Abstract
Adverse outcomes in coronavirus infection disease‐19 (COVID‐19) patients are not always due to the direct effects of the viral infection, but often due to bacterial coinfection. However, the risk factors for such bacterial coinfection are hitherto unknown. A case‐control study was conducted to determine risk factors for bacterial infection in moderate to critical COVID‐19. Out of a total of 50 cases and 50 controls, the proportion of cases with severe/critical disease at presentation was 80% in cases compared to 30% in controls (p < 0.001). The predominant site was hospital‐acquired pneumonia (72%) and the majority were Gram‐negative organisms (82%). The overall mortality was 30%, with comparatively higher mortality among cases (42% vs. 18%; p = 0.009). There was no difference between procalcitonin levels in both groups (p = 0.883). In multivariable logistic regression analysis, significant independent association was found with severe/critical COVID‐19 at presentation (AOR: 4.42 times; 95% CI: 1.63–11.9) and use of steroids (AOR: 4.60; 95% CI: 1.24–17.05). Notably, 64% of controls were administered antibiotics despite the absence of bacterial coinfection or secondary infection. Risk factors for bacterial infections in moderate to critically ill patients with COVID‐19 include critical illness at presentation and use of steroids. There is widespread empiric antibiotic utilization in those without bacterial infection.
Keywords: bacteria, coinfection, COVID‐19, nosocomial infections
Abbreviations
- COVID‐19
coronavirus infection disease‐19
- C‐RP
C‐reactive protein
- ERC
Ethics Review Committee
- IQR
interquartile range
- MDR
multi‐drug resistant
- MRSA
methicillin‐resistant Staphylococcus aureus
1. INTRODUCTION
Coronavirus infection disease‐19 (COVID‐19) pandemic has claimed more than 1 000 000 lives to date and its long‐term impact is yet to be determined. Respiratory viral infections have been well known to predispose patients to coinfections and these lead to increased disease severity and mortality as was observed in the 1918 influenza outbreak, where most mortalities were due to simultaneous bacterial infection. 1 Bacterial coinfection also led to poor outcomes in the 2009 H1N1 influenza pandemic. 2
The incidence of bacterial coinfection in COVID‐19 ranges from 3% to 30%. 3 , 4 Zhou et al. 5 showed that in the current COVID‐19 pandemic, 50% of patients who died, had secondary bacterial infections, while another study showed the presence of both bacterial and fungal infection. 6 Due to similar clinical phenotype and difficulties in identifying COVID‐19 disease from atypical bacterial pneumonia or nosocomial pneumonia some guidelines advise empirical antibiotics. 7 In a study conducted at 38 hospitals in Michigan 56.6% patients received empirical antibiotics therapy. 8
Studies indicate that bacterial coinfection and secondary infection complicate COVID‐19 though regional data is scarce. 6 Moreover, risk factors for these infections need to be better elucidated. Antimicrobials are used empirically which may lead to antimicrobial resistance in the long term. Hence, it is imperative to conduct comparative studies to identify those COVID‐19 patients who are candidates for empirical antibiotic therapy and curtail the widespread injudicious use of antibiotics. The objective of this study is was to determine the risk factors of bacterial infections in COVID‐19 patients. Secondary objectives were to determine differences in the organisms and sensitivity patterns in moderate versus severe/critical COVID‐19 and risk factors for mortality.
2. METHODS
A case‐control study was conducted from February 2020 to June 2020 at a tertiary care center in Karachi, Pakistan. Cases were defined as patients who had polymerase chain reaction confirmed, graded as moderate to severe/critical COVID‐19 as per World Health Organization (WHO) criteria for severity, 9 and had evidence of bacterial infection based on isolation of bacteria in any of the culture specimens collected during admission along with symptoms and signs consistent with infection. Controls were defined as patients who had graded as moderate to severe/critical COVID‐19 as per WHO criteria for severity 9 but who did not develop a bacterial infection during admission.
All adult patients (age ≥ 18 years) hospitalized with moderate and severe/critical COVID‐19 as per WHO definition for severity 10 at Aga Khan University Hospital, Karachi were assessed for the presence of bacterial coinfection at admission as well as followed for secondary bacterial infection during hospitalization. Patients with signs and symptoms consistent with bacterial infection during hospitalization with COVID‐19 at various sites such as urinary tract infection, hospital‐acquired pneumonia, ventilator‐associated pneumonia, and central line‐associated bloodstream infection, and so forth. As defined by Centers for Disease Control and Prevention (CDC) 11 and who were identified to have growth of significant bacteria in the respective appropriate culture specimens (e.g., urine, tracheal aspirate, and blood from the central line, etc.) collected during admission were included whereas those patients who had evidence of bacterial infection on culture but did not have signs and symptoms consistent with infection at any site as defined by CDC 11 were considered to be colonized and excluded from study and patients with mono‐microbial fungal infections were also excluded.
Patients who had any positive bacterial culture were identified from infection control records and were screened for eligibility criteria for cases. Controls were identified from medical records among all moderate to severe/critical COVID‐19 admissions. A sample size of 50 cases was obtained. For each case patient, 1:1 control patient(s) was also obtained (50 controls). The main outcome variable was the presence of bacterial infection defined as infection with clinically significant bacteria which is identified from a culture specimen among those with moderate or severe/critical COVID‐19. Exposure variables included patient‐related factors such as age in years, gender, comorbidities, type of ward/unit to which patient was admitted, presence of invasive devices, and immunosuppression received. We collected data on potential confounders including previous comorbid and severity of illness. Data were collected on structured proforma which was pretested for 1 week at the start of the study. Patients were further stratified as having community‐onset infection if they had culture specimen positive within 72 h of admission and hospital‐onset infection if they were found to have a bacterial infection after 72 h of hospitalization. Bacteriological identification was performed in the College of American Pathologists certified Clinical Microbiology Laboratory at Aga Khan University by conventional methods. Antibiotic susceptibility of isolated bacteria was evaluated by the standard disc diffusion method in accordance with the Clinical and Laboratory Standards Institute recommendations.
2.1. Statistical analysis
Frequencies with percentages were reported for categorical variables such as gender, type of infection by site, and so forth. According to case or control status. For continuous variables such as age, length of stay, and so forth. Median and interquartile range (IQR) were reported by case and control status. Logistic regression analysis was performed to determine the association between risk factors and bacterial infection in moderate and severe/critical COVID‐19 patients and the results were reported as adjusted odds ratios (AOR) with a 95% confidence interval (CI). p Value 0.05 was considered significant. Data were analyzed using Stata® Version 12.
The study received an exemption from ethical approval from the Aga Khan University Ethics Review Committee (ERC reference number: 2020‐5178‐14123).
3. RESULTS
3.1. Baseline characteristics
A total of 50 cases and 50 controls were included in the analysis after making relevant exclusions over a period of 4 months from February 2020 to June 2020. The median age of cases was 58 years (IQR) and of controls was 62 years (IQR). The overall male to female ratio was 2.8 and gender distribution was similar between the groups. The most frequent comorbid were diabetes and hypertension in both groups. The severity of illness was significantly different in both the groups with a high proportion (80%) of severe/critical COVID‐19 patients among cases compared to 30% in controls (p < 0.001).
3.2. Risk factors for bacterial infections in patients with COVID‐19
In the univariable analysis, the median C‐reactive protein (C‐RP) and median neutrophil to lymphocyte ratio (NLR) were significantly higher in cases compared to controls (Table 1). However, there was no statistically significant difference between procalcitonin levels in COVID‐19 patients with bacterial infection compared to those without bacterial infection (p = 0.883). With regard to the unit of admission, COVID‐19 patients with bacterial infections were more frequently admitted to the intensive care unit (ICU) (56%) compared to patients without bacterial infections who were mostly admitted to the ward (38%) (p < 0.001). The use of invasive devices such as endotracheal tube and central venous catheters were also more frequent among cases compared to controls (p < 0.001) (Table 1). Patients with bacterial infections were managed with invasive ventilation in 56% of cases compared to 14% controls (p < 0.001) and with noninvasive ventilation in 64% of cases compared to 34% controls (p = 0.003). A comparatively higher proportion of patients who had bacterial infections had received treatment with systemic steroids (92%) (p = 0.001). All patients with bacterial infections had received antibiotics and among controls (32/50) had received antibiotics. The choice of empiric antibiotics was based on local antibiogram and institutional guidelines for community‐acquired pneumonia and definitive antibiotic treatment was decided based on the identification and sensitivity pattern of the isolated organism. Multivariable logistic regression showed that patients who were severe to critically ill at the time of admission with COVID‐19 were 4.42 times (95% CI: 1.63–11.9) at risk for bacterial infection and treatment with steroids was also a significant risk factor (AOR: 4.60; 95% CI: 1.24–17.05). Admission to the ward unit was found to be protective (odds ratio [OR]: 0.15; 95% CI: 0.02–0.75).
Table 1.
Comparison of moderate and severe COVID‐19 patients with and without bacterial infections
| Variables | All (N = 100) | COVID‐19 with bacterial infection (n = 50) | COVID‐19 without bacterial infection (n = 50) | p Value |
|---|---|---|---|---|
| Median age in years (IQR) | 60 (52–70) | 58 (49–67) | 62 (54–70) | 0.512 |
| Sex | ||||
| Male | 74 (74%) | 39 (78%) | 35 (70%) | 0.362 |
| Female | 26 (26%) | 11 (22%) | 15 (30%) | |
| Comorbids | ||||
| Diabetes | 56 (56%) | 27 (54%) | 29 (58%) | 0.687 |
| Hypertension | 55 (55%) | 25 (50%) | 30 (60%) | 0.315 |
| Ischemic heart disease | 28 (28%) | 13 (26%) | 15 (30%) | 0.656 |
| Chronic kidney disease | 17 (17%) | 9 (18%) | 8 (16%) | 0.79 |
| COPD | 3 (3%) | 2 (4%) | 1 (2%) | |
| Severity of illness | <0.001 | |||
| Moderate | 43 (43%) | 10 (20%) | 33 (66%) | |
| Severe | 57 (57%) | 40 (80%) | 17 (34%) | |
| Laboratory investigations | ||||
| Median C‐reactive protein mg/L (IQR) | 147.47 (48.5–199.64) | 169.34 (95.86–231.19) | 81.57 (34.36–197.19) | 0.009 |
| Median neutrophil to lymphocyte ratio (IQR) | 6.20 (3.29–11.12) | 8.58 (5.41–14.66) | 4.28 (2.96–7.89) | 0.001 |
| Median procalcitonin ng/ml (IQR) | 0.25 (0.107–0.6) | 0.369 (0.15–1.78) | 0.14 (0.07–0.44) | 0.883 |
| Type of ward | ||||
| ICU admission | 37 (37%) | 28 (56%) | 9 (18%) | <0.001 |
| SCU admission | 42 (42%) | 20 (40%) | 22 (44%) | 0.685 |
| Ward admission | 21 (21%) | 2 (4%) | 19 (38%) | <0.001 |
| Presence of invasive devices | ||||
| Endotracheal tube | 37 (37%) | 28 (56%) | 7 (14%) | <0.001 |
| CVP line | 39 (39%) | 29 (58%) | 10 (20%) | <0.001 |
| Treatment | ||||
| Invasive ventilation | 35 (35%) | 28 (56%) | 7 (14%) | <0.001 |
| Noninvasive ventilation | 49 (49) | 32 (64%) | 17 (34%) | 0.003 |
| Antibiotics | 82 (82%) | 50 (100%) | 32 (64%) | <0.001 |
| Tocilizumab | 29 (29%) | 18 (36%) | 11 (22%) | 0.123 |
| Systemic steroids | 77 (77%) | 46 (92%) | 31 (62%) | 0.001 |
| Outcomes | ||||
| Length of stay, median (IQR) days | 9 (6–14) | 12.5 (7–18) | 7.5 (4–11) | 0.001 |
| Dead | 30 (30%) | 21 (42%) | 9 (18%) | 0.009 |
| Discharged | 70 (70%) | 29 (58%) | 41 (82%) | |
Abbreviations: COPD, chronic obstructive pulmonary disease; COVID‐19, coronavirus infection disease‐19; CVP, central venous pressure; ICU, intensive care unit; IQR, interquartile range; SCU, special care unit.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Site of infection and type of organisms
Among those COVID‐19 patients with bacterial infections, the majority (72%) were hospital‐acquired and 28% were community‐acquired at the onset. The commonest source of infection was hospital‐acquired pneumonia in n = 28 patients, followed by community‐acquired pneumonia in n = 8 patients, central line‐associated bloodstream infection in n = 7 patients, urinary tract infection in n = 6 patients, and skin and soft tissue infection in one patient. Seven out of fifty patients had a simultaneous infection at two sites. Gram‐negative organisms were more common (82%) than Gram‐positive organisms (Figure 1). The majority of patients had a mono‐microbial infection (70%). Eight out of fifty patients had bacteremia and one had candidemia. The most frequently isolated organism from blood was multi‐drug resistant (MDR) Acinetobacter in (3/10) patients, followed by ceftriaxone resistant. Escherichia coli in two patients, vancomycin‐resistant Enterococcus in two patients and ceftriaxone‐resistant Klebsiella pneumonia in one patient. In patients who developed hospital‐acquired pneumonia, the most commonly isolated pathogen was MDR Acinetobacter species in (13/28), followed by MDR Pseudomonas aeruginosa in 24% (5/28), Stenotrophomonas maltophilia in (5/28), methicillin‐resistant Staphylococcus aureus in 3/28, K. pneumoniae in 2/28 patients, and E. coli in 2/28 patients. Among patients with community‐acquired pneumonia, the most common organism was Staphylococcus aureus (4/8) in which two were methicillin‐resistant and P. aeruginosa in 3/8 ceftriaxone resistant K. pneumoniae in (1/8).
Figure 1.
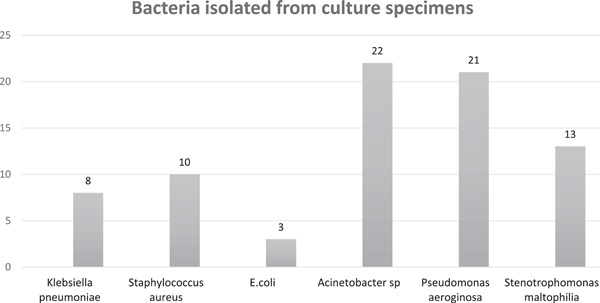
Clinically significant isolates from culture specimens in patients with moderate and severe coronavirus infection disease‐19
3.4. Subgroup analysis for mortality and length of stay
The overall mortality was 30%. Patients with COVID‐19 having bacterial coinfections or secondary infections had a comparatively greater proportion of deaths compared to controls (42% vs. 18%) OR = 3.29; 95% CI: 1.32–8.23 (p = 0.011). Other factors associated with overall mortality included severe/critical illness at presentation OR = 19.79; 95% CI: 4.36–89.7 (p < 0.001), use of antibiotics OR = 9.3 (95% CI: 1.17–73.4; p = 0.034), unit increase in NLR OR = 1.14; 95% CI: 1.06–1.24 (p = 0.001), hospital‐onset of infection OR = 4.33; 95% CI: 1.44–10.7, presence of central venous pressure line OR = 11.08; 95% CI: 4.02–30.5 (p < 0.001), and mechanical ventilation OR = 7.33; 95% CI: 2.83–18.9 (p < 0.001). Out of 21 patients with bacterial infections who died, 18 had an infection with Gram‐negative organisms with Acinetobacter sp (n = 9) and P. aeruginosa (n = 7) being the most common isolates. The median length of stay was also significantly longer among cases compared to controls (12.5 vs. 7.5 days (p = 0.001).
4. DISCUSSION
Our study found that risk factors for bacterial infections in COVID‐19 patients were severity of disease and use of steroids. A higher C‐RP and NLR was present in those with bacterial infection but did not come out to be statistically significant as independent risk factors on logistic regression analysis. Our patient cohort had moderate to severe or critical COVID‐19 at the time of hospitalization and we had excluded patients who had specimen positive with contaminants or who were presumed to be colonized and without clinically significant infection. There was a high male to the female ratio and several studies have reported a greater risk of severe illness with COVID‐19 in males compared to females. 12 , 13 We had very few community‐acquired infections and the majority were hospital‐acquired. This is similar to a study conducted by Hughes et al. 14 from the UK where only 3.2% coinfections were reported and a study from Spain describing the incidence of coinfections. 15 This was also confirmed in a meta‐analysis where the reported prevalence of coinfections was 3.5%. 16 Majority of the patients in our cohort developed hospital‐acquired infections. This is consistent with other reported literature. 16 , 17 The most common site of infection was pneumonia and this is in concordance with initial reports from China. 17 Gram‐negative infections have dominated as far as the type of organisms is concerned and this is similarly seen in studies reported from other parts of the world describing super‐infections or secondary bacterial infections. 15 , 17 There is a lack of reported literature on the sensitivity patterns of organisms isolated. In our study, we found MDR Acinetobacter as the predominant pathogen causing hospital‐acquired infections and MRSA as the main cause of coinfection in COVID‐19 in our patients. A case series of 19 patients from Iran has described Acinetobacter baumanii in the majority of patients requiring ICU admission 18 which reflects epidemiological similarity to Pakistan. In our study, the overall mortality was 30% and was higher in those who had bacterial infections. This is also consistent with data reported from Europe and parts of Asia. 16 , 19 Moreover, we found that overall antibiotic utilization was 82% and was 64% in patients who had no evidence of bacterial infection. This has also been similarly reported 19 indicating widespread injudicious use of antibiotics which can potentiate the problem of antimicrobial resistance. In our study, we did not find procalcitonin to be a reliable marker of distinguishing patients who had bacterial infections and those who did not have bacterial infections. Although studies have described procalcitonin levels to remain normal in severe COVID‐19, whether they increase in bacterial infections has not been reported and indicates the need of large comparative studies exploring this relationship. 20 Our study is the first comparative study with regard to bacterial infections in the setting of moderate and severe to critical COVID‐19 and describes the sensitivity pattern of organisms in addition to clearly classifying them into the community and hospital‐acquired infections.
Our study is limited as it's a single‐center experience which might affect generalizability. Moreover, we did not have molecular methods and genotyping available for our bacterial isolates and information on the isolation rate of the prevalent Gram‐negative and MRSA in non‐COVID‐19 patients over the study period was not available to draw comparisons with rates of infection seen in COVID‐19 patients.
5. CONCLUSIONS
Our study highlights the need for improving antibiotic stewardship practices and reserving antibiotics for those who are severely ill with COVID‐19 at presentation and require treatment with systemic steroids and provides insight into the lack of utility of serum procalcitonin as a marker of bacterial sepsis in this setting.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICS STATEMENT
The study was submitted for ethical approval to the Aga Khan University Ethics Review Committee and received exemption (ERC reference number: 2020‐5178‐14123). Consent to participate was not required because there was no direct interaction with participants. The study was eligible for exemption as data was collected from records and no personal identifiers were recorded or used.
AUTHOR CONTRIBUTIONS
Nosheen Nasir conceived the idea, major contributor to the manuscript, collected and analyzed data. Fazal Rehman conceived idea, contributed to manuscript, and collected the data. Syed Furrukh Omair collected data and contributed to the manuscript. All authors read and approved the manuscript.
Nasir N, Rehman F, Omair SF. Risk factors for bacterial infections in patients with moderate to severe COVID‐19: a case‐control study. J Med Virol. 2021;93:4564–4569. 10.1002/jmv.27000
REFERENCES
- 1. Taubenberger JK, Morens DM. The 1918 influenza pandemic and its legacy. Cold Spring Harb Perspect Med. 2020;10(10):a038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rice TW, Rubinson L, Uyeki TM, et al. Critical illness from 2009 pandemic influenza A (H1N1) virus and bacterial co‐infection in the United States. Crit Care Med. 2012;40(5):1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clancy CJ, Nguyen MH. COVID‐19, superinfections and antimicrobial development: what can we expect? Clin Infect Dis. 2020;71(10):2736‐2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lehmann CJ, Pho MT, Pitrak D, Ridgway JP, Pettit NN. Community acquired co‐infection in COVID‐19: a retrospective observational experience. [published online ahead of print July 1, 2020]. Clin Infect Dis. 2020:ciaa902. 10.1093/cid/ciaa902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawson TM, Moore LS, Zhu N, et al. Bacterial and fungal co‐infection in individuals with coronavirus: a rapid review to support COVID‐19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459‐2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lansbury L, Lim B, Baskaran V, Lim WS. Co‐infections in people with COVID‐19: a systematic review and meta‐analysis. J Infect. 2020;81(2):266‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaughn VM, Gandhi T, Petty LA, et al. Empiric antibacterial therapy and community‐onset bacterial co‐infection in patients hospitalized with COVID‐19: a multi‐hospital cohort study [published online ahead of print August 21, 2020]. Clin Infect Dis. 2020:ciaa1239. 10.1093/cid/ciaa1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. World Health Organization . Clinical management of COVID‐19. 2020. https://www.who.int/publications/i/item/clinical-management-of-covid-19 [PubMed]
- 10. World Health Organization . Clinical management of COVID‐19: interim guidance. Clinical management of COVID‐19: interim guidance. 2020.
- 11. Control CfD . CDC/NHSN Surveillance Definitions for Specific Types of Infections. 2020. https://www.cdc.gov/nhsn/pdfs/pscmanual/17pscnosinfdef_current.pdf
- 12. Stall NM, Wu W, Lapointe‐Shaw L, et al. Sex‐ and age‐specific differences in COVID‐19 testing, cases, and outcomes: a population‐wide study in Ontario, Canada. J Am Geriatr Soc. 2020;68(10):2188‐2191. [DOI] [PubMed] [Google Scholar]
- 13. Alkhouli M, Nanjundappa A, Annie F, Bates MC, Bhatt DL. Sex differences in case fatality rate of COVID‐19: insights from a multinational registry. Mayo Clin Proc. 2020;95(8):1613‐1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID‐19: a retrospective cohort study in a UK secondary‐care setting. Clin Microbiol Infect. 2020;26(10):1395‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcia‐Vidal C, Sanjuan G, Moreno‐García E, et al. Incidence of co‐infections and superinfections in hospitalized patients with COVID‐19: a retrospective cohort study. Clin Microbiol Infect, 27(1):83‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langford BJ, So M, Raybardhan S, et al. Bacterial co‐infection and secondary infection in patients with COVID‐19: a living rapid review and meta‐analysis. Clin Microbiol Infect. 2020;26(12):1622‐1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He Y, Li W, Wang Z, Chen H, Tian L, Liu D. Nosocomial infection among patients with COVID‐19: a retrospective data analysis of 918 cases from a single center in Wuhan, China. Infect Control Hosp Epidemiol. 2020;41(8):982‐983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co‐infections of the respiratory tract in COVID‐19 patients admitted to ICU. BMC Infect Dis. 2020;20(1):646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goncalves Mendes Neto A, Lo KB, Wattoo A, et al. Bacterial infections and patterns of antibiotic use in patients with COVID‐19. J Med Virol, 93(3):1489‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID‐19): a meta‐analysis. Clin Chim Acta. 2020;505:190‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]


