Abstract
The outbreak of atypical pneumonia (coronavirus disease 2019 [COVID‐19]) has been a global pandemic and has caused severe losses to the global economy. The virus responsible for COVID‐9, severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), has a spike glycoprotein (S protein) that binds angiotensin‐converting enzyme 2 (ACE2) present on host cell membranes to gain entry. Based on the full‐length human ACE2 cryo‐EM structure, we generated homology models of full‐length ACE2 proteins from various species (gorilla, monkey, pig, bovine, sheep, cat, dog, mouse, and rat). Although these ACE2 molecules were found to share similar overall structures, their S–ACE2 interface residues differed. These differences likely result in variations in the ACE2 binding affinities to the SARS‐CoV‐2 S protein. The highest affinities are predicted for human, gorilla, and monkey, while mouse and rat ACE2 are predicted to have the lowest affinities. Cat ACE2 is predicted to have a lower S protein affinity than dog ACE2. Although affinity is not the only factor that affects viral susceptibility, it is one of the most important factors. Thus, we believe that care should be taken with these animals to prevent the spread of SARS‐CoV‐2 among animal and human populations.
Keywords: ACE2, frequently contacted animals, S–ACE2 affinity, virus susceptibility
Research Highlights
We provide, for the first time, homology models of the full‐length ACE2 structures from several species that humans may have frequent contact with, either in daily life or in laboratory research.
S‐ACE2 interface residues differences likely result in variations in the ACE2 binding affinities to the SARS‐CoV‐2 S protein
The highest affinities are predicted for human, gorilla, and monkey, while mouse and rat ACE2 are predicted to have the lowest affinities.
CatACE2 is predicted to have a lower S protein affinity than dogACE2.
1. INTRODUCTION
The first documented outbreak of severe acute respiratory syndrome (SARS) was caused by SARS‐CoV in 2002. Today, we are experiencing a more severe situation with the outbreak of atypical pneumonia (coronavirus disease 2019 [COVID‐19]) caused by a new coronavirus, SARS‐coronavirus 2 (SARS‐CoV‐2). At the time around March/April, 2020, the COVID‐19 has spread throughout more than 170 countries, with more than seven million confirmed infections and over 200,000 confirmed deaths. Now (January, 2021), this global pandemic has become a much more serious issue affects the daily life of billions of people from almost all the countries. Research institutes, companies, and hospitals have exerted much effort to develop therapeutic drugs and vaccines for this virus, only few vaccines have been developed worldwide very recently.
It is now known that SARS‐CoV‐2 is a new member of the betacoronavirus genus, 1 sharing a genome sequence that is very similar to SARS‐CoV. Like SARS‐CoV and Middle East respiratory syndrome coronavirus, they can cross the species barriers and cause severe respiratory infections in human. 2 Zhou et al. 3 have showed that the origin of SARS‐CoV‐2 is probably bats, there are likely unknown intermediate hosts for SARS‐CoV‐2 before it can affect human. CoVs usually have a relative high mutation and recombination rates which facilities their adaptation to new host. 4 To processes the ability for cross‐species transmission, the crucial step is to evolve an ability to engage with receptors in the new host, 4 which occurs through the spike glycoprotein (S protein) in SARS‐CoV‐2.
Like the other known coronaviruses, SARS‐CoV‐2 has an S protein on its envelope that binds to receptors on host cell membranes. The S protein forms as homotrimer in which each S monomer is composed of two subunits, S1 and S2. S1 is involved in receptor binding, while S2 is responsible for fusion between the viral and cellular membranes. 5 , 6 , 7 , 8 , 9 In the S1 subunit, a receptor‐binding domain (RBD) is the region of the S protein that binds to its receptor, which is angiotensin‐converting enzyme 2 (ACE2). 10 Binding to ACE2 is the critical step by which SARS‐CoV‐2 enters the host cells. Recent studies have confirmed that ACE2 mediates the entry of SARS‐CoV‐2 into the host cell. 3 , 11 , 12 , 13 Furthermore, an in vitro study showed that SARS‐CoV‐2 could not infect cells that did not express ACE2, while the virus easily infected cells that overexpressed ACE2. 3 Therefore, whether the SARS‐CoV‐2 S protein can bind to ACE2 on the host cell reflects, to some degree, the susceptibility of the host to SARS‐CoV‐2 infection.
The cryo‐EM structures of full‐length human ACE2 and the SARS‐CoV‐2 S protein have been published at the early beginning of the pandemic. 1 , 14 These structures provide detailed information regarding the mechanisms of receptor binding and function. However, we lack ACE2 structures from other animals. These structures are very important for us to understand whether SARS‐CoV‐2 shares similar infection mechanisms among other animals and whether certain animals may play a prominent role as virus reservoirs, especially when the intermediate hosts of SARS‐CoV‐2 are still unknown.
Currently, most research is being focused on humans. Furthermore, it is impossible to solve ACE2 structures from hundreds of different species in a short time given that structure determination is time‐and labor‐intensive and is expensive. Therefore, in our study, we used homology modeling to generate hundreds of full‐length ACE2 models of animals that humans may have frequent contact with, either in daily life or in laboratory research; namely, gorilla, monkey, pig, bovine, sheep, cat, dog, mouse, and rat. We selected the optimal model for each of these species. By comparing the ACE2 structures and interface regions, we predict the SARS‐CoV‐2 S protein to have nearly the same affinity for gorilla, monkey and dog ACE2 as it does for human ACE2, while it has a lower affinity to ACE2 from pig, bovine, sheep and cat; these were further confirmed by molecular dynamic (MD) simulation studies. We believe that all of these animals are likely to be susceptible to SARS‐CoV‐2 infection. In contrast, the SARS‐CoV‐2 S protein might have a much lower affinity to mouse and rat ACE2 than to human ACE2. We suggested that care should be taken regarding these likely susceptible animals to prevent passing the virus to other animal populations or to humans.
2. MATERIALS AND METHODS
2.1. Sequence alignments
ACE2 sequences of the following species were obtained from UniProt (https://www.uniprot.org/): human (Q9BYF1), gorilla (G3QWX4), monkey (F7AH40), pig (K7GLM4), bovine (Q58DD0), sheep (W5PSB6), cat (Q56H28), dog (J9P7Y2), mouse (Q8R0I0), and rat (Q5EGZ1). The human ACE2 structure models (6M18, 6M17) and the SARS‐CoV‐2 spike protein structure model (6VSB) were obtained from the PDB (http://www.rcsb.org/).
Multiple sequence alignment (MSA) was performed using Clustal Omega. 15 All protein sequences combined in one file were loaded into the Clustal Omega web server in FASTA format. Where necessary, BioEdit 16 was used to open the MSA files generated by Clustal Omega and to manually adjust the alignments. All alignments were then viewed in Jalview. 17
2.2. Model building and validation
Optimized pairwise alignments between the target ACE2 and the full‐length human ACE2 structure (6M17) were used for model construction using Modeller9.19. 18 For each alignment, 100 models were generated. The “optimal” models for each species with the lowest energies, as indicated by the modeler objective function, were selected. These models were then validated with MolProbity, 19 the percentage of residues in the favored regions of the Ramachandran plot and the number (or percentage) of residues in the disallowed region of the plot that calculated based on our models and based on the cryo‐EM structure 6M17 were compared, to show reliability of our models to some extent. The entire SARS‐CoV‐2 S protein or the RBD of the S protein was rigid body fitted on these ACE2 models based on 6M17. All structure figures were generated with PyMOL 2.3.2. 20
2.3. Molecular dynamic simulations
MD calculations were performed using the GROMACS 21 software suite. The general OPLS‐AA/L all‐atom force field was used for the protein complexes. The protein complex was solvated with SPC water molecules in a periodically repeating cubic box. The net charge of the system was brought to neutrality by addition of dissociated NaCl. The structure was then relaxed through energy minimization (EM), the EM will stop when the maximum force less than 1000.0 kJ/mol/nm. After minimization, each system was gradually heated in the canonical ensemble from 0 to 300 K over a period of 100 ps, this stabilized the temperature of the system. Then the system were subjected to a 100 ps “NPT” equilibration (with a target pressure of 1 atm) to stabilize the pressure (and the density). All bonds were constrained. The time step was set to 2 fs. The PMEMD program was used for the molecular mechanics (MM) optimization and MD simulations. A 10 ns data production run was performed for each of the protein complexes. The coordinates were saved every 10 ps during the MD sampling process. For analysis, protein “backbone” was chosen for both the least‐squares fit and the group for root mean square deviation (RMSD) calculation.
3. RESULTS
3.1. Full‐length ACE2 models from frequently contacted animals share similar overall structures
Gorilla, monkey, pig, bovine, sheep, cat, dog, mouse, and rat are animals that humans frequently come into contact with either in daily life or through laboratory research. This study provides molecular‐level insights into the seriousness of the role these animals may play in the current COVID‐19 pandemic. There has been an increasing number of studies regarding SARS‐CoV‐2, and the 3D structures of the SARS‐CoV‐2 S protein and human ACE2 are available. Nevertheless, we still lack the ACE2 structures of other animal species.
Here, we obtained the ACE2 sequences of gorilla, monkey, pig, bovine, sheep, cat, dog, mouse, and rat from UniProt, performed MSA, and manually adjusted the alignments when necessary. Since these ACE2 proteins share a high amino acid sequence identity with human ACE2, we constructed homology models of these full‐length ACE2 structures that were based on the full‐length human ACE2 structure.
In total, 100 initial ACE2 models were generated for each of the abovementioned species. The optimal models with the lowest energies, as indicated by the modeler objective function, were selected. The RBD of the SARS‐CoV‐2 S protein or the full‐length SARS‐CoV‐2 S protein were rigid body docked onto these ACE2 models based on 6M17. This provided a general idea of how these ACE2 structures look and how the full‐length SARS‐CoV‐2 S protein makes contact with ACE2 (Figure 1). To validate the reliability of our models, we calculated the percentage of residues in the favored regions of the Ramachandran plot, the percentage of residues in the allowed regions and the number (or percentage) of residues in the disallowed region of the plot. By comparing those data of our “optimal” models of each animal species chosen in this study and the cryo‐EM structure of human ACE2–RBD complex (6M17), we could tell that our models share an over 98% of residues in the Ramachandran plot allowed regions, similar to that of 6M17; and in some cases, with even fewer Ramachandran outliers than 6M17 does (Table S2). These validation data, together with the high sequence similarity among those ACE2s (>90%), and low RMSD difference among those models, to some extent, prove that our models are reliable. The overall structures of the ACE2 models were similar to that of human ACE2 (RMSD approximately 0.25–0.31, Table S1), the residues at the binding interface between the SARS‐CoV‐2 S protein and ACE2 varied, thereby affecting the binding affinity of the S protein to the different ACE2 molecules.
Figure 1.
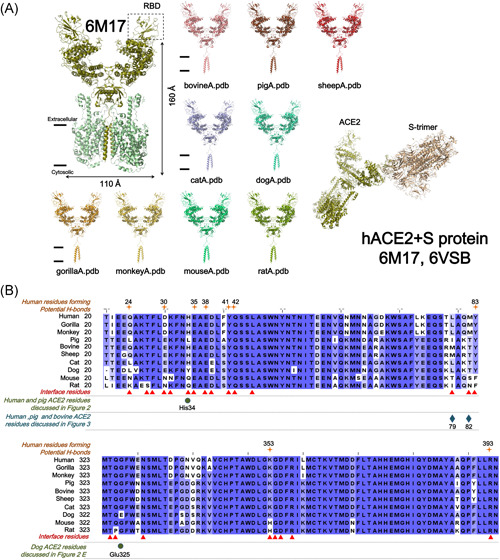
Angiotensin‐converting enzyme 2 (ACE2) structures from different species and the interface residues discussed in this study. (A) Upper left: overall cryo‐EM structure of the ACE2–RBD‐B0AT1 complex (6M17). ACE2 has a width of 110 Å and a height of 160 Å. Bottom right: rigid body fitting shows how the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) S protein trimer docks on human ACE2. The remaining images show the full‐length ACE2 structure models of the nine indicated species. All possess a similar overall structure. The assigned colors are used throughout all figures for clarity. (B) The interface residues discussed in this study
3.2. Gorilla and monkey ACE2 share the same features as human ACE2, while mouse and rat ACE2 might have a much lower affinity to SARS‐CoV‐2 S protein
The study by Renhong Yan 1 identified seven hACE2 residues that were involved in binding the S protein based on their full‐length cryo‐EM structure (6M17). Lan et al. 22 expanded the number to 20 hACE2 residues that were involved in S binding based on their partial hACE2 2.45 Å crystal structure (6M0J). The more curved T20‐N53 helix in 6M0J binds to the S protein RBD, similar to that of 6M17, revealing additional interaction residues (Figure 2A). We applied the same method as in the Jun Lan study and identified the same hACE2 residues in the S–hACE2 interface when using the full‐length cryo‐EM structure (6M17). This method was then applied to all ACE2 models in our study.
Figure 2.
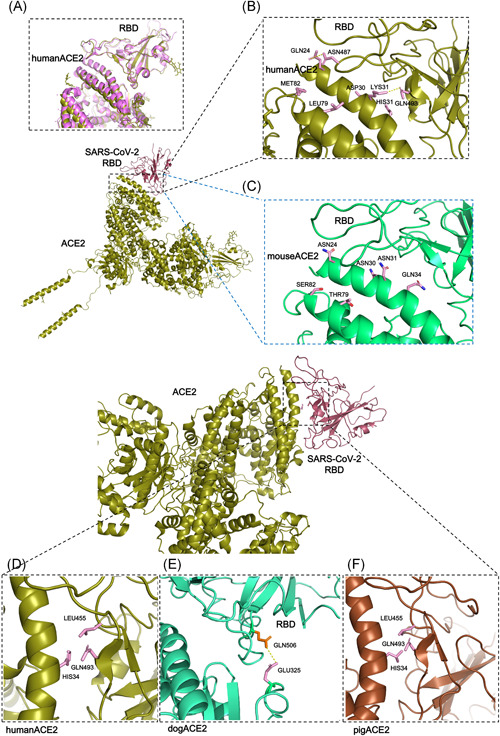
Interface comparison showing highlighted regions. (A) Structure alignment of 6M0J (purple) and 6M17 (deep olive). (B, C) Six corresponding interface residues of human ACE2 and mouse ACE2 shown in detail. (D, F) Position 34 of ACE2 from human and pig; their residues that interact with the SARS‐CoV‐2 S protein are shown in detail. (E) Position 325 of dog ACE2 forming a potential hydrogen bonds with position 506 of SARS‐COV‐2 S protein. ACE2, angiotensin‐converting enzyme 2; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
Among the S–ACE2 interface residues (Q24, T27, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L45, L79, M82, Y83, T324, Q325, N330, K353, G354, D355, R357, and R393; residue number based on hACE2) 13 are highly conserved (Q24, F28, E35, E37, Y41, Q42, L45, T324, N330, G354, D355, R357, and R393, Figure S2). The remaining residues are also conserved but in different patterns, which will be discussed below.
For clarity, we divide the ACE2 structures in our study into several groups. The first group consisted of gorilla and monkey. The residues forming the binding interface between the SARS‐CoV‐2 S protein and the gorilla or monkey ACE2 were no different than those of the human ACE2 binding interface. All of these structures were predicted to form a network of 10 hydrogen bonds and one salt bridge (Table S3). Although human ACE2–RBD interface calculated to exits 13 potential hydrogen bonds, these hydrogen bonds forming residues are exactly the same as in gorilla or monkey ACE2, which means gorilla or monkey ACE2 also possess the same hydrogen bonds forming ability to SARS‐CoV‐2 S–RBD protein. Therefore, the gorilla and monkey ACE2 likely have a similar affinity as human ACE2 to SARS‐CoV‐2 S.
Mouse and rat were in the second group, and the interface residues of this group showed the most variation. In mouse ACE2, seven of the S–ACE2 interface residues were different than for human ACE2; the majority of them had changed to uncharged polar amino acids (N24, N30, N31, Q34, T79, and S82 in mouse ACE2). These changes not only altered the surface characteristics in this region but also eliminated several hydrogen bonds (Figure 2B,C) and the electrostatic interactions in position 30. By calculating their interface properties (Table S3), mouse and rat ACE2–RBD complex have a much less solvation free energy gain upon formation of the interface than that of human ACE2–RBD complex, and a roughly 2 kcal/mol lesser contribution into the free energy of protein binding due to lost of several hydrogen bonds. Therefore, these mutations should result in a much lower affinity between SARS‐CoV‐2 S protein and mouse and rat ACE2. This might be why mice are not easily infected by SARS‐CoV‐2. In the study of SARS‐CoV, hACE2 transgenic mice are normally required. 23
3.3. CatACE2 might have a lower SARS‐CoV‐2 S protein affinity than dog ACE2
The next group investigated whether there were any differences between dogs and cats. Compared to human ACE2, only four residues were changed in the catACE2 interface (L24, S30, E38, and T82), while six dog ACE2 interface residues were different (L23, S29, Y33, E37, T81, and E326). Position 24 of human ACE2 is Q. This corresponds to L24 in cat ACE2 and L23 in dog ACE2. Changing from Q to L results in the loss of one hydrogen bond in the S–cat ACE2 and S–dog ACE2 interfaces and decreases the S–ACE2 affinity. D30 and M82 of human ACE2 are changed to S and to T in dogs, respectively. These polar uncharged amino acids often interact with water molecules in the environment and are therefore likely to interfere with the binding between S protein and ACE2. Based on this evidence, we predict that the SARS‐CoV‐2 S protein can bind to cat ACE2 and dog ACE2 albeit with a different affinity compared to human ACE2.
Compared to cat ACE2, dog ACE2 has two additional interface residues that are different. Y33 in dog ACE2 corresponds to H34 in human ACE2 and cat ACE2. In dog ACE2, the tail of the Y33 side chain protrudes closely to S494 of the SARS‐CoV‐2 S protein, likely respels the RBD away in this position. By contrast, in human and cat, H34 of ACE2 contacts L455 and Q493 of the SARS‐CoV‐2 S protein, forming a stable “triangle” plane (Figure 2D,E). While, E325 of dog ACE2 is notable—the corresponding residue in the other ACE2 structures is G326. Changing from a small residue, such as G, to E introduces an additional hydrogen bond forming with the NE2 group of Q506 of the SARS‐CoV‐2 S protein. We further examined in details the residue conservation (Figure 1B), the number of potential hydrogen bonds and salt bridges forming in the cat, dog, and human ACE2–RBD interface (Table S3), we believe that dog ACE2 could bind the SARS‐CoV‐2 S protein even with a similar affinity to that of human ACE2, but with a higher affinity than that of catACE2.
3.4. Sheep, bovine, and pig ACE2 should have a lower affinity to SARS‐CoV‐2 S protein than human ACE2
The ACE2 sequences of bovine, sheep, and pig have many residues that are changed compared to human ACE2; however, few of these changes are in the S–ACE2 interface region. M82 of human ACE2 is changed to T in bovine, sheep, and pig. L79 of human ACE2 is changed to M in bovine and sheep and to I in pig. Because the side chains of M82 and T82 likely point towards the external environment (Figure 3A,B), this M to T mutation may have little impact on its interaction with F486 of the SARS‐CoV‐2 S protein. In position 79, the L to I mutation likely has very little effect on the interaction; while the L to M mutation causes the methyl group of the M side chain to be placed much closer (<3 Å) to F486 in the SARS‐CoV‐2 S protein, resulting in a repulsion force (Figure 3C‐E). The calculation of solvation free energy gain upon formation of the interface and potential H‐bond numbers (Table S3) also point to a similar hypothesis. Therefore, we believe that the SARS‐CoV‐2 S protein has a slightly lower affinity to sheep and bovine ACE2 than to human ACE2.
Figure 3.
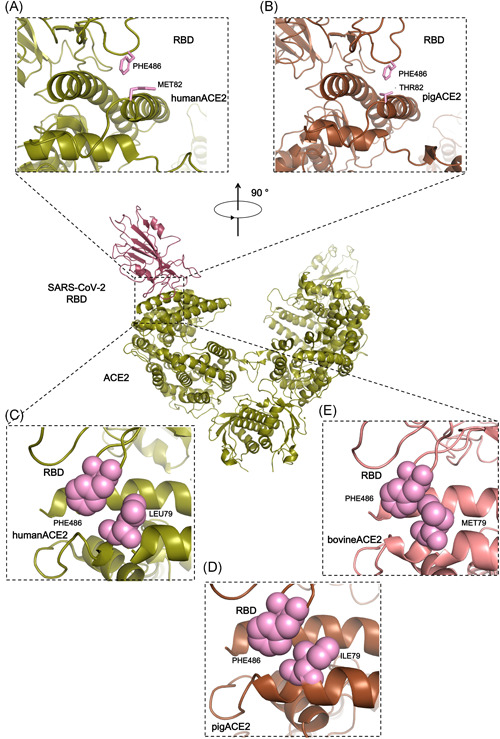
Comparison of highlighted residues from human, bovine, and pig ACE2. (A, B) Relative side chain positions of M82 in human ACE2 and T82 in pig ACE2, respectively. (C–E) Sphere model of position 79 of human, pig, and bovine ACE2 to show their relative distances. ACE2, angiotensin‐converting enzyme 2
In addition to residues in positions 82 and 79, pig ACE2 has two further differences in the interface region, namely L24 (corresponding to Q24 in human, bovine, and sheep) and L34 (corresponding to H34 in human, bovine, and sheep). In position 24, OE1 of Q24 from human/bovine/sheep ACE2 forms a hydrogen bond with ND2 of N487 in the SARS‐CoV‐2 S protein. This hydrogen bond is not present in pig ACE2. The H to L mutation in position 34 occurs in pig ACE2, where the side chain of L34 moves further away from the S protein L455 residue compared to H34 (Figure 2F). All these changes should weaken the interaction between pig ACE2 and the SARS‐CoV‐2 S protein.
3.5. MD simulation of the ten ACE2–RBD complexes
To further test our hypothesis, we performed MD simulations based on our models and the cryo‐EM structure 6M17, in hope we could tell the affinity difference of ACE2s binding to the SARS‐CoV‐2 S protein. We followed these logics: (1) to tell the affinity difference, there's no need to perform MD simulation on the whole ACE2 dimer and S protein trimer, one ACE2 monomer (actually, its N‐domain which involves in binding) and one S protein RBD domain are enough. Therefore we subtracted one ACE2 N‐domain (residues 18‐611) and its corresponding S protein RBD domain from 6M17 and from our models, performed MD simulation on the model of these nine ACE2(N‐domain)–RBD protein complexes. This strategy helps us saved a huge amount of computational resources and gpu time. (2) As the ACE2 from the nine species discussed here share a very high sequences identity (over 85%) to that of human, their N‐domain (residues 18‐611) share an even higher sequence identity and similarity (over 95%), the stability of their ACE2 N‐domain are very much alike. The affinity differences of S protein RBD domain binding to N‐domain of ACE2s from 10 species could be easily reflected by the stability of whole ACE2(N‐domain)–RBD complexes. Therefore, an easy MD simulation indicating the RMSD levels against simulation time is probably able to tell the differences.
In brief, we performed a 10 ns simulation for each of the ACE2–RBD complex exactly as described in Materials and Methods, and plotted the backbone RMSD levels against simulation time (Figure 4). Almost all the 10 ACE2–RBD complexes achieved equilibrium in less than 5 ns, indicating that 10 ns simulation is more than enough. Since the overall structures are quite stable, a small but consist difference will reflect the stability difference. The RMSD of human, gorilla and monkey ACE2–RBD complexes level off to approximately 0.22–0.26 nm during 10 ns (Figures 4A,E), indicating that the structure of the complexes are stable. The distribution of RMSD levels indicated that 50% RMSD values of human, monkey, gorilla are falling in the range of 0.221, 0.226, 0.215 nm, respectively, which means the stability of them are almost the same.
Figure 4.
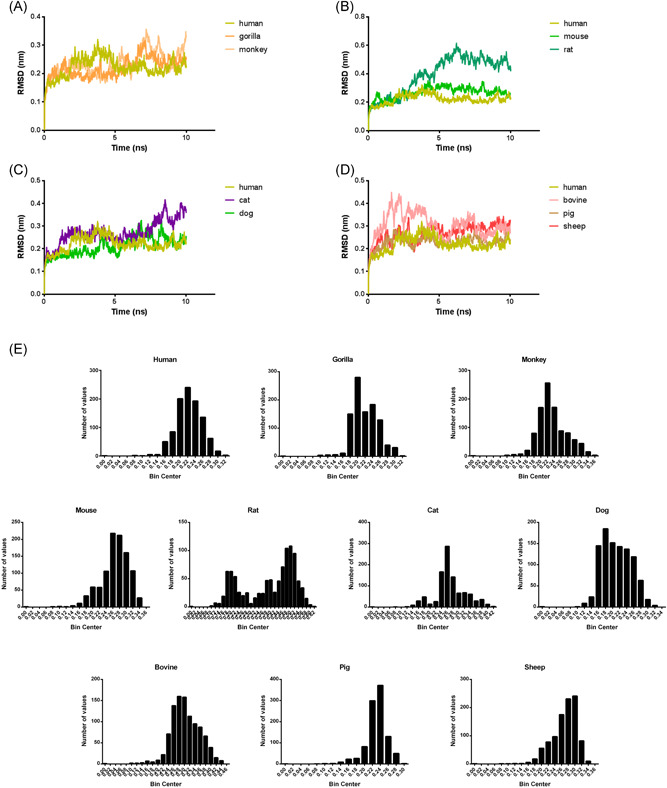
MD simulation of the ACE2 (N‐domain, residues 18‐611) from ten different species in complex with SARS‐CoV‐2 S protein RBD domain. (A–D) the root mean square deviation (RMSD) of 10 ACE2–RBD complexes discussed in our study. (E) the distribution of RMSD levels for each simulation to get a quantitative data for comparisons. ACE2, angiotensin‐converting enzyme 2; MD, molecular dynamic; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
Similarly, over 50% of the RMSD values of mouse and rat are shift to 0.270 and 0.441 nm, indicating that both mouse and rat ACE2–RBD complex are less stable than human ACE2–RBD complex, with rat processes the lowest complex stability (Figures 4B,E). As for cat and dog, over 50% of RMSD values are moved to 0.264 and 0.208 nm, respectively. This suggests that dog ACE2–RBD complex might processes a much higher stability than cat ACE2–RBD complex, or even higher than that of human (Figures 4C,E). Farm animals, such as bovine, pig, and sheep in this case have a RMSD level of 0.299, 0.233, and 0.275 nm, respectively, suggesting that their ACE2–RBD complexes are much less stable than human's (Figure 4D,E). To be noted, except the rat ACE2–RBD complex, the RMSD shifts of the remaining nine complexes are less than 3 Å during the 10 ns simulation, this suggests that ACE2–RBD complex of these nine species (including human) are reasonability stable.
As discussed above, the affinity differences between S protein and ACE2s from different species could be demonstrated by the stability of the whole ACE2–‐RBD complexes. If the stability of the ACE2–RBD complex is higher, it means this ACE2 should have a higher affinity to SARS‐CoV‐2 S protein. Therefore, our MD simulation agrees well with our main hypothesis that (1) monkey, gorilla ACE2 should have a similar affinity to SARS‐CoV‐2 S protein like human ACE2; (2) farm animals, mouse and rat ACE2s should have a lower affinity to SARS‐CoV‐2 S protein than human ACE2; (3) dog ACE2 probably have a higher affinity to SARS‐CoV‐2 S protein than cat ACE2.
4. DISCUSSION
In this study, we conducted an in silico analysis to answer two questions: (1) What are the structures of the ACE2 proteins from frequently contacted animals? (2) Can the SARS‐CoV‐2 S protein bind to these ACE2 molecules? We also predicted the susceptibilities of these species to SARS‐CoV‐2 infection based on their ACE2 structures. The 3D structures of ACE2 contain valuable information regarding their function, and the task for us is to find a reliable way to decode it. We used homology modeling to generate hundreds of full‐length ACE2 models from frequently contacted animals, including gorilla, monkey, pig, bovine, sheep, cat, dog, mouse, and rat. The optimal model for each species was selected for further analysis. The high sequence identity of ACE2 across species results in an overall structure that is very similar to human ACE2, and this high sequence identity also indicates that the homology models should be very similar to the real 3D structures solved by cryo‐EM or other methods (validations also proved this conclusion, Table S3). Therefore, analyses and hypothesis based on such homology models should be as reliable as hypothesis made based on atomic structures.
Previous studies have demonstrated that the domain located in the C‐terminus of SARS‐CoV‐2 S protein specifically interacts with the human ACE2, and therefore designated as RBD. 10 , 24 , 25 Some early in silico studies and studies performed roughly at the similar time suggest that some animals, such as cat, dog, and bovine, their ACE2s also involved in recognition of RBD of SARS‐CoV‐2 S protein. 26 , 27 , 28 They deducted that some key residues, such as those corresponds to K31 and E35 of human ACE2, involved in RBD binding. This could also be drawn from the molecular models built in this study, which docked with the RBD of SARS‐CoV‐2 S protein, they all possess the overall structures and stabilities similar to that of 6M17 (Figures 1 and 4, Tables S1 and S3). In March/April, 2020, based on our models, several hypothesis were raised and were latter confirmed by MD simulations. During this manuscripts were revising, there are other similar independent in silico studies published. 28 The species they and we investigated have some overlaps and we both raised similar conclusion that these animals are likely to be infected by or carrying SARS‐CoV‐2. Their studies and our studies could be supplements to each other. To be noted, current study is the only one to date which provides such amount of homology models of ACE2 from nine species. Our analysis and MD simulation are “bonus” results of these structural models. We could obtain more information based on structural analysis than only based on sequence alignments. These structural models are good supplements for X‐ray or Cryo‐EM structures and could be readily used for further detailed investigations.
Based on our analysis, we therefore believe that the SARS‐CoV‐2 S protein has almost the same affinity for gorilla and monkey ACE2 as for human ACE2. Wu et al. 29 recently demonstrated using both flow cytometry and SPR methods, that monkey ACE2 and human ACE2 indeed have the same affinity to SARS‐CoV‐2 RBD. Although affinity is not the only factor that affects viral susceptibility, it is one of the most important factors. Accordingly, gorilla and monkey are highly likely to be susceptible to SARS‐CoV‐2 infection and thus could be used as animal models for COVID‐19.
In contrast, the S–ACE2 interface residues of mouse and rat differ from those of human. The majority residues are changed to uncharged polar amino acids compared to human ACE2, and several hydrogen bonds are not present. These changes likely dramatically interfere with the interaction between the SARS‐CoV‐2 S protein and mouse/rat ACE2. Not surprisingly, our MD simulation found that rat ACE2 is likely to have the lowest affinity to SARS‐CoV‐2 S protein among all the ACE2s we investigated in this study; while, mouse might not be the second lowest one. Mouse ACE2 might still have a reasonably high binding potential to SARS‐CoV‐2 S protein. This might explain why only one residue substitution in SARS‐CoV‐2 S protein (N501Y) result in a detectable affinity between Mouse ACE2 to the new S protein mutants. 30 , 31 , 32 Based on the structural modeling, the N501Y substitution in the SARS‐CoV S protein increased the binding affinity of the protein to mouse ACE2, the binding energy changed from −14.32 kcal/mol to −14.96 kcal/mol. 30
Because four residues of the S‐catACE2 interface are different from that of human, we believe that the affinity of SARS‐CoV‐2 S protein to cat ACE2 is only slightly reduced. In dog ACE2, the additional hydrogen bond formed by E325 of dog ACE2 and Q506 of the SARS‐CoV‐2 S protein results in an incensement of the affinity. Our MD simulation results agree well with this hypothesis. And in Wu et al.'s study, 29 they detected by SPR method that the interactive affinity of SARS‐CoV‐2 RBD with dog ACE2 were even stronger than its interaction with human ACE2. As mentioned above, “affinity is not the only factor that affects viral susceptibility,” since dog encodes soluble ACE2 isoforms which may compete with full‐length ACE2 binding to SARS‐CoV‐2 S protein, 33 and therefore may have a lower susceptibility. A lower susceptibility to SARS‐CoV‐2 of dogs than that of cat was confirmed by Shi et al. 34
By the time of revising our manuscript, a cat ACE2 (N‐domain, residues 18‐614)‐RBD structure was solved using cryo‐electron microscopy. 29 We compared our cat ACE2–RBD model with the cat ACE2–RBD Cryo‐EM structure (7C8D), and found that both models share a very similar structure (Table S4, Figure S2). We could raise similar observations based on our cat ACE2 model as based on 7C8D discussed in their study. 29 This further supports the reliability of our models. Furthermore, Wuhan cat samples were detected containing the antibodies against SARS‐CoV‐2. 35 , 36 , 37 This could serve as evidence supporting the susceptibility of cats to SARS‐CoV‐2. Their study highlights the importance of solving ACE2 structures from difference species to get detailed and trustworthy ideas of whether and how SARS‐CoV‐2 infects these animals. Until now, research groups only solved two of these ACE2 structures (human and cat's), not because we don′t want to solve more, it is only because solving each of these structures costs almost the same resources (funds, time, labor‐force etc) as for solving the human's. Therefore, the strategies (i.e., homology modeling and in silico analysis), the models and the conclusions discussed in our study could serve as very good supplements.
Farm animals. such as sheep and bovine have only two residues in the S–ACE2 interface region that are different from that of human. The primary difference is in a side chain residue that causes repulsion in the interaction; therefore, the sheep and bovine ACE2 proteins might have a slightly lower affinity to the SARS‐CoV‐2 S protein compared to human ACE2. Similarly, based on the same strategy, we predict that pig ACE2 should also have a low S protein affinity. This was further confirmed by our MD simulation analysis and reports from other groups. 28 , 29 Regardless of the differences in affinities, care should be taken with these animals, as they are also likely to carry SARS‐CoV‐2 and spread the virus to other animal populations.
Our study provides, for the first time, models of the full‐length ACE2 structures from several species. These models can serve as useful tools in understanding the mechanisms by which SARS‐CoV‐2 S protein binds host cells. These full‐length models could also be applied in detailed analyses of other ACE2 regions in and future studies. By analyzing the S–ACE2 interfaces, we predicted the differences in affinity of the SARS‐CoV‐2 S protein to ten different ACE2 molecules. Although affinity is not the only factor that affects viral susceptibility, it is one of the most important factors. Regardless of the ranks describing that which of them should have the highest affinity to SARS‐CoV‐2 S protein and which of them might have the lowest affinity, the overall stability of the ACE2–RBD complex from most of the species are reasonably high. Also, during the evolution, ACE2s from all these species possessed a very high sequence and structure similarity especially in the N‐domain which involve in binding to SARS‐CoV‐2 S protein. Hence, care should be taken regarding these species that humans are in daily contact with, especially cats, dogs, bovine, sheep, and pigs, to prevent the spread of SARS‐CoV‐2.
Structural modeling and in silico analysis are highly beneficial in structure‐guided drug development and in other areas. We believe that these methods can also be powerful tools for providing important clues in the study of viruses, providing us with guidance for viral protection in the early stages of disease spread.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Figure S1. Sequence alignment of ACE2 from 10 indicated species. SARS‐CoV‐2 S protein interface residues are marked with red triangles. Human (Q9BYF1), gorilla (G3QWX4), monkey (F7AH40), pig (K7GLM4), bovine (Q58DD0), sheep (W5PSB6), cat (Q56H28), dog (J9P7Y2), mouse (Q8R0I0), and rat (Q5EGZ1).
Figure S2 Alignment of Cryo‐EM structure of catACE2 (residues 18‐614)‐RBD complex recently solved (7C8D, palegreen) and our catACE2 (residues 18‐611)‐RBD model (violet).
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGMENTS
The authors would like to thank Prof. Xing Zhang from Zhejiang University School of Medicine for his kind support on MD simulation. They would also like to thank Prof. Vincent Postis from Leeds Beckett University for providing constructive advices during this study and writing, and Katherine Thieltges from Liwen Bianji, Edanz Editing China for editing the English text of a draft of this manuscript. This study was supported by grants from the Zhejiang Province Science and Technology Plan of Traditional Chinese Medicine [grant number 2020ZQ031], the Natural Science Foundation of China [grant number 32000707], the Key Discipline of Traditional Chinese Medicine in Zhejiang Province [grant number 2017‐XK‐A31], and Zhejiang University [grant number SJS202014].
AUTHOR CONTRIBUTIONS
Caixia Gong collected the data. Caixia Gong and Cheng Ma performed the analysis. Cheng Ma drafted the manuscript.
Ma C, Gong C. ACE2 models of frequently contacted animals provide clues of their SARS‐CoV‐2 S protein affinity and viral susceptibility. J Med Virol. 2021;93:4469‐4479. 10.1002/jmv.26953
Contributor Information
Cheng Ma, Email: mac4@zju.edu.cn.
Caixia Gong, Email: gongcaixia@zju.edu.cn.
DATA AVAILABILITY STATEMENT
The PDB file of models used or analyzed during the current study are available from the corresponding author on reasonable request. All the remaining data are included in this article and its supplementary information files.
REFERENCES
- 1. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science. 2020;367(6485):1444‐1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He WT, Ji X, He W, et al. Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Mol Biol Evol. 2020;37(9):2641‐2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau SKP, Wong EYM, Tsang CC, et al. Discovery and sequence analysis of four deltacoronaviruses from birds in the middle east reveal interspecies jumping with recombination as a potential mechanism for avian‐to‐avian and avian‐to‐mammalian transmission. J Virol. 2018;92(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106(14):5871‐5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801‐8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burkard C, Verheije MH, Wicht O, et al. Coronavirus cell entry occurs through the endo‐/lysosomal pathway in a proteolysis‐dependent manner. PLOS Pathog. 2014;10(11):e1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kirchdoerfer RN, Cottrell CA, Wang N, et al. Pre‐fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two‐step, furin‐mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111(42):15214‐15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science. 2005;309(5742):1864‐1868. [DOI] [PubMed] [Google Scholar]
- 11. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS‐CoV‐2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5(4):562‐569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105‐116. [DOI] [PubMed] [Google Scholar]
- 16. Hall TA. BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95‐98. [Google Scholar]
- 17. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fiser A, Sali A. Modeller: generation and refinement of homology‐based protein structure models. Methods Enzymol. 2003;374:461‐491. [DOI] [PubMed] [Google Scholar]
- 19. Chen VB, Arendall WB 3rd, Headd JJ, et al. MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janson G, Zhang C, Prado MG, Paiardini A. PyMod 2.0: improvements in protein sequence‐structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33(3):444‐446. [DOI] [PubMed] [Google Scholar]
- 21. Pronk S, Páll S, Schulz R, et al. GROMACS 4.5: a high‐throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845‐854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 23. Yang XH, Deng W, Tong Z, et al. Mice transgenic for human angiotensin‐converting enzyme 2 provide a model for SARS coronavirus infection. Comp Med. 2007;57(5):450‐459. [PubMed] [Google Scholar]
- 24. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581(7807):221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Q, Zhang Y, Wu L, et al. Structural and Functional Basis of SARS‐CoV‐2 Entry by Using Human ACE2. Cell. 2020;181(4):894‐904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brooke GN, Prischi F. Structural and functional modelling of SARS‐CoV‐2 entry in animal models. Sci Rep. 2020;10(1):15917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS‐CoV‐2 infection. Biochem Biophys Res Commun. 2020;526(1):165‐169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhai X, Sun J, Yan Z, et al. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J Virol. 2020;94(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu L, Chen Q, Liu K, et al. Broad host range of SARS‐CoV‐2 and the molecular basis for SARS‐CoV‐2 binding to cat ACE2. Cell Discov. 2020;6:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu H, Chen Q, Yang G, et al. Adaptation of SARS‐CoV‐2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie X, Zou J, Fontes‐Garfias CR, et al. Neutralization of N501Y mutant SARS‐CoV‐2 by BNT162b2 vaccine‐elicited sera. bioRxiv. 2021. [DOI] [PubMed] [Google Scholar]
- 33. Gao S, Zhang L. ACE2 partially dictates the host range and tropism of SARS‐CoV‐2. Comput Struct Biotechnol J. 2020;18:4040‐4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science. 2020;368(6494):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Q, Zhang H, Gao J, et al. A serological survey of SARS‐CoV‐2 in cat in Wuhan. Emerg Microbes Infect. 2020;9(1):2013‐2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halfmann PJ, Hatta M, Chiba S, et al. Transmission of SARS‐CoV‐2 in domestic cats. N Engl J Med. 2020;383(6):592‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bosco‐Lauth AM, Hartwig AE, Porter SM, et al. Experimental infection of domestic dogs and cats with SARS‐CoV‐2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA. 2020;117(42):26382‐26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of ACE2 from 10 indicated species. SARS‐CoV‐2 S protein interface residues are marked with red triangles. Human (Q9BYF1), gorilla (G3QWX4), monkey (F7AH40), pig (K7GLM4), bovine (Q58DD0), sheep (W5PSB6), cat (Q56H28), dog (J9P7Y2), mouse (Q8R0I0), and rat (Q5EGZ1).
Figure S2 Alignment of Cryo‐EM structure of catACE2 (residues 18‐614)‐RBD complex recently solved (7C8D, palegreen) and our catACE2 (residues 18‐611)‐RBD model (violet).
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
The PDB file of models used or analyzed during the current study are available from the corresponding author on reasonable request. All the remaining data are included in this article and its supplementary information files.


