Abstract
3q29 deletion syndrome (3q29del) is a recurrent deletion syndrome associated with neuropsychiatric disorders and congenital anomalies. Dysmorphic facial features have been described but not systematically characterized. This study aims to detail the 3q29del craniofacial phenotype and use a machine learning approach to categorize individuals with 3q29del through analysis of 2D photos. Detailed dysmorphology exam and 2D facial photos were ascertained from 31 individuals with 3q29del. Photos were used to train the next‐generation phenotyping algorithm DeepGestalt (Face2Gene by FDNA, Inc, Boston, MA) to distinguish 3q29del cases from controls and all other recognized syndromes. Area under the curve of receiver operating characteristic curves (AUC‐ROC) was used to determine the capacity of Face2Gene to identify 3q29del cases against controls. In this cohort, the most common observed craniofacial features were prominent forehead (48.4%), prominent nose tip (35.5%), and thin upper lip vermillion (25.8%). The FDNA technology showed an ability to distinguish cases from controls with an AUC‐ROC value of 0.873 (p = 0.006) and led to the inclusion of 3q29del as one of the supported syndromes. This study found a recognizable facial pattern in 3q29del, as observed by trained clinical geneticists and next‐generation phenotyping technology. These results expand the potential application of automated technology such as FDNA in identifying rare genetic syndromes, even when facial dysmorphology is subtle.
Keywords: 3q29 deletion syndrome, craniofacial features, Face2Gene, facial dysmorphism
1. INTRODUCTION
3q29 deletion syndrome (OMIM #609425, hg19, chr3:195725000–197,350,000) is caused by a recurrent 1.6 megabase (Mb) deletion with an estimated prevalence of 1/30,000 births (Stefansson et al., 2013). The deletion is often de novo though up to 30% of cases may be inherited (Cox & Butler, 2015). The driver genes for the syndromic phenotypes are not known, though attention has been focused on the genes PAK2, DLG1, and NCBP2 (Grice et al., 2015; Mulle 2010; Singh et al., 2020). Data from mouse models suggest that haploinsufficiency of DLG1 alone cannot account for the phenotypes seen in 3q29 deletion syndrome (Rutkowski et al., 2019). Details about the phenotypic spectrum of the syndrome are emerging from both case reports and series (Ballif et al., 2008; Cox & Butler, 2015) and systematic studies (Glassford et al., 2016; Sanchez Russo et al., 2021).
The developmental and neuropsychiatric phenotypes associated with 3q29 deletion syndrome are well recognized. A previous Emory 3q29 Project study by Glassford et al. (2016) using self‐reported data from 44 study participants observed that 98% of 3q29 deletion carriers had developmental delay, and 89% of deletion carriers were symptomatic within the first year of life with feeding problems and failure to gain weight. A high incidence of dental problems, recurrent ear infections, and gastrointestinal disorders was also reported (Glassford et al., 2016). Individuals with the 3q29 deletion are at a 40‐fold increased risk for developing schizophrenia (Mulle et al., 2015) and are at increased risk for autism spectrum disorder (ASD) and social disability in even the absence of autism (Sanders et al., 2015; Pollack, 2019). These reports have included limited data about physical and craniofacial characteristics, highlighting the need for further delineation of dysmorphic features in the syndrome.
Most case reports and case series have documented some craniofacial abnormalities from retrospective reviews. A case series of 14 participants with 3q29 deletion syndrome described four participants with high nasal bridge, four with abnormal ear morphology, and five with microcephaly (Ballif et al., 2008). In a registry‐based study of 44 study individuals, self‐report data revealed a high proportion of participants with dental abnormalities including wide spaced teeth and dental crowding (Glassford et al., 2016). To date, a distinguishing facial phenotype associated with 3q29 deletion is not well‐described in the medical literature.
The use of microarrays and next‐generation phenotyping technology in the clinical setting by both genetics and non‐genetics providers is increasing (Michelson & Clark, 2020; Zarate et al., 2019). As one example, the DeepGestalt algorithm used in the Face2Gene (FDNA Inc, MA, USA) platform uses deep learning technologies to identify facial phenotypes in rare disorders. In addition to clinical data, this platform provides a list of potential differential diagnoses that can aid in the diagnosis of various genetic disorders. In the context of 3q29 syndrome, if a subtle, but recognizable, facial phenotype exists and can be detected by next‐generation phenotyping technology, it could be leveraged in the clinic to streamline genetic testing and identify individuals with this, shortening the time to diagnosis and allowing patients faster access to tailored interventions.
We therefore sought to delineate the craniofacial features of 3q29 deletion syndrome and ask whether next‐generation phenotyping technology could be used to identify a characteristic facial dysmorphology associated with 3q29 deletion syndrome. At the time this study was conducted, 3q29 deletion was uncharacterized by Face2Gene, thus the present research study is the first to train the system to recognize this copy number variant (CNV). This study will allow us to add to the growing body of knowledge about 3q29 deletion syndrome and to evaluate the clinical application of next‐generation phenotyping technology.
2. METHODS
Study participant recruitment followed the criteria outlined in the study protocol for the Emory 3q29 Project (Murphy et al., 2018). Study participants were recruited from the 3q29 Deletion Registry (3q29deletion.org), a voluntary registry database housed at Emory University (Glassford et al., 2016). Eligibility criteria are as follows: age of at least 6 years, molecular diagnosis of 3q29 deletion syndrome, English fluency, and ability to travel to Emory University (Atlanta, GA, USA) for a full evaluation (Murphy et al., 2018). One exception to the age criterion was made; a 4.85 year old who was part of a previously described multiplex family was included in the study (Murphy et al., 2020). At the evaluation, a trained clinical geneticist obtained a medical history and completed a detailed physical examination with attention to craniofacial features. 2D frontal and side photos were obtained during the physical examination.
After the visit, two clinical geneticists who conducted the medical history and physical exam discussed and evaluated all captured clinical data and 2D facial photographs for each participant. To standardize this process, each dysmorphic feature captured by the clinical geneticists was mapped to its associated Human Phenotype Ontology (HPO) term (Köhler et al., 2018), the standardized terms for phenotypic abnormalities. Descriptive statistics were used to describe the frequency of each phenotypic abnormality.
Face2Gene has two independent applications: the clinical application, which leverages the Face2Gene database to provide up to 30 suggested differential diagnoses based on craniofacial features, and a research application, that can be used to identify the presence of distinct facial patterns in a syndromic cohort not yet characterized by Face2Gene. We uploaded a total of 31 2D frontal photos, one for each participant in our cohort, into the research application in a de‐identified manner. Using the proprietary DeepGestalt (v.19.1.3) algorithm described by Gurovich et al. (2019), a mathematical representation, the descriptor, of the face was created for 3q29 syndrome. This facial descriptor can be shown as a two‐dimensional mask or composite image. Moreover, this mathematical descriptor can be readily compared to other such descriptors for other syndromes which yields a ranked list of possible differential diagnoses. When comparing the descriptor of the 3q29 syndrome with the descriptor of a particular patient's photo or sets of photos, this comparison can be visualized as a graphical heatmap laid over a composite image to show the degree of similarity between the two facial descriptors being compared.
The descriptor from the 2D facial photos of our subjects was then compared to an equally sized age‐, ethnicity‐, and gender‐matched control set that comprised typically developing individuals with no known genetic syndromes provided by FDNA. To protect anonymity of the control cohort, only the composite photo can be viewed. Both the 3q29 deletion cohort and control cohorts are randomly divided into two sets, a training set, used to train the algorithm, and a test set, used to test the training. This random split is repeated ten times and the results are provided as mean and aggregate results in the form of the area under a receiver operating characteristic curve (AUC‐ROC curve). The results of the binary comparisons are reported both numerically and graphically. A SD is reported for the AUC and a p value is calculated for the aggregate results from the ROC and score distribution. We performed three comparisons and labeled each comparison as a group. Group A compared all cases (n = 31) to all controls (n = 31), Group B compared cases 13 years and over (n = 14) to controls 13 years and over (n = 14), and lastly, Group C compared cases 12 years and under (n = 17) to controls 12 years and under (n = 17). Training of the DeepGestalt algorithm happens periodically, and in light of this study, the current version of the DeepGestalt algorithm does support the 3q29 syndrome.
3. RESULTS
3.1. Demographics
Demographic features for 31 study participants can be seen in Table 1. Twenty (65%) were male; ages ranged from 4 to 39 years (median = 11 years). The cohort was stratified into four age brackets used by Face2Gene (stata in years: 4–6, 7–12, 13–18, 19–40). All four age brackets were represented in our cohort with most participants aged between 7 years and 12 years of age (11 participants, 35.5%). Twenty‐eight (90.3%) of participants identified as Caucasian. The remaining three participants were denoted as mixed race, identifying as Caucasian and one other ethnicity.
TABLE 1.
Demographics (N = 31)
| n | % | ||
|---|---|---|---|
| Sex | |||
| Male | 20 | 64.5% | |
| Female | 11 | 35.5% | |
| Age | |||
| 4y‐6y | 6 | 19.4% | |
| 7y‐12y | 11 | 35.5% | |
| 13y‐18y | 9 | 29.0% | |
| 19y‐40y | 5 | 16.1% | |
| Ethnicity | |||
| Caucasian | 28 | 90.3% | |
| Mixed Race a | 3 | 9.70% |
Participants of mixed race identified as Caucasian and one other ethnicity.
3.2. Dysmorphic craniofacial features
Results from our clinical analysis of observed craniofacial features and 2D are reported in Table 2. In each facial region, dysmorphic features were seen in the majority of study participants as follows: 70.9% of participants had at least one forehead or cranium abnormality, 56.8% had an eye or eyebrow abnormality, 74.2% had a nose or midface abnormality, 61.3% had a lip or oral abnormality, and 67.7% had a mandible or face shape abnormality. Within these regions, the most commonly seen dysmorphic features were prominent forehead (48.4%), wide nose (48.4%), and thin upper lip vermillion (25.8%). Low‐hanging columella (22.6%), prominent nasal bridge (22.6%), and incisor macrodontia (22.6%) were also common (Figure 1). Other observed features were rare and only observed once or twice, particularly other eye, mouth, and ear features (Table S1).
TABLE 2.
Most commonly described craniofacial features of 3q29 deletion cohort (N = 31)
| HPO ID | HPO term | Totals (n) | % (N = 31) |
|---|---|---|---|
| HP:0000252 | Microcephaly | 5 | 16.1% |
| HP:0011220 | Prominent forehead | 15 | 48.4% |
| HP:0000582 | Upslanted palpebral fissure | 4 | 12.9% |
| HP:0000490 | Deeply set eyes | 7 | 22.6% |
| HP:0000368 | Low‐set, posteriorly rotated ears | 4 | 12.9% |
| HP:0000272 | Malar flattening | 5 | 16.1% |
| HP:0000445 | Wide nose | 15 | 48.4% |
| HP:0000426 | Prominent nasal bridge | 7 | 22.6% |
| HP:0005274 | Prominent nasal tip | 11 | 35.5% |
| HP:0009765 | Low hanging columella | 7 | 22.6% |
| HP:0000322 | Short philtrum | 5 | 16.1% |
| HP:0000219 | Thin upper lip vermilion | 8 | 25.8% |
| HP:0011081 | Incisor macrodontia | 7 | 22.6% |
| HP:0000308 | Microretrognathia | 5 | 16.1% |
| HP:0000324 | Facial asymmetry | 5 | 16.1% |
FIGURE 1.

The 3q29 deletion cohort
3.3. Face2Gene analysis
Results from the Face2Gene Research application's analysis can be found in Figure 2 and Table 3. Group A consisted of all 31 subjects in one cohort and the matched control cohort. The AUC‐ROC curves show that the algorithm successfully differentiated between a 3q29 deletion subject and a control patient 87.3% of the time with a p value of 0.006. The score distribution shows that there is a subtle but clear difference between 3q29 deletion facial features and the control feature, indicating that Face2Gene is moderately able to differentiate between a 3q29 deletion subject and control. When comparing subjects 13 and older to their matched controls (n‐14) in Group B, Face2Gene was only able to categorize them correctly 77.8% of the time at a p value of 0.079. Group C compared subjects 12 years and younger to a matched control cohort. Face2Gene was able to accurately categorize the pictures 92.3% of the time (AUC = 0.923) at a p value of 0.005.
FIGURE 2.
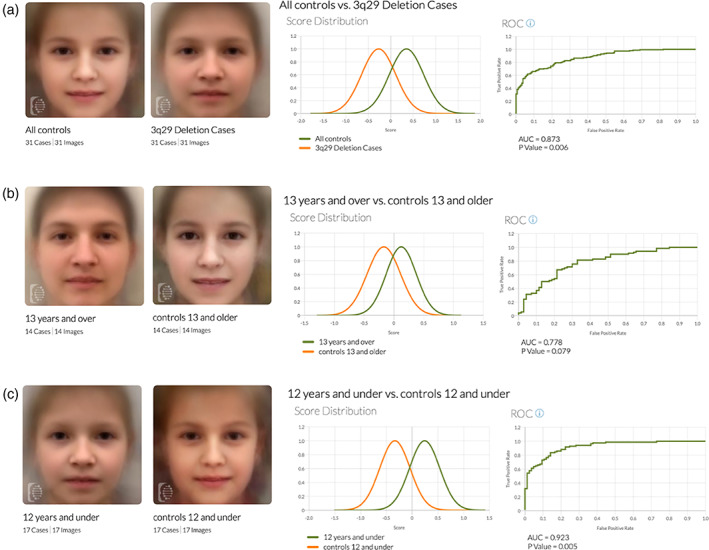
Face2Gene composites and binary comparisons by age comparison of control vs case composites and the binary comparison graphs. (A) Comparison of age‐, gender‐, and ethnicity‐matched controls to the whole 3q29 deletion cohort. (B) Comparison of cases 13 and older and age‐, gender, and ethnicity‐matched controls. (C) Comparison of cases 12 and under and age‐, gender, and ethnicity‐matched controls. Green line: Score distribution curve for case groups. (A) All 3q29 cases (B) Cases 13 years and over (C) Cases 12 years and under. Orange line: Score distribution curve for all control groups. (A) All controls (B) Controls 13 years and over (C) Controls 12 years and under
TABLE 3.
ROC results for age stratification
| Comparison groups | AUC‐ROC | p value |
|---|---|---|
| All cases vs. All controls | 0.873 | 0.006 |
| 13 years and over vs. Controls 13 years and over | 0.778 | 0.079 |
| 12 and under vs. Controls 12 years and under | 0.923 | 0.005 |
3.4. Common differentials
The DeepGestalt algorithm lists 30 differential diagnoses, each participant independent of the research algorithm. Each of these possible differentials are listed as a high, medium, or low match. How high a participant matches to a syndrome depends on how similar the participant's face matches DeepGestlat's model for that given syndrome. In version 19.1.3, this did not include a facial gestalt for 3q29 deletion syndrome. For 13 out of 31 patients, Marfan syndrome was listed as one of the top three differentials, making it the most common differential offered for 3q29 deletion syndrome cohort by Face2Gene. One participant was listed as a high match for Marfan syndrome, nine participants were listed as medium matches for Marfan, and three participants were listed as low matches for Marfan based on gestalt alone.
4. DISCUSSION
This is the first and largest study to systematically describe the craniofacial phenotype of 3q29 deletion syndrome. The focus of this study on the craniofacial phenotype builds upon and refines previously reported data. A literature summary of 40 previous case reports by Cox and Butler (2015), which includes a case series from Ballif et al. (2008), and the previous 3q29 Project study by Glassford et al. (2016) reported craniofacial findings that overlap with our study participants. Although our description of the 3q29 facial phenotypes is similar to previous studies, notable differences are the frequencies at which some features were observed. For example, Ballif et al. (2008) reported on 14 individuals with 3q29 deletion syndrome and found 72% with high nasal bridge 33% with posteriorly rotated ears, 40% with short philtrum, and 46.7% with microcephaly. In contrast, we found none of these features in greater than 25% of our cohort (Table S1). Our participant sample was larger than previous studies, likely making the observed frequencies in this study more representative of the true frequencies. In addition, previous studies were culled reviews of the literature and may not have used the same terms to describe similar features. This study explicitly looked at craniofacial features and used a standardized nomenclature. The true frequencies of these features may be somewhere between the observed frequencies of these different studies.
By documenting common features seen in 3q29 deletion participants, we are able to aid clinicians in recognizing this rare copy number variant. In our experience with the 3q29 project (Glassford et al. (2016)) and in the Ballif et al. (2008) study, patients with 3q29 deletion syndrome are often referred to genetics clinics primarily for developmental delays and autism diagnoses. Chromosomal microarray (CMA) is still the first‐tier clinical diagnostic test in individuals with developmental delays, autism, or congenital anomalies including dysmorphic features (Miller et al., 2010). By following this tiered testing schema and recognizing the dysmorphic features of 3q29 deletion carriers, it may be possible for patients to obtain a molecular diagnosis in a timely manner and prior to the onset of some of the more severe neuropsychiatric phenotypes (Sanchez Russo et al., 2021). Since the phenotype may be too subtle and too variable for clinicians to easily recognize 3q29 deletion syndrome as may be the case for other syndromes with characteristic facial phenotypes such as Cornelia‐de‐Lange or Down syndrome, using next‐generation phenotyping in primary care clinics may aid in the recognition of this rare disorder (Latorre‐Pellicer et al., 2020).
Research cohorts for facial models should reach an AUC‐ROC of 0.80–0.85 or more, as this is the range where results are both sensitive and specific enough to have clinical utility. In the non‐stratified 3q29 deletion cohort, Face2Gene was able to reach an area under the curve of 0.873 (p value = 0.006). AUC‐ROC curves of 0.8 to 0.85 or more have been described in the literature for conditions that presently have facial models within Face2Gene's “Clinic” application, like mucolipidosis type IV, where the AUC is 0.822 (p < 0.01) (Pode‐Shakked et al., 2020). When comparing individuals with mucolipidosis type IV to a control group made up of 100 other genetic syndromes, Face2Gene reached an AUC of 0.885 (p < 0.001) (Pode‐Shakked et al., 2020). By comparing individuals with mucolipidosis type IV to both an unaffected control group and a syndromic control group, authors were able to demonstrate Face2Gene's reliability in distinguishing facial features associated with a rare syndrome. Thus, a facial model for 3q29 deletion syndrome is feasible and currently in use in Version 20.1.1 This fact further strengthens the hypothesis that there is a non‐random pattern to the subtle facial dysmorphology associated with 3q29 deletion syndrome and that this pattern is different from any of the 313 other syndromes recognized by DeepGestalt's current model.
Stratifying the participant and control groups by age yielded interesting results. In Group B, participants aged 13 years and older were compared to unaffected controls. While the DeepGestalt algorithm could detect a difference 77.8% of the time, Group B did not reach significance (p = 0.079). However, Group C did reach significance with an AUC of 0.923 and a p value of 0.005 when comparing participants age 12 years and under to age‐ and gender‐matched controls. These observed groups suggest that dysmorphic features may be more obvious in childhood and becomes less discernable as the patient becomes older. It is important to note that this cohort was small. There were only 14 participants aged 13 or older and only 17 patients aged 12 or younger. Future studies looking at the facial phenotype of 3q29 deletion syndrome would benefit from a larger cohort. Additionally, observing the 3q29 deletion syndrome facial phenotype across the lifespan may provide further insights.
Marfan syndrome was the most commonly offered differential by the DeepGestlat algorithm within the Face2Gene “Clinic” application. Marfan syndrome was a top three differential for 13 out of 31 participants ranging from low to high facial match. While Marfan syndrome and 3q29 deletion are clinically distinct, they share some dysmorphic facial features including deep‐set eyes and malar flattening. Interestingly, previously published data show other overlapping features between 3q29 deletion syndrome and connective tissue disorders including myopia, scoliosis, chest cavity deformity, long/slender fingers, foot deformity, and ligamentous laxity (Cox & Butler, 2015; Sanchez Russo et al., 2021). These data suggest the hypothesis that there may be an underlying mechanism within the 3q29 deletion syndrome leading to this connective tissue‐like phenotype.
A limitation and consideration of our study is that our study is 90% Caucasian which has the ability to impact the Face2Gene facial recognition model as a whole. Males were disproportionately represented in our cohort which may have contributed to the stratified Face2Gene analysis not reaching significance. Lastly, our cohort had a very wide age gap with ages ranging between 4 and 40 years of age. This large age range may have an effect on clinical evaluation and the appearance of certain facial features. These limitations can all be ameliorated with a larger sample size of 3q29 deletion cases upon which to build the facial recognition model, which is a possible future direction. Regardless, this is the still the largest cohort of 3q29 deletion patients ascertained for systematic characterization of the 3q29 deletion facial phenotype.
5. CONCLUSION
The medical community continues to move forward in better understanding 3q29 deletion syndrome. This study is the largest to systematically describe the craniofacial phenotype of 3q29 deletion syndrome. Our analyses suggest there may be a non‐random pattern to the subtle facial dysmorphology associated with 3q29 deletion syndrome. This notion is supported by the application of machine‐learning algorithms to 2D photos, where 3q29 deletion study subjects can be distinguished from control subjects with 87% accuracy from 2D photos alone. Future studies looking at the natural history of 3q29 deletion syndrome will provide invaluable information on how to better diagnose and treat these individuals.
CONFLICT OF INTEREST
NF is a full time employee of FDNA, Inc (Boston, MA). All other authors declare there are no conflicts of interest. The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.
AUTHOR'S CONTRIBUTIONS
Bryan C Mak wrote the manuscript, analyzed the demographic and craniofacial data, and generated Tables 1, 2, and 3 and Figure 1. Rossana Sanchez Russo, Michael J. Gambello, Emily D. Black, Jennifer G. Mulle, and Melissa M. Murphy collected and provided clinical data and facial photos. Nicole Fleischer and Face2Gene provided technical support, the Face2Gene analysis, graphs, and controls. All authors reviewed the final manuscript.
Supporting information
Appendix S1: Supporting information
ACKNOWLEDGMENTS
First, we would like to thank the participants and their families that made this study possible. They have taught us so much and without their willingness, our research would not have been possible. This research was completed as part of the Emory 3q29 Project. We would like to acknowledge the members: Jennifer Gladys Mulle, Gary J. Bassell, Katrina Aberizk, T. Lindsey Burrell, Shanthi Cambala, Tamara Caspary, Joseph F. Cubells, David Culter, Paul A. Dawson, Michael P. Epstein, Ryan Guest, Michael J. Gambello, Sandra M. Goulding, Henry R. Johnston, Cheryl Klaiman, Sookyoung Koh,Elizabeth J. Leslie, Bryan C. Mak, Michale Mortillo, Trenell Mosley, Melissa M. Murphy, Becky Pollak, Ryan Purcell, Tim Rutkowski, Rossana Sanchez, Celine Saulnier, Jason Schroeder, Esra Sefik, Sarah Shultz, Elaine F. Walker, Stephen T. Warren, David Weinshenker, Zhexing Wen, and Michael E. Zwick. We would like to thank FDNA for their contributions to this project. This manuscript was the final outcome of Bryan C. Mak's capstone project, a graduation requirement of Emory University Genetic Counseling Training Program. As such, we would like to express our gratitude to the leadership, faculty, and everyone involved with the Genetic Counseling Training Program.
Mak, B. C. , Sanchez Russo, R. , Gambello, M. J. , Fleischer, N. , Black, E. D. , Leslie, E. , Murphy, M. M. , The Emory 3q29 Project, & Mulle, J. G. (2021). Craniofacial features of 3q29 deletion syndrome: Application of next‐generation phenotyping technology. American Journal of Medical Genetics Part A Part A, 185A:2094–2101. 10.1002/ajmg.a.62227
Funding information National Institute of Mental Health, Grant/Award Numbers: 1R01MH110701‐01A1, MH110701‐01A1; NIH T32, Grant/Award Number: GM0008490; Emory University
Contributor Information
Rossana Sanchez Russo, Email: rossana.sanchez@emory.edu.
Jennifer Gladys Mulle, Email: jmulle@emory.edu.
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.
REFERENCES
- Ballif, B. , Theisen, A. , Coppinger, J. , Gowans, G. , Hersh, J. , Madan‐Khetarpal, S. , Schmidt, K. R. , Tervo, R. , Escobar, L. F. , Friedrich, C. A. , McDonald, M. , Campbell, L. , Ming, J. E. , Zackai, E. H. , Bejjani, B. A. , & Shaffer, L. G. (2008). Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Molecular Cytogenetics, 1(1), 8. 10.1186/1755-8166-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D. , & Butler, M. (2015). A clinical case report and literature review of the 3q29 microdeletion syndrome. Clinical Dysmorphology, 24(3), 89–94. 10.1097/mcd.0000000000000077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford, M. , Rosenfeld, J. , Freedman, A. , Zwick, M. , & Mulle, J. (2016). Novel features of 3q29 deletion syndrome: Results from the 3q29 registry. American Journal of Medical Genetics Part A, 170(4), 999–1006. 10.1002/ajmg.a.37537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, S. J. , Liu, J. L. , & Webber, C. (2015). Synergistic interactions between drosophila orthologues of genes spanned by de novo human CNVs support multiple‐hit models of autism. PLoS Genetics, 11(3), e1004998. 10.1371/journal.pgen.1004998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurovich, Y. , Hanani, Y. , Bar, O. , Nadav, G. , Fleischer, N. , Gelbman, D. , Basel‐Salmon, L. , Krawitz, P. M. , Kamphausen, S. B. , Zenker, M. , Bird, L. M. , & Gripp, K. W. (2019). Identifying facial phenotypes of genetic disorders using deep learning. Nature Medicine, 25(1), 60–64. 10.1038/s41591-018-0279-0 [DOI] [PubMed] [Google Scholar]
- Köhler, S. , Carmody, L. , Vasilevsky, N. , Jacobsen, J. , Danis, D. , Gourdine, J. , Gargano, M. , Harris, N. L. , Matentzoglu, N. , McMurry, J. A. , Osumi‐Sutherland, D. , Cipriani, V. , Balhoff, J. P. , Conlin, T. , Blau, H. , Baynam, G. , Palmer, R. , Gratian, D. , Dawkins, H. , Segal, M. , … Robinson, P. N. (2019). Expansion of the human phenotype ontology (HPO) knowledge base and resources. Nucleic Acids Research, 47(D1), D1018–D1027. 10.1093/nar/gky1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre‐Pellicer, A. , Ascaso, Á. , Trujillano, L. , Gil‐Salvador, M. , Arnedo, M. , Lucia‐Campos, C. , Antoñanzas‐Pérez, R. , Marcos‐Alcalde, I. , Parenti, I. , Bueno‐Lozano, G. , Musio, A. , Puisac, B. , Kaiser, F. J. , Ramos, F. J. , Gómez‐Puertas, P. , & Pié, J. (2020). Evaluating Face2Gene as a tool to identify Cornelia de Lange syndrome by facial phenotypes. International Journal of Molecular Sciences, 21(3), 1042. 10.3390/ijms21031042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson, D. , & Clark, R. (2020). Optimizing genetic diagnosis of neurodevelopmental disorders in the clinical setting. Clinics in Laboratory Medicine, 40(3), 231–256. 10.1016/j.cll.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Miller, D. T. , Adam, M. P. , Aradhya, S. , Biesecker, L. G. , Brothman, A. R. , Carter, N. P. , Church, D. M. , Crolla, J. A. , Eichler, E. E. , Epstein, C. J. , Faucett, W. A. , Feuk, L. , Friedman, J. M. , Hamosh, A. , Jackson, L. , Kaminsky, E. B. , Kok, K. , Krantz, I. D. , Kuhn, R. M. , … Ledbetter, D. H. (2010). Consensus statement: Chromosomal microarray is a first‐tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American Journal of Human Genetics, 86(5), 749–764. 10.1016/j.ajhg.2010.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle, J. (2015). The 3q29 deletion confers >40‐fold increase in risk for schizophrenia. Molecular Psychiatry, 20(9), 1028–1029. 10.1038/mp.2015.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle, J. G. , Dodd, A. F. , McGrath, J. A. , Wolyniec, P. S. , Mitchell, A. A. , Shetty, A. C. , Sobreira, N. L. , Valle, D. , Rudd, M. K. , Satten, G. , Cutler, D. J. , Pulver, A. E. , & Warren, S. T. (2010). Microdeletions of 3q29 confer high risk for schizophrenia. American Journal of Human Genetics, 87(2), 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, M. , Lindsey Burrell, T. , Cubells, J. , España, R. , Gambello, M. , Goines, K. , Klaiman, C. , Li, L. , Novacek, D. M. , Papetti, A. , Russo, R. L. S. , Saulnier, C. A. , Shultz, S. , Walker, E. , & Mulle, J. G. (2018). Study protocol for the Emory 3q29 Project: Evaluation of neurodevelopmental, psychiatric, and medical symptoms in 3q29 deletion syndrome. BMC Psychiatry, 18(1), 183. 10.1186/s12888-018-1760-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, M. M. , Burrell, T. L. , Cubells, J. F. , Epstein, M. T. , Espana, R. , Gambello, M. J. , Goines, K. , Klaiman, C. , Koh, S. , Russo, R. S. , Saulnier, C. A. , Walker, E. , & Mulle, J. G. (2020). Comprehensive phenotyping of neuropsychiatric traits in a multiplex 3q29 deletion family: A case report. BMC Psychiatry, 20(1), 184. 10.1186/s12888-020-02598-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pode‐Shakked, B. , Finezilber, Y. , Levi, Y. , Putter, S. , Fleischer, N. , Greenbaum, L. , & Raas Rothschild, A. (2020). Shared facial phenotype of patients with mucolipidosis type IV: A clinical observation reaffirmed by next generation phenotyping. European journal of medical genetics, 63, 103927. 10.1016/j.ejmg.2020.103927 [DOI] [PubMed] [Google Scholar]
- Pollak, R. M. , Murphy, M. M. , Epstein, M. P. , Zwick, M. E. , Klaiman, C. , Saulnier, C. A. , Emory 3q29 Project , & Mulle, J. G. (2019). Neuropsychiatric phenotypes and a distinct constellation of ASD features in 3q29 deletion syndrome: Results from the 3q29 registry. Molecular Autism, 10, 30. 10.1186/s13229-019-0281-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski, T. P. , Purcell, R. H. , Pollak, R. M. , Grewenow, S. M. , Gafford, G. M. , Malone, T. , Khan, U. A. , Schroeder, J. P. , Epstein, M. P. , Bassell, G. J. , Warren, S. T. , Weinshenker, D. , Caspary, T. , & Mulle, J. G. (2019). Behavioral changes and growth deficits in a CRISPR engineered mouse model of the schizophrenia‐associated 3q29 deletion. Molecular psychiatry, 26, 772–783. 10.1038/s41380-019-0413-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Russo, R. , Gambello, M. J. , Murphy, M. M. , Aberizk, K. , Black, E. , Burrell, T. L. , Carlock, G. , Cubells, J. F. , Epstein, M. T. , Espana, R. , Goines, K. , Guest, R. M. , Klaiman, C. , Koh, S. , Leslie, E. J. , Li, L. , Novacek, D. M. , Saulnier, C. A. , Sefik, E. , Schultz, S. , … Mulle, J. G. (2021). Deep phenotyping in 3q29 deletion syndrome: recommendations for clinical care. Genetics in Medicine: official journal of the American College of Medical Genetics. 10.1038/s41436-020-01053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S. J. , He, X. , Willsey, A. J. , Ercan‐Sencicek, A. G. , Samocha, K. E. , Cicek, A. E. , Murtha, M. T. , Bal, V. H. , Bishop, S. L. , Dong, S. , Goldberg, A. P. , Jinlu, C. , Keaney, J. F., 3rd , Klei, L. , Mandell, J. D. , Moreno‐De‐Luca, D. , Poultney, C. S. , Robinson, E. B. , Smith, L. , … State, M. W. (2015). Insights into autism Spectrum disorder genomic architecture and biology from 71 risk loci. Neuron, 87(6), 1215–1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, M. D. , Jensen, M. , Lasser, M. , Huber, E. , Yusuff, T. , Pizzo, L. , Lifschutz, B. , Desai, I. , Kubina, A. , Yennawar, S. , Kim, S. , Iyer, J. , Rincon‐Limas, D. E. , Lowery, L. A. , & Girirajan, S. (2020). NCBP2 modulates neurodevelopmental defects of the 3q29 deletion in drosophila and Xenopus laevis models. PLoS Genetics, 16(2), e1008590. 10.1371/journal.pgen.1008590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson, H. , Meyer‐Lindenberg, A. , Steinberg, S. , Magnusdottir, B. , Morgen, K. , Arnarsdottir, S. , Bjornsdottir, G. , Walters, G. B. , Jonsdottir, G. A. , Doyle, O. M. , Tost, H. , Grimm, O. , Kristjansdottir, S. , Snorrason, H. , Davidsdottir, S. R. , Gudmundsson, L. J. , Jonsson, G. F. , Stefansdottir, B. , Helgadottir, I. , … Stefansson, K. (2013). CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature, 505(7483), 361–366. 10.1038/nature12818 [DOI] [PubMed] [Google Scholar]
- Zarate, Y. , Bosanko, K. , & Gripp, K. (2019). Using facial analysis technology in a typical genetic clinic: Experience from 30 individuals from a single institution. Journal of Human Genetics, 64(12), 1243–1245. 10.1038/s10038-019-0673-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.


