Abstract
The receptor‐binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 spike (S) protein plays a central role in mediating the first step of virus infection to cause disease: virus binding to angiotensin‐converting enzyme 2 (ACE2) receptors on human host cells. Therefore, S/RBD is an ideal target for blocking and neutralization therapies to prevent and treat coronavirus disease 2019 (COVID‐19). Using a target‐based selection approach, we developed oligonucleotide aptamers containing a conserved sequence motif that specifically targets S/RBD. Synthetic aptamers had high binding affinity for S/RBD‐coated virus mimics (K D≈7 nM) and also blocked interaction of S/RBD with ACE2 receptors (IC50≈5 nM). Importantly, aptamers were able to neutralize S protein‐expressing viral particles and prevent host cell infection, suggesting a promising COVID‐19 therapy strategy.
Keywords: aptamers, COVID-19, receptor-binding domain (RBD), SARS-CoV-2, virus neutralization
Synthetic ssDNA aptamers containing a conserved sequence motif that specifically targets the receptor‐binding domain (RBD) of the SARS‐CoV‐2 spike have the capacity to neutralize the virus and prevent host cell infection in vitro, suggesting a new therapeutic approach to treat COVID‐19.
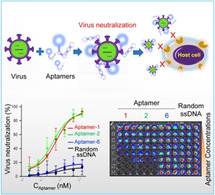
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a novel coronavirus recently identified as the causative agent of coronavirus disease 2019 (COVID‐19), [1] a respiratory disease exhibiting a wide range of clinical outcomes from mild disease to severe viral pneumonia and acute respiratory distress syndrome.[ 1b , 2 ] Trimeric spike (S) proteins are densely glycosylated molecules on the surface of SARS‐CoV‐2. [3] The receptor‐binding domain (RBD) of S protein mediates binding of the virus to angiotensin‐converting enzyme 2 (ACE2) receptors on host cells, which is the first step in cell entry and host infection.[ 3 , 4 ] Interaction of S/RBD with host cell ACE2 receptors involves dramatic conformational changes in S protein.[ 4a , 5 ] Because of its critical function in host cell entry and virus dissemination, S/RBD is an ideal target for the development of vaccines, [6] neutralizing antibodies, [7] and blocking inhibitors.[ 4b , 8 ] Aptamers are small‐molecule ligands comprised of short, single‐stranded oligonucleotides. [9] Target‐specific aptamers can be developed from synthetic ssRNA/ssDNA libraries via Systematic Evolution of Ligands by Exponential (SELEX) enrichment. [10] Through their unique three‐dimensional structures, aptamers specifically recognize and bind to a variety of targets with high affinity, similar to antigen‐antibody interactions. [11] Recently, aptamers that target S/RBD with high affinity have been described. [12] However, the extent to which aptamers may effectively neutralize SARS‐CoV‐2 has not yet been explored.
Herein we report the development of ssDNA aptamers specific for viral S/RBD, which have capacity to neutralize SARS‐CoV‐2 virus and prevent host cell infection in vitro.
Results and Discussion
Characterization of aptamer sequences specific for viral S/RBD. To develop neutralizing aptamers against SARS‐CoV‐2, virus mimics were generated by conjugating purified His‐tagged S/RBD proteins to Ni‐Sepharose beads. SELEX was performed using S/RBD‐coated virus mimics as targets (Figure S1a) and an ssDNA library consisting of a 40‐mer random core sequence and 18‐mer constant arm sequences at both ends (Figure 1 b). [13] Six rounds of combined enrichment and counter‐selection steps were performed (Figure S1b). To monitor SELEX progression, aptamer pools derived from each SELEX round were PCR‐amplified and the evolving capacity to bind virus mimics was quantified (Figure S1c–e). The final aptamer pool products were assessed by next‐generation sequencing and the enrichment progression from SELEX rounds 4 to 6 (R4 to R6) was calculated (Table S1). Phylogenetic trees of aptamers were generated from the 50 most predominant sequences in the final aptamer pool (R6) and potential motif sequences in each cluster of aptamer trees were determined (Figure 1 a). From over 100 000 sequencing reads, three distinct sequence motifs accounting for 90 %, 3 %, and 1 % of the total were identified. All motifs were found within central core sequences without involvement of constant arm sequences (Figure S2). For functional validation, two representative aptamer sequences from the Motif 1 cluster (aptamers‐1 and ‐2) and one sequence from the Motif 2 cluster (aptamer‐6) were selected and listed along with the ssDNA library (Figure 1 b). Minimum free energy secondary structures were predicted for selected aptamer sequences (Figure 1 c). Further analysis revealed that the core sequences of Motif 1‐derived aptamers‐1 and ‐2 had very similar secondary structures while the core sequence of Motif 2‐derived aptamer‐6 differed significantly from these two (Figure 1 d).
Figure 1.
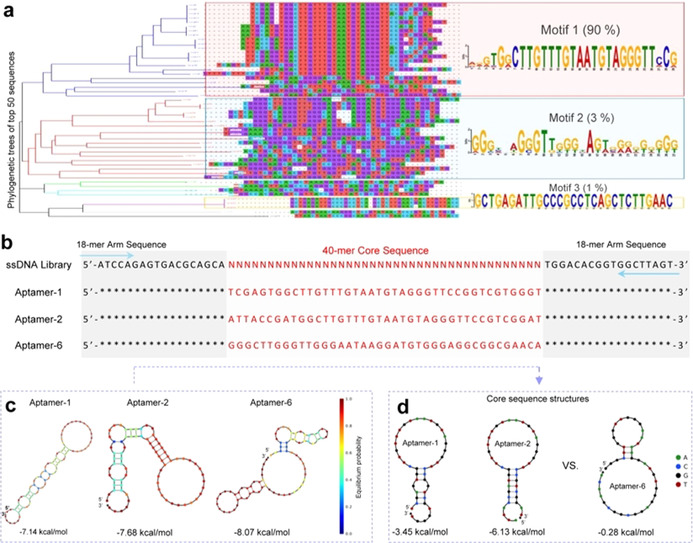
Aptamer sequences specific for viral S/RBD. a) Aptamers were developed using a target‐based enrichment process and final products were sequenced. The top 50 predominant aptamer sequences were selected from over 100 000 reads. Phylogenetic tree analysis identified three well‐preserved sequence motifs. b) Aptamers‐1 and ‐2 were derived from the Motif 1 sequence and aptamer‐6 from the Motif 2 sequence. Sequences of the ssDNA library used for aptamer development are also shown. Blue arrows indicate 18‐mer consistent arm sequences at both ends for primer annealing of PCR amplification. Red lettering indicates central cores composed of 40‐mer specific sequences of the aptamers or random sequences in the Random ssDNA library. c) Minimum free energy secondary structures of aptamers‐1, ‐2, and ‐6. d) Core sequence comparison confirms different secondary structures for aptamers derived from Motif 1 and 2 sequences.
Functional analysis of aptamers to bind S/RBD and spike proteins. For functional assessment, selected aptamer sequences were synthesized and labeled with a Cy3 fluorescent reporter at 5′ end. Individual synthetic aptamers were incubated with virus mimics at room temperature (RT) for 25 min and resultant binding was quantified by flow cytometry. Motif 1‐derived aptamers‐1 and ‐2 targeted S/RBD‐virus mimics with high affinities, K D=6.05±2.05 nM and 6.95±1.10 nM, respectively (Figure 2 a). Notably, Motif 2‐derived aptamer‐6 had a similar K D value, 7.52±3.20 nM, but significantly lower maximal binding capacity (Bmax) about one third of aptamers‐1 and ‐2. All aptamers also targeted S protein‐coated virus mimics (Figure 2 b) but did not bind to control His‐tag beads (Figure 2 c). To determine whether the aptamers targeted the same site/epitope on S/RBD, competition binding assays were performed using individual aptamer probes in the presence of equal amounts of unlabeled competitive aptamer sequences. Motif 1‐derived aptamers‐1 and ‐2 competed for binding sites on virus mimics. However, only small competitive effects were noted between these aptamers and Motif 2‐derived aptamer‐6, indicating that sequence motifs likely drive aptamer target specificity or Bmax (Figure 2 d). In addition, aptamers were tested under variable conditions to confirm biocompatibility. As functional oligonucleotides, aptamers depend on magnesium (Mg2+) ions for target binding activity. Aptamer binding to virus mimics was fully functional within the physiologic Mg2+ concentration range, 0.65–1.10 mmol L−1 (Figure 2 e). Further, aptamers similarly targeted S/RBD at temperatures ranging from 4–37 °C (Figure 2 f), indicating that they can function both in vitro and in vivo. Moreover, aptamers did not react with mixed culture cells or blood cells (Figure S3a, S3b) and were stable in human serum at 37 °C for 24 h (Figure S5), suitable for clinical use.
Figure 2.
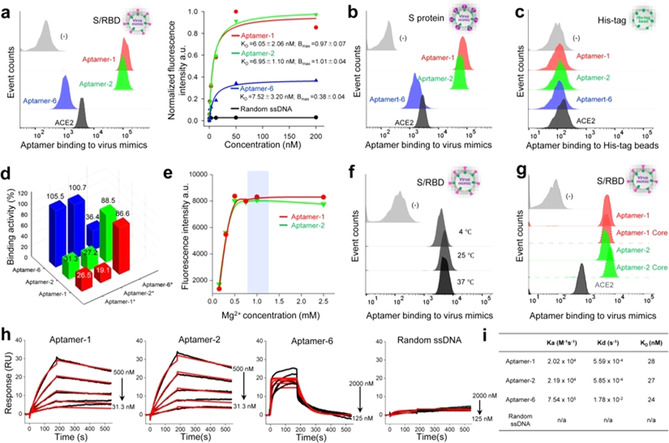
Functional characterization of aptamers. a) Binding assays of selected aptamers to S/RBD‐virus mimics, which were used for target‐based SELEX. Flow cytometry reveals that aptamers‐1 and ‐2 containing Motif 1 sequences bound virus mimics with high capacity, while aptamer‐6, containing the Motif 2 sequence, had significantly lower binding capacity. b) Aptamers targeted S protein‐virus mimics with identical pattern. c) No aptamers reacted with control His‐tag beads under the same experimental conditions. d) Competition assays demonstrated that aptamers‐1 and ‐2 competed with each other for virus mimic binding but not with aptamer‐6, suggesting that aptamers‐1 and ‐2 target different S/RBD sites/epitopes than aptamer‐6. e) Aptamers‐1 and ‐2 were fully functional within the physiologic concentration range of magnesium. f) Aptamer‐1 had the same target binding capacity at 4 °C, 25 °C, and 37 °C. g) The central core sequences of aptamers‐1 and ‐2 possessed full S/RBD binding capacity. h) Sensorgrams and kinetic binding parameters of aptamer binding to immobilized SARS‐CoV‐2 S proteins. Black lines: raw data; red lines: 1:1 Langmuir fitting. i) SPR studies reveal high binding capacity of aptamers‐1 and ‐2 with very similar kinetics of association and dissociation constants (k a and k d, respectively). In contrast, aptamer‐6 had faster k a and k d kinetics. Although aptamers showed significantly different k a and k d, they had similar K D values ranging from 24–28 nM.
To determine the functional sequences of the aptamers, 40‐mer core sequences containing consensus Motif 1 structures were synthesized (Figure 1 d). Binding assays revealed that the core sequences had the same capacity to bind virus mimics as the full‐length aptamers (Figure 2 g). To characterize target binding kinetics, surface plasmon resonance (SPR) studies were performed with purified and immobilized SARS‐CoV‐2 S proteins. Aptamers‐1 and ‐2 had similar association and dissociation constants (k a and k d) (Figure 2 h). In contrast, aptamer‐6 showed faster k a and k d kinetics although its K D value was nearly identical to aptamers‐1 and ‐2 (Figure 2 i).
Aptamers block S/RBD‐ACE2 interaction and neutralize viral particles. To effectively neutralize SARS‐CoV‐2, aptamers need not only to specifically target S/RBD but also block its interaction with ACE2 receptors. To determine the ability of the aptamers to block S/RBD‐ACE2 interactions, enzyme‐linked immunosorbent assays (ELISA) were performed. First, biotinylated S/RBD proteins were mixed with serial dilutions of aptamers and then added into microplates pre‐coated with purified ACE2 receptor proteins to mimic the host cell surface (Figure 3 a). After incubation at RT for 30 min, resultant S/RBD‐ACE2 binding was quantified by measuring reaction color intensity with a microplate reader. Aptamers‐1 and ‐2 blocked S/RBD binding to ACE2 in a dose‐dependent manner. In contrast, aptamer‐6 did not affect S/RBD‐ACE2 interactions. Similar findings were observed in binding assays using microplates pre‐coated with S/RBD or S protein to mimic virus surface (Figure 3 b,c). Notably, aptamer‐mediated blocking effects occurred at 4 °C, 25 °C, and 37 °C (Figure S3c), indicating that aptamers are suitable for in vivo use. Next, HEK293T cells stably expressing ACE2 (ACE‐293T) were used as host cells in blocking assays. Biotinylated S/RBD proteins were mixed with serially diluted aptamers and then incubated with cultured ACE‐ 293T host cells at RT for 1 h (Figure 3 d). For reporting purposes, treated cells were stained with Cy3‐labeled streptavidin and resultant changes in host cell binding of S/RBD were quantified by flow cytometry. Treatments with aptamers‐1 and ‐2 blocked S/RBD protein binding to host cells with IC50=5.2 nM and 4.4 nM, respectively. Aptamer‐6 had minimal blocking effects and control random ssDNA sequences had no effect. Aptamer‐induced blockage of S/RBD‐host cell interaction was also confirmed by fluorescent microscopic examination of treated cells. To assess the ability of aptamers to target native S proteins, we generated pseudoviruses which are recombinant lentiviral particles expressing surface SARS‐CoV‐2 S proteins and carrying a firefly luciferase reporter gene. Serially diluted pseudovirus was pre‐immobilized on microplates and then treated with biotinylated aptamers (Figure 3 e). Biotinylated ACE2 receptor proteins were used as a positive control probe. Quantitative ELISA analysis demonstrated that aptamers bound viral particles in a virus dose‐dependent manner, similar to results achieved with ACE2.
Figure 3.
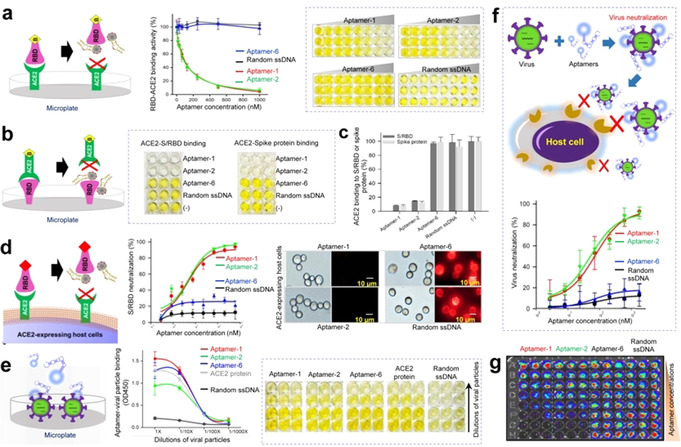
Aptamers block S/RBD‐ACE2 interaction and neutralize viral particles to prevent host cell infection. a) Schematic depicting aptamer‐mediated blockade of S/RBD‐ACE2 interaction. ELISA reveals that aptamers‐1 and ‐2 block S/RBD binding to ACE2 receptor proteins precoated on microplates (mimicking host cell surface). In contrast, aptamer‐6 and random ssDNA sequences have no blocking effects under the same conditions. b) Aptamers‐1 and ‐2 prevent ACE2 binding to S/RBD and c) S proteins precoated on microplates (mimicking virus surface). d) Flow cytometry and fluorescent microscopy demonstrate that aptamers‐1 and ‐2 interrupt S/RBD binding to ACE2‐expressing host cells. Aptamer‐6 shows minimal blocking effects and random ssDNA sequences have no effect. e) Aptamers specifically target S protein‐expressing viral particles in a virus dose‐dependent manner, similar to the pattern observed with ACE2 protein. f) Virus neutralization assays. Aptamers‐1 and ‐2 effectively neutralize viral particles and prevent host cell infection, while aptamer‐6 and control random ssDNA sequences do not. g) Aptamer virus neutralization effects were also confirmed post‐treatment using bioluminescent imaging of microplates, which contained intact host cells in the presence of luciferin for signal development.
Finally, to interrogate aptamer therapeutic potential, virus neutralization assays were performed using a paired S protein‐expressing viral particle and ACE‐293T host cell system. Because the viral particles carry the luciferase reporter gene, viral infection results in intracellular expression of luciferase in host cells. Accordingly, the activity of cellular luciferase is proportional to viral infection intensity and the number of infected cells. ACE‐293T host cells were pre‐seeded into microplates overnight. Viral particles mixed with serial dilutions of aptamers were added to microplates containing host cells. After a short spinoculation and incubation at 37 °C for 2 h to allow virus infection, old medium was replaced with fresh and cells were further cultured at 37 °C for 72 h. To evaluate viral infection rates, treated host cells were lysed in microplates and cellular luciferase activity was evaluated using a Luciferin assay kit. Resultant signals were detected using a microplate luminometer. Quantitative analysis revealed that aptamers‐1 and ‐2 neutralized viral particles and prevented host cell infection with neutralization IC50=76.9 nM and 53.0 nM, respectively (Figure 3 f). Aptamer‐6 and random ssDNA sequences had little or no neutralization effects under the same experimental conditions. These findings were also confirmed by post‐treatment bioluminescent imaging of microplates, which contained intact host cells in the presence of luciferin for signal development (Figure 3 g).
To assess clinical potential of the aptamers, we conducted a pilot study using the conventional microneutralization assay system composed of primary SARS‐CoV‐2 strain USA‐WA1/2020 and host Vero E6 cells. [14] Figure S6 reveals that treatments of aptamers‐1 and ‐2 resulted in virus neutralization and efficiently protected host cells from virus infection. In contract, control aptamer‐6 and random ssDNA sequences had no effect on host cell infection by primary virus under the same condition. To understand whether our aptamers also neutralize other coronaviruses, additional studies are undergoing currently to determine binding specificity and affinity of the aptamers to SARS and MERS, as well as the variants of SARS‐CoV‐2. Resultant findings will be reported in near future separately.
For virus neutralization, aptamers need not only to specifically target S/RBD but also block its interaction with ACE2 receptors. Functional assays revealed that, although aptamer‐6 possesses very similar K D to aptamers‐1 and ‐2 to bind S/RBD, it failed to block the S/RBD‐ACE2 interaction. This failure may be due to its fast k a and k d kinetics that result in lower B max to bind S/RBD and/or variation in targeting sites or epitopes on S/SRBD. For detection of SARS‐CoV‐2, our lab developed aptamers using S protein‐coated virus mimics for target‐based SELEX (Figure S4a, S4b). The selected S aptamers specifically targeted S protein with high binding affinity (Figure S4c) and also bound to S/RBD‐virus mimics with similar pattern to those achieved by ACE2 protein control (Figure S4d). Notably, although S aptamers have the ability to bind to S/RBD, they failed to block the interaction of S/RBD with ACE2‐expressing host cells (Figure S4e). A likely explanation for this is that the S/RBD site(s) targeted by S aptamers is/are not involved in the S/RBD‐ACE2 interaction. In addition, we have observed that 40‐mer core sequences of aptamers‐1 and ‐2 can neutralize S/RBD and prevent its binding to viral particles, indicating that failure to block S/RBD function is not due to the small size of aptamers (unpublished data). Therefore, future studies aiming to identify neutralizing aptamers against SARS‐CoV‐2 should evaluate binding kinetic features, B max, and S/RBD targeting sites or epitopes.
To enhance the degree of effective virus neutralization, aptamer cocktails that allow targeting of multiple S/RBD epitopes may show promise. To this end, identification of additional aptamers containing different motif sequences specific for S/RBD with favorable binding kinetics and blocking capacity is required. Because of their synthetic oligonucleotide properties, multivalent aptamer nanostructures can be formulated through covalent or non‐covalent methods. Such aptamer polymer nanostructures can simultaneously target multiple S/RBD molecules on individual viruses and, thus, may augment binding affinity to achieve higher virus neutralization efficacy.
Conclusion
In summary, this proof‐of‐concept study demonstrates that synthetic ssDNA aptamers can block S/RBD‐ACE2 interactions, neutralize SARS‐CoV‐2 virus, and prevent host cell infection in vitro. The development of neutralizing aptamers provides a new promising therapeutic approach to treat COVID‐19, in addition to neutralization antibodies and molecular blockers. To validate the clinical utility of neutralizing aptamers, our lab is pursuing preclinical studies with authentic SARS‐CoV‐2 virus and primary natural host cells is indispensable. Notably, while this submission was under review, a related study was published. [15]
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We would like to thank Dr. Sasha M. Pejerrey and Adrienne Winston for their scientific editing of the manuscript.
X. Liu, Y.-l. Wang, J. Wu, J. Qi, Z. Zeng, Q. Wan, Z. Chen, P. Manandhar, V. S. Cavener, N. R. Boyle, X. Fu, E. Salazar, S. V. Kuchipudi, V. Kapur, X. Zhang, M. Umetani, M. Sen, R. C. Willson, S.-h. Chen, Y. Zu, Angew. Chem. Int. Ed. 2021, 60, 10273.
References
- 1.
- 1a. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270–273; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Zhu N., Zhang D. Y., Wang W. L., Li X. W., Yang B., Song J. D., Zhao X., Huang B. Y., Shi W. F., Lu R. J., Niu P. H., Zhan F. X., Ma X. J., Wang D. Y., Xu W. B., Wu G. Z., Gao G. G. F., Tan W. J., N. Engl. J. Med. 2020, 382, 727–733; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1c. Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z., Nature 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Huang C. L., Wang Y. M., Li X. W., Ren L. L., Zhao J. P., Hu Y., Zhang L., Fan G. H., Xu J. Y., Gu X. Y., Cheng Z. S., Yu T., Xia J. A., Wei Y., Wu W. J., Xie X. L., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J. G., Wang G. F., Jiang R. M., Gao Z. C., Jin Q., Wang J. W., Cao B., Lancet 2020, 395, 497–506; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2b. Dong E. S., Du H. R., Gardner L., Lancet Infect. Dis. 2020, 20, 533–534.32087114 [Google Scholar]
- 3. Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Cell 2020, 181, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Yan R. H., Zhang Y. Y., Li Y. N., Xia L., Guo Y. Y., Zhou Q., Science 2020, 367, 1444–1448; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b. Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Muller M. A., Drosten C., Pohlmann S., Cell 2020, 181, 271–280; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Lan J., Ge J. W., Yu J. F., Shan S. S., Zhou H., Fan S. L., Zhang Q., Shi X. L., Wang Q. S., Zhang L. Q., Wang X. Q., Nature 2020, 581, 215–220. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Wrapp D., Wang N. S., Corbett K. S., Goldsmith J. A., Hsieh C. L., Abiona O., Graham B. S., McLellan J. S., Science 2020, 367, 1260–1263; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5b. Walls A. C., Xiong X. L., Park Y. J., Tortorici M. A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., Zambon M., Rey F. A., Corti D., Veesler D., Cell 2019, 176, 1026–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Yang J. Y., Wang W., Chen Z. M., Lu S. Y., Yang F. L., Bi Z. F., Bao L. L., Mo F., Li X., Huang Y., Hong W. Q., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z. Q., Luo M., Gao H., Zheng Y., Gong Y. Q., Jiang X. H., Xu Y. F., Lv Q., Li D., Wang M. N., Li F. D., Wang S. Y., Wang G. P., Yu P., Qu Y. J., Yang L., Deng H. X., Tong A. P., Li J., Wang Z. L., Yang J. L., Shen G. B., Zhao Z. W., Li Y. H., Luo J. W., Liu H. Q., Yu W. H., Yang M. L., Xu J. W., Wang J. B., Li H. Y., Wang H. X., Kuang D. X., Lin P. P., Hu Z. T., Guo W., Cheng W., He Y. L., Song X. R., Chen C., Xue Z. H., Yao S. H., Chen L., Ma X. L., Chen S. Y., Gou M. L., Huang W. J., Wang Y. C., Fan C. F., Tian Z. X., Shi M., Wang F. S., Dai L. Z., Wu M., Li G., Wang G. Y., Peng Y., Qian Z. Y., Huang C. H., Lau J. Y. N., Yang Z. L., Wei Y. Q., Cen X. B., Peng X. Z., Qin C., Zhang K., Lu G. W., Wei X. W., Nature 2020, 586, 572–577; [DOI] [PubMed] [Google Scholar]
- 6b. Yu J. Y., Tostanoski L. H., Peter L., Mercado N. B., McMahan K., Mahrokhian S. H., Nkolola J. P., Liu J. Y., Li Z. F., Chandrashekar A., Martinez D. R., Loos C., Atyeo C., Fischinger S., Burke J. S., Slein M. D., Chen Y. Z., Zuiani A., Lelis F. J. N., Travers M., Habibi S., Pessaint L., Van Ry A., Blade K., Brown R., Cook A., Finneyfrock B., Dodson A., Teow E., Velasco J., Zahn R., Wegmann F., Bondzie E. A., Dagotto G., Gebre M. S., He X., Jacob-Dolan C., Kirilova M., Kordana N., Lin Z. J., Maxfield L. F., Nampanya F., Nityanandam R., Ventura J. D., Wan H. H., Cai Y. F., Chen B., Schmidt A. G., Wesemann D. R., Baric R. S., Alter G., Andersen H., Lewis M. G., Barouch D. H., Science 2020, 369, 806–811; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Tai W. B., He L., Zhang X. J., Pu J., Voronin D., Jiang S. B., Zhou Y. S., Du L. Y., Cell. Mol. Immunol. 2020, 17, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.
- 7a. Huo J. D., Le Bas A., Ruza R. R., Duyvesteyn H. M. E., Mikolajek H., Malinauskas T., Tan T. K., Rijal P., Dumoux M., Ward P. N., Ren J. S., Zhou D. M., Harrison P. J., Weckener M., Clare D. K., Vogirala V. K., Radecke J., Moynie L., Zhao Y. G., Gilbert-Jaramillo J., Knight M. L., Tree J. A., Buttigieg K. R., Coombes N., Elmore M. J., Carroll M. W., Carrique L., Shah P. N. M., James W., Townsend A. R., Stuart D. I., Owens R. J., Naismith J. H., Nat. Struct. Mol. Biol. 2020, 27, 846–854; [DOI] [PubMed] [Google Scholar]
- 7b. Liu L. H., Wang P. F., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F. W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T. Q., Chen Z. W., Yuen K. Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z. Z., Huang Y. X., Shapiro L., Ho D. D., Nature 2020, 584, 450–456; [DOI] [PubMed] [Google Scholar]
- 7c. Chi X. Y., Yan R. H., Zhang J., Zhang G. Y., Zhang Y. Y., Hao M., Zhang Z., Fan P. F., Dong Y. Z., Yang Y. L., Chen Z. S., Guo Y. Y., Zhang J. L., Li Y. N., Song X. H., Chen Y., Xia L., Fu L., Hou L. H., Xu J. J., Yu C. M., Li J. M., Zhou Q., Chen W., Science 2020, 369, 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.
- 8a. Su H., Zhou F., Huang Z., Ma X., Natarajan K., Zhang M., Huang Y., Su H., Angew. Chem. Int. Ed. 2021, https:doi/full/10.1002/anie.202008835; Angew. Chem. 2021, https:doi/full/10.1002/ange.202008835; [Google Scholar]
- 8b. de Oliveira O. V., Rocha G. B., Paluch A. S., Costa L. T., J. Biomol. Struct. Dyn. 2020, 10.1080/07391102.2020.1772885; [DOI] [Google Scholar]
- 8c. Huo J. D., Zhao Y. G., Ren J. S., Zhou D. M., Duyvesteyn H. M. E., Ginn H. M., Carrique L., Malinauskas T., Ruza R. R., Shah P. N. M., Tan T. K., Rijal P., Coombes N., Bewley K. R., Tree J. A., Radecke J., Paterson N. G., Supasa P., Mongkolsapaya J., Screaton G. R., Carroll M., Townsend A., Fry E. E., Owens R. J., Stuart D. I., Cell Host Microbe 2020, 28, 497–497; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8d. Sacco M. D., Ma C., Lagarias P., Gao A., Townsend J. A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M. T., Kolocouris A., Xiang Y., Chen Y., Wang J., Sci. Adv. 2020, 6, eabc0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robertson D. L., Joyce G. F., Nature 1990, 344, 467–468. [DOI] [PubMed] [Google Scholar]
- 10.
- 10a. Tuerk C., Gold L., Science 1990, 249, 505–510; [DOI] [PubMed] [Google Scholar]
- 10b. Ellington A. D., Szostak J. W., Nature 1990, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Cerchia L., Esposito C. L., Camorani S., Rienzo A., Stasio L., Insabato L., Affuso A., de Franciscis V., Mol. Ther. 2012, 20, 2291–2303; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Tran P. H. L., Xiang D. X., Nguyen T. N. G., Tran T. T. D., Chen Q., Yin W., Zhang Y. M., Kong L. X., Duan A., Chen K. S., Sun M. M., li Y., Hou Y. C., Zhu Y. M., Ma Y. C., Jiang G. Q., Duan W., Theranostics 2020, 10, 3849–3866; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11c. Muharemagic D., Zamay A., Ghobadloo S. M., Evgin L., Savitskaya A., Bell J. C., Berezovski M. V., Mol. Ther. Nucl. Acids 2014, 3, e167; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11d. He W. G., Elizondo-Riojas M. A., Li X., Lokesh G. L. R., Somasunderam A., Thiviyanathan V., Volk D. E., Durland R. H., Englehardt J., Cavasotto C. N., Gorenstein D. G., Biochemistry 2012, 51, 9592–9592; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11e. Zhou J. H., Rossi J., Nat. Rev. Drug Discovery 2017, 16, 181–202; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11f. Culver H. R., Clegg J. R., Peppas N. A., Acc. Chem. Res. 2017, 50, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Song Y. L., Song J., Wei X. Y., Huang M. J., Sun M., Zhu L., Lin B. Q., Shen H. C., Zhu Z., Yang C. Y., Anal. Chem. 2020, 92, 9895–9900; [DOI] [PubMed] [Google Scholar]
- 12b. Zhang L. Y., Fang X. N., Liu X. B., Ou H. C., Zhang H. Y., Wang J. J., Li Q., Cheng H. Y., Zhang W. Y., Luo Z. F., Chem. Commun. 2020, 56, 10235–10238. [DOI] [PubMed] [Google Scholar]
- 13. Sefah K., Shangguan D., Xiong X. L., O'Donoghue M. B., Tan W. H., Nat. Protoc. 2010, 5, 1169–1185. [DOI] [PubMed] [Google Scholar]
- 14. Salazar E., Kuchipudi S. V., Christensen P. A., Eagar T., Yi X., Zhao P., Jin Z., Long S. W., Olsen R. J., Chen J., Castillo B., Leveque C., Towers D., Lavinder J., Gollihar J., Cardona J., Ippolito G., Nissly R., Bird I., Greenawalt D., Rossi R. M., Gontu A., Srinivasan S., Poojary I., Cattadori I. M., Hudson P. J., Josleyn N. M., Prugar L., Huie K., Herbert A., Bernard D. W., Dye J. M., Kapur V., Musser J. M., J. Clin. Invest. 2020, 130, 6728–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun M., Liu S., Wei X., Wan S., Huang M., Song T., Lu Y., Weng X., Lin Z., Chen H., Song Y., Yang C., Angew. Chem. Int. Ed. 2021, 10.1002/anie.202100225; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 10.1002/ange.202100225. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


