized patients with Covid‐19 ‐ preliminary report. New Engl J Med. 2021; 384: 693‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Axfors C, Schmitt AM, Janiaud P, et al. Mortality outcomes with hydroxychloroquine and chloroquine in COVID‐19: an international collaborative meta‐analysis of randomized trials. Nat Commun. 2021. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen HH, Shaw DM, Petty LE, et al. Host genetic effects in pneumonia. Am J Hum Genet. 2021;108(1):194‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ioannidis JPA, Axfors CA, Contopoulos‐Ioannidis DG. Second versus first wave of COVID‐19 deaths: shifts in age distribution and in nursing home fatalities. Environ Res. 2021;195:110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Veiga ABGD, Martins LG, Riediger I, Mazetto A, Debur MDC, Gregianini TS. More than just a common cold: endemic coronaviruses OC43, HKU1, NL63, and 229E associated with severe acute respiratory infection and fatality cases among healthy adults. J Med Virol. 2021;93(2):1002‐1007. [DOI] [PubMed] [Google Scholar]
- 80. Ioannidis JPA. Meta‐research: the art of getting it wrong. Res Synth Methods. 2010;1:169‐184. [DOI] [PubMed] [Google Scholar]
- 81. Campbell J, Bellamy N, Gee T. Differences between systematic reviews/meta‐analyses of hyaluronic acid/hyaluronan/hylan in osteoarthritis of the knee. Osteoarthritis Cartilage. 2007;15:1424‐1436. [DOI] [PubMed] [Google Scholar]
- 82. Jadad AR, Cook DJ, Browman GP. A guide to interpreting discordant systematic reviews. Can Med Assoc J. 1997;156:1411‐1416. [PMC free article] [PubMed] [Google Scholar]
- 83. Moayyedi P, Deeks J, Talley NJ, Delaney B, Forman D. An update of the Cochrane systematic review of Helicobacter pylori eradication therapy in nonulcer dyspepsia: resolving the discrepancy between systematic reviews. Am J Gastroenterol. 2003;98:2621‐2626. [DOI] [PubMed] [Google Scholar]
- 84. Linde K, Willich SN. How objective are systematic reviews? Differences between reviews on complementary medicine. J Royal Soc Med. 2003;96:17‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott Architect SARS‐CoV‐2 IgG assay and seroprevalence in Boise. Idaho. J Clin Microbiol. 2020;58(8):e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendices


