Abstract
The recent coronavirus disease 2019 (COVID‐19), causing a global pandemic with devastating effects on healthcare and social‐economic systems, has no special antiviral therapies available for human coronaviruses (CoVs). The severe acute respiratory syndrome coronavirus 2 (SARS‐Cov‐2) possesses a nonstructural protein (nsp14), with amino‐terminal domain coding for proofreading exoribonuclease (ExoN) that is required for high‐fidelity replication. The ability of CoVs during genome replication and transcription to proofread and exclude mismatched nucleotides has long hindered the development of anti‐CoV drugs. The resistance of SARS‐CoV‐2 to antivirals, especially nucleoside analogs (NAs), shows the need to identify new CoV inhibition targets. Therefore, this review highlights the importance of nsp14‐ExoN as a target for inhibition. Also, nucleoside analogs could be used in combination with existing anti‐CoV therapeutics to target the proofreading mechanism.
Keywords: coronavirus, exoribonuclease, nonstructural protein 14, proofreading, RNA recombination
Abbreviations
- COVID‐19
coronavirus disease 2019
- CoVs
coronaviruses
- ExoN
exoribonuclease
- MERS‐CoV
Middle East Respiratory Syndrome coronavirus
- MHV
murine hepatitis virus
- N7‐MTase
N7‐methyltransferase
- NAs
nucleoside analogs
- nsp14
nonstructural protein
- ORFs
open reading frames
- RdRp
RNA‐dependent RNA polymerase
- RTC
replication‐transcription complex
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐Cov‐2
severe acute respiratory syndrome coronavirus 2
- sgmRNAs
subgenomic mRNAs
- TRSs
transcription regulatory sequences
1. INTRODUCTION
Coronaviruses (CoVs) are enveloped, single‐stranded positive‐sense RNA viruses with some of the largest identified RNA viral genomes, about 30 kb. CoVs are a set of related viruses that belong to the Coronaviridae family in the order Nidovirales. 2 Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the causative agent of coronavirus disease 2019 (COVID‐19) and belongs to the genus Betacoronavirus along with the severe acute respiratory syndrome coronavirus (SARS‐CoV) and the Middle East Respiratory Syndrome coronavirus (MERS‐CoV). All three of these viruses raised as novel coronaviruses, are causing respiratory diseases and transmitted from animals to humans (zoonotic viruses), and cause respiratory diseases. 3 SARS‐CoV‐2 is closest (88%) to two SARS‐like bat‐derived CoVs (bat‐SL‐CoVZC45 and bat‐SL‐CoVZXC21), with (79%) overall SARS‐CoV sequence identity and (50%) MERS‐CoV identity 4
The SARS‐CoV polycistronic RNA genome encodes 14 open reading frames (ORFs), some of which overlap. 1 The genomes 5′ end is capped and has a 3′ poly(A) tail with short 5′ and 3′ untranslated region sequences forming regulatory stem‐loop structures and a single leader sequence proximal to the 5′ end. Two‐thirds of the genome encodes two large polyproteins, pp1a and pp1ab, that are cleaved into 16 nonstructural proteins. The last one‐third of the genome encodes structure and additional proteins (Figure 1). The bifunctional nsp14 contains an N‐terminal 3′‐to‐5′ exoribonuclease (ExoN) and a C‐terminal N7‐methyltransferase (N7‐MTase) domain. In the 3′‐to‐5′ direction, nsp14 catalyzes nucleoside monophosphate excision from nucleic acids, using a mechanism that relies on two divalent metal ions and a reactive water molecule. This exonucleolytic activity is crucial for the Coronavirus replication proofreading activity, a property lacking in other RNA viruses that improve its fidelity replication and has played an essential role in nidoviral evolution and genome expansion. 1 The first proofreading protein identified in an RNA virus is the coronavirus ExoN, and targeting ExoN is a promising strategy to develop new coronavirus inhibitors and antiviral sensitizers. 5 The development of anti‐CoV drugs has long been hampered by the ability of CoVs during genome replication and transcription to proofread and eliminate mismatched nucleotides (Table 1).
Figure 1.
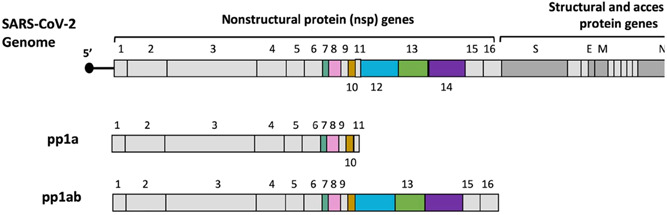
Single‐stranded RNA genome of severe acute respiratory syndrome coronavirus 2. Two‐thirds of the genome encodes two major polyproteins, pp1a and pp1ab, which are divided into 16 nonstructural proteins 40
Table 1.
Role of nonstructural proteins of SARS‐CoV‐2
| Protein | Function | Reference |
|---|---|---|
| Nsp1 | It plays a role in host‐range restriction, countering the innate antiviral response of the host, and suppressing apoptosis induction during the early stages of infection to promote viral growth. | Jauregui et al. 6 and Narayanan et al. 7 |
| Nsp2 | It is involved in disrupting intracellular host signaling. | Cornillez‐Ty et al. 8 |
| Nsp3 | It is suggested that it facilitates mRNA transcript translation while suppressing host protein synthesis. | Lu et al. 9 , Anand et al. 10 , Stobart et al. 11 , Cottam et al. 12 , and Benvenuto et al. 13 |
| Nsp4 | Vital role is replication and the assembly of the replicative structures. | Oostra et al. 14 |
| Nsp5 | Protease activity | Mielech et al.15 |
| Nsp6 | Autophagosome formation for immunomodulatory protein degradation | Cottam et al. 12 |
| Nsp7 | Primer‐Independent RNA polymerase Activity | Xiao et al. 16 |
| Nsp8 | Primase activity | Tan et al. 17 |
| Nsp9 | It has been suggested that it works as a single‐stranded RNA binding protein. | Bouvet et al. 18 |
| Nsp10 | It serves as a cofactor for both nsp16's 2′O‐methyltransferase and nsp14's N7‐guaninemethyltransferase/exoribonuclease activities. | Bouvet et al.19, Deming et al. 20 |
| Nsp11 | Necessary for replication. | Fang et al. 21 |
| Nsp12 | RNA‐dependent RNA polymerase. | te Velthuis et al. 22 |
| Nsp13 | RTPase activity hydrolyzes the RNA strand's at 5′ end. | Bouvet et al. 18 |
| Nsp14 | Exoribonuclease activity and Methyl transferase. | Minskaia et al. 23 and Chen et al. 24 |
| Nsp15 | Endoribonuclease | Bhardwaj et al. 25 |
| Nsp16 | 2′‐O‐methyltransferase methylates the RNA strand at position 2′. | Chen et al. 26 and Bouvet et al. 18 |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
2. CORONAVIRUS PROOFREADING MECHANISM
The replication of RNA virus has a high error rate (or low viral fidelity), resulting in the virus living as diverse genome mutant populations or “quasispecies.” RNA viruses can adapt to various replicative environments and selective stresses due to low replicative fidelity, however, a disastrous error could lead to viral extinction. This factor threatens the development of antivirals against CoVs and other RNA viruses as they can easily establish drug resistance while preserving viral replicative fitness. 27 The distinctive 3′ to 5′ exoribonuclease (ExoN) proofreading feature is an additional obstacle to the production of nucleoside analogs (NAs) as antivirals against CoVs. Proofreading activities that protect the virus from mutagenesis are provided by nonstructural protein 14. 5 Within the family Coronaviridae, nsp14 is strongly conserved. 28 In Group 2 coronaviruses and possibly all coronaviruses, nsp14‐ExoN is involved in RNA proofreading, and the acquisition of ExoN function is necessary for the achievement and preservation of broad genomes of coronaviruses. 29
3. BIFUNCTIONAL NSP14 PROTEIN
Nsp14 is a 60 kDa bifunctional enzyme involved in replication fidelity with an N‐terminal ExoN domain and N7‐methyltransferase (N7‐MTase) C‐terminal domain that is involved in messenger RNA (mRNA) capping. 26 SARS‐CoV‐2 and SARS‐CoV nsp14 share more than 95% similarity in amino acid sequence. 30 Also, nsp14 is involved in many other virus life cycles and pathogenicity processes, including recombination of the viral genome and control of innate immune responses. 31 The exoribonucleolytic activity of nsp14 3ʹ−5ʹ relies on metal ions, preferably Mg2+ over Mn2+, Co2+, and Zn2+ promote residual activity, while catalysis was not supported by Ca2+, Ni2+, and Cu2+. 32
3.1. N7‐Methyltransferase domain
The C‐terminal N7‐MTase domain contains a DxG S‐adenosyl‐l‐methionine (SAM)‐a binding motif conserved among CoVs and is involved in mRNA capping and host immune response evasion. 33 In pre‐mRNA splicing, mRNA export, RNA stability, this cap structure plays essential roles by blocking degradation by the 5ʹ−3ʹ exoribonuclease. Translational initiation by promoting the 4E (eIF4E) binding initiation factor of eukaryotic translation, 34 and escapes from the cellular innate immune system recognition. 35 In cytoplasmic granular compartments, host and viral RNA molecules missing the 5ʹ cap structure are degraded. 36 In SARS‐CoV nsp14, mutations in the DxG motif specifically eliminate the activity of N7‐MTase that is essential for the formation of mRNA caps, and they have an exact detrimental effect on replication without disrupting the activity of ExoN, demonstrating the importance of this enzymatic function of nsp14. 33
3.2. Exoribonuclease domain
The activity of nsp14 exoribonuclease resides in the N terminal. 37 By removing mismatched nucleotides from the 3ʹ end of the rising RNA strand, the ExoN domain is proposed to correct errors produced by the RdRp. 38 CoV nsp14‐ExoN is a member of the DEDD superfamily of DNA and RNA exonucleases, similar to the proofreading subunit (ε) of Escherichia coli DNA polymerase III. 23 This superfamily contains four preserved D‐E‐D‐D acidic residues necessary for enzymatic action, and mutation in these residues within CoV ExoN significantly decreases the activity or ablates the ExoN. 23 A recent study found that the ExoN domain plays a role in the frequency of subgenomic microRNA (sgmRNA) recombination and recombination patterns in virally infected cells and virions in nsp14. Inactivating nsp14‐ExoN in murine hepatitis virus (MHV‐CoV), significantly altered recombination patterns and decreased recombination frequency compared with wild‐type MHV‐CoV. 31 In both MHV‐CoV model and SARS‐CoV model, ExoN activity is vital for high‐fidelity replication. 29
4. DEDDH SUPERFAMILY
The protein's ExoN domain was first described by comparative sequence analysis that demonstrated a distant association between the N‐terminal portion of the protein and the cellular exonuclease DEDD protein superfamily. 1 DEDD refers to four preserved catalytic aspartic acid (D)/glutamic acid (E) (Asp/Glu) amino acids that are needed by the protein to form two metal‐binding sites that drive nucleotide excision through a two‐metal ion‐assisted process. 39 X‐ray crystallography of the nsp14/nsp10 complex and its functional ligands were able to identify a range of main factors that highlighted distinct differences between nsp14 and its DEDD superfamily homologs and shed light on several characteristics of the structure and function of the protein. 28
Ma et al. 28 compared the nsp14 ExoN active sites with proofreading homologs of E. coli DNA polymerase I Klenow fragment and the polymerase III subunit ε. They reported that Asp90, Glu92, Glu191, His268, and Asp2733 were the catalytic residues in nsp14. Glu191 replaced the Asp424 of the Klenow fragment and Asp103 of the ε subunit. This makes nsp14 a “DEED outlier.” Furthermore, Asp243, a catalytic center, is highly preserved and mutating it inhibit nsp14 activity, although its function has yet to be deciphered. 40 nsp14 has exoribonuclease activity and is capable of degrading ssRNA in 3ʹ–5ʹ directions. 23 The nsp14 activity requires divalent metal ions as a cofactor and it is essential to have a secondary structure of the RNA substrate. Furthermore, mutating the conserved D/E residues showed severe defects in viral RNA synthesis, and no viable virus could be recovered. 23 Two Mg2+ ions are expected to coordinate the five catalytic DEEDh residues that aid in removing misincorporated nucleotides. To activate a nucleophilic attack on the phosphate of the substrate nucleotide, MgA must activate a water molecule, and MgB would then promote the product's exit. 40
5. THE INTERACTION BETWEEN NSP14‐EXON AND THE NSP10
In many processes during the viral infection cycle, interactions between viral proteins play crucial roles. A yeast two‐hybrid screen of intraviral protein‐protein interactions revealed nsp10 as an interacting partner of nsp14. 41 Experiments in vitro in E. coli has identified that dsRNA with a 30 mismatch is the preferred substrate of the nsp10‐bound nsp14 that mimics a faulty replication product. Although nsp14 alone has exoribonuclease activity, this study also showed more than a 35‐fold increase in nsp14 activity when bound to nsp10, indicating that nsp10 has a role in stabilizing the active site of ExoN in the correct conformation for catalysis of the substrate. 18 The interaction between nsp10 and nsp14 is figuratively equivalent to a hand (nsp14) over a fist (nsp10) (Figure 2). During exoribonuclease assays, the addition of nsp10 to the complex increased the ExoN activity, indicating that nsp10 improves ExoN activity in situ in the active complex. 42 In the presence of nsp10, ExoN active site residues are adequately positioned and form a highly active ExoN, as determined by the distance observed between catalytic residues. 38 Mutations in the nsp10 domain that interact with nsp14 resulted in a decrease in viral replication fidelity. The complexed protein structure showed that nsp10 confers structural integrity and stability to nsp14's ExoN domain. 18 In the absence of nsp10, the ExoN catalytic pocket partly collapses, accounting for the poor ExoN behavior of nsp14 alone. 38
Figure 2.
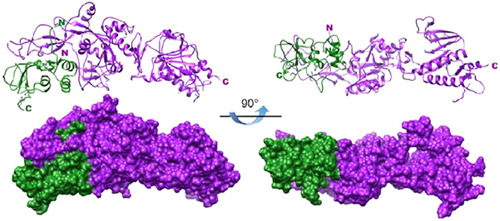
Structure model of severe acute respiratory syndrome coronavirus nsp10/nsp14 complex. From a side view and 90° rotation concerning the side view, ribbon structures of nsp10 (green) and nsp14 (purple) Each protein's amino‐terminal and carboxyl‐terminal extremities are indicated by the letters N and C with their respective colors 38
6. THE ROLE OF ZINC FINGER MOTIFS DURING VIRAL REPLICATION
A study revealed significant differences between nsp14 ExoN and other members of the DEDD exonuclease superfamily in addition to the revised catalytic motif. 39 Although the core structural elements are similar, nsp14 ExoN has two zinc fingers essential for exoribonuclease activity and a site of interaction with nsp10. Zinc finger 2 is located near the catalytic core, and the domain is disrupted by the action of the enzyme via C261A or H264R mutations, meaning that it has a role in catalysis. On the other hand, zinc finger 1 mutation result in insoluble proteins of nsp14 when expressed in E. coli, and this, together with its composition, indicated the role for SARS‐CoV nsp14 in structural stability. 40
The proximity of zinc finger 1 to the interaction site of nsp10 could influence the interaction between nsp10 and nsp14 and thus decrease the catalytic activity of ExoN. Zinc finger 2 mutations have eliminated viral viability, consistent with previous studies. 30 The induction of a particular mutation in zinc finger 1 of the nsp14‐ExoN domain alphacoronavirus created a viable virus that induced a decrease in dsRNA intermediate aggregation and decreased antiviral response and apoptosis compared with the wild‐type virus. 43
7. THE ACTIVITY OF SARS‐COV‐2 NSP14 EXON IS METAL DEPENDENT
It is also recognized that the nsp14 ExoN domain activity relies on divalent cations. 19 In the presence of both Mg2+ and Mn2+, SARS‐CoV‐2 ExoN nsp14 is active, with more noticeable activity in the presence of Mg2+. In agreement with the data obtained for SARS‐CoV nsp14 and other proteins from the DEDD family, Ca2+, Ni2+, and Cu+2 did not help catalysis. 19 nsp14 residual activity also depends on the presence of Zn2+ ion. 39 However, the addition of the chelating agent EDTA to the reaction entirely blocks nsp14 ExoN activity, confirming the significance for this activity of divalent ions, namely Mg2+ and Mn2+, close to that described for the SARS‐CoV counterpart. 19
8. THE ROLE OF NSP14 IN RNA RECOMBINATION
Studies comparing CoV strains closely related to SARS‐CoV‐2 have indicated that SARS‐CoV‐2 has acquired the capacity to infect human cells by recombining within the spike protein series. 44 CoV recombination has been reported to be associated with the increase in spread and severe illness, resulting in multiple livestock CoVs vaccine failures. 45 On virus‐specific transcription regulatory sequences (TRSs), the CoV replication‐transcription complex (RTC) performs intramolecular recombination to produce a set of subgenomic mRNAs (sgmRNAs) with typical ends of 5ʹ and 3ʹ. 46 Therefore, as a natural part of their replication, CoVs, perform recombination, creating complex populations of recombined RNA molecules. 31
Gribble et al. 31 reveal that the CoVs nsp14‐ExoN is necessary for native recombination, and inactivation of ExoN results in decreased recombination frequency and altered recombination products. The viral genome, notably DNA viruses, had shown that exonucleases are essential for recombination. To test this hypothesis, Gribble et al. 31 infected Murine DBT cells with wild‐type (Exon+) or ExoN‐deleted (ExoN−) murine hepatitis virus (MHV). Indeed, infected cells (MHV‐ExoN−) had significantly decreased unique recombination junctions compared to wild‐type (MHV‐Exon+). Thus, relative to (MHV‐Exon+), (MHV‐ExoN−) had reduced recombination junction frequency and number of specific junctions, suggesting that lack of operation of nsp14‐ExoN resulted in considerably less recombination throughout infection. Further research requires to clarify the function of SARS‐CoV‐2 nsp14‐ExoN interaction in RNA recombination. The role of nsp14‐ExoN in recombination, combined with the several essential integrated functions, make it vulnerable, and highly selective target for antiviral treatment inhibition and viral attenuation.
9. NSP14‐EXON AS A TARGET FOR COMBINATION COV INHIBITORS
Since the beginning of the COVID‐19 trials, the class of antivirals that are RNA‐dependent RNA polymerase (RdRp) inhibitors such as Favirpiravir, Remdesivir, Ribavirin, and Galidesivir have been of high importance. These drugs, which are nucleoside analogs, work either by introducing viral RNA mutations or terminating the chain during replication. These medications had a strong effect on viruses that do not have proofreading enzymes. 47 However, in SARS‐CoV‐2, Due to its 3ʹ−5ʹ exoribonuclease proofreading operation, ExoN is capable of excising integrated nucleoside analogs. This results in negating these medicines' function to varying extents, depending on the kind of nucleoside analog chemistry (Ribavirin, 5FU, and Remdesivir). Compounds with affinity to the catalytic subunit nsp14‐ExoN or those with allosteric effects at critical sites with retained residues cause the entire viral RNA repair to alter conformationally. Therefore, finding an inhibitor for this viral nuclease should be the researchers' priority. 48 ExoN inactivation was shown to create a “mutator phenotype,” which was apparent from a 15‐ to 21‐fold rise in the incidence of mutation during replication and passage in cell culture compared to the wild‐type control. 29 There is evidence that Remdesivir has 4.5 times more efficacy in the ExoN mutant background, and Ribavirin has 200 times higher efficacy relative to the wild‐type ExoN viral genome. 4 , 38 This provides a clear justification for a mixture of Favirpiravir/Remdesivir/Ribavirin/Galidesivir and SARS‐CoV‐2 ExoN inhibitor combinations to be used. There is currently no available drug for inhibiting nsp14. To identify small molecules/peptides/natural molecules that have the potential to inhibit nsp14 ExoN, comprehensive molecular docking studies are urgently needed. 48 The crystal structure of SARS‐CoV nsp14‐nsp10 offers the possibility of molecular docking of the nsp14 ExoN domain to various available drugs. Three known antivirals, Conivaptan, Hesperidin, and Glycyrrhizic acid, show promise based on docking results and their known inhibitory effects on in vitro beta coronaviruses 49 and in patients. 50 As potential proofreading inhibitors of ExoN (nsp14), dexamethasone metasulfobenzoate, conivaptan, hesperidin, and glycyrrhizic acid, and their use in combination with RdRp inhibitors could lead to potentially high levels of antiviral activity and promising COVID‐19 therapy 51 (Figure 3). According to Smith et al., 5 while inhibitors of ExoN alone might be useful, Combining it with an RNA mutagen would increase the ExoN inhibition's intrinsic fidelity defect and drive high‐level mutagenesis. A possible benefit of such an approach would be to quickly drive the virus to extinction while limiting or preventing the virus's ability to resolve reversion inhibition.
Figure 3.
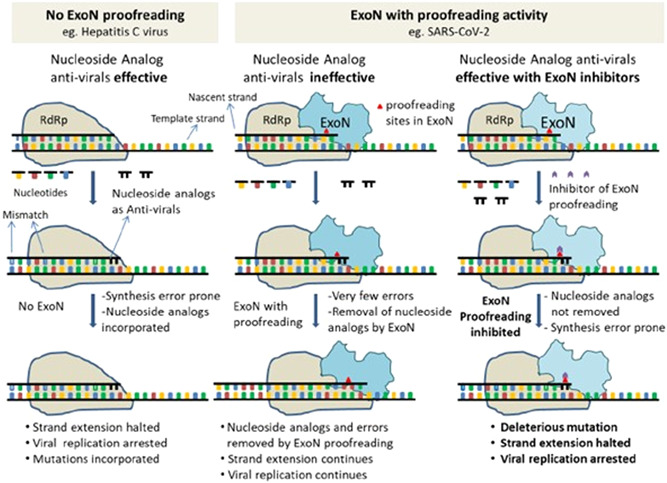
Schematic describing ExoN proofreading activity and mode of action of inhibitors. (Left Panel) Replication in viruses like Hepatitis C Virus (HCV), with no proofreading mechanism. (Mid panel) Replication in viruses like SARS‐CoV with proofreading Exoribonuclease. (Right panel) It is the same as a mid‐panel but in an ExoN inhibitor 51
10. CONCLUSION
There have been many outbreaks of major zoonotic CoV diseases in the 21st century: SARS‐CoV in 2002/3, the ongoing MERS‐CoV that began in 2012, and the current worldwide SARS‐CoV‐2 pandemic. However, the SARS‐CoV‐2 outbreak has emerged when postgenomics technologies are increasingly evolving in molecular biology in terms of sophistication and target specificity. CoVs have an nsp14, bifunctional enzyme with an amino‐terminal domain coding for proofreading exoribonuclease and N7‐methyltransferase domain to methylate the viral RNA cap. The exoribonuclease function of ExoN 3ʹ–5ʹ is a central player in a variety of essential life‐cycle processes for coronaviruses and has made many previously active antiviral compounds ineffective against CoVs. ExoN is a logical therapeutic target for new technologies in genomics.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
Thanks to Dr. Nawroz O. Kareem (Keele University, UK) and Dr. Anwar O. Mohammad (University of Sulaimani, Iraq) for their critical reviews of the manuscript.
Tahir M. Coronavirus genomic nsp14‐ExoN, structure, role, mechanism, and potential application as a drug target. J Med Virol. 2021;93:4258–4264. 10.1002/jmv.27009
REFERENCES
- 1. Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Cambridge, UK: Elsevier; 2012. [Google Scholar]
- 3. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2020;19:141‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shannon A, Le NTT, Selisko B, et al. Remdesivir and SARS‐CoV‐2: structural requirements at both nsp12 RdRp and nsp14 exonuclease active‐sites. Antiviral Res. 2020;178:104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith EC, Blanc H, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9(8):e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jauregui AR, Savalia D, Lowry VK, Farrell CM, Wathelet MG. Identification of residues of SARS‐CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One. 2013;8(4):e62416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narayanan K, Ramirez SI, Lokugamage KG, Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cornillez‐Ty CT, Liao L, Yates JR, Kuhn P, Buchmeier MJ. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol. 2009;83(19):10314‐10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu J‐H, Zhang DM, Wang GL, et al. Sequence analysis and structural prediction of the severe acute respiratory syndrome coronavirus nsp5. Acta Biochim Biophys Sin (Shanghai). 2005;37(7):473‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anand K. Coronavirus main proteinase (3CLpro) structure: basis for design of anti‐SARS drugs. Science. 2003;300(5626):1763‐1767. [DOI] [PubMed] [Google Scholar]
- 11. Stobart CC, Sexton NR, Munjal H, et al. Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol. 2013;87(23):12611‐12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benvenuto D, Angeletti S, Giovanetti M, et al. Evolutionary analysis of SARS‐CoV‐2: how mutation of non‐structural protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81(1):e24‐e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJM, de Haan CAM. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol. 2007;81(22):12323‐12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mielech AM, Chen Y, Mesecar AD, Baker SC. Nidovirus papain‐like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao Y, Ma Q, Restle T, et al. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer‐independent RNA polymerase activity. J Virol. 2012;86(8):4444‐4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan YW, Fung TS, Shen H, Huang M, Liu DX. Coronavirus infectious bronchitis virus non‐structural proteins 8 and 12 form stable complex independent of the non‐translated regions of viral RNA and other viral proteins. Virology. 2018;513:75‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bouvet M, Lugari A, Posthuma CC, et al. Coronavirus Nsp10, a critical co‐factor for activation of multiple replicative enzymes. J Biol Chem. 2014;289(37):25783‐25796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3'‐end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc Natl Acad Sci USA. 2012;109(24):9372‐9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deming DJ, Graham RL, Denison MR, Baric RS. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol. 2007;81(19):10280‐10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fang SG, Shen H, Wang J, Tay FPL, Liu DX. Proteolytic processing of polyproteins 1a and 1ab between non‐structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology. 2008;379(2):175‐180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. te Velthuis AJW, Arnold JJ, Cameron CE, van den Worm SHE, Snijder EJ. The RNA polymerase activity of SARS‐coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38(1):203‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103(13):5108‐5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen P, Jiang M, Hu T, et al. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol. 2007;40(5):649‐655. [DOI] [PubMed] [Google Scholar]
- 25. Bhardwaj K, Guarino L, Kao CC. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol. 2004;78(22):12218‐12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Cai H, Pan J, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106(9):3484‐3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perales C, Domingo E. Antiviral strategies based on lethal mutagenesis and error threshold. Curr Top Microbiol Immunol. 2015:392‐339. [DOI] [PubMed] [Google Scholar]
- 28. Ma Y., Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. 2015;112(30):9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eckerle LD, Becker MM, Halpin RA, et al. Infidelity of SARS‐CoV Nsp14‐exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6(5):e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ogando NS, Zevenhoven‐Dobbe JC, van der Meer Y, Bredenbeek PJ, Posthuma CC, Snijder EJ. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS‐CoV and SARS‐CoV‐2. J Virol. 2020;94(23):e01246‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gribble J, Stevens LJ, Agostini ML, et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17(1):e1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saramago M, Bárria C, Costa VG, et al. New targets for drug design: importance of nsp14/nsp10 complex formation for the 3'‐5' exoribonucleolytic activity on SARS‐CoV‐2. FEBS J. 2021. 10.1111/febs.15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Case JB, Ashbrook AW, Dermody TS, Denison MR. Mutagenesis of S‐adenosyl‐l‐methionine‐binding residues in coronavirus nsp14 N7‐methyltransferase demonstrates differing requirements for genome translation and resistance to innate immunity. J Virol. 2016;90(16):7248‐7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furuichi Y, Shatkin AJ. Viral and Cellular mRNA Capping: Past and Prospects, in Advances in Virus Research. Cambridge, MA: Academic Press; 2000:135‐184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nallagatla SR, Toroney R, Bevilacqua PC. A brilliant disguise for self RNA: 5'‐end and internal modifications of primary transcripts suppress elements of innate immunity. RNA Biol. 2008;5(3):140‐144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H, Kiledjian M. Decapping the message: a beginning or an end. Biochem Soc Trans. 2006;34(1):35‐38. [DOI] [PubMed] [Google Scholar]
- 37. Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses. RNA Biol. 2011;8(2):270‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. 2018;115(2):E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen P, Jiang M, Hu T, Liu Q, Chen XS, Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol. 2007;40(5):649‐655. [DOI] [PubMed] [Google Scholar]
- 40. Robson F, Khan KS, Le TK, et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79(5):710‐727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan J, Peng X, Gao Y, et al. Genome‐wide analysis of protein‐protein interactions and involvement of viral proteins in SARS‐CoV replication. PLoS One. 2008;3(10):e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Subissi L, Imbert I, Ferron F, et al. SARS‐CoV ORF1b‐encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 2014;101:122‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Becares M, Pascual‐Iglesias A, Nogales A, Sola I, Enjuanes L, Zuñiga S. Mutagenesis of Coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J Virol. 2016;90(11):5399‐5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patiño‐Galindo JÁ, Filip I, AlQuraishi M, Rabadan R. Recombination and lineage‐specific mutations led to the emergence of SARS‐CoV‐2. bioRxiv. 2020:2020. 10.1101/2020.02.10.942748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen N, Li S, Zhou R, et al. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017;242:90‐95. [DOI] [PubMed] [Google Scholar]
- 46. Sola I, Almazán A, Zúñiga S, Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2015;2(1):265‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Warren TK, Jordan R, Lo MK, et al. Therapeutic efficacy of the small molecule GS‐5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Senanayake SL. Drug repurposing strategies for COVID‐19. Future Drug Discov. 2020;2(2):fdd‐2020‐0010. [Google Scholar]
- 49. De Clercq E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev Anti Infect Ther. 2006;4(2):291‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoever G, Baltina L, Michaelis M, et al. Antiviral activity of glycyrrhizic acid derivatives against SARS−coronavirus. J Med Chem. 2005;48(4):1256‐1259. [DOI] [PubMed] [Google Scholar]
- 51. Khater S, Dasgupta N, Das G. Combining SARS‐CoV‐2 proofreading exonuclease and RNA‐dependent RNA polymerase inhibitors as a strategy to combat COVID‐19: a high‐throughput in silico screen. OSF Reprints. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]


