Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) was discovered 27 years ago and its link to several pathologies – Kaposi's sarcoma, primary effusion lymphoma, and the B cell variant of Multicentric Castleman disease – is now well established. However, many questions remain about how KSHV causes tumors. Here, I will review studies from the last few years (primarily 2019–2021) that report new information about KSHV biology and tumorigenesis, including new results about KSHV proteins implicated in tumorigenesis, genetic and environmental variability in KSHV-related tumor development, and potential vulnerabilities of KSHV-caused tumors that could be novel therapeutic targets.
Keywords: KSHV, HHV-8, Kaposi's sarcoma, PEL, Viral oncogenesis
1. Introduction
The gamma-herpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) was discovered 27 years ago in a Kaposi's sarcoma (KS) lesion of an AIDS patient [1]. Since then, the connection between infections with KSHV and the tumors it causes – KS and primary effusion lymphoma (PEL) – has become the best established among the human tumor viruses. KSHV is found in all KS and PEL samples. KS is a tumor of endothelial cells that is characterized by highly vascularized lesions on the skin and mucosal tissue, with varying degree of aggressive presentations. PEL is a rare but highly fatal B-cell lymphoma that is usually found in body cavities and is composed of plasmablastic cells. In addition to KS and PEL, KSHV also causes the B cell variant of Multicentric Castleman Disease, a lymphoproliferative disease, and an inflammatory syndrome termed KSHV-induced cytokine syndrome (KICS). KSHV-induced diseases are predominantly found in immunocompromised individuals. AIDS-associated KS (also known as epidemic KS) and iatrogenic KS arise as a consequence of immunosuppression from AIDS or immunosuppressant therapy after transplants, respectively. However, there are subtypes of KS that appear to occur in the absence of overt immunosuppression, such as the classical KS that occurs predominantly in older men of Mediterranean descent, and the endemic KS that is common in African adults and children. Because of the association with immunosuppression, KS became very prominent during the AIDS epidemics, which led to the discovery of KSHV. Since the advent of anti-retroviral therapy (ART), cases of KS in the US have drastically decreased. However, in the past 10 years they have leveled to ~900 cases a year [2]. KS remains the second most common AIDS-associated cancer in the US and the second most common cancer in sub-Saharan Africa. For reasons that remain unclear, ART has also been less successful in reducing KS incidence in sub-Saharan Africa, possibly because more patients in this region tend to start therapy when they have already developed advanced KS [3]. To date, approved therapies for KS and PEL treatment remain very limited and have limited efficacy. KS is mostly managed by counteracting the immune suppression, while PEL and MCD prognosis remains very poor, with patients typically surviving for only a few months to a year. However, since it is discovery in 1994, we have learned a lot about the biology and pathogenesis of KSHV and its interaction with the infected host, which will hopefully lead to new avenues to therapy. In this review, I will cover advances on some major questions in the KSHV field, focusing particularly on studies from the last few years.
2. How does KSHV infection lead to tumorigenesis?
Although the epidemiological connection between KSHV and KS, PEL, and MCD is well-established, the process of KSHV-induced transformation is still poorly understood. Like all herpesviruses, KSHV can infect cells in a lytic or latent fashion (Fig. 1). Many of the KS and PEL tumor cells are latently infected, although lytic replication is prominent in MCD. During latent infection, only a few viral genes are expressed: latent nuclear antigen (LANA/ORF73), viral cyclin (vCyclin/ORF72), viral FLIP (vFLIP/K13), the Kaposins (K12), 12 viral micro RNAs (K-miRNAs) and, in PEL cells, viral interferon regulatory factor 3 (vIRF3/K10.5/LANA2). LANA in particular is key for maintenance of latent infection, because it tethers the viral genome to the host chromosomes, preventing virus loss during cell division [4]. While the role of LANA in KSHV latency has been well established for many years, recently there have been increasing efforts to target LANA for therapy, with the idea of reducing viral persistence. Two studies have shown that in an experimental setting, CRISPR-Cas9 could be used to damage the LANA gene and thus reduce latent virus carriage [5,6]. Other groups have sought to identify small molecules that interfere with LANA-DNA interactions [7,8]. However, these approaches are still at an early experimental stage, and no in vivo studies have been performed. Therefore, it is unclear whether they can be adapted for therapy and whether there would be a significant impact on tumorigenesis in vivo. Studies over the last 27 years have also uncovered roles for LANA, the other latent proteins, and the K-miRNAs in cell survival, proliferation and/or angiogenesis. These activities likely contribute to tumorigenesis. Nonetheless, new aspects of the function of these proteins and RNAs continue to be reported. For example, in the last couple of years LANA has been shown to drive cleavage of the Aurora B kinase [9]. LANA also increases expression of the pro-survival factor MCL-1 by sequestering the E3 ubiquitin ligase FBW7, which normally drives MCL-1 degradation [10]. vCyclin was recently shown to help KSHV-infected primary lymphatic endothelial cells overcome senescence, a growth arrest that functions as anti-oncogenic mechanism [11]. KSHV may also be able to overcome cell senescence by activating an alternative pathway of telomere maintenance through homologous recombination, since telomere shortening leads to senescence [12]. Whether a specific KSHV factor is important in telomere maintenance by homologous recombination remains unknown. The function of viral K-miRNAs and other non-coding RNAs in tumorigenesis was recently reviewed by Tagawa et al. [13], therefore I will not discuss it more in detail here.
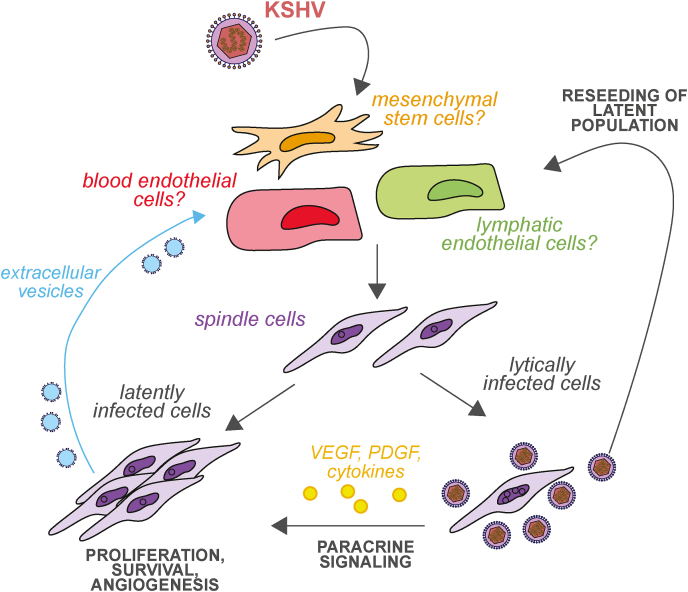
Fig. 1.
Current model of transformation by KSHV. After KSHV infects cells, they undergo a morphological, metabolic and gene expression change that turns them into “spindle cells”. Latently infected cells contribute to tumors through increased proliferation and survival, and angiogenesis. Lytically infected cells produce virus that contributes to reseeding and maintaining the latent pool of cells. They also secrete various cytokines, angiogenic factors and other signals that contribute to growth and transformation of the tumor. Latently infected cells also produce extracellular vesicles that can affect cellular phenotypes.
One theme that has consistently emerged from studies over the last 27 years is that, despite the pro-oncogenic functions of latent genes, simply infecting cells with KSHV does not transform them, unlike infection with the related gammaherpesvirus Epstein-Barr virus (EBV). For example, whereas vCyclin can propel cells past senescence, the infected cells still undergo crisis, a second proliferative block during which the cells cease to grow and die [11]. Paradoxically, transformation has been seen more often with single KSHV gene expression or in cell types that are not considered relevant for human tumors. For example, overexpression of the viral protein kinase vPK/ORF36 alone induces lymphomas in aged mice [14]. Also, rat mesenchymal precursor cells become transformed after KSHV infection and can give rise to tumors in nude mice [15]. Consistent with the poor transforming potential of KSHV, substantial evidence points to a heavy involvement of paracrine signaling in the development and maintenance of KS, PEL and MCD. In particular, cytokines like interleukin 6 (IL-6) and angiogenic factors like vascular endothelial growth (VEGF) have been implicated in KSHV-dependent tumorigenesis and are in some cases produced by KSHV lytically infected cells (see section 3) (Fig. 1). New potential therapies based on altering cytokine signaling are being explored for these diseases, as recently reviewed in Alomari and Totonchy [16]. VEGF-A targeting antibodies have also shown some promise in in vivo models, especially antibody fragments that may have better penetration in tumors [17]. Yet an additional wrinkle to the role of inter-cellular communication in tumorigenesis was added by the discovery that extracellular vesicles released from KSHV-infected cells may also alter the phenotype of nearby uninfected cells [18] (Fig. 1). In general, tumorigenesis by KSHV appears to rely on a complex set of interactions between infected cells and the microenvironment that we are still in the process of deciphering.
3. What is the contribution of lytic replication to tumorigenesis?
Another key component of the tumorigenesis process is viral lytic replication. This is at first glance surprising, as replicating cells are expected to die and thus cannot directly contribute to tumor mass. The contribution of lytic replication to tumorigenesis was initially revealed in the late '90s by the unexpected regression of Kaposi's sarcoma in AIDS patients under treatment with anti-herpesviral drugs for CMV retinitis [19]. New findings over the years have provided additional support for this model. For example, recent longitudinal studies have shown that antibodies against lytic proteins can be detected in patients that develop Kaposi's sarcoma prior to the onset of disease [20]. The presence of these antibodies suggests that high levels of reactivation from latency occurred before disease development.
Lytic replication is thought to support tumorigenesis in several ways. First of all, the KSHV genome is routinely lost during passaging of KSHV-infected cells, including endothelial cells (although it is retained in cell lines derived from PEL tumors). Therefore, in vivo lytic reactivation and virus production is needed to maintain the population of latently infected cells (Fig. 1). Interestingly, evidence from studies using 3D tissue culture models indicate that in a tissue there is either more spontaneous lytic reactivation and/or more efficient reinfection of cells, which enables better genome maintenance relative to cells grown in 2D culture [21,22]. In addition to reseeding the latent population, lytic genes may also serve oncogenic functions, in some cases because they promote pro-tumorigenic and angiogenic paracrine signaling (Fig. 1). Viral IL-6 homolog (vIL-6/K2), viral G-protein coupled receptor (vGPCR/ORF74), viral interferon regulatory factor 1 (vIRF1/K9), viral protein kinase (vPK/ORF36), and the membrane proteins K1 and K15 are all genes that are classically considered lytic but have oncogenic functions. vIL-6 is a homolog of the human cytokine and binds the human IL-6 receptor [23]. Among its many functions, recent studies have shown that vIL-6 increases expression of integrin β3 [24] and decreases expression of caveolin 1 [25] through its activation of STAT3. Both changes contribute to angiogenic-like behavior in cells expressing vIL-6. vGPCR is a constitutively active receptor that resembles the chemokine receptor CXCR-1/2, activates MAP kinase pathways, and stimulates secretion of the angiogenic factor VEGF [26]. Recent reports have shown that vGPCR signaling also induces expression and secretion of platelet-derived growth factor (PDGF), which activates its receptor PDGFRA and has oncogenic and angiogenic effects [27]. Moreover, VEGF induction is in part mediated by vGPCR stimulation of the expression of cyclooxygenase-2 (COX2), an enzyme that synthesizes prostaglandins [28]. Interestingly, both of these effects have potential as therapeutic targets, as PDGFRA inhibitors like sunitinib and COX2 inhibitors like celecoxib and NS398 can reduce vGPCR-induced tumorigenesis [27,28]. However, these agents have only been tested in experimental models, and more investigation is needed to translate these findings to the clinic. K1 and K15 are transmembrane proteins that control intracellular signaling and promote proliferation and angiogenesis [26]. The KSHV-encoded kinase vPK contributes to transformation via mimicry of ribosomal S6 kinase, a downstream effector of the mTOR pathway. vPK phosphorylation of S6 kinase targets leads to an increase in global translation and angiogenic-like phenotypes [29]. Moreover, transgenic mice expressing this protein develop lymphomas that resemble PEL and have elevated levels of cytokines in their plasma [14]. vIRF1 is an immune evasion factor [30], but it also contributes to angiogenesis by upregulating the expression of multiple genes with oncogenic functions, including CUB-domain containing protein 1 [31], sperm-associated antigen 9 [32] and glutaredoxin 3 [33]. Upregulation of most of these genes requires interactions between vIRF1 and the cellular transcription factor lymphoid enhancer-binding factor 1 [[31], [32], [33]]. Upregulation of glutaredoxin 3 is indirectly mediated through regulation of a circular RNA and a cellular miRNA [33].
There is some debate as to whether expression of these oncogenic lytic genes requires the full lytic cycle, or if they are expressed in a “relaxed” latency state without completion of the viral replication cycle. In support of a full lytic cycle induction, several other genes considered to be canonically lytic genes, like the glycoprotein K8.1 and the immune evasion factor K5, as well as K1 and K15, have been detected in KS biopsies over the years [26,[34], [35], [36]]. Recent advances in transcriptome-wide RNA sequencing technologies have allowed researchers to examine gene expression during KS and in PEL cells more in detail. These transcriptome-wide studies have revealed some level of lytic gene expression in tumors. Expression profiling of KS lesions suggests that lytic reactivation may vary substantially between patients and/or lesions, although some expression of genes classically considered to be lytic is found in most lesions [37,38]. Sequencing of the lymphoma cell line BCP-1 showed high expression of a subset of lytic genes, including PAN and a cluster of genes involved in immune regulation (K2, K4, K5) [39]. Single-cell RNA sequencing of the BC1 PEL line also detected many lytic genes in up to 50% of the cells, but at very low levels per cell [40].
In any case, there is considerable interest in understanding whether lytic replication or its effects can be targeted for therapy. This has also coincided with the development of new reagents that have improved our ability to study the lytic cycle and generate KSHV mutants. These reagents include systems in which reactivation of the lytic cycle can be induced more efficiently through doxycycline-inducible exogenous expression of the KSHV master lytic regulator, RTA/ORF50 [[41], [42], [43]]. Moreover, bacterial artificial chromosome-based systems have been developed to generate KSHV mutants, in particular the now widely used BAC16 system [44,45].
4. What is the origin of the KS spindle cells?
Another unresolved question is the origin of the cells that are infected by KSHV and give rise to KS. The long-standing model is that the most likely targets are endothelial cells, since KS cells express both panendothelial and lymphatic endothelial markers [46]. However, KS cells also appear poorly differentiated and express markers of other cell types. Indeed, KSHV infection causes reprogramming of the cellular gene expression, as well as a morphological change of the cells to KS “spindle cells” [47,48] (Fig. 1). In 2004, several studies showed that KSHV can infect both blood and lymphatic endothelial cells, and alter gene expression in both cell types to an intermediate state between the two [[49], [50], [51]]. Since then, it has become apparent that lymphatic endothelial cells can support spontaneous reactivation by KSHV, which is thought to be important for tumorigenesis (see section 3) and maintenance of the latent cell pool (Fig. 1). Two recent studies have revealed that this is because these cells express the PROX1 transcription factor, which is a key protein for lymphatic endothelium development [34,52]. PROX1 directly stimulates expression of RTA/ORF50, the master regulator of lytic reactivation, by binding to its promoter [34,52]. Gramolelli et al. also showed a role for another lymphatic endothelium transcription factor, SOX18, in KSHV persistence [34]. SOX18 promotes an increase in the number of latent genomes by binding near the origin of replication [34]. Interestingly, whereas PROX1 and SOX18 regulate each other during lymphatic endothelium development, KSHV decouples this reciprocal regulation, presumably to facilitate the continued expression of both transcription factors [34]. Since KSHV infection also induces higher levels of PROX1 and SOX18 expression in blood endothelial cells [34], these results suggest that KSHV infection drives cells to return to an embryonic stage of lymphatic endothelial development, when PROX1 and SOX18 are highly expressed. The two studies on PROX1 also suggest that the spontaneous reactivation in lymphatic endothelial cells is key to maintaining and expanding the population of infected cells in the tumor [34,52].
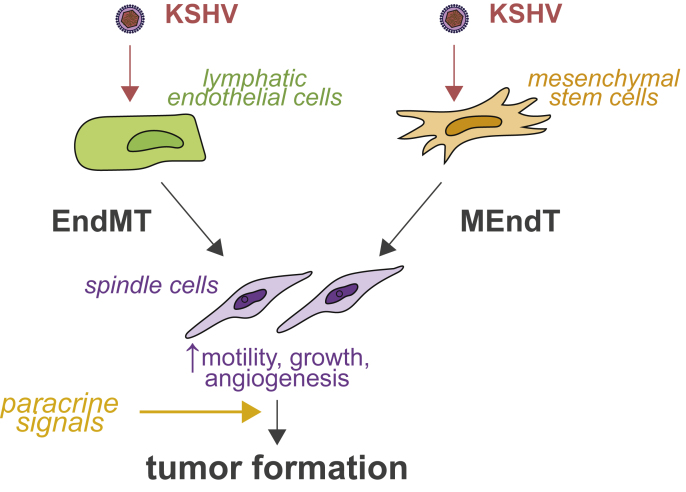
Notwithstanding these results in endothelial cells, some groups have advanced the idea that mesenchymal stem cells may be the source, or at least one of the sources, of the KS spindle cells [[53], [54], [55], [56], [57], [58]]. Indeed, KS spindle cells also express mesenchymal markers like nestin or PDGFRA [54]. Some studies have suggested that this could be due to an endothelial-to-mesenchymal (EndMT) transition that occurs in response to KSHV infection of endothelial cells [21,54,59] (Fig. 2). EndMT is a transdifferentiation process similar to epithelial-to-mesenchymal transition (EMT), which is implicated in tumorigenesis and metastasis [60,61]. EndMT may be related to LANA's ability to turn on expression of mesenchymal markers [62]. However, several studies over the years have shown that KSHV can also latently infect both rodent and human mesenchymal precursor/stem cells in cell culture [15,[53], [54], [55], [56], [57], [58]]. KSHV-infected mesenchymal stem cells express a mixed set of mesenchymal and endothelial markers, like KS tumor cells [53,54,57]. They also have increased angiogenic properties, including secretion of pro-angiogenic factors like basic fibroblast-growth factor [53,54,57] and an invasive and transformed phenotype in cell culture-based assays [15,53,58]. Recent studies have also shown that KSHV-infected mesenchymal cells transplanted into mice migrate to sites of wound healing [58]. This mimics a process called the Koebner phenomenon whereby KS lesions form at wound healing sites [63,64]. In addition, KSHV-infected rat mesenchymal precursor cells form tumors in nude mice [15]. Conversely, infection of murine mesenchymal cells is not sufficient for transformation [55]. Murine cells require growth in “KS-like” media with heparin and endothelial cell growth factors to become transformed, again emphasizing the role of soluble cues in the transformation of KSHV-infected cells [55] (Fig. 2). Treatment with the KS-like media induces epigenetic changes that alter host and viral gene expression and activate key paracrine pathways, most notably PDGFRA, which promotes cell growth [55]. While these data on infection of mesenchymal stem cells seem at odds with the prevalent model that KS arises from endothelial cells, Li et al. found that while KSHV infection of endothelial cells leads to EndMT, KSHV infection of mesenchymal cells leads to the opposite transdifferentiation, mesenchymal-to-endothelial transition (MEndT) [54]. These results suggests that KSHV may infect both types of cells and drive their transdifferentiation to an intermediate state (Fig. 2).
Fig. 2.
Potential origins of spindle cells from endothelial and mesenchymal cells. KSHV infection may lead to de-differentiation of both endothelial and mesenchymal cells through endothelial-to-mesenchymal (EndMT) and mesenchymal-to-endothelial transition (MEndT), respectively. The infected cells thus because more motile, angiogenic and proliferative. Nonetheless, paracrine signals are likely needed for full transformation of the cells.
In further support of an important physiological role for mesenchymal cell infection by KSHV, a potential connection between KSHV and osteosarcoma, a cancer of mesenchymal origin, was recently described [65]. Analysis of a group of osteosarcoma patients from the Xinjiang Uyghur autonomous region revealed much higher KSHV seropositivity than in a matched control group, and KSHV DNA and expression of LANA were detected in the osteosarcoma tumors of KSHV-positive patients [65]. Moreover, gene expression profiling suggested a qualitative difference between the KSHV-positive and the KSHV-negative osteosarcomas [65]. It remains to be seen if this new connection stands the test of time. The connection between KSHV and osteosarcoma may have been missed due to heterogeneity in osteosarcomas and low KSHV positivity in many populations. In contrast, KSHV seropositivity is relatively high among the Xinjiang Uyghur population [66]. Alternatively, this relationship may be specific to the Uyghur population. The latter would be similar to nasopharyngeal carcinoma and Epstein-Barr virus, which are linked predominantly in specific Asian ethnic groups [67].
5. Why do only some people develop KSHV-induced tumors?
While KSHV is required for KS and PEL tumorigenesis, it is clearly not sufficient. There is a lot of individual variability in disease development and presentation within both AIDS-associated KS and endemic KS. Several studies have tried to decipher factors that underlie this disease variability, as well as the variability in KSHV prevalence among different populations. For example, KSHV seropositivity is very high in Africa but low in North America [68]. Although one model has been that the risk of KS development may depend on the strength of the immune response of different individuals, recent studies have argued against this idea. For example, KS patients do not have lower levels of anti-KSHV neutralizing antibodies or antibodies mediating antibody-dependent cellular cytotoxicity relative to disease-free KSHV seropositive individuals [69,70]. Moreover, T cell responses are similarly weak in both groups [71]. Another possibility is that there are genetic differences between populations that underlie differences in risk. This is an attractive model since there is clear familial clustering of KS cases (for example [72]). Thus far, targeted studies or studies of early-onset KS cases have predominantly been used to identify genetic determinants of KSHV susceptibility or KS and MCD development. These studies have identified several interesting candidate polymorphisms, reviewed more extensively Blumenthal et al. [73]. In particular, polymorphism in specific HLA subtypes, in genes involved in regulation of immune responses like NFκB, TRIM31, LY6G6C, and in subtypes of the natural killer cell receptor KIR have been connected to susceptibility to KSHV infection or development of KS or MCD [73]. A genetic association with both susceptibility to infection and disease development was also found for Ephrin A, a potential entry receptor for KSHV [73,74]. However, most of these associations were found in a single study and their wider importance as genetic determinants of KSHV infection and disease remains unknown. Moreover, there has only been one genome-wide association study so far of any phenotype related to KSHV infection and disease [75]. In this study, polymorphisms in HLA genes were linked to levels of antibodies against KSHV proteins, a proxy for KSHV infection and disease progression [75]. How these polymorphisms modify KSHV infection and disease is currently unclear.
In addition to genetic or acquired immune differences, some of the differences in epidemiology and presentation may be due to the timing of KSHV infection. For example, pediatric KS patients in Malawi have a KS presentation that partially resembles MCD, with lymphadenopathy, viral replication and high levels of IL-6 and IL-10 [76]. This may be connected to the fact that these children acquire KSHV early in life, which is only common in areas where KSHV is endemic. Another influential factor for disease progression may be co-infections (recently reviewed by Thakker and Verma [77]). In particular, since PEL often develops in cells that are both KSHV and EBV positive, Faure et al. [78] set up a system of dual infection in primary peripheral B cells, and found that EBV infection shortly after KSHV infection promoted transformation of the B cells and higher KSHV infection. This reinforces that idea that dual KSHV/EBV infection is key for tumorigenesis in most cases of PEL. Lastly, the possibility that specific mutations need to arise in KSHV genomes to promote tumorigenesis has been considered. To test this hypothesis, a recent study sequenced several KSHV isolates from both oral swabs and tumors of patients [79]. While they found very little intra-host variability at single nucleotide level, consistent with the proof-reading activity of both the host and viral DNA polymerases, they did see that in several of the tumors the KSHV genome had major rearrangements with translocations and duplications [79]. However, it is unclear if these rearrangements contributed to tumorigenesis or were simply a consequence of genomic instability in the tumor tissue. Altogether, it is clear that multiple factors including viral genetics, host genetics and environmental factors contribute to the development of KS and PEL tumors.
6. What host activities are KSHV tumors addicted to that could be used as therapeutic targets?
Many tumors are “addicted” to specific cellular pathways and oncogenes, and these addictions represent potential vulnerabilities and drug targets for therapy [80]. While over the years many targeted studies have uncovered roles of individual pathways in KS and KSHV biology, with the advent of CRISPR screening it has become possible to comprehensively interrogate the landscape of genes and pathways necessary for KSHV tumorigenesis [81]. Three recent studies have sought to use genome-wide CRISPR screens to identify pathways that KSHV-induced tumors are “addicted” to. These screens looked for genes required for cell survival and growth in three different KSHV latent infection systems. Manzano et al. used BCBL1 cells originally derived from a PEL patient [82], Holmes et al. employed a widely used system of KSHV-infected human endothelial cells [83], and Gruffaz et al. used KSHV-infected rat embryonic mesenchymal stem cells [84]. Manzano et al. [82] found 210 genes that are specifically required for PEL growth and presumably oncogenesis, including the apoptosis regulators MCL1 and cFLIP, and the cyclin CCND2. cFLIP and CCND2 were unexpected, as the virus encodes homologs of these proteins (vFLIP and vCyclin). MCL1 and CCND2 were particularly interesting because they can be targeted using existing drugs. Indeed, treatment of multiple PEL cell lines with the MCL1 inhibitor S63845 or the CCND2 inhibitor palbociclib led to cell death or cell cycle arrest, respectively [82]. Another key regulator of PEL growth identified in this study was IRF4, which, together with its cofactor BATF and KSHV vIRF3, binds “super-enhancer” sequences [85]. These super-enhancers in turn regulate many of the key genes that PEL cells require for growth, including MCL1, cMYC and IRF4 itself [86]. The importance of super-enhancers for PEL growth explains why PEL are sensitive to BET domain inhibitors, as BET domain-containing proteins bind super-enhancers [87]. Holmes et al. [83] identified mitochondrial translation as essential for the growth of KSHV-infected endothelial cells. They also found that mitochondrial size and numbers were altered by KSHV latent infection in these cells [83]. Moreover, treatment with antibiotics that affect mitochondrial translation, such as chloramphenicol and tigecycline, reduced the growth of KSHV-infected endothelial cells and triggered cell death in PEL cell lines, pointing to these molecules as potential new anti-KSHV agents [83]. Gruffaz et al. [84] found a requirement for the nuclear export machinery for proliferation and growth in soft agar of KSHV-transformed rat embryonic mesenchymal stem cells. Pharmacological inhibition of the export protein XPO1 by KPT-8602 led to growth inhibition, which was likely due to activation of p53 [84]. Although these studies have focused on different vulnerabilities, they did find a few shared genes. For example, the screen hits from Manzano et al. [82] also include several subunits of the mitochondrial ribosome, like the ones identified in the Holmes et al. [83] study. Of note, these studies have revealed interesting new potential targets for treatment. However, these potential treatments have so far only been tested in tissue culture and more research is needed to determine whether they can truly be used for therapy.
Another aspect of cellular physiology that could constitute a therapeutic target is cellular metabolism. Cellular metabolism is often remodeled in cancers, with increased dependency on aerobic glycolysis and glutamine metabolism for growth, also known as the Warburg effect [88]. Since 2010, it has been apparent that latent KSHV infection changes many aspects of metabolism in a way that is similar to the Warburg effect. KSHV infection increases the cell's reliance on glycolysis and glutaminolysis and upregulates fatty acid metabolism and peroxisome number (reviewed in Lagunoff [89] and Dai et al. [90]). However, there has been some controversy surrounding these ideas, because some of the results differ between the latently infected endothelial cell model and the rat mesenchymal transformation model. In the KSHV-transformed rat mesenchymal precursor cells, glycolysis is not required for growth and glutaminolysis provides nucleotides for DNA replication rather than energy [91,92]. It remains to be seen whether the transformed phenotype or the difference in the source of the cells explains this discrepancy. RNAseq data from KS tumor lesions present yet another picture. Gene expression changes detected in the KS samples also suggest a shift towards glycolysis, but instead point to a decrease in lipid metabolism [38]. Again, it is unclear if the discrepancy between these results and the studies in KSHV latently infected cells is simply due to the system. It is also possible that lipid metabolism is reduced in these KS samples because they are from HIV-positive patients undergoing ART, which could also affect lipid metabolism [38]. An additional KSHV-associated change that is also commonly found in cancer cells is increased proline metabolism [93]. Mediated by the K1 protein, this metabolic change is required for high levels of proliferation of KSHV-infected cells in 3D culture [93]. In principle, many of the KSHV-associated metabolic changes could be exploited for targeted therapy, although at present the details remain to be worked out.
In addition to these pathways, it is possible that many genes involved in processes such as control of lytic gene expression and endothelial cell infection, including those described in other sections of this review, may serve as targets for KSHV treatment. For example, as mentioned in section 4, Gramolelli et al. reported that SOX18, an endothelial transcription factor, is important for genome maintenance during latent infection. The authors also showed that disrupting SOX18 dimerization and function with the small molecules SM4 or R-(+)-propanolol reduces genome copies in latently-infected cells [34], suggesting these treatments could be used as a therapeutic strategy against KS and KSHV infections. This and other findings described in this section exemplify how basic studies on the biology of KSHV during latent and lytic infection may inform novel treatment strategies.
7. Conclusions
27 years after its discovery, we have made progress on understanding the biology and tumorigenesis of KSHV, but many questions remain partly unresolved. In the last ten years, the range of tools to study KSHV has been expanded thanks to the work of multiple laboratories, which has allowed us to better dissect the function of proteins and to study the lytic cycle more in depth. However, we still lack a good transformation model that recapitulates KS or PEL development in human cells, and an animal model to study the interactions between the tumors and the physiology of the host organism. Nonetheless, many of the advances in the basic understanding of KSHV biology and its role in tumorigenesis provide hope that better treatments will be coming in the near future.
Author statement
Marta Maria Gaglia: Funding; Writing – original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
I apologize for not referencing all relevant studies because of length limitations. I thank Rachel Lent for critical reading of the manuscript. Work on KSHV in the Gaglia laboratory is supported by American Cancer Society Research Scholar Grant 131320-RSG-17-189-01-MPC.
References
- 1.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994 Dec 16;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Robbins H.A., Pfeiffer R.M., Shiels M.S., Li J., Hall H.I., Engels E.A. Excess Cancers Among HIV-Infected People in the United States. JNCI J Natl Cancer Inst [Internet] 2015 Apr 1;107(4) doi: 10.1093/jnci/dju503. [cited 2021 Mar 21]. (dju503). Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngalamika O., Munsaka S., Lidenge S.J., West J.T., Wood C. AIDS Res Hum Retroviruses [Internet]; 2021. Antiretroviral Therapy for HIV-Associated Cutaneous Kaposi's Sarcoma: Clinical, HIV-Related, and Sociodemographic Predictors of Outcome.https://www.liebertpub.com/doi/10.1089/aid.2020.0099 Jan 1 [cited 2021 Mar 23]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Leo A., Calderon A., Lieberman P.M. Control of viral latency by episome maintenance proteins. Trends Microbiol. 2020 Feb 1;28(2):150–162. doi: 10.1016/j.tim.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haddad C.O., Kalt I., Shovman Y., Xia L., Schlesinger Y., Sarid R. Targeting the Kaposi's sarcoma-associated herpesvirus genome with the CRISPR-Cas9 platform in latently infected cells. Virol. J. 2021 Mar 17;18(1):56. doi: 10.1186/s12985-021-01527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tso F.Y., West J.T., Wood C. Reduction of Kaposi’s sarcoma-associated herpesvirus latency using CRISPR-cas9 to edit the latency-associated nuclear antigen gene. J Virol [Internet] 2019 Apr 1;93(7):e02183–18. doi: 10.1128/JVI.02183-18. http://jvi.asm.org/content/93/7/e02183-18 [cited 2021 Mar 23] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderon A., Soldan S.S., Leo A.D., Deng Z., Frase D.M., Anderson E.M. Identification of Mubritinib (TAK 165) as an inhibitor of KSHV driven primary effusion lymphoma via disruption of mitochondrial OXPHOS metabolism. Oncotarget. 2020 Nov 17;11(46):4224–4242. doi: 10.18632/oncotarget.27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirsch P., Jakob V., Oberhausen K., Stein S.C., Cucarro I., Schulz T.F. Fragment-based discovery of a qualified hit targeting the latency-associated nuclear antigen of the oncogenic Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Med. Chem. 2019 Apr 25;62(8):3924–3939. doi: 10.1021/acs.jmedchem.8b01827. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Q., Ding L., Zi Z., Gao S., Wang C., Wang Y. Viral-mediated AURKB cleavage promotes cell segregation and tumorigenesis. Cell Rep. 2019 Mar 26;26(13):3657–3671. doi: 10.1016/j.celrep.2019.02.106. e5. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y.J., Kim Y., Kumar A., Kim C.W., Toth Z., Cho N.H. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen dysregulates expression of MCL-1 by targeting FBW7. PLoS Pathog. 2021 Jan 20;17(1) doi: 10.1371/journal.ppat.1009179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMaio T.A., Vogt D.T., Lagunoff M. KSHV requires vCyclin to overcome replicative senescence in primary human lymphatic endothelial cells. PLoS Pathog. 2020 Jun 18;16(6) doi: 10.1371/journal.ppat.1008634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lippert T.P., Marzec P., Idilli A.I., Sarek G., Vancevska A., Bower M. Oncogenic herpesvirus KSHV triggers hallmarks of alternative lengthening of telomeres. Nat. Commun. 2021 Jan 21;12(1):512. doi: 10.1038/s41467-020-20819-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tagawa T., Serquiña A., Kook I., Ziegelbauer J. Viral non-coding RNAs: stealth strategies in the tug-of-war between humans and herpesviruses. Semin. Cell Dev. Biol. 2021 Mar 1;111:135–147. doi: 10.1016/j.semcdb.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders P.M., Montgomery N.D., Montgomery S.A., Bhatt A.P., Dittmer D.P., Damania B. Human herpesvirus–encoded kinase induces B cell lymphomas in vivo. J. Clin. Invest. 2018 Jun 1;128(6):2519–2534. doi: 10.1172/JCI97053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones T., Ye F., Bedolla R., Huang Y., Meng J., Qian L. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest. 2012 Mar 1;122(3):1076–1081. doi: 10.1172/JCI58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alomari N., Totonchy J. Cytokine-targeted therapeutics for KSHV-associated disease. Viruses. 2020 Oct;12(10):1097. doi: 10.3390/v12101097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eason A.B., Sin S.-H., Shah M., Yuan H., Phillips D.J., Droste M. DLX1008 (brolucizumab), a single-chain anti-VEGF-A antibody fragment with low picomolar affinity, leads to tumor involution in an in vivo model of Kaposi Sarcoma. PloS One. 2020 May 14;15(5) doi: 10.1371/journal.pone.0233116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNamara R.P., Chugh P.E., Bailey A., Costantini L.M., Ma Z., Bigi R. Extracellular vesicles from Kaposi Sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog. 2019 Feb 4;15(2) doi: 10.1371/journal.ppat.1007536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin D.F., Kuppermann B.D., Wolitz R.A., Palestine A.G., Li H., Robinson C.A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N. Engl. J. Med. 1999 Apr 8;340(14):1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 20.Wakeham K., Thomas Johnston W., Angela Nalwoga, Webb Emily L., Mayanja Billy N., Wendell Miley. Trends in Kaposi's sarcoma‐associated Herpesvirus antibodies prior to the development of HIV‐associated Kaposi's sarcoma: a nested case‐control study. Int. J. Canc. 2015 Apr 11;136(12):2822–2830. doi: 10.1002/ijc.29329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F., Pekkonen P., Laurinavicius S., Sugiyama N., Henderson S., Günther T. KSHV-initiated notch activation leads to membrane-type-1 matrix metalloproteinase-dependent lymphatic endothelial-to-mesenchymal transition. Cell Host Microbe. 2011 Dec 15;10(6):577–590. doi: 10.1016/j.chom.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Dubich T., Dittrich A., Bousset K., Geffers R., Büsche G., Köster M. 3D culture conditions support Kaposi's sarcoma herpesvirus (KSHV) maintenance and viral spread in endothelial cells. J. Mol. Med. 2021 Mar 1;99(3):425–438. doi: 10.1007/s00109-020-02020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakakibara S., Tosato G. Viral interleukin-6: role in Kaposi's sarcoma-associated herpesvirus–associated malignancies. J. Interferon Cytokine Res. 2011 Jul 18;31(11):791–801. doi: 10.1089/jir.2011.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivera-Soto R., Dissinger N.J., Damania B. Kaposi’s sarcoma-associated herpesvirus viral interleukin-6 signaling upregulates integrin β3 levels and is dependent on STAT3. J Virol [Internet] 2020 Feb 14;94(5):e01384–19. doi: 10.1128/JVI.01384-19. http://jvi.asm.org/content/94/5/e01384-19 [cited 2021 Mar 29]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Wang Q., Qi X., Guo Y., Lu H., Chen Y. Viral interleukin-6 encoded by an oncogenic virus promotes angiogenesis and cellular transformation by enhancing STAT3-mediated epigenetic silencing of caveolin 1. Oncogene. 2020 Jun;39(23):4603–4618. doi: 10.1038/s41388-020-1317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abere B., Schulz T.F. KSHV non-structural membrane proteins involved in the activation of intracellular signaling pathways and the pathogenesis of Kaposi's sarcoma. Curr Opin Virol. 2016 Oct 1;20:11–19. doi: 10.1016/j.coviro.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Cavallin L.E., Ma Q., Naipauer J., Gupta S., Kurian M., Locatelli P. KSHV-induced ligand mediated activation of PDGF receptor-alpha drives Kaposi's sarcomagenesis. PLoS Pathog. 2018 Jul 9;14(7) doi: 10.1371/journal.ppat.1007175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina M.V., D'Agostino A., Ma Q., Eroles P., Cavallin L., Chiozzini C. KSHV G-protein coupled receptor vGPCR oncogenic signaling upregulation of Cyclooxygenase-2 expression mediates angiogenesis and tumorigenesis in Kaposi's sarcoma. PLoS Pathog. 2020 Oct 15;16(10) doi: 10.1371/journal.ppat.1009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatt A.P., Wong J.P., Weinberg M.S., Host K.M., Giffin L.C., Buijnink J. A viral kinase mimics S6 kinase to enhance cell proliferation. Proc. Natl. Acad. Sci. Unit. States Am. 2016 Jul 12;113(28):7876–7881. doi: 10.1073/pnas.1600587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z., Jacobs S.R., West J.A., Stopford C., Zhang Z., Davis Z. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. U. S. A. 2015 Aug 4;112(31):E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., Wang Q., Qi X., Lu H., Chen Y., Shi J. An oncogenic viral interferon regulatory factor upregulates CUB domain-containing protein 1 to promote angiogenesis by hijacking transcription factor lymphoid enhancer-binding factor 1 and metastasis suppressor CD82. Cell Death Differ. 2020 Dec;27(12):3289–3306. doi: 10.1038/s41418-020-0578-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Wang F., Shi J., Feng Q., Chen Y., Qi X. Sperm associated antigen 9 promotes oncogenic KSHV-encoded interferon regulatory factor-induced cellular transformation and angiogenesis by activating the JNK/VEGFA pathway. PLoS Pathog. 2020 Aug 10;16(8) doi: 10.1371/journal.ppat.1008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao S., Jia X., Wang F., Sheng L., Song P., Cao Y. CircRNA ARFGEF1 functions as a ceRNA to promote oncogenic KSHV-encoded viral interferon regulatory factor induction of cell invasion and angiogenesis by upregulating glutaredoxin 3. PLoS Pathog. 2021 Feb 4;17(2) doi: 10.1371/journal.ppat.1009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gramolelli S., Elbasani E., Tuohinto K., Nurminen V., Günther T., Kallinen R.E. Oncogenic herpesvirus engages endothelial transcription factors SOX18 and PROX1 to increase viral genome copies and virus production. Canc. Res. 2020 Aug 1;80(15):3116–3129. doi: 10.1158/0008-5472.CAN-19-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L., Dittmer D.P., Tomlinson C.C., Fakhari F.D., Damania B. Immortalization of primary endothelial cells by the K1 protein of Kaposi's sarcoma–associated herpesvirus. Canc. Res. 2006 Apr 1;66(7):3658–3666. doi: 10.1158/0008-5472.CAN-05-3680. [DOI] [PubMed] [Google Scholar]
- 36.Pantanowitz L., Früh K., Marconi S., Moses A.V., Dezube B.J. Pathology of rituximab-induced Kaposi sarcoma flare. BMC Clin. Pathol. 2008 Dec;8(1):1–5. doi: 10.1186/1472-6890-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosseinipour M.C., Sweet K.M., Xiong J., Namarika D., Mwafongo A., Nyirenda M. Viral profiling identifies multiple subtypes of Kaposi's sarcoma. mBio. 2014 Oct 31;5(5) doi: 10.1128/mBio.01633-14. e01633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tso F.Y., Kossenkov A.V., Lidenge S.J., Ngalamika O., Ngowi J.R., Mwaiselage J. RNA-Seq of Kaposi's sarcoma reveals alterations in glucose and lipid metabolism. PLoS Pathog. 2018 Jan 19;14(1) doi: 10.1371/journal.ppat.1006844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao S., Strong M.J., Wang X., Moss W.N., Concha M., Lin Z. High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the cancer cell line encyclopedia project. J. Virol. 2015 Jan 1;89(1):713–729. doi: 10.1128/JVI.02570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rondeau N.C., Finlayson M.O., Miranda J.L. Widespread traces of lytic Kaposi sarcoma-associated herpesvirus in primary effusion lymphoma at single-cell resolution. Microbiol Resour Announc [Internet] 2020 Nov 5;9(45):e00851–20. doi: 10.1128/MRA.00851-20. https://mra.asm.org/content/9/45/e00851-20 Available from: [cited 2021 Mar 21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura H., Lu M., Gwack Y., Souvlis J., Zeichner S.L., Jung J.U. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 2003 Apr;77(7):4205–4220. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myoung J., Ganem D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods. 2011 Jun;174(1–2):12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dollery S.J., Maldonado T.D., Brenner E.A., Berger E.A. iTIME.219: an immortalized KSHV infected endothelial cell line inducible by a KSHV-specific stimulus to transition from latency to lytic replication and infectious virus release. Front Cell Infect Microbiol [Internet] 2021;11:264. doi: 10.3389/fcimb.2021.654396. http://www.frontiersin.org/articles/10.3389/fcimb.2021.654396/full Available from: [cited 2021 May 16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brulois K.F., Chang H., Lee A.S.-Y., Ensser A., Wong L.-Y., Toth Z. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012 Sep;86(18):9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou F.-C., Zhang Y.-J., Deng J.-H., Wang X.-P., Pan H.-Y., Hettler E. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002 Jun;76(12):6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beckstead J.H., Wood G.S., Fletcher V. Evidence for the origin of Kaposi's sarcoma from lymphatic endothelium. Am. J. Pathol. 1985 May;119(2):294–300. [PMC free article] [PubMed] [Google Scholar]
- 47.Cancian L., Hansen A., Boshoff C. Cellular origin of Kaposi's sarcoma and Kaposi's sarcoma-associated herpesvirus-induced cell reprogramming. Trends Cell Biol. 2013 Sep 1;23(9):421–432. doi: 10.1016/j.tcb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Gramolelli S., Ojala P.M. Kaposi's sarcoma herpesvirus-induced endothelial cell reprogramming supports viral persistence and contributes to Kaposi's sarcoma tumorigenesis. Curr Opin Virol. 2017 Oct 1;26:156–162. doi: 10.1016/j.coviro.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Carroll P.A., Brazeau E., Lagunoff M. Kaposi's sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology. 2004 Oct 15;328(1):7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong Y.-K., Foreman K., Shin J.W., Hirakawa S., Curry C.L., Sage D.R. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma–associated herpesvirus. Nat. Genet. 2004 Jul;36(7):683–685. doi: 10.1038/ng1383. [DOI] [PubMed] [Google Scholar]
- 51.Wang H.-W., Trotter M.W.B., Lagos D., Bourboulia D., Henderson S., Mäkinen T. Kaposi sarcoma herpesvirus–induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet. 2004 Jul;36(7):687–693. doi: 10.1038/ng1384. [DOI] [PubMed] [Google Scholar]
- 52.Choi D., Park E., Kim K.E., Jung E., Seong Y.J., Zhao L. The lymphatic cell environment promotes Kaposi sarcoma development by prox1-enhanced productive lytic replication of Kaposi sarcoma herpes virus. Canc. Res. 2020 Aug 1;80(15):3130–3144. doi: 10.1158/0008-5472.CAN-19-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee M.-S., Yuan H., Jeon H., Zhu Y., Yoo S., Shi S. Human mesenchymal stem cells of diverse origins support persistent infection with Kaposi’s sarcoma-associated herpesvirus and manifest distinct angiogenic, invasive, and transforming phenotypes. mBio [Internet] 2016 Mar 2;7(1):e02109–15. doi: 10.1128/mBio.02109-15. https://mbio.asm.org/content/7/1/e02109-15 Available from: [cited 2021 Mar 27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y., Zhong C., Liu D., Yu W., Chen W., Wang Y. Evidence for Kaposi sarcoma originating from mesenchymal stem cell through KSHV-induced mesenchymal-to-endothelial transition. Canc. Res. 2018 Jan 1;78(1):230–245. doi: 10.1158/0008-5472.CAN-17-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naipauer J., Rosario S., Gupta S., Premer C., Méndez-Solís O., Schlesinger M. PDGFRA defines the mesenchymal stem cell Kaposi's sarcoma progenitors by enabling KSHV oncogenesis in an angiogenic environment. PLoS Pathog. 2019 Dec 27;15(12) doi: 10.1371/journal.ppat.1008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parsons C.H., Szomju B., Kedes D.H. Susceptibility of human fetal mesencyhmal stem cells to Kaposi sarcoma-associated herpesvirus. Blood. 2004 Nov 1;104(9):2736–2738. doi: 10.1182/blood-2004-02-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo S., Jang J., Yoo C., Lee M.-S. Kaposi's sarcoma-associated herpesvirus infection of human bone-marrow-derived mesenchymal stem cells and their angiogenic potential. Arch. Virol. 2014 Sep 1;159(9):2377–2386. doi: 10.1007/s00705-014-2094-3. [DOI] [PubMed] [Google Scholar]
- 58.Wang X., Chen W., Yuan Y. KSHV enhances mesenchymal stem cell homing and promotes KS-like pathogenesis. Virology. 2020 Oct 1;549:5–12. doi: 10.1016/j.virol.2020.07.012. [DOI] [PubMed] [Google Scholar]
- 59.Gasperini P., Espigol-Frigole G., McCormick P.J., Salvucci O., Maric D., Uldrick T.S. Kaposi sarcoma herpesvirus promotes endothelial-to-mesenchymal transition through notch-dependent signaling. Canc. Res. 2012 Mar 1;72(5):1157–1169. doi: 10.1158/0008-5472.CAN-11-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009 Jun 1;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Potenta S., Zeisberg E., Kalluri R. The role of endothelial-to-mesenchymal transition in cancer progression. Br. J. Canc. 2008 Nov;99(9):1375–1379. doi: 10.1038/sj.bjc.6604662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaur N., Tikla T., Kaul R. Kaposi sarcoma-associated herpes virus (KSHV) latent protein LANA modulates cellular genes associated with epithelial-to-mesenchymal transition. Arch. Virol. 2019 Jan 1;164(1):91–104. doi: 10.1007/s00705-018-4060-y. [DOI] [PubMed] [Google Scholar]
- 63.Janier M., Morel P., Civatte J. The Koebner phenomenon in AIDS-related Kaposi's sarcoma. J. Am. Acad. Dermatol. 1990 Jan;22(1):125–126. doi: 10.1016/s0190-9622(08)80011-4. [DOI] [PubMed] [Google Scholar]
- 64.Potouridou I., Katsambas A., Pantazi V., Armenaka M., Vareltzidis A., Stratigos J. Koebner phenomenon in classic Kaposi's sarcoma. Acta Derm. Venereol. 1997 Nov;77(6):481. doi: 10.2340/0001555577481. [DOI] [PubMed] [Google Scholar]
- 65.Chen Q., Chen J., Li Y., Liu D., Zeng Y., Tian Z. Kaposi’s sarcoma herpesvirus is associated with osteosarcoma in Xinjiang populations. Proc. Natl. Acad. Sci. India. 2021;118(10) doi: 10.1073/pnas.2016653118. http://www.pnas.org/content/118/10/e2016653118 Available from: Mar 9 [cited 2021 Mar 24] e2016653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He N., Shao X., Chen Y., Zhang T., Minhas V., Wood C. Human Herpesvirus 8 Seroprevalence, China - Volume 18, Number 1-January 2012-Emerging Infectious Diseases journal - CDC. https://wwwnc.cdc.gov/eid/article/18/1/10-2070_article [cited 2021 Jun 14]; 150-152, Available from: [DOI] [PMC free article] [PubMed]
- 67.Tsao S.W., Tsang C.M., Lo K.W. Epstein–Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc B Biol Sci. 2017 Oct 19;372(1732) doi: 10.1098/rstb.2016.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cesarman E., Damania B., Krown S.E., Martin J., Bower M., Whitby D. Kaposi sarcoma. Nat Rev Dis Primer. 2019 Jan 31;5(1):1–21. doi: 10.1038/s41572-019-0060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar P., Kuwa N.Y., Minhas V., Marimo C., Shea D.M., Kankasa C. Higher levels of neutralizing antibodies against KSHV in KS patients compared to asymptomatic individuals from Zambia. PloS One. 2013 Aug 14;8(8) doi: 10.1371/journal.pone.0071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poppe L.K., Wood C., West J.T. The presence of antibody-dependent cell cytotoxicity–mediating antibodies in Kaposi sarcoma–associated herpesvirus–seropositive individuals does not correlate with disease pathogenesis or progression. J. Immunol. 2020 Nov 15;205(10):2742–2749. doi: 10.4049/jimmunol.2000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roshan R., Labo N., Trivett M., Miley W., Marshall V., Coren L. T-cell responses to KSHV infection: a systematic approach. Oncotarget. 2017 Nov 25;8(65):109402–109416. doi: 10.18632/oncotarget.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaasinen E., Aavikko M., Vahteristo P., Patama T., Li Y., Saarinen S. Nationwide registry-based analysis of cancer clustering detects strong familial occurrence of Kaposi sarcoma. PloS One. 2013 Jan 24;8(1) doi: 10.1371/journal.pone.0055209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blumenthal M.J., Castro E.M.C., Whitby D., Katz A.A., Schäfer G. Evidence for altered host genetic factors in KSHV infection and KSHV-related disease development. Rev. Med. Virol. 2021 Mar;31(2) doi: 10.1002/rmv.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blumenthal M.J., Schutz C., Meintjes G., Mohamed Z., Mendelson M., Ambler J.M. EPHA2 sequence variants are associated with susceptibility to Kaposi's sarcoma-associated herpesvirus infection and Kaposi's sarcoma prevalence in HIV-infected patients. Cancer Epidemiol. 2018 Oct 1;56:133–139. doi: 10.1016/j.canep.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sallah N., Miley W., Labo N., Carstensen T., Fatumo S., Gurdasani D. Distinct genetic architectures and environmental factors associate with host response to the γ 2-herpesvirus infections. Nat. Commun. 2020 Jul 31;11(1):3849. doi: 10.1038/s41467-020-17696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El‐Mallawany N.K., Mehta P.S., Kamiyango W., Villiera J., Peckham‐Gregory E.C., Kampani C. KSHV viral load and Interleukin-6 in HIV-associated pediatric Kaposi sarcoma—exploring the role of lytic activation in driving the unique clinical features seen in endemic regions. Int. J. Canc. 2019;144(1):110–116. doi: 10.1002/ijc.31863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thakker S., Verma S.C. Co-infections and pathogenesis of KSHV-associated malignancies. Front Microbiol [Internet] 2016;7:151. doi: 10.3389/fmicb.2016.00151. http://www.frontiersin.org/articles/10.3389/fmicb.2016.00151/full Available from: [cited 2021 Mar 15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faure A., Hayes M., Sugden B. How Kaposi's sarcoma-associated herpesvirus stably transforms peripheral B cells towards lymphomagenesis. Proc. Natl. Acad. Sci. Unit. States Am. 2019 Aug 13;116(33):16519–16528. doi: 10.1073/pnas.1905025116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Santiago J.C., Goldman J.D., Zhao H., Pankow A.P., Okuku F., Schmitt M.W. Intra-host changes in Kaposi sarcoma-associated herpesvirus genomes in Ugandan adults with Kaposi sarcoma. PLoS Pathog. 2021 Jan 19;17(1) doi: 10.1371/journal.ppat.1008594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weinstein I.B. Addiction to oncogenes--the achilles heal of cancer. Science. 2002 Jul 5;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 81.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-cas9 system. Science. 2014 Jan 3;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manzano M., Patil A., Waldrop A., Dave S.S., Behdad A., Gottwein E. Gene essentiality landscape and druggable oncogenic dependencies in herpesviral primary effusion lymphoma. Nat. Commun. 2018 Aug 15;9(1):3263. doi: 10.1038/s41467-018-05506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holmes D.L., Vogt D.T., Lagunoff M. A CRISPR-Cas9 screen identifies mitochondrial translation as an essential process in latent KSHV infection of human endothelial cells. Proc. Natl. Acad. Sci. Unit. States Am. 2020 Nov 10;117(45):28384–28392. doi: 10.1073/pnas.2011645117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruffaz M., Yuan H., Meng W., Liu H., Bae S., Kim J.-S. CRISPR-Cas9 screening of Kaposi’s sarcoma-associated herpesvirus-transformed cells identifies XPO1 as a vulnerable target of cancer cells. mBio [Internet] 2019 Jun 25;10(3):e00866–19. doi: 10.1128/mBio.00866-19. https://mbio.asm.org/content/10/3/e00866-19 Available from: [cited 2021 Mar 21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Manzano M., Günther T., Ju H., Nicholas J., Bartom E.T., Grundhoff A. Kaposi’s sarcoma-associated herpesvirus drives a super-enhancer-mediated survival gene expression program in primary effusion lymphoma. mBio [Internet] 2020 Aug 25;11(4):e01457–20. doi: 10.1128/mBio.01457-20. https://mbio.asm.org/content/11/4/e01457-20 Available from: [cited 2021 Mar 29] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C., Zhang L., Ke L., Ding W., Jiang S., Li D. Primary effusion lymphoma enhancer connectome links super-enhancers to dependency factors. Nat. Commun. 2020 Dec 9;11(1):6318. doi: 10.1038/s41467-020-20136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tolani B., Gopalakrishnan R., Punj V., Matta H., Chaudhary P.M. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. 2014 May;33(22):2928–2937. doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Canc. 2011 Feb;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 89.Lagunoff M. Activation of cellular metabolism during latent Kaposi's Sarcoma herpesvirus infection. Curr Opin Virol. 2016 Aug 1;19:45–49. doi: 10.1016/j.coviro.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dai L., Lin Z., Jiang W., Flemington E.K., Qin Z. Lipids, lipid metabolism and Kaposi's sarcoma-associatedherpesvirus pathogenesis. Virol. Sin. 2017 Oct 1;32(5):369–375. doi: 10.1007/s12250-017-4027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu Y., Li T., Silva SR da, Lee J.-J., Lu C., Eoh H. A critical role of glutamine and asparagine γ-nitrogen in nucleotide biosynthesis in cancer cells hijacked by an oncogenic virus. mBio [Internet] 2017 Sep 6;8(4):e01179–17. doi: 10.1128/mBio.01179-17. https://mbio.asm.org/content/8/4/e01179-17 Available from: [cited 2021 Mar 30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhu Y., Silva SR da, He M., Liang Q., Lu C., Feng P. An oncogenic virus promotes cell survival and cellular transformation by suppressing glycolysis. PLoS Pathog. 2016 May 17;12(5) doi: 10.1371/journal.ppat.1005648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choi U.Y., Lee J.J., Park A., Zhu W., Lee H.-R., Choi Y.J. Oncogenic human herpesvirus hijacks proline metabolism for tumorigenesis. Proc. Natl. Acad. Sci. Unit. States Am. 2020 Apr 7;117(14):8083–8093. doi: 10.1073/pnas.1918607117. [DOI] [PMC free article] [PubMed] [Google Scholar]