Abstract
Novel coronary pneumonia (COVID‐19) is a respiratory distress syndrome caused by a new type of coronavirus. Understanding the genetic basis of susceptibility and prognosis to COVID‐19 is of great significance to disease prevention, molecular typing, prognosis, and treatment. However, so far, there have been only two genome‐wide association studies (GWASs) on the susceptibility of COVID‐19. Starting with these reported DNA variants, we found the genes regulated by these variants through cis‐eQTL and cis‐meQTL acting. We further did a series of bioinformatics analysis on these potential risk genes. The analysis shows that the genetic variants on EHF regulate the expression of its neighbor CAT gene via cis‐eQTL. There was significant evidence that CAT and the SARS‐CoV‐2‐related S protein binding protein ACE2 interact with each other. Intracellular localization results showed that CAT and ACE2 proteins both exists in the cell membrane and extracellular area and their interaction could have an impact on the cell invasion ability of S protein. In addition, the expression of these three genes showed a significant positive correlation in the lungs. Based on these results, we propose that CAT plays a crucial intermediary role in binding effectiveness of ACE2, thereby affecting the susceptibility to COVID‐19.
Keywords: COVID‐19, bioinformatics, protein interaction, candidate gene, eQTL
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- COVID‐19
coronavirus disease 2019
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
Novel coronary pneumonia (COVID‐19) is a respiratory distress syndrome caused by a new type of coronavirus. The disease began to break out in Wuhan, China, at the end of 2019, and officially entered the global pandemic phase in April 2020. Until now, it is still in a pandemic stage in a number of countries. 1 , 2 , 3 So far, the cumulative number of deaths due to COVID‐19 worldwide has reached 1 870 000. The COVID‐19 is the direct cause of the current global political and economic turmoil.
From an epidemiological perspective, an important feature of COVID‐19 is that a considerable proportion of population can turn into a state of asymptomatic or mild illness for a long time after being infected with the virus, while a small proportion quickly enter the moderate or severe stage. 4 This phenomenon of population differences strongly suggests that the susceptibility of COVID‐19 is influenced by genetic factors. Understanding the genetic basis of susceptibility to COVID‐19 is of great significance to disease prevention, molecular typing, prognosis and treatment.
However, so far, there have been only two genome‐wide association studies (GWASs) on COVID‐19. 5 , 6 One of them by Kachuri et al compared the genotypes of 676 individuals with positive SARS‐CoV‐2 tests and 1334 with negative tests at the genome‐wide level, and found the genetic variation on the EHF gene showing significant association with the tests results. Another GWAS by Ellinghaus et al considered 1610 severe patients as cases and 2205 mild patients or healthy people as controls, and found that genetic variants in the LZTFL1 and ABO genes were significantly associated with the severity of COVID‐19. The designs of the two studies were in fact very different in that the first one by Kachuri et al focused on the susceptibility to COVID‐19 and the second one by Ellinghaus et al focused on the prognosis of COVID‐19. Although these two studies were not very strict in the selection of control samples, the positive findings have provided an important reference for our understanding of COVID‐19 susceptibility and prognosis. As for the mechanism by which these positive locus affect the susceptibility and prognosis of COVID‐19, it is yet unclear. We next bioinformatically analyzed the potential risk genes associated with COVID‐19 susceptibility and prognosis.
We found the genes regulated by these variants through cis‐eQTL and cis‐meQTL acting. We further performed protein interaction network analysis, intracellular location analysis, and gene expression correlation analysis for these potentially functionally related genes. The results showed that the genetic variants on EHF regulate the expression of its neighbor CAT gene via cis‐eQTL acting. There was significant evidence that CAT and the SARS‐CoV‐2‐related S protein binding protein interact with each other. Intracellular localization results showed that CAT protein and ACE2 both exists in the cell membrane and extracellular area. In addition, the expression of these three genes showed a significant positive correlation in the lungs. Based on these results, we propose a hypothesis that CAT plays a crucial intermediary role in binding effectiveness of ACE2, thereby affecting the susceptibility to COVID‐19.
2. SUPPORTIVE EVIDENCE
We started with the functional annotations of the positive findings of the two GWASs. These included the EHF rs286914 significantly associated with the susceptibility of COVID‐19 6 and the LZTFL1 rs11385942 and ABO rs657152 significantly associated with the prognosis of COVID‐19. 5 Although one of the GWAS 6 is a preprint, it also laid foundation for follow‐up research. The rs286914 is an intronic variant of EHF on chromosome 11 and the A allele of this SNP was associated with an increased risk of positive respiratory virus test (OR = 1.52, Table S1). The A allele from the 1000 genomes project showed a large frequency difference between major continents with patterns showing higher frequencies in Africa (AFR 38.20%), America (AME 33.72%), and Europe (EUR 30.82%), but lower frequencies in East Asia (EAS 12.20% ) and South Asia (SAS 15.24%, Figure 1A). Kachuri GWAS were conducted in UK Biobank participants. As a result, the genomic variants with high frequency in these populations are more likely to be detected. In this case, the variant happened to be rarer in Asians. However, this is merely a reflection of the Winner's curse. We further investigated the existing eQTL 7 and meQTL databases, 8 and found that rs286914 (EHF) is a cis‐eQTL of the CAT gene about 140kbp upstream and a cis‐meQTL of cg18414381 on the EHF gene in peripheral blood (Tables 1 and 2, NeQTL = 5311, NmeQTL = 4170). Rs286914 also may be the eQTL of CAT in kidney tissue (P = .08, http://mulinlab.org/qtlbase/). This insignificant result might be partially explained by an insufficient sample size (N = 166). Until now, no significant correlation between rs286914 and CAT has been observed in other tissues including the lung. In the future, the sample size needs to be expanded to further verify their correlation in other tissues, especially in lung. This finding suggested a functional role of rs286914 on the expression of CAT while it does not necessarily directly affect the expression of EHF because rs286914 is not a significant cis‐eQTL of EHF and cis‐meQTL is very common over the genome. 9 , 10
FIGURE 1.
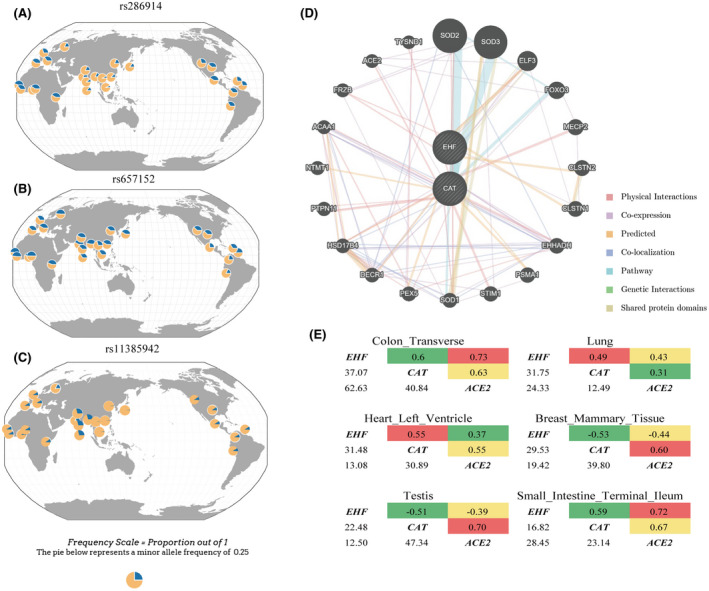
A, Global genotype frequency distribution of rs286914 in EHF; (B) Global genotype frequency distribution of rs657152 in ABO; (C): Global genotype frequency distribution of rs11385942 in LZTFL1; (D) Gene interaction networks created in GeneMania for EHF and CAT. The query genes was analyzed (striped circles) and additionally automatically generated their interacting genes (non‐striped circles). The color of the lines connecting the genes denotes the interaction types (supplementary materials). The size of the circle indicates the importance of the gene in the specific interactions while the width of lines indicates the weight of interaction between genes (Table S2). E, Correlation of CAT, ACE2, and EHF gene expression among top 6 tissues from the GTEx database (Table S6)
TABLE 1.
Cis‐eQTL of SNPs associated with COVID‐19 susceptibility and prognosis in peripheral blood
| SNP | EA | CHR | Gene | eQTL | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Distance (kbp) | Beta | P value | FDR | ||||
| rs286914 | A | 11p13 | EHF | CAT | 140 | −4.78 | 1.74E‐06 | 0 |
| rs11385942 | GA | 3p21.31 | LZTFL1 | — | — | — | — | — |
| rs657152 | A | 9q34.2 | ABO | GBGT1 | 100 | −5.00 | 5.72E‐07 | 0 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
TABLE 2.
Cis‐meQTL of SNPs associated with COVID‐19 susceptibility and prognosis in peripheral blood
| SNP | EA | CHR | Gene | cis‐meQTL | |||||
|---|---|---|---|---|---|---|---|---|---|
| CpG | Gene | Distance (kbp) | Beta | se | P value | ||||
| rs286914 | A | 11p13 | EHF | cg18414381 | EHF | — | 0.013 | 0.001 | 4.88E‐113 |
| rs11385942 | GA | 3p21.31 | LZTFL1 | — | — | — | — | — | — |
| rs657152 | A | 9q34.2 | ABO | cg11879188 | ABO | — | −0.059 | 0.001 | 0 |
| cg13506600 | ABO | — | −0.035 | 0.001 | 0 | ||||
| cg21160290 | ABO | — | −0.150 | 0.002 | 0 | ||||
| cg22535403 | ABO | — | −0.117 | 0.001 | 0 | ||||
| cg24267699 | ABO | — | −0.037 | 0.001 | 0 | ||||
| cg07241568 | ABO | — | −0.010 | 0.000 | 3.13E‐176 | ||||
| cg12020464 | ABO | — | 0.031 | 0.001 | 1.46E‐133 | ||||
| cg06818865 | ABO | — | −0.019 | 0.001 | 8.20E‐119 | ||||
| cg13531387 | — | — | 0.019 | 0.001 | 2.11E‐111 | ||||
| cg14271713 | — | — | 0.008 | 0.000 | 4.66E‐101 | ||||
| cg00878953 | — | — | 0.007 | 0.000 | 2.26E‐79 | ||||
| cg13952840 | — | — | −0.009 | 0.001 | 2.14E‐40 | ||||
| cg13963044 | C9orf7 | 190 | −0.034 | 0.003 | 2.03E‐39 | ||||
| cg14440550 | ABO | — | 0.019 | 0.001 | 4.15E‐38 | ||||
| cg13660174 | SURF4 | 90 | −0.010 | 0.001 | 1.16E‐36 | ||||
| cg03474926 | RALGDS | 100 | −0.004 | 0.000 | 6.19E‐30 | ||||
| cg18089000 | GBGT1 | 80 | −0.007 | 0.001 | 1.45E‐27 | ||||
| cg14653977 | GBGT1 | 80 | −0.009 | 0.001 | 1.64E‐27 | ||||
| cg01169778 | GBGT1 | 80 | −0.005 | 0.001 | 2.69E‐18 | ||||
| cg13980863 | — | — | 0.003 | 0.000 | 1.54E‐17 | ||||
| cg13850847 | ABO | — | −0.006 | 0.001 | 1.84E‐15 | ||||
| cg00339415 | OBP2B | 40 | 0.007 | 0.001 | 5.95E‐15 | ||||
| cg13040392 | — | — | 0.011 | 0.002 | 1.36E‐11 | ||||
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
We reviewed literature for evidence supporting functional links between these two genes (CAT and EHF) and respiratory diseases. CAT is downregulated in lung cancer 11 and asthma, 12 and promote the apoptosis of non‐small cell lung cancer cells by accelerating the degradation of caveolin‐1. 13 At the same time, CAT had a protective effect on pulmonary fibrosis 14 and protected lung epithelial cells from hydrogen peroxide‐induced apoptosis, 15 supporting that CAT plays an important role in the development of respiratory system diseases. CAT also has a significant anti‐inflammatory effect that regulates the production of cytokines in white blood cells, thereby protecting alveolar cells from oxidative damage and inhibiting the replication of SARS‐CoV‐2, 16 emphasizing its potential utility in the treatment of COVID‐19 or other severe inflammation‐related diseases in respiratory system. The observation that the EHF variant rs286914 is an eQTL of CAT, together with the lack of literature support for a functional role of EHF in lung diseases including COVID‐19, further supports our hypothesis that rs286914 on the EHF may affect COVID‐19 via regulatory effects on the expression of CAT.
The GWAS study of Ellinghaus et al found that the individuals carrying the ABO rs657152 A allele showed more severe respiratory symptoms after infection (OR = 1.39, Table S1). 5 There is no obvious difference in the frequency of rs657152 between continents (Figure 1B). In addition, we found that ABO rs657152 is a significant cis‐eQTL regulating the expression of GBGT1 in peripheral blood, located about 80 kb upstream of ABO (P = 5.72e‐7, Table 1). Meanwhile, rs657152, as a cis‐meQTL in peripheral blood, is significantly associated with the methylation level of 23 nearby methylation sites (Table 2). This region covers multiple genes (ABO, C9orf7, SURF4, RALGDS, GBGT1 and OBP2B). So far, there is no literature evidence supporting that these genes play an important role in respiratory system diseases. Besides, rs657152 was not observed as eQTL or meQTL in lung.
Ellinghaus et al also found that rs11385942, located in the intron region of the LZTFL1, was significantly associated with the prognosis of COVID‐19 5 (OR = 2.11, Table S1). The risk allele from the 1000 genomes project showed a large frequency difference between major continents with patterns showing higher frequencies in South Asia (SAS 29.55%), but lower frequencies in Europe (EUR 8.05%), Africa (AFR 5.30%), America (AME 4.61%), and East Asia (EAS 0.5%, Figure 1C). The rs11385942 was not a significant eQTL or meQTL in the peripheral blood and lung. Given the existing literature and databases, we have not sorted out a clear chain of evidence explaining the observed genetic associations (rs657152 and rs11385942) with the prognosis of the COVD‐19.
Next, we used GeneMANIA 17 to investigate the protein‐protein interactions between EHF and CAT. This analysis identified a functional network consisting of 22 genes. Interestingly, the ACE2 gene, encoding the SARS‐CoV‐2 S protein receptor, 18 was identified in this network having a protein level interaction with CAT (Figure 1D, Table S2). Noteworthy, among the 163 599 genes from 9 organisms, ACE2 had direct protein‐protein interactions with only 6 genes, including CAT (Figure S1, Table S3). Intracellular localization analysis 19 , 20 showed that CAT and ACE2 proteins both exist in the cell membrane and extracellular area which make it clear that the interaction could have an impact on the cell invasion ability of S protein (Table S5). No direct protein level interaction was found between EHF and ACE2. This result further support our hypothesis that rs286914 (EHF) as an eQTL of CAT may affect the binding efficiency of ACE2 and SARS‐CoV‐2 S protein through CAT, thereby affecting the susceptibility of COVID‐19.
There is no protein‐protein interaction between the ABO and other genes where the COVID‐19 prognosis‐associated variants and methylation site are located (GBGT1, C9orf7, SURF4, RALGDS, OBP2B). As illustrated in the network diagram, several genes had the same protein domain, possibly performing same functions (Figure S2, Table S4). It remains unclear why blood type A individuals had more severe COVID‐19 prognosis than those with blood type O. 21
We performed a correlation analysis for the expression of EHF, CAT and ACE2 based on the gene expression data of 49 tissues from the GTEx database (https://commonfund.nih.gov/GTEx/) 22 . These 3 genes were simultaneously expressed in 40 different tissues including the lung (Table S6). In 30 different tissues including the lung, the expression levels of these 3 genes showed significant correlations for at least one pairs after Bonferroni correction (P < 4.16e‐4). The majority of these correlations were positive (80%). The most significant correlation was observed in transverse colon, where the expressions of the three genes were all strongly positively correlated (0.60 < r < 0.73, 2.37e‐63 < P < 8.47e‐38, Figure 1E). The 2th significant finding was observed in lung tissue, where the expression of the three genes all showed a significant positive correlation (0.31 < r < 0.49, 1.76e‐32 < P < 3.24e‐13), which was consistent with that observed in the transverse colon, although the correlation and significance levels were reduced. These correlation patterns indicate that there are intrinsic connections within these three genes and they may regulate the progression of lung‐related diseases together. Here, a colocalization analysis would be preferred to test whether the GWAS and eQTL signals are overlapping. However, this analysis could not be done due to the failure to obtain COVID‐19 GWAS data. When the GWAS data are available in the future, this analysis can be supplemented to verify the association between CAT and COVID‐19.
3. CONCLUSION
In summary, EHF rs286914 functionally regulates the expression of CAT via cis‐eQTL acting. The expressions of EHF, CAT, and ACE2 are positively correlated with each other in lung. Moreover, there was a significant protein‐protein interaction between CAT and ACE2 while no significant interactions were found between EHF and ACE2. Intracellular location analysis showed that both CAT protein and ACE2 exist in cell membrane and extracellular area and their interaction could have an impact on the cell invasion ability of S protein. These multiple lines of evidence support the hypothesis that EHF may as an intermediary to affect the binding efficiency of ACE2 to SARS‐CoV‐2 S protein through CAT, thereby affecting the susceptibility of COVID‐19.
CONFLICT OF INTEREST
The authors declare that no conflicts of interest exist.
AUTHOR CONTRIBUTIONS
Fan Liu and Yu Qian conceived the study; Yu Qian analyzed the data, prepared the figures and supporting information. All authors took part in writing the article.
Supporting information
Fig S1
Fig S2
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Text S1
ACKNOWLEDGEMENTS
This project was supported by the Strategic Priority Research Program of Chinese Academy of Sciences, Grant No. XDB38010400.
Qian Y, Li Y, Liu X, et al. Evidence for CAT gene being functionally involved in the susceptibility of COVID‐19. The FASEB Journal. 2021;35:e21384. 10.1096/fj.202100008
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun J, He WT, Wang L, et al. COVID‐19: epidemiology, evolution, and cross‐disciplinary perspectives. Trends Mol Med. 2020;26:483‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shi Y, Wang G, Cai XP, et al. An overview of COVID‐19. J Zhejiang Univ Sci B. 2020;21:343‐360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide association study of severe COVID‐19 with respiratory failure. N Engl J Med. 2020;383(16):1522‐1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kachuri L, Francis SS, Morrison M, et al. (2020) The landscape of host genetic factors involved in infection to common viruses and SARS‐CoV‐2. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westra H‐J, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huan T, Joehanes R, Song C, Peng F, Guo Y. Genome‐wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gong J, Wan H, Mei S, et al. Pancan‐meQTL: a database to systematically evaluate the effects of genetic variants on methylation in human cancer. Nucleic Acids Res. 2019;47:D1066‐D1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McClay JL, Shabalin AA, Dozmorov MG, et al. High density methylation QTL analysis in human blood via next‐generation sequencing of the methylated genomic DNA fraction. Genome Biol. 2015;16:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coursin DB, Cihla HP, Sempf J, Oberley TD, Oberley LW. An immunohistochemical analysis of antioxidant and glutathione S‐transferase enzyme levels in normal and neoplastic human lung. Histol Histopathol. 1996;11:851‐860. [PubMed] [Google Scholar]
- 12. Ghosh S, Janocha AJ, Aronica MA, et al. Nitrotyrosine proteome survey in asthma identifies oxidative mechanism of catalase inactivation. J Immunol. 2006;176:5587–5597. [DOI] [PubMed] [Google Scholar]
- 13. Rungtabnapa P, Nimmannit U, Halim H, Rojanasakul Y, Chanvorachote P. Hydrogen peroxide inhibits non‐small cell lung cancer cell anoikis through the inhibition of caveolin‐1 degradation. Am J Physiol Cell Physiol. 2011;300:C235‐C245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Odajima N, Betsuyaku T, Nagai K, et al. The role of catalase in pulmonary fibrosis. Respir Res. 2010;11:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arita Y, Harkness SH, Kazzaz JA, et al. Mitochondrial localization of catalase provides optimal protection from H2O2‐induced cell death in lung epithelial cells. Am J Physiol. Lung cell Mol Physiol. 2006;290:L978‐L986. [DOI] [PubMed] [Google Scholar]
- 16. Qin M, Cao Z, Wen J, et al. An antioxidant enzyme therapeutic for COVID‐19. Adv Mater. 2020;32:e2004901. [DOI] [PubMed] [Google Scholar]
- 17. Warde‐Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214‐W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158‐D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204‐D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Latz CA, DeCarlo C, Boitano L, et al. Blood type and outcomes in patients with COVID‐19. Ann Hematol. 2020;99:2113‐2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Consortium, G. T . The genotype‐tissue expression (GTEx) project. Nat Genet. 2013;45:580‐585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Text S1


