Abstract
Seminal vesicles are an integral part of the male reproductive accessory gland system. They produce a complex array of secretions containing bioactive constituents that support gamete function and promote reproductive success, with emerging evidence suggesting these secretions are influenced by our environment. Despite their significance, the biology of seminal vesicles remains poorly defined. Here, we complete the first proteomic assessment of mouse seminal vesicles and assess the impact of the reproductive toxicant acrylamide. Mice were administered acrylamide (25 mg/kg bw/day) or control daily for five consecutive days prior to collecting seminal vesicle tissue. A total of 5013 proteins were identified in the seminal vesicle proteome with bioinformatic analyses identifying cell proliferation, protein synthesis, cellular death, and survival pathways as prominent biological processes. Secreted proteins were among the most abundant, and several proteins are linked with seminal vesicle phenotypes. Analysis of the effect of acrylamide on the seminal vesicle proteome revealed 311 differentially regulated (FC ± 1.5, p ≤ 0.05, 205 up-regulated, 106 downregulated) proteins, orthogonally validated via immunoblotting and immunohistochemistry. Pathways that initiate protein synthesis to promote cellular survival were prominent among the dysregulated pathways, and rapamycin-insensitive companion of mTOR (RICTOR, p = 6.69E-07) was a top-ranked upstream driver. Oxidative stress was implicated as contributing to protein changes, with acrylamide causing an increase in 8-OHdG in seminal vesicle epithelial cells (fivefold increase, p = 0.016) and the surrounding smooth muscle layer (twofold increase, p = 0.043). Additionally, acrylamide treatment caused a reduction in seminal vesicle secretion weight (36% reduction, p = 0.009) and total protein content (25% reduction, p = 0.017). Together these findings support the interpretation that toxicant exposure influences male accessory gland physiology and highlights the need to consider the response of all male reproductive tract tissues when interpreting the impact of environmental stressors on male reproductive function.
Keywords: seminal vesicle, proteomics, mass spectrometry, acrylamide, mouse models, oxidative stress, mTOR, toxicant, physiology, male reproductive tract
Abbreviations: 8-OHdG, 8-hydroxy-2' -deoxyguanosine; αSMA, alpha smooth muscle actin; ARHGAP10, Rho GTPase-activating protein 10; B2M, Beta-2-microglobulin; BCAP29, B-cell receptor-associated protein 29; CAR1, carbonic anhydrase 2; CASP6, caspase-6; CD9, cluster of differentiation 9 antigen; CD38, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1; CDK4/6, cyclin-dependent kinase 4/6; CEACAM10, carcinoembryonic antigen-related cell adhesion molecule 10; DDA, Data-dependent acquisition; DKKL1, dickkopf-like protein 1; DNAJB5, DnaJ homolog subfamily B member 5; EIF2, eukaryotic initiation factor; EIF4EBP1, eukaryotic translation initiation factor 4E-binding protein 1; FDR, false discovery rate; HILIC, hydrophilic interaction liquid chromatography; HNRNPC, heterogeneous nuclear ribonucleoproteins C1/C2; IRAK4, interleukin-1 receptor-associated kinase 4; LARP1, La-related protein 1; LCN2, neutrophil gelatinase-associated lipocalin; LFQ, Label-free quantification; LIPA, lysosomal acid lipase/cholesteryl ester hydrolase; MFGE8, lactadherin; MLXIPL, MLX-interacting protein-like; MPZ, myelin protein P0; MSI2, RNA-binding protein Musashi homolog 2; MYC, Myc proto-oncogene protein; MYCN, N-myc proto-oncogene protein; nLC-MS/MS, nanoliquid chromatography–tandem mass spectrometry; NPC2, NPC intracellular cholesterol transporter 2; NRF2, nuclear factor erythroid 2-related factor-2; PATE4, prostate and testis expressed protein 4; PLAU, urokinase-type plasminogen activator; PSAP, prosaposin; QSOX1, sulfhydryl oxidase 1; RHO, Rho family of GTPases; RICTOR, rapamycin-insensitive companion of mTOR; RPL28, 60S ribosomal protein L28; SERPINA1D, alpha-1-antitrypsin 1–4; SERPINA3K, serine protease inhibitor A3K; SERPINE2, glia-derived nexin; SPINK1, serine protease inhibitor kazal-type 1; SPINKL, serine protease inhibitor kazal-like protein, minor form; SVS, seminal vesicle secretory protein; TBS, tris-buffered saline; TBST, TBS supplemented with 0.1% (v/v) Tween-20; TGM4, protein-glutamine gamma-glutamyltransferase 4; TP53, transcriptional regulator tumor protein p53; UGT1A7C, UDP-glucuronosyltransferase 1A7; V/V, percent volume/volume; W/V, percent weight/volume; WDR46, WD repeat-containing protein 46
Graphical abstract
Highlights
-
•
First proteomic characterization of the mouse seminal vesicle tissue.
-
•
Secreted proteins are among the most abundant proteins in the seminal vesicle tissue.
-
•
Paternal exposure to reproductive toxicant acrylamide alters seminal vesicle proteome.
-
•
Acrylamide treatment results in reduced seminal vesicle secretory capacity.
In Brief
Seminal vesicles produce bioactive constituents that support gamete function and promote reproductive success. Despite their significance, seminal vesicle biology remains poorly defined. Here, we have exploited proteomics to generate the first mechanistic insights into mouse seminal vesicles in normal physiology and pathology. Our collective data affirm the hypothesis that seminal vesicles are sensitive to environmental factors and exposures in a manner that may have consequences for fetal development and later offspring health.
The male accessory sex glands comprise the prostate, seminal vesicles, and bulbourethral glands (1). The combined secretory activity of these tissues gives rise to seminal plasma, a highly viscous fluid containing a complex array of bioactive molecules that support male gamete function after ejaculation and stimulate gene expression and immune response changes in the female reproductive tract (2, 3). In the majority of mammalian species, including humans, the androgen-dependent seminal vesicles are the dominant contributor to seminal plasma (4, 5). Indeed, elegant experiments to surgically excise the accessory glands have clearly established seminal vesicles as being the most influential tissue that contributes to the rodent ejaculate. In rodents, seminal vesicle secretions are directly implicated in regulating the conception environment within the female reproductive tract to elicit an immune response that promotes optimal fetal development and supports long-term offspring health (1, 2, 4, 6, 7, 8, 9, 10, 11). In view of the mounting evidence that such roles are conserved among mammals (12, 13, 14), it is likely that the complex molecular signals contained within seminal plasma are of critical importance for promoting reproductive success and maximizing progeny fitness.
The regulatory influence exerted by seminal vesicle secretions at conception (1) provides impetus to understand whether this tissue is affected by lifestyle and/or environmental exposures (3, 15). While genetic and epigenetic alterations to the male germline are conventionally thought to account for the transmission of paternal environmental exposures to subsequent generations, emerging evidence demonstrates that altered function of seminal vesicle tissue may exert an important influential role (3, 16, 17). An impact of the paternal environment on seminal vesicle function has been observed in rams, where seminal vesicle proteins were altered in response to paternal heat stress (18), while neonatal exposure to the endocrine-disrupting compound, diethylstilbestrol, leads to toxicity in mouse seminal vesicle from early developmental stages (19). Compelling data comes from rodent models, where paternal diet has been demonstrated to shape offspring health through mechanisms involving both the fertilizing spermatozoon and the seminal plasma in which they are delivered (16, 20). Together, these data highlight that altered seminal vesicle fluid composition not only has potential to impair fertility but may also affect fetal development and impart long-term health consequences for offspring (3, 16, 17, 21).
To develop an understanding of the impact of environmental exposures on male reproductive function, we have previously used acrylamide as a reproductive toxicant to assess its impact on germ cell populations in the testes and in epididymis (22, 23, 24). In these former studies, we demonstrated that acrylamide exposure leads to elevated levels of DNA damage throughout sperm development and maturation, with subtle changes in Sertoli cell structure following long-term acrylamide exposure. Specifically, exposure of testicular germ cells to acrylamide leads to enhanced levels of DNA damage that fail to be resolved during sperm maturation, yet do not appear to affect fertility or fetal development. Comparable levels of DNA damage are also characteristic of sperm exclusively exposed to acrylamide during epididymal transit. However, exposure timed to coincide with the developmental window of epididymal transit and ejaculation leads to substantially increased rates of fetal loss. These data indicate that the male transfers the burden of acrylamide stress to the female through mechanisms that are independent of sperm DNA damage. These results gave us the impetus to utilize this acute exposure model to assess acrylamide impact on the seminal vesicles, to better understand the contribution of this understudied tissue to a male’s response to reproductive insults.
To begin to address this challenge, we have completed the first proteomic assessment of mouse seminal vesicle tissues using offline peptide fractionation via hydrophilic interaction liquid chromatography coupled with high-resolution reverse-phase nanoflow liquid chromatography–tandem mass spectrometry (nLC-MS/MS). Additionally, we have exploited an established model involving acute exposure to the reproductive toxicant acrylamide that we have previously demonstrated alters male gamete quality and increases fetal loss (22), to explore the impact of this environmental exposure on seminal vesicle function. The results highlight cellular mechanisms altered in the seminal vesicles in response to acrylamide and thus provide evidence that this crucial reproductive tissue is sensitive to environmental factors and exposures.
Experimental Procedures
Ethics Approval
All experimental procedures involving mice were conducted with the approval of the University of Newcastle Animal Care and Ethics Committee (approval number A-2017-726). Male outbred Swiss mice (8–12 weeks of age) were obtained from a breeding colony held at the University of Newcastle central animal facility and maintained according to the recommendations prescribed by the Animal Care and Ethics Committee. Mice were housed under a controlled lighting regimen (12-h light: 12 h dark) at 21 to 22 °C and supplied with food and water ad libitum.
Chemicals and Reagents
All reagents were purchased from Merck, unless otherwise specified.
Acrylamide Treatment Regimen and Tissue Collection
Mice received an intraperitoneal injection of acrylamide (25 mg/kg bw/day) or control (phosphate buffered saline (PBS): 0.01 M phosphate buffer, 0.0027 M KCl and 0.137 M NaCl, pH 7.4) each morning for five consecutive days (injection volume 100 μl) according to an established acute exposure protocol described in Katen et al. (22), which results in fetal loss in females sired by acrylamide-treated males. In accordance with this established exposure regimen, mice were euthanized 72 h following the final acrylamide injection, and seminal vesicles were dissected and weighed, ensuring that the anterior prostate (coagulating gland) was left in situ. Seminal vesicle fluid was expelled from one gland by gently squeezing seminal vesicle tissue using forceps and then weighed. Tissues were washed thoroughly in Tris-buffered saline (TBS) comprising 50 mM Tris-HCl, 150 mM NaCl, in mass spectrometry grade H2O, pH 7.6. In separate tubes, tissue and fluid were snap frozen in liquid nitrogen and stored at −80 °C. For histological analysis, excised seminal vesicles from the gland containing seminal vesicle fluid were fixed for 8 h in Bouin’s solution (5% v/v acetic acid, 25% v/v formaldehyde, 70% v/v picric acid). Bouin’s solution was removed with daily changes of 75% (v/v) ethanol for at least 5 days. Tissues were embedded in paraffin and 5 μm sections were cut prior to staining with hematoxylin and eosin. Stained sections were examined and photographed on a Zeiss Axio A.2 fluorescence microscope (Carl Zeiss AG) and epithelial cell height and epithelial mucosal folding assessed using OLYMPUS cellSens standard software (Olympus Corporation), as described (25).
Proteomic Sample Preparation of Mouse Seminal Vesicles
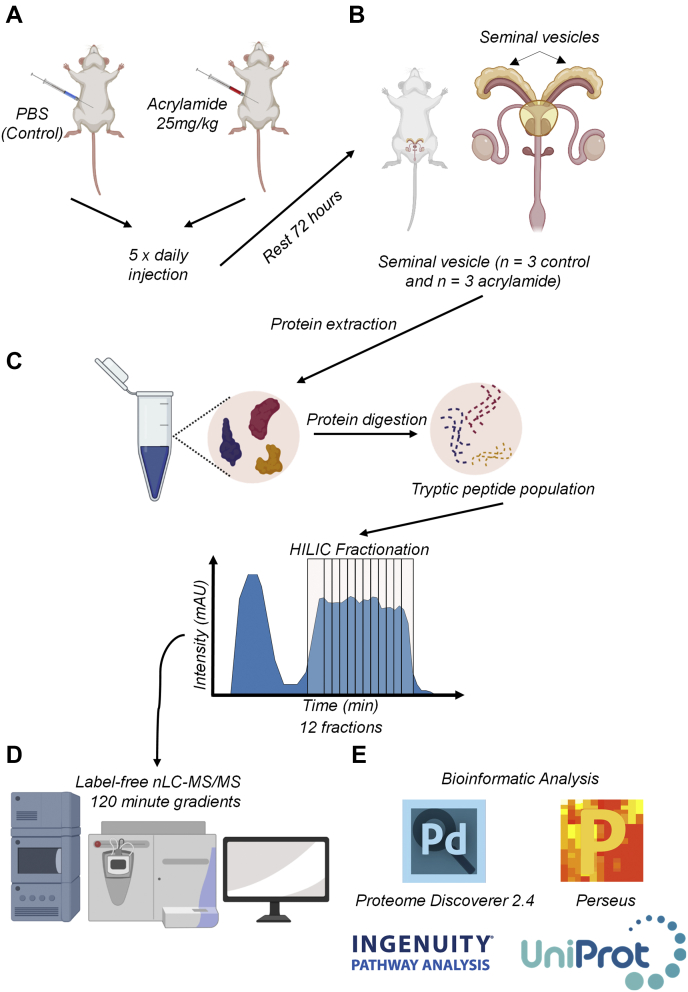
Seminal vesicle tissues were cut into small segments with a scalpel and homogenized in ice-cold 0.1 M Na2CO3 containing protease and complete EDTA-free phosphatase inhibitors (Roche Holding AG) using a FastPrep-24TM 5G homogenizer (MP Biomedicals) with a Cool Prep Adaptor (2 × 1 min, 4.0 m/s, at 4 °C). Homogenized samples were then sonicated (3 × 10 s cycles, 100% output power). Protein concentration was determined using a bicinchoninic acid assay (Thermo Fisher Scientific) and diluted to a final concentration of 1.4 mg in a solution of 6 M urea and 2 M thiourea. Samples underwent reduction and alkylation with 10 mM dithiothreitol (30 min at room temperature) and 20 mM iodoacetamide (30 min protected from light at room temperature) respectively. Peptide populations were digested with 1:30 Lys-C/Trypsin Mix (Promega) (26, 27, 28, 29, 30) at room temperature for 3 h. Using 50 mM triethylammonium bicarbonate, the concentration of urea was brought below 1 M and digested overnight at 37 °C. Lipids were precipitated from peptide suspensions using formic acid (2% v/v final concentration), and the resultant peptides were purified using Oasis 1 cc desalting columns (Waters Corp). Peptides were quantified using the Qubit protein assay (Thermo Fisher Scientific), and 48 μg of each sample was subjected to offline fractionation via hydrophilic interaction liquid chromatography (HILIC) (Fig. 1, supplemental Fig. S1, A and B).
Fig. 1.
Overview of the proteomics workflow.A, male Swiss mice (8–12 weeks of age, n = 3 per treatment group) were injected with acrylamide (25 mg/kg bw/day) or PBS (control) following the methods stated in Katen et al., (22). B, following the final injection, mice were rested for 72 h prior to collection of seminal vesicles. C, protein was extracted, reduced, alkylated, and digested, prior to off-line fractionation of the tryptic peptide populations using hydrophilic interaction liquid chromatography (HILIC). D, HILIC fractions were then run on high-resolution nano liquid chromatography–tandem MS (nLC-MS/MS), before being, (E) analyzed with Proteome Discoverer 2.4, Perseus, Ingenuity Pathway Analysis, and UniProt. Some components of this figure created with BioRender.com.
nLC-MS/MS Analysis
Reverse-phase nLC-MS/MS was performed on control and acrylamide-treated HILIC fractions (12 from each seminal vesicle) (supplemental Fig. S1, A and B) using a Q-Exactive Plus hybrid quadrupole-Orbitrap MS coupled to a Dionex Ultimate 3000RSLC nanoflow high-performance LC system (Thermo Fisher Scientific). Samples were loaded onto an Acclaim PepMap 100 C18 75 μm × 20 mm trap column (Thermo Fisher Scientific) for preconcentration and online desalting. Separation was then achieved using an EASY-Spray PepMap C18 75 μm × 250 mm column (Thermo Fisher Scientific), employing a linear gradient of acetonitrile (2–40%, 300 nl/min, 120 min). The MS was operated in data-dependent acquisition (DDA) mode. The Orbitrap mass analyzer was used at a resolution of 35,000, to acquire full MS with an m/z range of 380 to 2000, incorporating a target automatic gain control value of 1 × 106 and maximum fill times of 50 ms. The 20 most intense multiple charged precursors were selected for higher-energy collision dissociation fragmentation with stepped collisional energies of 25%, 28%, and 30%. MS/MS fragments were measured at an Orbitrap resolution of 17,500 using an automatic gain control target of 5 × 105 and maximum fill time of 120 ms.
Data Processing and Analysis
Database searching of all raw files was performed using Proteome Discoverer 2.4 (Thermo Fisher Scientific). SEQUEST HT was used to search against the UniProt Mus musculus database (25,260 sequences, downloaded November 12, 2019). Database searching parameters included up to two missed cleavages, a precursor mass tolerance set to 10 ppm, fragment mass tolerance of 0.02 Da, and trypsin designated as the digestion enzyme. Cysteine carbamidomethylation was set as a fixed modification while dynamic modifications included acetylation (N-terminus), oxidation (M), and phosphorylation (S/T and Y). Interrogation of the corresponding reversed database was also performed to evaluate the false discovery rate (FDR) of peptide identification using Percolator based on q-values, which were estimated from the target-decoy search approach. To filter out target peptide spectrum matches over the decoy-peptide spectrum matches, a fixed FDR of 1% was set at the peptide level. Mass recalibration and label-free quantification (LFQ) of area under the curve MS1 peaks were carried out using Proteome Discoverer nodes, Spectrum Files RC, Minora Feature Detector, and Feature Mapper (31, 32, 33, 34, 35, 36, 37). Minora Feature Detector is the dominant algorithm behind the LFQ whereby it first detects chromatographic peaks in the raw spectral data and maps them to identified peptide spectrum matches. A feature is created when a peak fits the mapping of chromatographic peaks to the calculated theoretical isotope patterns of the PSM. A similar process occurs for peaks that have no association with any features; this is calculated by accruing all chromatographic peaks within a short retention time range and searching for peaks that fit isotope patterning. To be accepted, intensities and mass deviations of these peaks must fit a theoretical pattern of an Averagine peptide. The Feature Mapper node builds on the Minora output and carries out a retention time alignment and feature/peptide linking within groups. Fold changes and significance between control and acrylamide-treated groups were calculated by Proteome Discoverer 2.4 using a nonnested pairwise ratio approach, whereby the program calculates the peptide group ratios as the geometric median of all combinations of ratios from all of the replicates for each group. Protein ratios were subsequently calculated as the geometric median of the peptide group ratios, and statistical testing was completed using a Student’s t test. The protein list was exported as an Excel file and further refined to include only those with a quantitative value in all three replicates within at least one group (i.e., control and/or acrylamide treated) and a minimum of two unique peptides. Using Perseus, version 1.6.10.43 (38), scatter plots of log-transformed normalized abundances (supplemental Fig. S1, C and D) were generated, and Pearson correlation (r2) was calculated, while global analysis of the data was undertaken to generate a principal component analysis plot. For a global illustration of protein abundance compared with fold-change, an MA plot was generated using Microsoft Excel. To determine the ranking order of proteins in the seminal vesicle tissue proteome by abundance, normalized abundance quants of the three biological replicates (prior to scaling) were averaged and ordered based on high to low.
Ingenuity Pathway Analysis
Ingenuity Pathway Analysis software (Qiagen) was used to analyze refined seminal vesicle tissue proteomic lists as previously described (26). Briefly, canonical pathways, upstream regulators, and disease and function analyses were generated and assessed by generating a p-value and Z-score enrichment measurement of the overlapping proteins from the dataset in a particular pathway, function, or regulator (39).
Phenotype and Seminal Vesicle Fluid Comparisons
The curated comparative proteome was searched against an exported protein list consisting of the full complement of Mouse Genome Informatics (40) and the International Mouse Phenotyping Consortium (41) datasets to identify those proteins expressed in the seminal vesicle that were also associated with seminal vesicle phenotypes. To further complement these analyses, proteomic datasets from three seminal vesicle fluid proteomic characterization papers (42, 43, 44) were obtained and filtered for reviewed proteins identified in UniProt. Proteins that were identified in at least one manuscript were then used to form a list of proteins expressed in mouse seminal vesicle fluid. This list was then compared with our seminal vesicle proteome.
Immunohistochemistry
Sections from Bouin’s fixed seminal vesicle tissue were probed with antibodies using established methods for immunofluorescence and DAB immunohistochemistry analysis (22, 23, 27, 45). Sections were dewaxed in xylene and rehydrated. Antigen retrieval and blocking of nonspecific antibody binding were performed as described (supplemental Table S1). Sections were washed in PBS before incubation in primary antibodies (supplemental Table S1, in 1% w/v BSA diluted in PBS) overnight at 4 °C in a humidified chamber. Following washing in PBS (3 × 5 min), sections were incubated with appropriate secondary antibodies (supplemental Table S1, in 1% w/v BSA diluted in PBS) for 1 h at room temperature. Slides were washed in PBS (3 × 5 min) prior to mounting. For immunofluorescence analysis, slides were mounted in an antifade reagent comprising 10% v/v Mowiol 4 to 88, 30% v/v glycerol in 0.2 M Tris (pH 8.5), and 2.5% v/v 1,4-diazobicyclo-(2.2.2)-octane in 0.2 M Tris (pH 8.5). For immunohistochemistry analysis, detection was performed using diaminobenzidine tetrachloride, and sections were counterstained with hematoxylin and mounted in DPX mountant for histology. Slides were examined and photographed on a Zeiss Axio A.2 fluorescence microscope (Carl Zeiss AG). For 8-hydroxy-2’-deoxyguanosine (8-OHdG), ImageJ Fiji software (version 1.48v; National Institute of Health) (46, 47) was used to quantify the level of fluorescence normalized to DAPI labeling as described previously (23, 48). For alpha smooth muscle actin (αSMA) staining, OLYMPUS cellSens standard software (Olympus Corporation) was used to assess the depth of the smooth muscle cell layer as described previously (25).
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Immunoblotting
Equivalent amounts of protein (10 μg) were boiled in SDS-PAGE sample buffer (2% v/v β2-mercaptoethanol, 2% w/v SDS, and 10% w/v sucrose in 0.375 M Tris, pH 6.8, with bromophenol blue) at 100 °C for 5 min, prior to being resolved by SDS-PAGE (150 V, 1 h), and transferred to nitrocellulose membranes (350 mA, 1 h). To detect proteins of interest, membranes were blocked under optimized conditions for 1 h at room temperature (Supplemental Table S1) before being incubated in primary antibody overnight at 4 °C (Supplemental Table S1). Following washing in (3 × 5 min in TBS supplemented with 0.1% (v/v) Tween-20 (TBST, pH 7.4)), sections were incubated with appropriate secondary antibodies for 1 h at room temperature. After additional washes (3 × 5 min in TBST), labeled proteins were detected using an enhanced chemiluminescence kit (GE Healthcare). For quantification of protein expression, relevant bands were assessed by densitometry using Multi Gauge V3.0 (Fujifilm) and normalized against glyceraldehyde-3-phosphate dehydrogenase. Data were expressed relative to the amount of protein in control seminal vesicle tissue.
Seminal Vesicle Fluid Solubilization and Quantification
Seminal vesicle fluid was collected and solubilized using a modified version of a previously reported method (49). Briefly, 8 M guanidine hydrochloride was added to seminal vesicle fluid and samples were vortexed for 5 min to completely dissolve the coagulated material. Immediately after, samples were incubated at 95 °C for 5 min to deactivate endogenous phosphatases and proteases and sonicated (3 × 10 s cycles, 100% output power). For protein quantification, samples were diluted to 2 M guanidine hydrochloride, and protein concentration was determined by a bicinchoninic acid assay (Thermo Fisher Scientific).
Experimental Design and Statistical Rationale
Proteomic analyses were performed using seminal vesicle tissue (n = 3 biological replicates collected from three individual mice/group). Differentially expressed proteins were defined as those with a fold-change ≥1.5 or ≤−1.5 and p value ≤0.05. All other data were assessed for normality using the Shapiro–Wilk normality test. Normally distributed data were analyzed by unpaired Student’s t-tests to detect differences between treatment groups. Data not normally distributed were analyzed by a Mann–Whitney U test. Differences between groups were considered significant when p ≤ 0.05. The number of biological replicates used in each experiment are presented in the figure captions. Graphical data were prepared using GraphPad Prism version 8.2.1 for Windows (GraphPad Software) and are presented as mean values ± SEM.
Results
Analysis of the Global Mouse Seminal Vesicle Proteome
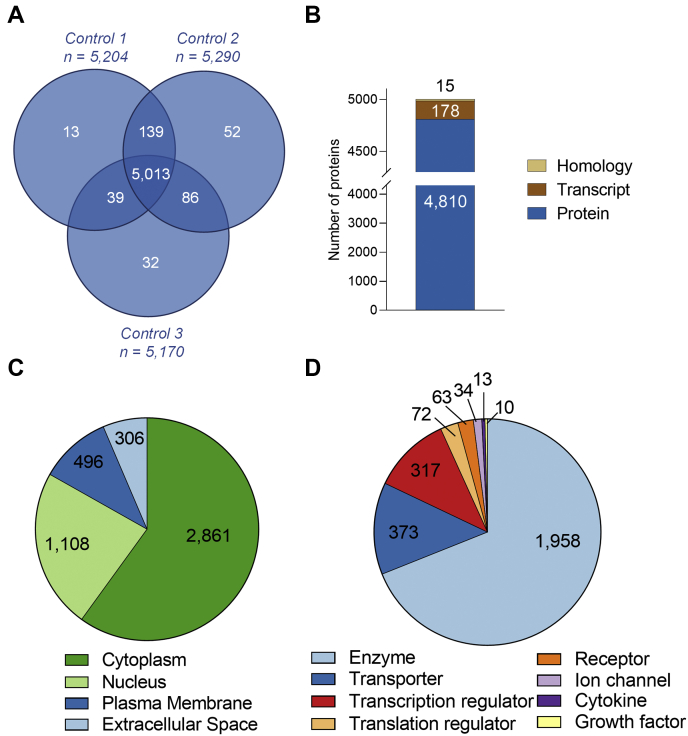
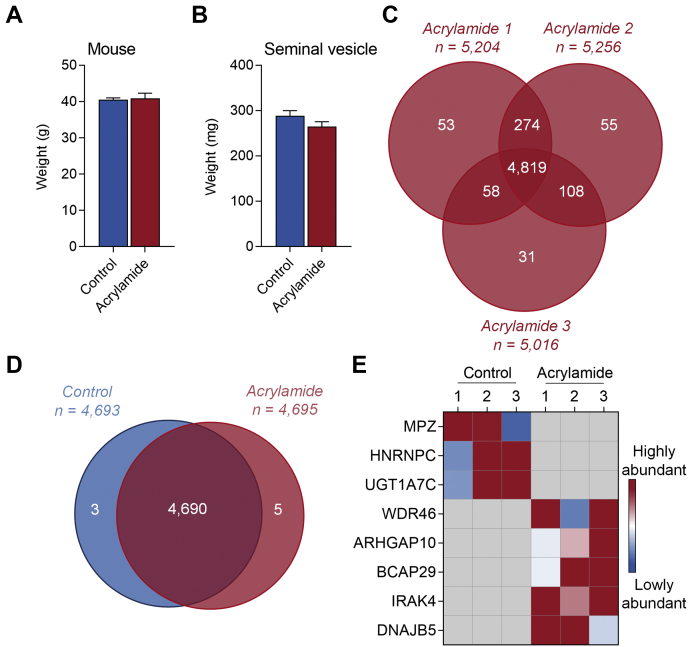
Secretions from the seminal vesicle exert substantial influence on male reproductive function (1, 2), but the biology of this tissue remains poorly defined. To address this, we have characterized the mouse seminal vesicle proteome using LFQ DDA high-resolution MS. Proteomic analysis of the control seminal vesicle tissue returned a complex core proteome comprising 5013 proteins (FDR ≤ 0.01, Fig. 2A, supplemental Table S2) with high Pearson correlation coefficients (0.932 average r2), across all biological replicates (supplemental Fig. S1C). An average of 16.4 peptides (14.9 unique peptides) were identified per seminal vesicle protein, representing a coverage of 39.8% per protein (Table 1). Further interrogation revealed that the majority (96.1%) of the proteome consists of known proteins with evidence at the protein level, but an additional 178 proteins (3.6%) had annotated evidence at the transcript level, and a further 15 proteins (0.3%) were identified that had only been inferred from homology (Fig. 2B, supplemental Table S2).
Fig. 2.
Characterization of the mouse seminal vesicle proteome.A, proteins identified from each biological replicate of control treated mice were compared, with proteins identified in all three replicates constituting the core seminal vesicle proteome. B, using UniProt, the core proteome was then assessed to determine whether the proteins had annotated evidence at protein or transcript level or had been inferred from homology. An initial characterization of the core seminal vesicle proteome using Ingenuity Pathway Analysis revealed: (C) cellular localization and (D) protein classification.
Table 1.
Seminal vesicle proteome summary
| Number of proteins | ||||
|---|---|---|---|---|
| Group | n = 3 FDR ≤ 0.01 |
Av. peptide hits/protein | Av. unique peptide hits/protein | Av. protein coverage (%) |
| Control | 5013 | 16.4 | 14.9 | 39.8 |
| Acrylamide | 4819 | 16.9 | 15.3 | 40.8 |
To gain an overview of the cellular localization and potential functions of the core seminal vesicle proteome, we utilized Ingenuity Pathway Analysis (39), which successfully mapped 4976/5013 (99.2%) of the identified proteome. Among the core seminal vesicle proteome, the majority of proteins were mapped to the cytoplasm (2861 proteins, 57.5%), followed by cellular localization within the nucleus (1108 proteins, 22.3%), plasma membrane (496 proteins, 10%), and extracellular space (306 proteins, 6.1%) (Fig. 2C), with the remaining 205 proteins (4.1%) mapped to the category “other.” Classification of protein type revealed that the most common category of proteins were enzymes (1958 proteins, 39.3%), followed by transporters (373 proteins, 7.5%), transcription regulators (317 proteins, 6.6%), translation regulators (72 proteins, 1.4%), and receptors (63 proteins, 1.3%) (Fig. 2D). Proteins classified as ion channels, cytokines, and growth factors constituted less than 1% of the total proteome (Fig. 2D), while 2136 (42.9%) proteins were assigned to the functional category “other” (data not presented).
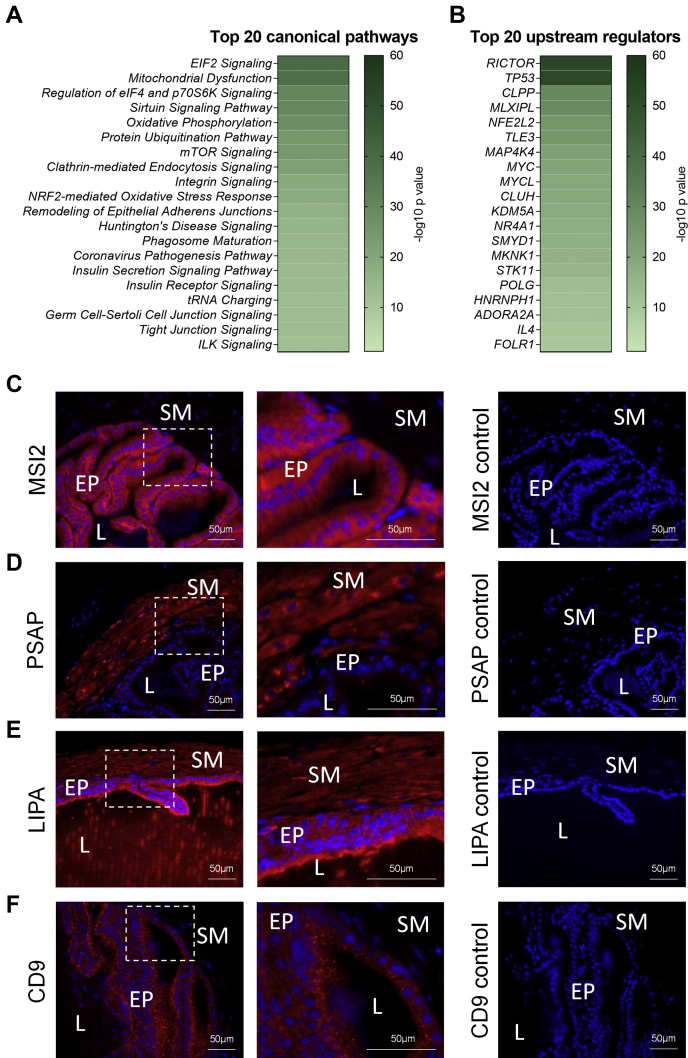
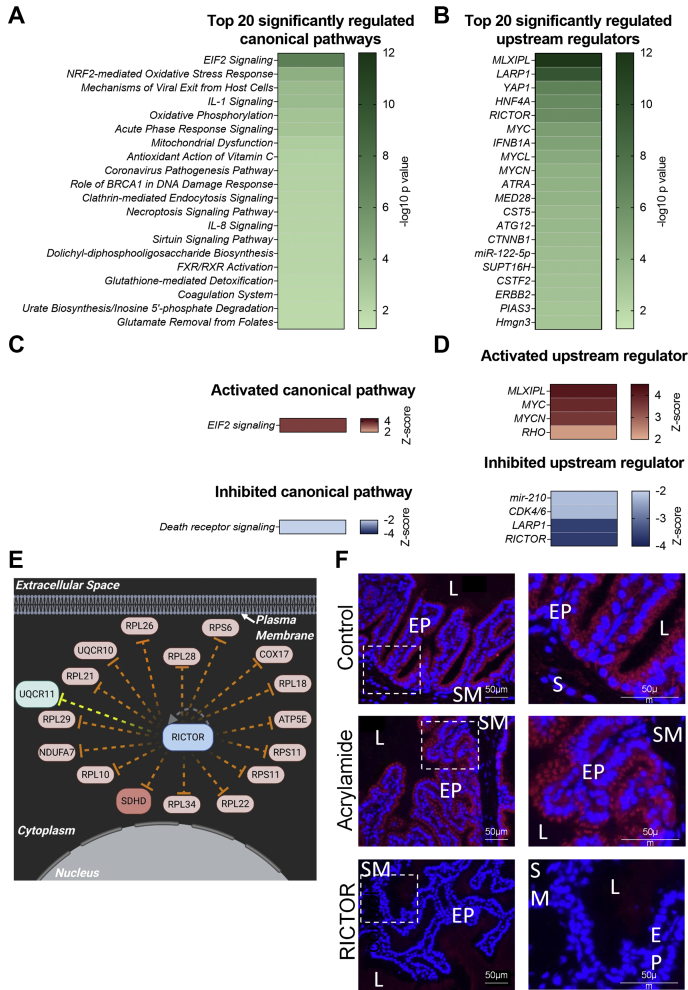
To assess the functional roles of proteins within the core mouse seminal vesicle proteome, Ingenuity Pathway Analysis was used to predict canonical pathways, downstream molecular, cellular, physiological system development, and disease functions associated with this proteome (Fig. 3, A and B, supplemental Tables S3 and S4). This strategy revealed that canonical pathways associated with cell proliferation, protein synthesis, and protein turnover were prominent (Fig. 3A and supplemental Table S3). Specifically, these signaling pathways included eukaryotic initiation factor (EIF)2 (cell proliferation and protein synthesis: 144 proteins, 70.2% coverage), regulation of EIF4 and P70S6K (cell proliferation and protein synthesis: 106 proteins, 71.6% coverage), protein ubiquitination (protein synthesis and protein turnover: 147 proteins, 56.1% coverage), and mTOR (cell proliferation, protein synthesis, and protein turnover: 120 proteins, 60.6% coverage). Among other relevant predicted pathways identified were cellular stress pathways, including mitochondrial dysfunction (120 proteins, 75% coverage), sirtuin signaling (158 proteins, 59% coverage), and nuclear factor erythroid 2-related factor-2 (NRF2)-mediated oxidative stress response (103 proteins, 57.5% coverage) (Fig. 3A and supplemental Table S3). Functions associated with both cell proliferation/protein production and cellular stress were also observed following assessment of molecular and physiological functions associated with the seminal vesicle proteome (supplemental Table S4).
Fig. 3.
Functions of proteins expressed in the mouse seminal vesicle. Functions of the mouse core seminal vesicle proteome were assessed by Ingenuity Pathway Analysis. This analysis was used to predict canonical signaling pathways and upstream regulators of the proteins expressed that were highly (p ≤ 0.05) predicted to be involved in mouse seminal vesicle function. Data depicted represents the top 20: (A) canonical signaling pathways and (B) upstream regulators, presented as a heat map based on –log10 p values. A subset of proteins from the mouse seminal vesicle proteome were selected for immunofluorescence confirmation (n = 4). Candidate proteins were selected based on their association with functions of the seminal vesicle, including cellular proliferation/protein synthesis and cell death and survival. Representative images of low and high magnification and secondary antibody only controls for (C) RNA-binding protein Musashi homolog 2 (MSI2), (D) prosaposin (PSAP), (E) lysosomal acid lipase/cholesteryl ester hydrolase (LIPA), and (F) cluster of differentiation 9 antigen (CD9). White boxes indicate regions focused on for high power. EP, Epithelial cells; L, Seminal vesicle lumen; SM, Smooth muscle.
To identify the key regulators of the seminal vesicles, we then utilized the predictive upstream regulator function of Ingenuity Pathway Analysis. Among the curated regulators, those associated with cell proliferation and protein synthesis were highly predicted, including: RICTOR (p = 2.72E-53, 188 targets), transcriptional regulator tumor protein p53 (TP53) (p = 1.18E-52, 515 targets), MLX-interacting protein-like (MLXIPL, p =5.1E-30, 86 targets), and Myc proto-oncogene protein (MYC, p = 3.25E-21, 132 targets) (Fig. 3B, supplemental Table S5).
To provide a snapshot of the localization of proteins mapping to the proteome of seminal vesicles, a subset of proteins that were associated with seminal vesicle molecular functions identified in our in silico analysis were selected for validation by immunolocalization analysis (Fig. 3, C–F). Of these, RNA-binding protein Musashi homolog 2 (MSI2), which was associated with “cellular growth and proliferation” (p = 8.5E-10) and known to govern epithelial cell migration (50), was expressed in epithelial cells (predicted cytoplasm) (Fig. 3C). Additionally prosaposin (PSAP) (51), lysosomal acid lipase/cholesteryl ester hydrolase (LIPA) (52), and cluster of differentiation 9 antigen (CD9) (53), with revealed molecular and cellular functions related to cellular stress, “cell death and survival” (p = 6.5E-53), “cellular function and maintenance” (p = 2.6E-16), and “cellular compromise” (p = 2.5E-9), were investigated. Immunolocalization analysis revealed PSAP to be expressed in the smooth muscle layer (predicted extracellular space) (Fig. 3D), LIPA presents in the epithelium and smooth muscle layer (predicted cytoplasm) (Fig. 3E), and CD9 expression localized to the epithelium (predicted plasma membrane) (Fig. 3F).
Comparative Analysis of the Global Seminal Vesicle Proteome to Proteins Expressed in Seminal Vesicle Fluid
The primary function of the seminal vesicles is to synthesize and secrete a diversity of bioactive factors, including proteins that support gamete function and promote reproductive success (1, 2, 4). To compare our core proteome (supplemental Table S2) to proteins secreted in seminal vesicle fluid, we obtained seminal vesicle fluid proteomic data from Bayram et al., 2020 (42), Chang et al., 2010 (43), and Dean et al., 2009 (44). Filtering this data for proteins reviewed by UniProt identified 58 proteins in common with one or more of the previous studies, including 26, 47, and 26 in common with the Bayram, Chang, and Dean studies respectively. Of these, all were detected in our core seminal vesicle proteome (Table 2, supplemental Table S2), and these included the well-characterized seminal vesicle secretory (SVS) protein family members: 3a, 4, 5, 6, prostate and testis-expressed 4 (PATE4), glia-derived nexin (SERPINE2), protein-glutamine gamma-glutamyltransferase 4 (TGM4), and serine protease inhibitor kazal-like protein, minor form (SPINKL), which have established physiological functions in copulatory plug formation and the modulation of sperm fertilizing ability (2). Strikingly among the 58 proteins, 18/58 (31%) were in the top 1% and 34/58 (59%) were in the top 10% of the most abundant proteins detected in the core seminal vesicle proteome (Table 2).
Table 2.
Comparative analysis of the core seminal vesicle proteome to seminal vesicle fluid proteome
| Accession | Description | Gene symbol | Coverage [%] | # Peptides | # Unique peptides | Order of abundance in core proteome |
|---|---|---|---|---|---|---|
| P18419 | Seminal vesicle secretory protein 4 | Svs4 | 80 | 20 | 20 | 1 |
| P30933 | Seminal vesicle secretory protein 5 | Svs5 | 80 | 18 | 18 | 2 |
| Q64356 | Seminal vesicle secretory protein 6 | Svs6 | 72 | 21 | 21 | 5 |
| Q09098 | Prostate and testis expressed protein 4 | Pate4 | 64 | 13 | 13 | 6 |
| Q61400 | Carcinoembryonic antigen-related cell adhesion molecule 10 | Ceacam10 | 79 | 23 | 22 | 7 |
| Q8BZH1 | Protein-glutamine gamma-glutamyltransferase 4 | Tgm4 | 94 | 90 | 89 | 9 |
| F2Z472 | Seminal vesicle secretory protein 3A | Svs3a | 84 | 37 | 37 | 10 |
| P01887 | Beta-2-microglobulin | B2m | 70 | 11 | 11 | 11 |
| P20029 | Endoplasmic reticulum chaperone BiP | Hspa5 | 83 | 85 | 83 | 12 |
| P09036 | Serine protease inhibitor Kazal-type 1 | Spink3; Spink1 | 64 | 13 | 13 | 15 |
| P09103 | Protein disulfide-isomerase | P4hb | 91 | 76 | 76 | 18 |
| P10126 | Elongation factor 1-alpha 1 | Eef1a1 | 81 | 51 | 33 | 19 |
| P07724 | Serum albumin | Alb | 93 | 91 | 91 | 21 |
| P45376 | Aldo-keto reductase family 1 member B1 | Akr1b3 | 74 | 35 | 34 | 26 |
| P14211 | Calreticulin | Calr | 78 | 44 | 44 | 30 |
| Q07235 | Glia-derived nexin | Serpine2 | 74 | 33 | 33 | 33 |
| Q8BND5 | Sulfhydryl oxidase 1 | Qsox1 | 70 | 59 | 7 | 36 |
| P81117 | Nucleobindin-2 | Nucb2 | 85 | 69 | 69 | 46 |
| Q921I1 | Serotransferrin | Trf | 72 | 70 | 70 | 52 |
| P35700 | Peroxiredoxin-1 | Prdx1 | 95 | 32 | 30 | 62 |
| P17182 | Alpha-enolase | Eno1 | 91 | 50 | 43 | 74 |
| Q01853 | Transitional endoplasmic reticulum ATPase | Vcp | 86 | 107 | 106 | 75 |
| P24369 | Peptidyl-prolyl cis-trans isomerase B | Ppib | 75 | 30 | 30 | 76 |
| Q8CEK3 | Serine protease inhibitor kazal-like protein, minor form | Spinkl | 52 | 9 | 9 | 92 |
| P21460 | Cystatin-C | Cst3 | 66 | 14 | 14 | 105 |
| P18242 | Cathepsin D | Ctsd | 64 | 27 | 27 | 112 |
| P06869 | Urokinase-type plasminogen activator | Plau | 78 | 36 | 36 | 124 |
| P13020 | Gelsolin | Gsn | 68 | 49 | 49 | 164 |
| P07759 | Serine protease inhibitor A3K | Serpina3k | 54 | 29 | 27 | 251 |
| O09159 | Lysosomal alpha-mannosidase | Man2b1 | 51 | 47 | 47 | 252 |
| Q9QY48 | Deoxyribonuclease-2-beta | Dnase2b | 43 | 20 | 20 | 259 |
| P22599 | Alpha-1-antitrypsin 1–2 | Serpina1b | 65 | 31 | 12 | 285 |
| Q3UN54 | Secreted seminal-vesicle Ly-6 protein 1 | A630095E13Rik | 54 | 6 | 6 | 325 |
| P12032 | Metalloproteinase inhibitor 1 | Timp1 | 59 | 12 | 12 | 431 |
| Q61207 | Prosaposin | Psap | 57 | 24 | 24 | 509 |
| P28665 | Murinoglobulin-1 | Mug1 | 54 | 59 | 37 | 519 |
| P28798 | Progranulin | Grn | 71 | 32 | 32 | 523 |
| P32261 | Antithrombin-III | Serpinc1 | 69 | 33 | 33 | 583 |
| P01027 | Complement C3 | C3 | 69 | 103 | 103 | 669 |
| O88968 | Transcobalamin-2 | Tcn2 | 79 | 22 | 22 | 808 |
| Q9Z0J0 | NPC intracellular cholesterol transporter 2 | Npc2 | 59 | 17 | 17 | 846 |
| P06728 | Apolipoprotein A-IV | Apoa4 | 74 | 27 | 27 | 923 |
| Q06890 | Clusterin | Clu | 33 | 17 | 17 | 953 |
| E9Q557 | Desmoplakin | Dsp | 38 | 86 | 86 | 995 |
| Q9D309 | Protein FAM3B | Fam3b | 70 | 19 | 19 | 1028 |
| P11276 | Fibronectin | Fn1 | 40 | 61 | 61 | 1073 |
| P21614 | Vitamin D-binding protein | Gc | 81 | 33 | 33 | 1092 |
| P23953 | Carboxylesterase 1C | Ces1c | 35 | 17 | 14 | 1324 |
| P06797 | Cathepsin L1 | Ctsl | 56 | 16 | 16 | 1363 |
| Q01339 | Beta-2-glycoprotein 1 | Apoh | 48 | 17 | 17 | 1628 |
| P20918 | Plasminogen | Plg | 51 | 31 | 31 | 1864 |
| Q62181 | Semaphorin-3C | Sema3c | 39 | 20 | 20 | 1936 |
| P11672 | Neutrophil gelatinase-associated lipocalin | Lcn2 | 44 | 6 | 6 | 1982 |
| Q9QZL9 | Dickkopf-like protein 1 | Dkkl1 | 40 | 10 | 10 | 2154 |
| P13634 | Carbonic anhydrase 1 | Car1 | 58 | 11 | 11 | 2239 |
| P07309 | Transthyretin | Ttr | 71 | 8 | 8 | 2259 |
| Q9ESB3 | Histidine-rich glycoprotein | Hrg | 25 | 12 | 12 | 2997 |
| Q00897 | Alpha-1-antitrypsin 1–4 | Serpina1d | 48 | 23 | 2 | 3714 |
The core seminal vesicle fluid proteome was determined as described in supplemental Table S2 and compared with a high confidence mouse seminal vesicle fluid proteome. This high confidence proteome was generated by obtaining data from Bayram et al., 2020 (42), Chang et al., 2010 (43), and Dean et al., 2009 (44) and filtering this data to only include proteins reviewed by UniProt. This led to a final high confidence list of 58 proteins. Included in this dataset is the order of abundance in the core proteome, determined by the average of normalized abundance quants of the three biological replicates (prior to scaling), to demonstrate the relative level of expression of seminal vesicle secreted proteins within the proteome.
Acute Acrylamide Exposure Alters the Seminal Vesicle Proteome
We have previously demonstrated that paternal acrylamide exposure alters sperm quality in a manner that has consequences for fetal development (22). To examine whether acrylamide also dysregulates seminal vesicle function, male mice were subjected to an established acute acrylamide exposure protocol (22), whereby the animals received daily injections of either acrylamide or PBS (control) for five consecutive days prior to seminal vesicle tissue collection 72 h after the final injection (Fig. 1).
The treatment regimen did not substantially alter mouse body weight (Fig. 4A) or change seminal vesicle weight (Fig. 4B), but did elicit marked changes in the proteomic composition of the seminal vesicle tissue. A global analysis of the seminal vesicle tissue proteome from acrylamide-treated males returned high confidence identification of 4819 proteins (FDR ≤ 0.01, Fig. 4C, supplemental Table S6), with strong Pearson correlation coefficients (0.925 average r2) across all biological replicates (supplemental Fig. S1D). Consistent with the depth of coverage achieved in control samples, an average of 16.9 peptides (15.3 unique peptides), representing a coverage of 40.8% per protein (Table 1), were identified per protein in the seminal vesicle tissue sourced from acrylamide-treated animals. Combined analysis of acrylamide (4819 proteins; Fig. 4C, supplemental Table S6) and control seminal vesicle proteomes (5013 proteins; Fig. 2A, supplemental Table S2) identified 5478 proteins (FDR ≤ 0.01). Filtering for proteins with three quantitative values in at least one treatment group (i.e., control or acrylamide), and either 3 or 0 quantitative values in the opposing group, resulted in a final proteome of 4698 proteins (Fig. 4D, supplemental Table S7). Among these, three were exclusively identified in control seminal vesicle tissue: myelin protein P0 (MPZ), heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC), and UDP-glucuronosyltransferase 1A7 (UGT1A7C). Five proteins were uniquely identified in acrylamide-exposed seminal vesicle tissue: WD repeat-containing protein 46 (WDR46), Rho GTPase-activating protein 10 (ARHGAP10), B-cell receptor-associated protein 29 (BCAP29), interleukin-1 receptor-associated kinase 4 (IRAK4), and DnaJ homolog subfamily B member 5 (DNAJB5) (Fig. 4, D and E). Additional analysis of the 4698 identified proteins using the Mouse Genome Informatics Phenotypes/Alleles project as well as the International Mouse Phenotyping Consortium databases revealed that 60/4698 (1.3%) proteins are associated with phenotypes in seminal vesicle structure or function (60 proteins) (supplemental Table S7).
Fig. 4.
Characterization of the mouse seminal vesicle proteome following acrylamide exposure.A, mouse weight (g) (n = 7 individual mice per treatment group) and (B) seminal vesicle weight (mg) (n = 11 individual mice per treatment group) were initially assessed prior to isolation of proteins. Data are presented as a column graph with mean ± SEM values and analyzed by unpaired t test with significance indicated (∗) if p ≤ 0.05 when comparing the acrylamide to the control group. Using a Venn diagram, (C) proteins detected across the n = 3 replicates of the acrylamide group were determined, (D) then compared with the control group filtering for proteins with three quantitative values in at least one of control or acrylamide groups, and either 3 or 0 quantitative values in the other group. E, proteins identified as unique to control or acrylamide are presented as a heat map.
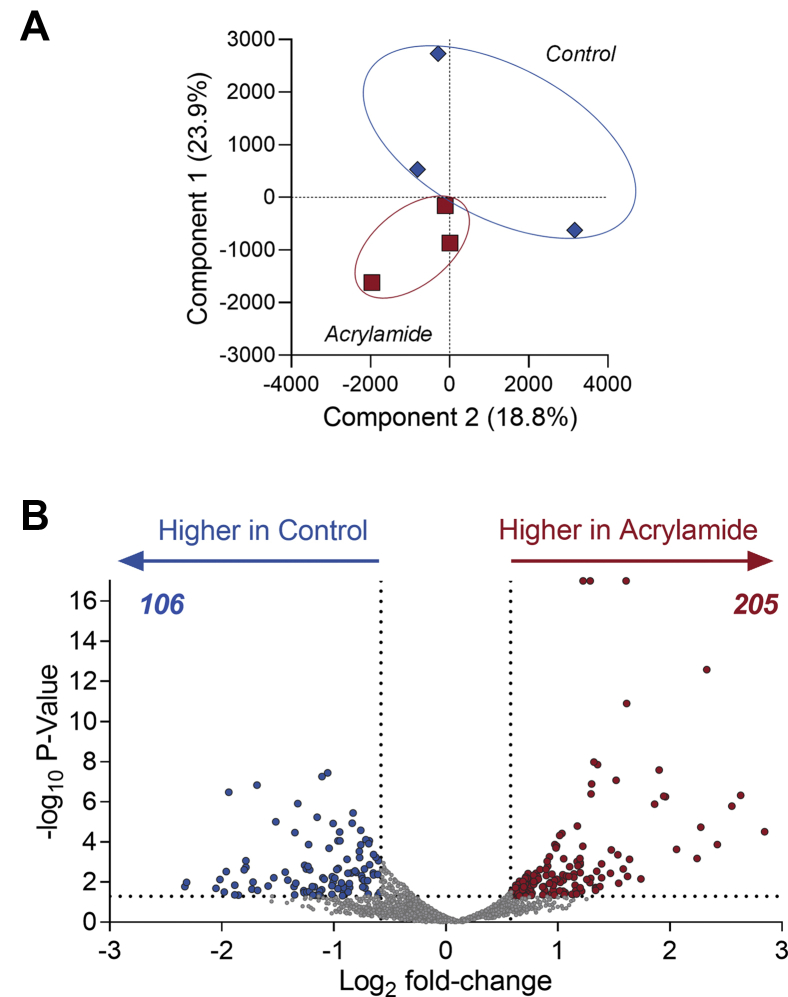
Assessment of differentially regulated proteins in mouse seminal vesicles revealed a subset of proteins that were altered in response to acrylamide exposure. Initially, global assessment of protein profiles using principal component analysis demonstrated biological variance among the control mice but notably a tighter clustering among the acrylamide-treated mice, indicative of a consistent response to this reproductive toxicant (Fig. 5A). Assessment of the regulation of individual proteins demonstrated that 205 proteins were upregulated and 106 downregulated (fold-change ≥1.5 or ≤−1.5 and p ≤ 0.05) (Fig. 5B, supplemental Table S7). These significantly regulated proteins ranged in abundances (supplemental Fig. S1E) and 8/311 (2.6%) were associated with seminal vesicle phenotypes (Table 3).
Fig. 5.
Acrylamide exposure alters the mouse seminal vesicle proteome. Seminal vesicle proteomic data from acrylamide and control mice was assessed (n = 3 biological replicates per group) to identify those proteins with a fold-change ≥ 1.5 or ≤ −1.5 and p ≤ 0.05 using Proteome Discoverer 2.4. A, principal component analysis of proteomic data. B, graphical depiction of differentially regulated proteins using a volcano plot.
Table 3.
Comparative analysis of the seminal vesicle proteome to seminal vesicle phenotypes identified from the Mouse Genome Informatics and The International Mouse Phenotyping Consortium datasets
| Accession | Protein names | Gene symbol | FC (acrylamide versus control) | p-value | Phenotype description |
|---|---|---|---|---|---|
| Q61103 | Zinc finger protein ubi-d4 | Dpf2 | 2.14 | 5.10E-03 | abnormal seminal vesicle morphology |
| Q9D1Q4 | Dolichol-phosphate mannosyltransferase subunit 3 | Dpm3 | 1.83 | 1.66E-02 | abnormal seminal vesicle morphology |
| Q9D142 | Uridine diphosphate glucose pyrophosphatase NUDT14 | Nudt14 | 1.62 | 1.71E-02 | abnormal seminal vesicle morphology |
| P63037 | DnaJ homolog subfamily A member 1 | Dnaja1 | 1.58 | 8.86E-03 | small seminal vesicle |
| Q61792 | LIM and SH3 domain protein 1 | Lasp1 | 1.57 | 1.32E-02 | small seminal vesicle |
| Q8C5P7 | Testis development-related protein | Tdrp | −1.52 | 4.10E-03 | abnormal seminal vesicle morphology |
| P06869 | Urokinase-type plasminogen activator | Plau | −1.56 | 4.37E-04 | herniated seminal vesicle |
| Q9Z0M9 | Interleukin-18-binding protein | Il18 bp | −2.35 | 1.66E-03 | abnormal seminal vesicle morphology |
Seminal vesicle phenotypes associated with proteins expressed in the mouse seminal vesicle were determined. These data were then filtered based upon those proteins that met our differential regulation criteria (−1.5 ≤ FC ≥ 1.5, p ≤ 0.05) and proteins that matched these criteria are presented along with their associated phenotypes.
Functional assessment of the differentially expressed proteins using Ingenuity Pathway Analysis (Fig. 6, supplemental Fig. S2, supplemental Tables S8–S10) revealed that canonical pathways and molecular functions dysregulated following acrylamide exposure were associated with the regulation of protein synthesis and cellular stress (Fig. 6, A and C, supplemental Fig. S2, A–C and supplemental Tables S8 and S9). Utilization of a Z-score algorithm to assign either pathway activation (Z-score ≥ 2) or inhibition (Z-score ≤ −2) revealed EIF2 signaling (Z-score = 3.46), which is associated with eukaryotic protein synthesis (54), as predicted to be activated in acrylamide-exposed seminal vesicles, while death receptor signaling (Z-score = −2), associated with cellular stress and apoptosis (55), was predicted to be inhibited (Fig. 6C). Interestingly, assessment of molecular and physiological functions revealed a large number of proteins associated with the terms “cell death and survival” and “cell proliferation/protein synthesis” (supplemental Fig. S2A), with overrepresentation of the functions “apoptosis” in the category “cell death and survival” and “translation of protein” in the category “protein synthesis” (supplemental Fig. S2, B and C, supplemental Table S9).
Fig. 6.
Signaling pathways and upstream regulators influenced by acrylamide exposure in the mouse seminal vesicles. Ingenuity Pathway Analysis was used to predict (p ≤ 0.05) canonical signaling pathways and upstream regulators of the mouse seminal vesicle proteins that were differentially regulated following acrylamide treatment. Data are represented as a heat map based on –log10 p values and show the top 20: (A) canonical signaling pathways and (B) upstream regulators. To gain a greater understanding of the regulation of these pathways, (C) canonical signaling pathways and (D) upstream regulators that were predicted to be activated (Z-score ≥ 2) or inhibited (Z-score ≤ −2) with p ≤ 0.05 are represented as heat maps based on Z-score value. E, proteins differentially expressed following acrylamide exposure (red = upregulated, green = downregulated) that are documented to be regulated by RICTOR are presented in an interaction network. Connecting lines indicate several predicted relationships that lead to activation (orange) and a relationship not consistent with that published in the literature (yellow). Image created with BioRender.com. F, representative images from immunofluorescence analysis of RICTOR in control and acrylamide samples at low and high power and secondary antibody only controls (n = 5/group). White boxes indicate regions focused on for high power. EP, Epithelial cells; L, Seminal vesicle lumen; SM, Smooth muscle.
To identify the upstream signaling pathways involved in transmitting the acrylamide-mediated dysregulation of mouse seminal vesicle function, we utilized the upstream regulator function of Ingenuity Pathway Analysis. A total of 206 upstream factors were predicted to be regulators of the seminal vesicle response to acute acrylamide exposure (p ≤ 0.05, supplemental Table S10). To assess the activation or inhibition status of these regulators, we first filtered to remove all chemical molecules other than those classified as endogenous mammalian chemicals, leaving 156 predicted upstream regulators (p ≤ 0.05, Fig. 6B, supplemental Table S10). Of these, four were predicted to be activated (Fig. 6D) including MLXIPL, MYC, N-myc proto-oncogene protein (MYCN), and Rho family of GTPases (RHO), while the four inhibited regulators included RICTOR, La-related protein 1 (LARP1), cyclin-dependent kinase 4/6 (CDK4/6), and miRNA-210 (Fig. 6D, supplemental Fig. S2, D–F, supplemental Table S10).
Notably RICTOR, a major component of the mTOR signaling pathway known to influence both protein synthesis (56) and epithelial cell function (57, 58), was a predicted upstream regulator identified in the core mouse seminal vesicle proteome (p = 2.52E-53, Fig. 3B and supplemental Table S5) and was also prominent among dysregulated upstream regulators following acrylamide exposure (Z-score = −3.64, p = 6.69E-07. Fig. 6, B, D–F and supplemental Table S10), despite not having altered expression (1.2 fold-change, p = 6.4E-01, supplemental Table S7). As a prelude for future studies focusing on defining the regulatory role of RICTOR in seminal vesicle tissues, we used immunofluorescence to assess the distribution of the protein in both control and acrylamide-treated tissue samples. This approach revealed intense RICTOR labeling that was primarily associated with the apical surface of seminal vesicle epithelial cells, yet failed to respond in terms of either abundance or localization upon acrylamide challenge (Fig. 6F).
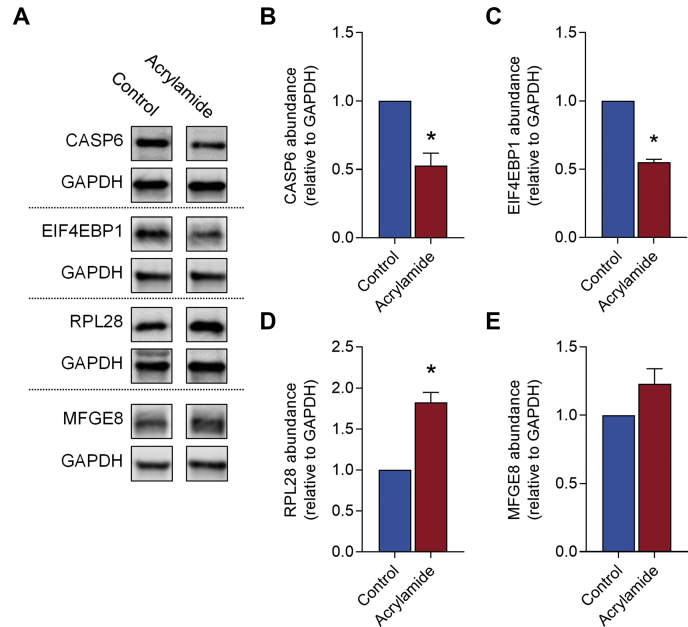
To validate differential protein expression, a subset of proteins whose expression was dysregulated by acrylamide treatment and that were associated with responsive cellular and molecular functions identified by bioinformatic analyses were selected for orthogonal quantification by immunoblotting (n = 4 per group). Specifically, these proteins were associated with “cell death and survival”; Caspase-6 (CASP6, −1.71-fold-change in acrylamide versus control, p = 2.91E-04), or “protein synthesis”; (60S ribosomal protein L28 (RPL28, 1.96-fold-change in acrylamide versus control, p = 1.33E-04), and eukaryotic translation initiation factor 4E-binding protein 1 (EIF4EBP1, −2.06-fold-change in acrylamide versus control, p = 2.06E-04)). Additionally, we also assessed Lactadherin (MFGE8, −1.2-fold-change in acrylamide versus control, p = 6.83E-01) as a protein that did not exhibit change in the proteomic dataset. Consistent with the proteomic data, CASP6 (Fig. 7, A and B, −1.9-fold-change in acrylamide versus control, p = 0.0021), and EIF4EBP1 (Fig. 7, A and C, −1.8-fold-change in acrylamide versus control, p < 0.0001) were suppressed, while RPL28 (Fig. 7, A and D, 1.8 fold-change in acrylamide versus control, p = 0.0005) was induced following acrylamide exposure. Additionally, MFGE8 (Fig. 7, A and E, 1.23 fold-change in acrylamide versus control, p = 0.1420) was not altered.
Fig. 7.
Orthogonal validation of proteins identified in the mouse seminal vesicle proteome. Quantitative mass spectrometry data were validated via immunoblotting of differentially regulated proteins (n = 4 individual mice per treatment group). Candidate proteins were selected based on those differentially regulated proteins that were associated with canonical pathways and upstream regulators with (A) Representative Immunoblotting images shown. Proteins assessed included (A and B) Caspase-6 (CASP6), (A and C) Eukaryotic translation initiation factor 4E-binding protein 1 (EIF4EBP1), (A and D) 60S ribosomal protein L28 (RPL28), and (A and E) lactadherin (MFGE8). Immunoblotting data were assessed using Multi Gauge software, normalized to the control group and presented as a column graph with mean ± SEM values. Data were analyzed by unpaired t test. ∗ p ≤ 0.05 indicates statistical significance when comparing acrylamide with control groups.
Acrylamide Treatment Leads to Increased Levels of Oxidative DNA Damage in the Mouse Seminal Vesicle
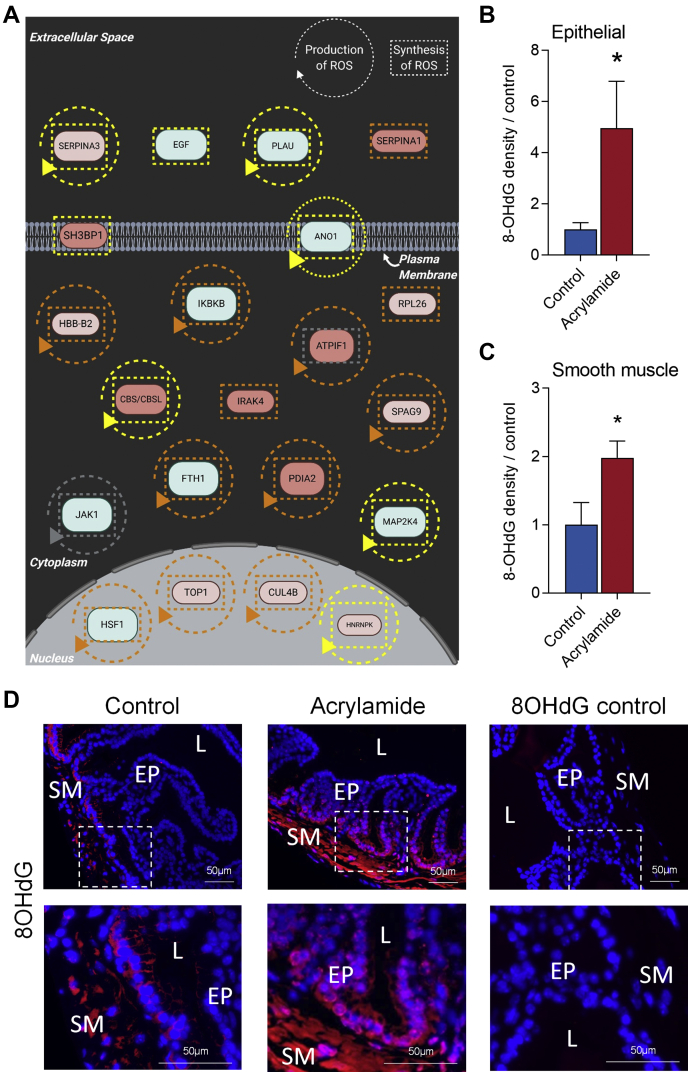
We have previously demonstrated that exposure to acrylamide leads to increased levels of oxidative DNA damage in epididymal sperm (22, 23, 24, 59). Consistent with these findings, we observed a number of canonical signaling pathways and functions associated with the cellular response to oxidative stress that were altered in the mouse seminal vesicles following acrylamide exposure (Fig. 6, supplemental Tables S8 and S9). These pathways included NRF2-mediated oxidative stress response (11 proteins, p = 6.92E-05), acute-phase response signaling (9 proteins, p = 9.12E-04), antioxidant actions of vitamin C (6 proteins, p = 4.17E-03), sirtuin signaling pathway (10 proteins, p = 7.59E-03), glutathione-mediated detoxification (3 proteins, p = 9.77E-03) (Fig. 6A and supplemental Table S8), and free radical scavenging (22 proteins, p = 3.69E-03) (Fig. 8A and supplemental Table S9). Given this evidence, we assessed whether acrylamide exposure causes oxidative stress in seminal vesicle tissues. Specifically, seminal vesicle tissue sections were assessed for their levels of oxidative DNA damage burden using an 8-OHdG probe. This analysis confirmed that acrylamide treatment elicits elevated 8-OHdG in epithelial cells (Fig. 8, B and D, fivefold increase, p = 0.016) and the smooth muscle cell layer (Fig. 8, C and D, twofold increase, p = 0.04).
Fig. 8.
Acrylamide induces oxidative stress in the mouse seminal vesicle.A, Ingenuity Pathway Analysis identified the production and synthesis of reactive oxygen species as a significant (p ≤ 0.05) cellular function associated with proteins that were differentially regulated following acrylamide exposure. These data are presented as interaction networks where a circular arrow surrounding the protein name indicates it is associated with the production of reactive oxygen species, and a square dotted line surrounding the protein indicates it is associated with the synthesis of reactive oxygen species. These lines indicate several predicted relationships that lead to activation (orange), relationship inconsistent with the literature (yellow), and relationship is known but effects on function is yet to be completely characterized (gray). Image created with BioRender.com. B–D, oxidative DNA damage was measured using 8-Hydroxy-2' -deoxyguanosine (8-OHdG) in (B) epithelial cells and (C) the smooth muscle layer, with (D) representative images at low and high magnification and secondary antibody only controls (n = 5/group). White boxes indicate regions focused on for high power. Data were analyzed by unpaired t test and are presented as a column graph with mean ± SEM values. ∗p ≤ 0.05 indicates statistical significance when comparing acrylamide with control groups. EP, Epithelial cells; L, Seminal vesicle lumen; SM, Smooth muscle.
Acrylamide Treatment Leads to Alterations of Seminal Vesicle Secretory Capacity
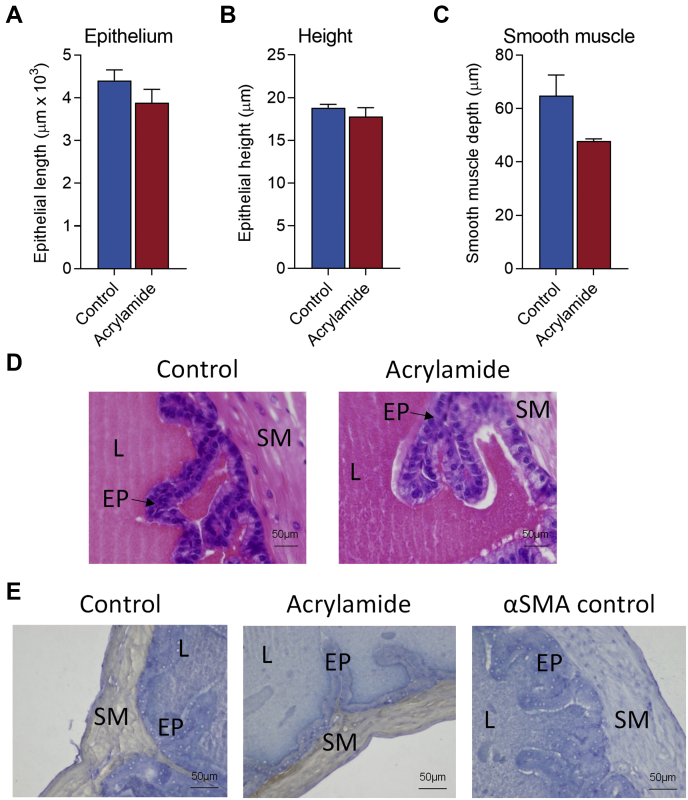
Collectively, these data demonstrate that seminal vesicle tissue is responsive to acute acrylamide exposure; however, whether these changes alter the secretory capacity of the seminal vesicles remained to be answered. To begin to address this question, we undertook histological analysis of seminal vesicles to examine the impact of acrylamide on epithelial cells and the smooth muscle layer, which play important roles in modulating secretory capacity (25). Analysis of seminal vesicle epithelial cells revealed no significant change in epithelial cell mucosal folding/branching (Fig. 9, A and D), nor any change in epithelial cell height, reflective of maintenance of a tall secretory columnar epithelium following acrylamide exposure (Fig. 9, B and D). Similarly, no significant change was observed in the depth of the smooth muscle cell layer (Fig. 9, C and E) in acrylamide-treated seminal vesicle tissue compared with the controls, suggesting that SVS capacity was unchanged.
Fig. 9.
Seminal vesicle cellular structure is not altered following acrylamide exposure.A and D, epithelial cell branching/folding (B and D) epithelial cell height, and (C and E) depth of stromal muscle layer wete assessed as surrogate measures of seminal vesicle secretory capacity with (D and E) representative histological images of (D) hematoxylin and eosin and (E) alpha smooth muscle actin (αSMA) presented. Data were analyzed by unpaired t test and are presented as column graphs with mean ± SEM values from n = 5 individual mice per group. EP, Epithelial cells; L, Seminal vesicle lumen; SM, Smooth muscle.
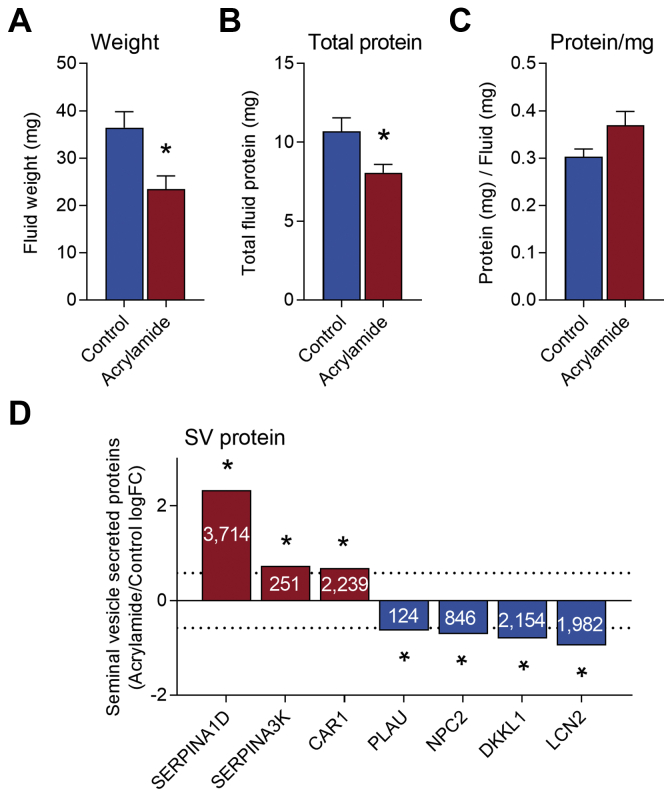
However, despite not overtly affecting gross seminal vesicle morphology or weight (Fig. 4B, Fig. 9, A–E), acrylamide treatment did significantly reduce the volume of seminal vesicle secretions harvested from treated animals (Fig. 10A, 36% reduction, p < 0.01). Notably, this response was also reflected in a reduction in the total amount of seminal vesicle fluid protein collected (Fig. 10B, 25% reduction, p = 0.02), with a small but nonsignificant increase in protein concentration per mg fluid collected observed (Fig. 10C, p = 0.055).
Fig. 10.
Seminal vesicle secretory function is perturbed following acrylamide exposure. Seminal vesicle fluid was collected and (A) seminal vesicle fluid weight (mg), (B) total seminal vesicle fluid protein (mg), and (C) seminal vesicle fluid protein/mg seminal vesicle fluid were assessed. Data are presented as column graph with mean ± SEM values and data from n = 11 individual mice per treatment group. Data were analyzed by unpaired t test. ∗p ≤ 0.05 indicates statistical significance. D, expression of seminal vesicle secreted proteins (n = 3 individual mice per treatment group) identified by proteomics of mouse seminal vesicle fluid (42, 43, 44) as statistically significant (p ≤ 0.05 indicated by ∗) and fold-change ≥ 1.5 or ≤ −1.5 in our proteomic analysis. Proteins assessed included alpha-1-antitrypsin 1–4 (SERPINA1D), serine protease inhibitor a3k (SERPINA3K), carbonic anhydrase 2 (CAR1), urokinase-type plasminogen activator (PLAU), NPC intracellular cholesterol transporter 2 (NPC2), dickkopf-like protein 1 (DKKL1), and neutrophil gelatinase-associated lipocalin (LCN2). Data are presented as column graphs of log fold-change (logFC).
Additionally, analysis of our proteomic data for seminal vesicle secreted proteins showed evidence of alterations to seminal vesicle secreted proteins (Table 2) (42, 43, 44). Seven of these, alpha-1-antitrypsin 1–4 (SERPINA1D, 5.0 FC, p = 2.56E-13), serine protease inhibitor A3K (SERPINA3K, 1.66 FC, p = 9.39E-03), carbonic anhydrase 2 (CAR1, 1.5 FC, p = 1.60E-2), urokinase-type plasminogen activator (PLAU, −1.56 FC, p = 4.4E-04), NPC intracellular cholesterol transporter 2 (NPC2, −1.6 FC, p = 8.0E-05), dickkopf-like protein 1 (DKKL1, −1.7 FC, p = 3.21E-03), and neutrophil gelatinase-associated lipocalin (LCN2, −1.9 FC, p = 9.0E-05) were significantly altered (FC ≥ 1.5 or ≤ −1.5 and p ≤ 0.05) in seminal vesicle tissue of acrylamide-treated males compared with control (Fig. 10D, Table 2). Notably, of the 12 other seminal vesicle secreted proteins (supplemental Fig. S3, Table 2) that did not reach the effect size threshold but did reach statistical significance, seven were in the top 1% most abundant seminal vesicle proteins. These included PATE4 (−1.25 FC, p = 3.54E-02), SVS3A (−1.30 FC, p = 1.98E-02), carcinoembryonic antigen-related cell adhesion molecule 10 (CEACAM10, −1.34FC, p = 1.07E-02), serine protease inhibitor kazal-type 1 (SPINK1, −1.36 FC, p = 8.3E-03), beta-2-microglobulin (B2M, −1.37 FC, p = 7.28E-03) SVS6 (−1.41 FC, p = 4.34E-03), and sulfhydryl oxidase 1 (QSOX1, −1.44 FC, p = 2.73E-03) (supplemental Fig. S3, Table 2).
Discussion
Supporting the delivery of spermatozoa to the site of fertilization has long been considered the sole function of seminal plasma, but this narrow delineation is now inadequate. Indeed, seminal plasma fulfills a more complex role in virtually all species that deposit gametes in the female reproductive tract at intromission (1). Thus, in addition to its primary function in sperm delivery, seminal plasma modifies the postcopulatory female reproductive tract environment in a manner that supports embryo implantation and exerts long-term consequences for offspring health (2, 3, 15, 21). In most species, the major contributor to seminal plasma volume is the seminal vesicle glands (2, 4). Although compelling evidence implicates these glands as crucial regulators of reproductive success (3, 15, 17), understanding of their physiology and proteomic constitution remains limited. This is illustrated through analysis of reviewed M. musculus proteins in UniProt (25,260 proteins) whereby only 26 proteins have been characterized as seminal vesicle expressed, despite substantially more proteins linked with seminal vesicle phenotypes (40, 41). Thus, in completing the first proteomic assessment of the mouse seminal vesicle, we have built greater understanding of this gland’s complex biology and demonstrated that the seminal vesicle tissue, akin to that of other segments of the male reproductive tract, is sensitive to environmental exposures.
The principal responsibility of the seminal vesicles is to synthesize and secrete a diversity of bioactive factors, including proteins that promote reproductive success by supporting gamete function or interacting with the female reproductive tract (2, 4, 5). Consistent with this primary function, proteins secreted by the seminal vesicle (42, 43, 44) were among the most abundant proteins detected in seminal vesicle tissue. These proteins play critical roles in semen coagulation (e.g., members of the SVS family and PATE4), and the regulation of male gamete function (e.g., members of the SVS family, SPINKL and SERPINE2) (1, 2, 3, 4). Consistent with other secretory mucosal surfaces, wherein equivalent cellular pathways mediate the cell fate decisions needed to maintain healthy tissue homeostasis and organ functionality (60, 61, 62), our in silico analysis of the seminal vesicle proteome identified cell proliferation, protein synthesis, and protein turnover as prominent biological processes. In addition, proteins linked to cellular death and survival pathways were abundantly represented within the seminal vesicle proteome.
Notably, the composition of seminal vesicle secretions is known to exhibit significant plasticity in response to changing paternal environments (63, 64, 65) or even cues produced by a female partner (66); functions that are postulated to maximize fertilization success and transmit evolutionary adaptations to offspring (63, 66, 67). These above studies raise the possibility that in addition to sperm, the seminal vesicles are sensitive to environmental signals and stressors (3, 15, 17). To assess the impact of environmental exposures on seminal vesicle function, we utilized a readily tractable in vivo acute acrylamide exposure model. This model, which utilizes doses of acrylamide that are substantially higher than human chronic exposure estimates (68), has been extensively utilized by us (22) and others (69, 70) to investigate adverse impacts of acrylamide exposure on sperm quality and fetal development. Specifically, exposure of testicular germ cells to acrylamide leads to enhanced levels of DNA damage that fail to be resolved during sperm maturation, yet do not appear to affect fetal development. Comparable levels of DNA damage are also characteristic of sperm exclusively exposed to acrylamide during epididymal transit. However, exposure during epididymal transit leads to substantially increased rates of fetal loss (22, 69, 71, 72). This raises the intriguing possibility that the male transfers the burden of acrylamide stress to the female through mechanisms that are independent of sperm DNA damage. While the nature of these paternal stress signals and the timing of their transfer to spermatozoa remain an area of active investigation, our data raise the prospect that other tissues in the male reproductive tract such as the seminal vesicles may be similarly affected by paternal stressors and therefore deserve consideration in terms of a holistic appraisal of their impact on male reproductive health and fertility.
Consistent with this possibility, paternal exposure to the reproductive toxicant, acrylamide altered the seminal vesicle proteome. In silico analysis of the dysregulated proteins provided evidence that the seminal vesicles initiate protein synthesis and promote cellular survival in response to the acrylamide insult, in order to return seminal vesicle tissue to normal homeostasis and functionality. The cellular responses we observed among dysregulated proteins share analogy with changes recorded following oxidative stress insults, wherein cells typically increase the expression of antioxidant genes and activate pathways that promote survival and adaptation to counter the cytotoxic effects of excessive reactive oxygen species production (73). Indeed, our studies demonstrate that treatment with the antioxidant resveratrol for 3 months rescues acrylamide-induced sperm DNA damage in mouse chronic exposure model (59), but this rescue is lost after a longer, 6 month exposure when other consequences of treatment have become apparent. Similarly, other studies have demonstrated that antioxidants can inhibit oxidative stress in rodent seminal vesicles (74, 75). Here we demonstrate that acrylamide exposure leads to the dysregulation of oxidative stress pathways accompanied by elevation of 8-OHdG lesions in the seminal vesicle epithelium, indicating that oxidative stress likely plays a leading role in the pathological response of this tissue to acute acrylamide.
Notably in our analysis, we have detected proteins uniquely expressed in the seminal vesicle proteome, with ARHGAP10, BCAP29, DNAJB5, IRAK4, and WDR46 present following acrylamide treatment and HNRNPC, MPZ, and UGT1A7C absent. In agreement with our in silico analysis, these uniquely acrylamide-expressed proteins are associated with driving cellular maintenance, proliferation, and repair in response to cellular stressors. Specifically, DNAJB5, a member of the heat shock protein 40 chaperone family (76), is a regulator of cellular protein homeostasis through its interaction with histone deacetylases and is activated in response to oxidative stress (77, 78). Moreover, the expression of DNAJB5 is known to increase in response to other cellular stressors such cancer (79) or infection (80).
Notwithstanding the paucity of studies focused on changes to the global proteomic landscape induced by acrylamide challenge in other tissues, we sought to explore the conservation of the proteomic response of the seminal vesicles to that of other tissues. We noted a 68% overlap (23/34) between proteins identified within our dataset and significantly dysregulated proteins identified in hippocampal tissue (81) and striatal synaptosomes (82) (supplemental Table S11). However, of the 23 detected proteins, only glutathione S-transferase omega-1 (upregulated) and alpha-soluble NSF attachment protein (downregulated) were also significantly dysregulated in seminal vesicle tissue. Although these studies suggest that the seminal vesicles respond to acrylamide in a tissue-specific manner compared with existing data (81, 82), further studies are required to confirm this, particularly through the examination of proteomic changes in response to acrylamide from other male reproductive tract tissues.
Much of our knowledge of seminal vesicle biology comes from studies examining the impact of androgens on this tissue. Indeed, androgens are pivotal for all aspects of seminal vesicle development and function through their capacity to trigger differentiation of epithelial cells into a secretory epithelial layer (4, 25, 83). This raises the possibility that dysregulated production of androgens may contribute to altered seminal vesicle function following acrylamide exposure. However, we did not observe the expected reduction in seminal vesicle size or tissue morphology (84), nor did we detect any in silico evidence of endocrine pathway disruption. These findings suggest that our subchronic acrylamide exposure regimen was below the threshold needed to influence serum testosterone levels (85, 86), and thus androgens are unlikely to be solely responsible for driving the seminal vesicle response to acrylamide.
Due to the lack of comprehensive assessment of the proteomic composition of seminal vesicles, the identity of other regulators of seminal vesicle function remains unknown, although estrogens (87) and sexual activity (88) are implicated. In an attempt to shed light on novel regulators, we utilized Ingenuity Pathway Analysis to identify regulators of central importance to the seminal vesicle. Notably, in silico analyses identified RICTOR, a component of the mTOR pathway with established roles in protein production, protein secretion, and epithelial cell function (56, 57, 58, 89, 90, 91, 92, 93, 94), as a putative regulator of the seminal vesicle tissue response to acrylamide exposure. While we have yet to directly assess how RICTOR influences the physiological responses of seminal vesicle tissue, we found no evidence of overt modulation of either RICTOR expression of intracellular distribution in response to acrylamide challenge. Despite this, it is notable that deficiency of the closely related mTORC1 molecule regulatory-associated protein of MTOR complex 1 (RPTOR) does dysregulate seminal vesicle function (95), thus reinforcing the tenet that components of the mTOR complex play an important role in seminal vesicle biology. It is also notable that impaired mTORC2 signaling, mediated by RICTOR, has been implicated in increased cellular ROS sensitivity (96, 97).
The altered protein composition of seminal vesicle tissue following acrylamide exposure raises the question of whether seminal vesicle secretory function is similarly influenced. While no change in the weight of seminal vesicle tissue was detected, a decrease in seminal vesicle secretion volume and overall protein concentration was observed. This change in secretory function is consistent with other studies of epithelial cells in which environmental perturbations or genetic deficiency has been linked to a hyposecretory phenotype (98, 99, 100), yet the exact mechanisms by which this change is enacted remain uncertain. Additionally, our data implicates the synthesis of prominent seminal vesicle secreted factors, including SVS3A, SVS6, and PATE4, as being dysregulated in seminal vesicle tissue following acrylamide exposure. These proteins have well-established roles in copulatory plug formation (2, 9, 44), with PATE4-deficient male mice unable to form a complete copulatory plug leading to subfertility (9). Additional proteins with altered synthesis included the serine protease inhibitors SERPINA1D (101), SERPINA3K (102), and PLAU (103), which may contribute to copulatory plug formation, modification of sperm function, or activation of seminal fluid signaling molecules, as has been shown for other protease inhibitors in seminal fluid (104).
Taken together, these findings raise the possibility that alterations to seminal vesicle function may contribute to the embryonic loss observed following acute acrylamide exposure in the sire, as well as epididymal sperm changes (22, 69, 70). However, the specific proteomic impacts of acrylamide exposure on seminal vesicle fluid and the biological consequences of these changes remain to be addressed—particularly as this fluid constitutes a highly biased proteomic sample with a small subset of highly dominant proteins that rapidly coagulate upon recovery, making proteomic analysis and biological assessment of their functions challenging (42, 63). Indeed, our combined analysis of published seminal vesicle fluid proteomes (42, 43, 44) only identified 58 reviewed proteins from the UniProt database as being secreted by the seminal vesicle. These data further emphasize the importance of gaining a comprehensive profile of the true protein composition of seminal vesicle fluid, as acknowledged by Bayram et al., (42). Our future studies aim to address this and increase the depth of the mouse seminal vesicle fluid proteome and explore the biological significance of altered seminal vesicle secretions on fertility and fetal development.
In summary, we have exploited an advanced proteomic platform to generate the first mechanistic insight into mouse seminal vesicles in normal physiology and pathology. These data identify how the seminal vesicle tissue responds to the reproductive toxicant, acrylamide, lending support to the emerging evidence that suggests we need to consider the responses of all male reproductive tract tissues when interpreting the potential impact of toxicants/environmental stressors on male fertility.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (105) with the dataset identifier PXD021998 and 10.6019/PXD021998.
Supplemental data
This article contains supplemental data (81, 82).
Conflict of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors gratefully acknowledge Nathan Smith from The University of Newcastle Analytical Biomolecular Research Facility (ABRF) and the Academic and Research Computing Support team at the University of Newcastle.
Funding and additional information
The preparation of this article was supported by the award of National Health and Medical Research Council (NHMRC) Project Grants APP1147932, awarded to B. N. and M. D. D., and APP1163319, awarded to B. N., R. J. A. and E. G. B. B. N. (APP1154837), M. D. D. (APP1173892), and E. G. B. (APP1138701) are recipients of NHMRC Research Fellowships.
Author contributions
D. A. S.-B., E. G. B., M. D. D., T. L., R. J. A., S. D. R., S. A. R., B. N., and J. E. S. conceptualization; D. A. S.-B. data curation; D. A. S.-B., I. R. B., A. L. A., S. J. S., L. A. M., and J. E. S. formal analysis; E. G. B., M. D. D., R. J. A., and B. N. funding acquisition; D. A. S.-B., N. A. T., and J. E. S. investigation; N. A. T., S. D. R., B. N., and J. E. S. methodology; B. N. and J. E. S. project administration; B. N. resources; S. A. R., B. N., and J. E. S. supervision; D. A. S.-B. and J. E. S. visualization; D. A. S.-B., I. R. B., A. L. A., S. J. S., and L. A. M. validation; D. A. S.-B., B. N., and J. E. S. writing—original draft; D. A. S.-B., N. A. T., E. G. B., M. D. D., I. R. B., A. L. A., S. J. S., L. A. M., T. L., R. J. A., S. D. R., S. A. R., B. N., and J. E. S. writing—review and editing.
Supplemental Data
References
- 1.Schjenken J.E., Sharkey D.J., Robertson S.A. Seminal vesicle—secretion. In: Skinner M.K., editor. Encyclopedia of Reproduction. 2nd Ed. Academic Press; Oxford: 2018. pp. 349–354. [Google Scholar]
- 2.Noda T., Ikawa M. Physiological function of seminal vesicle secretions on male fecundity. Reprod. Med. Biol. 2019;18:241–246. doi: 10.1002/rmb2.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schjenken J.E., Robertson S.A. The female response to seminal fluid. Physiol. Rev. 2020;100:1077–1117. doi: 10.1152/physrev.00013.2018. [DOI] [PubMed] [Google Scholar]
- 4.Bromfield J.J., Ibrahim L.A., Rizo J.A. Seminal vesicle gland—overview. In: Skinner M.K., editor. Encyclopedia of Reproduction. 2nd Ed. Academic Press; Oxford: 2018. pp. 341–343. [Google Scholar]
- 5.Davies D.C., Hall G., Hibbitt G., Moore H.D. The removal of the seminal vesicles from the boar and the effects on the semen characteristics. J. Reprod. Fertil. 1975;43:305–312. doi: 10.1530/jrf.0.0430305. [DOI] [PubMed] [Google Scholar]
- 6.Pang S.F., Chow P.H., Wong T.M. The role of the seminal vesicles, coagulating glands and prostate glands on the fertility and fecundity of mice. J. Reprod. Fertil. 1979;56:129–132. doi: 10.1530/jrf.0.0560129. [DOI] [PubMed] [Google Scholar]
- 7.Queen K., Dhabuwala C.B., Pierrepoint C.G. The effect of the removal of the various accessory sex glands on the fertility of male rats. J. Reprod. Fertil. 1981;62:423–426. doi: 10.1530/jrf.0.0620423. [DOI] [PubMed] [Google Scholar]
- 8.Kawano N., Araki N., Yoshida K., Hibino T., Ohnami N., Makino M., Kanai S., Hasuwa H., Yoshida M., Miyado K., Umezawa A. Seminal vesicle protein SVS2 is required for sperm survival in the uterus. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4145–4150. doi: 10.1073/pnas.1320715111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noda T., Fujihara Y., Matsumura T., Oura S., Kobayashi S., Ikawa M. Seminal vesicle secretory protein 7, PATE4, is not required for sperm function but for copulatory plug formation to ensure fecundity. Biol. Reprod. 2019;100:1035–1045. doi: 10.1093/biolre/ioy247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean M.D. Genetic disruption of the copulatory plug in mice leads to severely reduced fertility. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim B.J., Park D.R., Nam T.S., Lee S.H., Kim U.H. Seminal CD38 enhances human sperm capacitation through its interaction with CD31. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess E.A., Ludwick T.M., Martig R.C., Ely F. Influence of seminal vesiculectomy on certain physical and biochemical properties of bovine semen. J. Dairy Sci. 1960;43:256–265. [Google Scholar]
- 13.Ortiz W.G., Rizo J.A., Carvalheira L.R., Ahmed B.M.S., Estrada-Cortes E., Harstine B.R., Bromfield J.J., Hansen P.J. Effects of intrauterine infusion of seminal plasma at artificial insemination on fertility of lactating Holstein cows. J. Dairy Sci. 2019;102:6587–6594. doi: 10.3168/jds.2019-16251. [DOI] [PubMed] [Google Scholar]
- 14.Wong C.L., Lee K.H., Lo K.M., Chan O.C., Goggins W., O W.S., Chow P.H. Ablation of paternal accessory sex glands imparts physical and behavioural abnormalities to the progeny: An in vivo study in the golden hamster. Theriogenology. 2007;68:654–662. doi: 10.1016/j.theriogenology.2007.04.062. [DOI] [PubMed] [Google Scholar]
- 15.Morgan H.L., Watkins A.J. The influence of seminal plasma on offspring development and health. Semin. Cell Dev. Biol. 2020;97:131–137. doi: 10.1016/j.semcdb.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Watkins A.J., Dias I., Tsuro H., Allen D., Emes R.D., Moreton J., Wilson R., Ingram R.J.M., Sinclair K.D. Paternal diet programs offspring health through sperm- and seminal plasma-specific pathways in mice. Proc. Natl. Acad. Sci. U. S. A. 2018;115:10064–10069. doi: 10.1073/pnas.1806333115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane M., Robker R.L., Robertson S.A. Parenting from before conception. Science. 2014;345:756–760. doi: 10.1126/science.1254400. [DOI] [PubMed] [Google Scholar]
- 18.Rocha D.R., Martins J.A., van Tilburg M.F., Oliveira R.V., Moreno F.B., Monteiro-Moreira A.C., Moreira R.A., Araujo A.A., Moura A.A. Effect of increased testicular temperature on seminal plasma proteome of the ram. Theriogenology. 2015;84:1291–1305. doi: 10.1016/j.theriogenology.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Hamilton K.J., Wang T., Coons L.A., Jefferson W.N., Li R., Wang Y., Grimm S.A., Ramsey J.T., Liu L., Gerrish K.E., Williams C.J., Wade P.A., Korach K.S. DNA methylation and transcriptome aberrations mediated by ERα in mouse seminal vesicles following developmental DES exposure. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4189–E4198. doi: 10.1073/pnas.1719010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binder N.K., Sheedy J.R., Hannan N.J., Gardner D.K. Male obesity is associated with changed spermatozoa Cox4i1 mRNA level and altered seminal vesicle fluid composition in a mouse model. Mol. Hum. Reprod. 2015;21:424–434. doi: 10.1093/molehr/gav010. [DOI] [PubMed] [Google Scholar]
- 21.Bromfield J.J., Schjenken J.E., Chin P.Y., Care A.S., Jasper M.J., Robertson S.A. Maternal tract factors contribute to paternal seminal fluid impact on metabolic phenotype in offspring. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2200–2205. doi: 10.1073/pnas.1305609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katen A.L., Sipila P., Mitchell L.A., Stanger S.J., Nixon B., Roman S.D. Epididymal CYP2E1 plays a critical role in acrylamide-induced DNA damage in spermatozoa and paternally mediated embryonic resorptions. Biol. Reprod. 2017;96:921–935. doi: 10.1093/biolre/iox021. [DOI] [PubMed] [Google Scholar]
- 23.Katen A.L., Chambers C.G., Nixon B., Roman S.D. Chronic acrylamide exposure in male mice results in elevated DNA damage in the germline and heritable induction of CYP2E1 in the testes. Biol. Reprod. 2016;95:86. doi: 10.1095/biolreprod.116.139535. [DOI] [PubMed] [Google Scholar]
- 24.Nixon B.J., Stanger S.J., Nixon B., Roman S.D. Chronic exposure to acrylamide induces DNA damage in male germ cells of mice. Toxicol. Sci. 2012;129:135–145. doi: 10.1093/toxsci/kfs178. [DOI] [PubMed] [Google Scholar]
- 25.Welsh M., Moffat L., Jack L., McNeilly A., Brownstein D., Saunders P.T., Sharpe R.M., Smith L.B. Deletion of androgen receptor in the smooth muscle of the seminal vesicles impairs secretory function and alters its responsiveness to exogenous testosterone and estradiol. Endocrinology. 2010;151:3374–3385. doi: 10.1210/en.2009-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Degryse S., de Bock C.E., Demeyer S., Govaerts I., Bornschein S., Verbeke D., Jacobs K., Binos S., Skerrett-Byrne D.A., Murray H.C., Verrills N.M., Van Vlierberghe P., Cools J., Dun M.D. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia. 2018;32:788–800. doi: 10.1038/leu.2017.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nixon B., De Iuliis G.N., Hart H.M., Zhou W., Mathe A., Bernstein I.R., Anderson A.L., Stanger S.J., Skerrett-Byrne D.A., Jamaluddin M.F.B., Almazi J.G., Bromfield E.G., Larsen M.R., Dun M.D. Proteomic profiling of mouse epididymosomes reveals their contributions to post-testicular sperm maturation. Mol. Cell. Proteomics. 2019;18:S91–S108. doi: 10.1074/mcp.RA118.000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nixon B., Johnston S.D., Skerrett-Byrne D.A., Anderson A.L., Stanger S.J., Bromfield E.G., Martin J.H., Hansbro P.M., Dun M.D. Modification of crocodile spermatozoa refutes the tenet that post-testicular sperm maturation is restricted to mammals. Mol. Cell. Proteomics. 2019;18:S59–S76. doi: 10.1074/mcp.RA118.000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dun M.D., Chalkley R.J., Faulkner S., Keene S., Avery-Kiejda K.A., Scott R.J., Falkenby L.G., Cairns M.J., Larsen M.R., Bradshaw R.A., Hondermarck H. Proteotranscriptomic profiling of 231-BR breast cancer cells: Identification of potential biomarkers and therapeutic targets for brain metastasis. Mol. Cell. Proteomics. 2015;14:2316–2330. doi: 10.1074/mcp.M114.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dun M.D., Mannan A., Rigby C.J., Butler S., Toop H.D., Beck D., Connerty P., Sillar J., Kahl R.G.S., Duchatel R.J., Germon Z., Faulkner S., Chi M., Skerrett-Byrne D., Murray H.C. Shwachman-Bodian-Diamond syndrome (SBDS) protein is a direct inhibitor of protein phosphatase 2A (PP2A) activity and overexpressed in acute myeloid leukaemia. Leukemia. 2020;34:3393–3397. doi: 10.1038/s41375-020-0814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douse C.H., Tchasovnikarova I.A., Timms R.T., Protasio A.V., Seczynska M., Prigozhin D.M., Albecka A., Wagstaff J., Williamson J.C., Freund S.M.V., Lehner P.J., Modis Y. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020;11:4940. doi: 10.1038/s41467-020-18761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasch P., Schneider A., Blumenscheit C., Doellinger J. Identification of microorganisms by liquid chromatography-mass spectrometry (LC-MS(1)) and in silico peptide mass libraries. Mol. Cell. Proteomics. 2020;19:2125–2138. doi: 10.1074/mcp.TIR120.002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruckbauer S.T., Minkoff B.B., Yu D., Cryns V.L., Cox M.M., Sussman M.R. Ionizing radiation-induced proteomic oxidation in Escherichia coli. Mol. Cell. Proteomics. 2020;19:1375–1395. doi: 10.1074/mcp.RA120.002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartel N.G., Chew B., Qin J., Xu J., Graham N.A. Deep protein methylation profiling by combined chemical and immunoaffinity approaches reveals novel PRMT1 targets. Mol. Cell. Proteomics. 2019;18:2149–2164. doi: 10.1074/mcp.RA119.001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Federspiel J.D., Greco T.M., Lum K.K., Cristea I.M. Hdac4 interactions in Huntington's disease viewed through the prism of multiomics. Mol. Cell. Proteomics. 2019;18:S92–S113. doi: 10.1074/mcp.RA118.001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Herranz M., Nekulova M., Faktor J., Hernychova L., Kote S., Sinclair E.H., Nenutil R., Vojtesek B., Ball K.L., Hupp T.R. The effects of IFITM1 and IFITM3 gene deletion on IFNgamma stimulated protein synthesis. Cell Signal. 2019;60:39–56. doi: 10.1016/j.cellsig.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasci D., van Ingen H., Scheltema R.A., Heck A.J.R. Histone interaction landscapes visualized by crosslinking mass spectrometry in intact cell nuclei. Mol. Cell. Proteomics. 2018;17:2018–2033. doi: 10.1074/mcp.RA118.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 39.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith C.L., Eppig J.T. The mammalian phenotype ontology: Enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:390–399. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickinson M.E., Flenniken A.M., Ji X., Teboul L., Wong M.D., White J.K., Meehan T.F., Weninger W.J., Westerberg H., Adissu H., Baker C.N., Bower L., Brown J.M., Caddle L.B., Chiani F. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayram H.L., Franco C., Brownridge P., Claydon A.J., Koch N., Hurst J.L., Beynon R.J., Stockley P. Social status and ejaculate composition in the house mouse. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20200083. doi: 10.1098/rstb.2020.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang W.C., Chou C.K., Tsou C.C., Li S.H., Chen C.H., Zhuo Y.X., Hsu W.L., Chen C.H. Comparative proteomic analysis of proteins involved in the tumorigenic process of seminal vesicle carcinoma in transgenic mice. Int. J. Proteomics. 2010;2010:726968. doi: 10.1155/2010/726968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dean M.D., Clark N.L., Findlay G.D., Karn R.C., Yi X., Swanson W.J., MacCoss M.J., Nachman M.W. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Mol. Biol. Evol. 2009;26:1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schjenken J.E., Moldenhauer L.M., Zhang B., Care A.S., Groome H.M., Chan H.Y., Hope C.M., Barry S.C., Robertson S.A. MicroRNA miR-155 is required for expansion of regulatory T cells to mediate robust pregnancy tolerance in mice. Mucosal Immunol. 2020;13:609–625. doi: 10.1038/s41385-020-0255-0. [DOI] [PubMed] [Google Scholar]
- 46.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abràmoff M.D., Magalhães P.J., Ram S.J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 48.Houston B.J., Nixon B., Martin J.H., De Iuliis G.N., Trigg N.A., Bromfield E.G., McEwan K.E., Aitken R.J. Heat exposure induces oxidative stress and DNA damage in the male germ line. Biol. Reprod. 2018;98:593–606. doi: 10.1093/biolre/ioy009. [DOI] [PubMed] [Google Scholar]
- 49.Tremellen K.P., Seamark R.F., Robertson S.A. Seminal transforming growth factor beta1 stimulates granulocyte-macrophage colony-stimulating factor production and inflammatory cell recruitment in the murine uterus. Biol. Reprod. 1998;58:1217–1225. doi: 10.1095/biolreprod58.5.1217. [DOI] [PubMed] [Google Scholar]
- 50.Bennett C.G., Riemondy K., Chapnick D.A., Bunker E., Liu X., Kuersten S., Yi R. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res. 2016;44:3788–3800. doi: 10.1093/nar/gkw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochiai T., Takenaka Y., Kuramoto Y., Kasuya M., Fukuda K., Kimura M., Shimeno H., Misasi R., Hiraiwa M., Soeda S. Molecular mechanism for neuro-protective effect of prosaposin against oxidative stress: Its regulation of dimeric transcription factor formation. Biochim. Biophys. Acta. 2008;1780:1441–1447. doi: 10.1016/j.bbagen.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Zhao T., Ding X., Du H., Yan C. Myeloid-derived suppressor cells are involved in lysosomal acid lipase deficiency-induced endothelial cell dysfunctions. J. Immunol. 2014;193:1942–1953. doi: 10.4049/jimmunol.1301941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brosseau C., Colas L., Magnan A., Brouard S. CD9 tetraspanin: A new pathway for the regulation of inflammation? Front. Immunol. 2018;9:2316. doi: 10.3389/fimmu.2018.02316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adomavicius T., Guaita M., Zhou Y., Jennings M.D., Latif Z., Roseman A.M., Pavitt G.D. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 2019;10:2136. doi: 10.1038/s41467-019-10167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavrik I., Golks A., Krammer P.H. Death receptor signaling. J. Cell Sci. 2005;118:265–267. doi: 10.1242/jcs.01610. [DOI] [PubMed] [Google Scholar]
- 56.Wang X., Proud C.G. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 57.Lamouille S., Connolly E., Smyth J.W., Akhurst R.J., Derynck R. TGF-beta-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J. Cell Sci. 2012;125:1259–1273. doi: 10.1242/jcs.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sampson L.L., Davis A.K., Grogg M.W., Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katen A.L., Stanger S.J., Anderson A.L., Nixon B., Roman S.D. Chronic acrylamide exposure in male mice induces DNA damage to spermatozoa; potential for amelioration by resveratrol. Reprod. Toxicol. 2016;63:1–12. doi: 10.1016/j.reprotox.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Avivar-Valderas A., Wen H.C., Aguirre-Ghiso J.A. Stress signaling and the shaping of the mammary tissue in development and cancer. Oncogene. 2014;33:5483–5490. doi: 10.1038/onc.2013.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delgado M.E., Grabinger T., Brunner T. Cell death at the intestinal epithelial front line. FEBS J. 2016;283:2701–2719. doi: 10.1111/febs.13575. [DOI] [PubMed] [Google Scholar]
- 62.Gudipaty S.A., Conner C.M., Rosenblatt J., Montell D.J. Unconventional ways to live and die: Cell death and survival in development, homeostasis, and disease. Annu. Rev. Cell Dev. Biol. 2018;34:311–332. doi: 10.1146/annurev-cellbio-100616-060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Claydon A.J., Ramm S.A., Pennington A., Hurst J.L., Stockley P., Beynon R. Heterogenous turnover of sperm and seminal vesicle proteins in the mouse revealed by dynamic metabolic labeling. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014993. M111.014993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramm S.A., Edward D.A., Claydon A.J., Hammond D.E., Brownridge P., Hurst J.L., Beynon R.J., Stockley P. Sperm competition risk drives plasticity in seminal fluid composition. BMC Biol. 2015;13:87. doi: 10.1186/s12915-015-0197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iamsaard S., Tongpan S., Yannasithinon S., Arun S., Wu A.T.H., Sukhorum W. Effect of chronic stress on expression and secretion of seminal vesicle proteins in adult rats. Andrologia. 2020;53 doi: 10.1111/and.13800. [DOI] [PubMed] [Google Scholar]
- 66.Kerwin P., Yuan J., von Philipsborn A.C. Female copulation song is modulated by seminal fluid. Nat. Commun. 2020;11:1430. doi: 10.1038/s41467-020-15260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weigmann K. Lifestyle in the sperm: There is growing evidence that epigenetic marks can be inherited. But what is the nature of the information they store and over how many generations do they prevail? EMBO Rep. 2014;15:1233–1237. doi: 10.15252/embr.201439759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.JECFA . 2010. Joint FAO/WHO Expert Committee on Food Additives. Summary Report of the Seventy-Second Meeting, Expert Committee on Food Additives. Rome. [Google Scholar]
- 69.Ghanayem B.I., Witt K.L., El-Hadri L., Hoffler U., Kissling G.E., Shelby M.D., Bishop J.B. Comparison of germ cell mutagenicity in male CYP2E1-null and wild-type mice treated with acrylamide: Evidence supporting a glycidamide-mediated effect. Biol. Reprod. 2005;72:157–163. doi: 10.1095/biolreprod.104.033308. [DOI] [PubMed] [Google Scholar]
- 70.Shelby M.D., Cain K.T., Hughes L.A., Braden P.W., Generoso W.M. Dominant lethal effects of acrylamide in male mice. Mutat. Res. 1986;173:35–40. doi: 10.1016/0165-7992(86)90008-4. [DOI] [PubMed] [Google Scholar]
- 71.Holland N., Ahlborn T., Turteltaub K., Markee C., Moore D., 2nd, Wyrobek A.J., Smith M.T. Acrylamide causes preimplantation abnormalities in embryos and induces chromatin-adducts in male germ cells of mice. Reprod. Toxicol. 1999;13:167–178. doi: 10.1016/s0890-6238(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 72.Marchetti F., Bishop J., Lowe X., Wyrobek A.J. Chromosomal mosaicism in mouse two-cell embryos after paternal exposure to acrylamide. Toxicol. Sci. 2009;107:194–205. doi: 10.1093/toxsci/kfn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koromilas A.E. M(en)TORship lessons on life and death by the integrated stress response. Biochim. Biophys. Acta Gen. Subj. 2019;1863:644–649. doi: 10.1016/j.bbagen.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Tsounapi P., Honda M., Dimitriadis F., Kawamoto B., Hikita K., Muraoka K., Saito M., Sofikitis N., Takenaka A. Impact of antioxidants on seminal vesicles function and fertilizing potential in diabetic rats. Asian J. Androl. 2017;19:639–646. doi: 10.4103/1008-682X.186871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li R., Li H., Rao K., Liu K., Zhang Y., Liu X., Wang T., Wang S., Liu Z., Liu J. Curcumin ameliorates atrophy of seminal vesicle via reduction of oxidative stress in castrated mice. PeerJ. 2019;7 doi: 10.7717/peerj.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hageman J., van Waarde M.A., Zylicz A., Walerych D., Kampinga H.H. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 2011;435:127–142. doi: 10.1042/BJ20101247. [DOI] [PubMed] [Google Scholar]
- 77.Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J.D., Vatner S.F., Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 78.Hageman J., Rujano M.A., van Waarde M.A., Kakkar V., Dirks R.P., Govorukhina N., Oosterveld-Hut H.M., Lubsen N.H., Kampinga H.H. A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell. 2010;37:355–369. doi: 10.1016/j.molcel.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 79.Tong M., Chan K.W., Bao J.Y., Wong K.Y., Chen J.N., Kwan P.S., Tang K.H., Fu L., Qin Y.R., Lok S., Guan X.Y., Ma S. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012;72:6024–6035. doi: 10.1158/0008-5472.CAN-12-1269. [DOI] [PubMed] [Google Scholar]
- 80.Sims A.C., Tilton S.C., Menachery V.D., Gralinski L.E., Schäfer A., Matzke M.M., Webb-Robertson B.J., Chang J., Luna M.L., Long C.E., Shukla A.K., Bankhead A.R., 3rd, Burkett S.E., Zornetzer G., Tseng C.T. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 2013;87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagashima D., Zhang L., Kitamura Y., Ichihara S., Watanabe E., Zong C., Yamano Y., Sakurai T., Oikawa S., Ichihara G. Proteomic analysis of hippocampal proteins in acrylamide-exposed Wistar rats. Arch. Toxicol. 2019;93:1993–2006. doi: 10.1007/s00204-019-02484-9. [DOI] [PubMed] [Google Scholar]
- 82.Barber D.S., Stevens S., LoPachin R.M. Proteomic analysis of rat striatal synaptosomes during acrylamide intoxication at a low dose rate. Toxicol. Sci. 2007;100:156–167. doi: 10.1093/toxsci/kfm210. [DOI] [PubMed] [Google Scholar]
- 83.Simanainen U., McNamara K., Davey R.A., Zajac J.D., Handelsman D.J. Severe subfertility in mice with androgen receptor inactivation in sex accessory organs but not in testis. Endocrinology. 2008;149:3330–3338. doi: 10.1210/en.2007-1805. [DOI] [PubMed] [Google Scholar]
- 84.Deanesly R., Parkes A.S. Size changes in the seminal vesicles of the mouse during development and after castration. J. Physiol. 1933;78:442–450. doi: 10.1113/jphysiol.1933.sp003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Camacho L., Latendresse J.R., Muskhelishvili L., Patton R., Bowyer J.F., Thomas M., Doerge D.R. Effects of acrylamide exposure on serum hormones, gene expression, cell proliferation, and histopathology in male reproductive tissues of Fischer 344 rats. Toxicol. Lett. 2012;211:135–143. doi: 10.1016/j.toxlet.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anvari M., Talebi A.R., Mangoli E., Shahedi A., Ghasemi M.R., Pourentezari M. Effects of acrylamide in the presence of vitamin E on sperm parameters, chromatin quality, and testosterone levels in mice. Clin. Exp. Reprod. Med. 2020;47:101–107. doi: 10.5653/cerm.2019.03230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bianco J.J., Handelsman D.J., Pedersen J.S., Risbridger G.P. Direct response of the murine prostate gland and seminal vesicles to estradiol. Endocrinology. 2002;143:4922–4933. doi: 10.1210/en.2002-220493. [DOI] [PubMed] [Google Scholar]
- 88.Robertson S.A., Mau V.J., Tremellen K.P., Seamark R.F. Role of high molecular weight seminal vesicle proteins in eliciting the uterine inflammatory response to semen in mice. J. Reprod. Fertil. 1996;107:265–277. doi: 10.1530/jrf.0.1070265. [DOI] [PubMed] [Google Scholar]
- 89.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Le Bacquer O., Queniat G., Gmyr V., Kerr-Conte J., Lefebvre B., Pattou F. mTORC1 and mTORC2 regulate insulin secretion through Akt in INS-1 cells. J. Endocrinol. 2013;216:21–29. doi: 10.1530/JOE-12-0351. [DOI] [PubMed] [Google Scholar]
- 91.Morrison M.M., Young C.D., Wang S., Sobolik T., Sanchez V.M., Hicks D.J., Cook R.S., Brantley-Sieders D.M. mTOR directs breast morphogenesis through the PKC-alpha-Rac1 signaling axis. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jankiewicz M., Groner B., Desrivières S. Mammalian target of Rapamycin regulates the growth of mammary epithelial cells through the inhibitor of deoxyribonucleic acid binding Id1 and their functional differentiation through Id2. Mol. Endocrinol. 2006;20:2369–2381. doi: 10.1210/me.2006-0071. [DOI] [PubMed] [Google Scholar]
- 93.Pauloin A., Chanat E. Prolactin and epidermal growth factor stimulate adipophilin synthesis in HC11 mouse mammary epithelial cells via the PI3-kinase/Akt/mTOR pathway. Biochim. Biophys. Acta. 2012;1823:987–996. doi: 10.1016/j.bbamcr.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 94.Serrano I., McDonald P.C., Lock F.E., Dedhar S. Role of the integrin-linked kinase (ILK)/Rictor complex in TGFbeta-1-induced epithelial-mesenchymal transition (EMT) Oncogene. 2013;32:50–60. doi: 10.1038/onc.2012.30. [DOI] [PubMed] [Google Scholar]
- 95.Schell C., Kretz O., Liang W., Kiefer B., Schneider S., Sellung D., Bork T., Leiber C., Rüegg M.A., Mallidis C., Schlatt S., Mayerhofer A., Huber T.B., Grahammer F. The rapamycin-sensitive complex of mammalian target of Rapamycin is essential to maintain male fertility. Am. J. Pathol. 2016;186:324–336. doi: 10.1016/j.ajpath.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 96.Cai W., Andres D.A. mTORC2 is required for rit-mediated oxidative stress resistance. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang R.-H., Kim H.-S., Xiao C., Xu X., Gavrilova O., Deng C.-X. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang H.S., Sim H.J., Cho H., Bang W.Y., Kim H.E., Kwon T.K., Kwon T., Park T.J. Alpha-tocopherol exerts protective function against the mucotoxicity of particulate matter in amphibian and human goblet cells. Sci. Rep. 2020;10:6224. doi: 10.1038/s41598-020-63085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim S.C., Lee H.J., Joo J.H., Yoon J.H., Choi J.Y. Vitamin A deficiency induces fluid hyposecretion from the airway submucosal glands of mice. J. Nutr. 2012;142:739–743. doi: 10.3945/jn.111.154047. [DOI] [PubMed] [Google Scholar]
- 100.Shea-Donohue T., Notari L., Stiltz J., Sun R., Madden K.B., Urban J.F., Jr., Zhao A. Role of enteric nerves in immune-mediated changes in protease-activated receptor 2 effects on gut function. Neurogastroenterol. Motil. 2010;22:e1138–e1291. doi: 10.1111/j.1365-2982.2010.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Winkler I.G., Hendy J., Coughlin P., Horvath A., Levesque J.P. Serine protease inhibitors serpina1 and serpina3 are down-regulated in bone marrow during hematopoietic progenitor mobilization. J. Exp. Med. 2005;201:1077–1088. doi: 10.1084/jem.20042299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Takahara H., Sinohara H. Mouse plasma trypsin inhibitors: Inhibitory spectrum of contrapsin and alpha-1-antitrypsin. Thromb. Res. 1982;27:45–50. doi: 10.1016/0049-3848(82)90276-6. [DOI] [PubMed] [Google Scholar]
- 103.Vassalli J.-D., Huarte J., Bosco D., Sappino A.-P., Sappino N., Velardi A., Wohlwend A., Ernø H., Monard D., Belin D. Protease-nexin I as an androgen-dependent secretory product of the murine seminal vesicle. EMBO J. 1993;12:1871–1878. doi: 10.1002/j.1460-2075.1993.tb05835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laflamme B.A., Wolfner M.F. Identification and function of proteolysis regulators in seminal fluid. Mol. Reprod. Dev. 2013;80:80–101. doi: 10.1002/mrd.22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (105) with the dataset identifier PXD021998 and 10.6019/PXD021998.