Abstract
To assess the practicability (usability and satisfaction) and analytical performances of the VitaPCR™ SARS‐CoV‐2 Assay (Credo Diagnostics Biomedical Pte. Ltd.), a rapid point‐of‐care nucleic acid amplification test (NAAT), by reference to real‐time reverse‐transcription polymerase chain reaction (rRT‐PCR) for respiratory viruses. The practicability of the VitaPCR™ Assay and Instrument was assessed from usability evaluation and a satisfaction questionnaire. Nasopharyngeal swabs were collected from 239 patients with coronavirus disease 2019 (COVID‐19)‐like illness during the second epidemic wave, in Paris, France. Overall, the usability of the VitaPCR™ Instrument was high. The satisfaction questionnaire indicated a high appreciation of the VitaPCR™ NAAT mainly for the short duration of analysis in only 20 min. A total of 140 and 99 samples were positive and negative for SARS‐CoV‐2 RNA by rRT‐PCR, respectively. In the event of significant viral load (i.e., N gene C t values 33), the platform's analytical performances dropped significantly, with lower sensitivity, concordance, and accuracy, while its specificity remained high. The VitaPCR™ SARS‐CoV‐2 Assay is an accurate rapid point‐of‐care NAAT, suitable for clinical practice for the rapid diagnosis of COVID‐19, especially in patients with COVID‐19‐suspected symptoms.
Keywords: bedside testing, COVID‐19, innovative approach, NAAT, PCR, point‐of‐care, practicability, SARS‐CoV‐2, sensitivity, specificity, VitaPCR™
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), a novel coronavirus that causes coronavirus disease 2019 (COVID‐19), started in Wuhan province, China, in December 2019, and was declared by the World Health Organization as a global pandemic on March 11, 2020. 1 SARS‐CoV‐2 infection has caused a worldwide viral pandemic, with >1,670,455 deaths and >75,371,570 cases reported internationally, with France reporting 2,427,316 cases and 59,619 deaths, as of December 18, 2020. 2
Controlling the outbreak in the community and hospitals mainly relied on the availability of highly sensitive and specific nucleic acid amplification‐based molecular testing for SARS‐CoV‐2, which is the gold standard for the diagnosis of SARS‐CoV‐2 in respiratory samples. 3 , 4 Molecular assays are crucial for the rapid application of infection control measures, case identification, and contact tracing. 5 Several nucleic acid amplification test (NAAT) platforms are currently available to be used at central reference or hospital laboratory levels. These large platforms are not suitable for small laboratory structures such as district laboratory level, lacking high‐level facilities and instruments, clinics, medical offices, and in general for decentralized diagnostics, as in establishments for the elderly, or even airports. Indeed, one of the major drawbacks of these assays is the need for viral nucleic acid extraction from clinical specimens. Furthermore, the assay run times of 1–3 h are still too long for timely decision support in a variety of important clinical situations (e.g., bed assignments for patients being admitted from the emergency department who may require cohorting by their COVID‐19 status). Thus, diagnostic tests specific to SARS‐CoV‐2 infection are urgently required to confirm suspected cases, screen patients, and conduct virus surveillance. In this scenario, a point‐of‐care, rapid, robust, and cost‐efficient device is crucial and urgently needed for the detection of COVID‐19. 6 Indeed, point‐of‐care tests are used to diagnose patients without sending samples to centralized facilities, thereby enabling communities without laboratory infrastructure to detect infected patients.
In search for a platform with a shorter turnaround time, we sought to evaluate the recently released VitaPCR™ SARS‐CoV‐2 Assay and Instrument (Credo Diagnostics Biomedical Pte. Ltd.). We report herein our field experience on the practicability and analytical performances of the VitaPCR™ SARS‐CoV‐2 Assay during the second wave of COVID‐19 epidemic in Paris, France, from our continuous quality improvement program required by the accreditation of medical biology laboratories.
2. MATERIALS AND METHODS
2.1. Patients specimens and procedures
Patients who were suspected of COVID‐19 attending the Centre Cardiologique du Nord—CCN, Saint‐Denis, France, were prospectively included during the last second epidemic wave in France from October to November 2020. Patients were subjected to nasopharyngeal flocked swab (Copan) in one nostril, and to the dedicated nylon flocked swab of the VitaPCR™ SARS‐CoV‐2 kit in the other nostril. SARS‐CoV‐2 RNA detection was first carried out by our reference multiplex real‐time reverse‐transcription polymerase chain reaction (rRT‐PCR) on native nasopharyngeal sample specimens. The swab specimens for VitaPCR™ testing were kept frozen at −20°C before use.
2.2. Nucleic acid amplification testing
2.2.1. Reference multiplex molecular detection of SARS‐CoV‐2
Nucleic acid extraction was performed from a 300 μl elution volume of nasopharyngeal flocked swab sample discharged in 1000 μl of physiological serum (NaCl 0.9%), using EX3600 extractor (Liferiver and Shanghai ZJ Bio‐Tech Co., Ltd), according to the manufacturer's instructions, and finally eluted in 50 μl (final volume). SARS‐CoV‐2 was detected in 5 μl of extracted RNA using the multiplex real‐time PCR novel coronavirus (2019‐nCoV) Real‐Time Multiplex RT‐PCR Kit (detection for three genes) (Liferiver and Shanghai ZJ Bio‐Tech Co., Ltd), and constituted the reference multiplex rRT‐PCR for SARS‐CoV‐2 RNA detection. This assay can simultaneously detect three coronavirus target genes, including the envelope protein gene (E), the RNA‐dependent RNA polymerase gene (ORF1ab of RdRP gene) and the nucleocapsid protein gene (N), using reverse transcription followed by real‐time PCR, providing individual cyclic threshold (C t) values for each target gene. Real‐time PCR was carried out with CFX96™ Real‐Time PCR Detection System (Bio‐Rad Laboratories), according to the manufacturer's instructions. The experiment and result interpretation were carried out according to the manufacturer's protocol.
2.2.2. VitaPCR™ SARS‐CoV‐2 Assay
The VitaPCR™ SARS‐CoV‐2 Assay performed on the VitaPCR™ Instrument (Credo Diagnostics Biomedical Pte. Ltd.; distributed in France by Biosynex) is a rapid molecular in vitro diagnostic test utilizing an rRT‐PCR amplification technology, using a patented thermal control system and enzymes. The assay is used for the qualitative detection of SARS‐CoV‐2 viral RNA without extraction in nasopharyngeal and oropharyngeal swabs from patients with signs and symptoms of respiratory infection who are suspected of COVID‐19. Aiming at a real point‐of‐care solution, the sample preparation is simplified in a 2 min hands‐on time procedure. Additionally, the well‐established lyophilization technique allows the assay to be stored at room temperature (25°C) for 1 year.
The detection target for detection of SARS‐CoV‐2 RNA is in the region of the virus nucleocapsid (N) gene. The assay is designed for first‐line specific detection of the SARS‐CoV‐2 RNA. In addition, detection of SARS‐like conserved region in N genes of SARS‐CoV‐2, SARS‐CoV, and bat SARS‐like coronavirus is provided as confirmatory testing. 7 An additional primer/probe set to detect human β‐globin, which is also used as a sample adequacy control, is included in the reagent for ensuring adequate processing of sample and monitoring the presence of inhibition factors in the rRT‐PCR process. During the whole reaction process, it is possible to visualize the progress curves of the real‐time PCR for the target SARS‐CoV‐2 and SARS‐CoV‐2‐like genes as well as for the internal DNA control. At the end of the analysis, the C t of each amplification curve is given.
One by one thawed nasopharyngeal sample taken with the dedicated nylon flocked swab was unloaded into a tube containing 4 ml of lysis buffer, with final analysis carried out with 30 μl of lysis buffer containing the sample, which was taken with a calibrated pipette after shaking to be placed in the PCR reagent tube containing a lyophilized master mix, according to the manufacturer's instructions. The reagent tube loaded with the sample was then placed on the VitaPCR™ Instrument for analysis in approximately 20 min. The experiment and result interpretation were carried out according to the manufacturer's protocol. In addition, the internal quality control (IQC) consisted of a synthetic DNA of the SARS‐CoV‐2 N gene segment, which sequence is used for both universal primer/probe and specific primer/probe set target (control set for SARS‐CoV‐2 PCR produced by Biosynex)
Forty microlitres of IQC was added into the sample collection buffer tube of the VitaPCR™ SARS‐CoV‐2 Assay, to process the test procedure. IQC was carried out twice a week.
2.2.3. Practicability of the VitaPCR™ platform
The practicability evaluation of the study platform was divided into two sub‐studies carried out by trained health care professionals.
2.2.4. Substudy 1: Usability evaluation
The usability of the VitaPCR™ Instrument for the qualitative molecular diagnosis of SARS‐CoV‐2 was assessed among volunteer health workers from the laboratory, including six laboratory technicians and four biologists. The participants were first trained on both platforms, and every one carefully read the instructions for each kit. Afterwards, the volunteers performed at least five measurements with each system, then completed the usability grid comprising 11 items (Figure 1).
Figure 1.
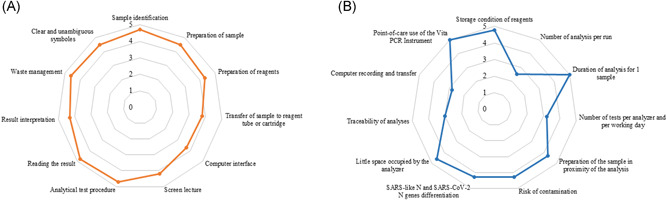
(A) Usability and (B) satisfaction. Evaluations of the VitaPCR™ SARS‐CoV‐2 Assay and Instrument (Credo Diagnostics Biomedical Pte. Ltd.) for the qualitative molecular diagnosis of SARS‐CoV‐2 RNA among six technicians and four biologists. Each usability and satisfaction item received a note using an arbitrary quantitative five‐point Likert's scale 8 ranging from 1 (very difficult), 2 (difficult), 3 (relatively easy) to 4 (easy) to 5 (very easy or comfortable). For each item, the mean note is indicated by a point
2.2.5. Substudy 2: Satisfaction questionnaire
Afterwards, the participants fulfilled the satisfaction questionnaire concerning their experiences with the VitaPCR™ platform, comprising 11 items (Figure 1).
Each usability and satisfaction item received a note using an arbitrary quantitative five‐point Likert's scale 8 ranging from 1 (very difficult), 2 (difficult), 3 (relatively easy) to 4 (easy) to 5 (very easy or comfortable).
2.2.6. Statistical analyzes
Data were entered into an Excel database and analyzed using IBM® SPSS® Statistics 20 software (IBM, SPSS Inc.). Means or medians were calculated for quantitative variables and proportions for categorical variables. The results were presented along with their 95% confidence interval (CI) using the Wilson score bounds for categorical variables. 9 The results of SARS‐CoV‐2 RNA detection by the multiplex rRT‐PCR (Liferiver and Shanghai ZJ Bio‐Tech Co., Ltd.) were used as the reference standard to estimate the sensitivity and specificity of the VitaPCR™ platform to detect SARS‐CoV‐2 RNA, with corresponding 95% CI. The concordance between the VitaPCR™ platform and multiplex molecular detection of SARS‐CoV‐2 RNA was assessed by percent agreement corresponding to the observed proportion of identical results between VitaPCR™ compared to rRT‐PCR detection. The reliability between the VitaPCR™ and the multiplex molecular detection of SARS‐CoV‐2 RNA was estimated by Cohen's κ coefficient, 10 and the degree of agreement was determined as ranked by Landlis and Koch. 11 The accuracy of the VitaPCR™ platform to correctly diagnose SARS‐CoV‐2 infection was estimated by Youden's J index (J = sensitivity + specificity − 1). 12 Positive predictive values (PPV) and negative predictive values (NPV) were calculated according to Bayes's formulae, by considering the official reported prevalence of SARS‐CoV‐2‐RNA positivity in symptomatic patients in the Paris area, France, on November 17, 2020 (Santé publique France, 2020).
2.2.7. Ethics statement
The study was used as a clinical evaluation of the continuous quality improvement program and COVID‐19 management measures performance evaluation, according to the national law on the accreditation of medical biology laboratories, 13 providing an exemption from informed consent application, according to the French public health code (Code de la Santé Publique, article L 1121‐1.1; https://www.legifrance.gouv.fr/). The data set was completely anonymous and did not contain any identifiable personal health information.
3. RESULTS
3.1. Practicability evaluation
Overall, the mean note of all usability items was 4.5 on the five‐point Likert's scale (Figure 1). The mean note of all satisfaction items was 4.3. The easy storage condition of reagents, the short duration of analysis in 20 min, the little place occupied by the VitaPCR™ Instrument and the possibility of point‐of‐care use of the VitaPCR™ Instrument were considered important strengths. However, only one analysis per run and the lack of connection with the laboratory's management computer system were seen as potential weaknesses.
3.2. Analytical performances using clinical samples
The analytical performances of the VitaPCR™ platform were assessed on clinical nasopharyngeal secretions samples from 239 patients attending the Centre Cardiologique du Nord, Saint‐Denis, France, for the diagnosis of COVID‐19, prospectively collected around the peak of the second wave of the COVID‐19 epidemic in France, using our routine multiplex rRT‐PCR for the detection of SARS‐CoV‐2 RNA, as a reference method. Among them, 140 and 99 samples were positive and negative for SARS‐CoV‐2 RNA by rRT‐PCR, respectively.
Overall, the VitaPCR™ platform showed high sensitivity, specificity, PPVs and NPVs of 90.0%, 99.0%, 94.6%, and 98.1%, respectively (Table 1), as well as high or almost perfect agreement (93.7%), reliability assessed by Cohen's κ coefficient (0.87), and accuracy assessed by Youden's J index (89%) to detect SARS‐CoV‐2 RNA. These analytical performances were further stratified according to the C t values of the N gene detected by reference rRT‐PCR, considering C t‐related criteria of very high (C t ≤ 20) and high (C t ≤ 33) SARS‐CoV‐2 RNA load. 14 , 15 In the event of a very high and high viral load, the analytical performances remained stable and excellent. However, in the event of moderate or very low viral load (C t > 33), the platform's analytical performances dropped significantly, with lower medium (60.0%) sensitivity, concordance, and accuracy, while its specificity remained high.
Table 1.
Analytical performances of the VitaPCR™ Assay and Instrument (Credo Diagnostics Biomedical Pte. Ltd.) for the qualitative molecular diagnosis of SARS‐CoV‐2 RNA using 140 positive and 99 negative nasopharyngeal samples by reference rRT‐PCR,a according to their N gene Ct values
| VitaPCR™ Assay and Instrumentb | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N gene Ct by rRT‐PCR, median (IQR)c | N | N gene Ct by VitaPCR™, median (IQR) | TP(n) | FN (n) | Sensitivity,d % [95% CI])a | Specificity,d % [95% CI] | Agreement,e % [95% CI] | Concordance,f % [95% CI] | Youden’ J index,g % [95% CI] | PPV,h % [95% CI] | NPV,h % [95% CI] | ||
| N gene C t i of positive samples by reference rRT‐PCR | ≤20 | 16 (10–20) | 34 | 22 (14–27) | 34 | 0 | 100 [97.2–100] | 99.0 [95.5–100] | 99.2 [95.8–100] | 0.97 [0.93–0.99] | 99.0 [95.5–100] | 95.1 [90.0–97.7] | 100 [97.1–100] |
| 21–33 | 28 (21–33) | 81 | 27 (0–35) | 77 | 4 | 95.1 [90.9–97.4] | 99.0 [96.2–99.7] | 97.2 [93.6–98.8] | 0.94 [0.89–0.96] | 94.1 [89.6–96.7] | 94.8 [90.5–97.2] | 99.1 [96.4–100] | |
| >33 | 35 (34–37) | 25 | 36 (0–37) | 15 | 10 | 60.0 [51.2–68.2] | 99.0 [95.3–100] | 91.1 [84.8–95.0] | 0.68 [0.59–0.76] | 59.0 [50.2–67.3] | 92.1 [86.0–95.7] | 92.8 [86.8–96.2] | |
| All C t values | 25 (10–37) | 140 | 22 (0–37) | 126 | 14 | 90.0 [85.5–93.2] | 99.0 [96.8–99.7] | 93.7 [89.9–96.1] | 0.87 [0.82–0.91] | 89.0 [84.4–92.4] | 94.6 [91.0–96.8] | 98.1 [95.5–99.2] | |
Abbreviations: C t, cycle threshold; FN, false negative; FP, false positive; IQR, interquartile range; NPV, negative predictive value; PPV, positive predictive value; rRT‐PCR, real‐time reverse‐transcription‐polymerase chain reaction; TP, true positive; TN, true negative.
95% confidence intervals in brackets were calculated by using the Wilson score bounds.
The CE IVD‐marked VitaPCR™ SARS‐CoV‐2 Assay uses a patented process of rRT‐PCR targeting the nucleocapsid (N) gene of SARS‐CoV‐2 and a SARS‐like N conserved region of SARS‐CoV‐2, SARS‐CoV, and bat SARS‐like coronavirus.
The CE IVD‐marked novel coronavirus (2019‐nCoV) Real Time Multiplex RT‐PCR Kit (detection for three genes) (Liferiver & Shanghai ZJ Bio‐Tech Co., Ltd.) constituted the reference multiplex rRT‐PCR for SARS‐CoV‐2 RNA detection.
Among the 99 negative samples by reference rRT‐PCR, one sample was positive by VitaPCR™ SARS‐CoV‐2 Assay, providing 98 TN and 1 FN samples by VitaPCR™ platform.
Agreement = TP + TN/TP + FP + TN + FN, expressed in percentage.
The Cohen's κ coefficient calculation was used to estimate the concordance 10 and interpreted according to the Landis and Koch scale, 11 as follows: <0 as indicating no agreement, 0–0.20 as slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1 as almost perfect concordance.
The accuracy of the VitaPCR™ platform to correctly diagnose SARS‐CoV‐2 infection was estimated by Youden's J index (J = sensitivity + specificity − 1). 12
PPV and NPV were calculated according to the Bayes's formulae, by taking into account the official reported prevalence of SARS‐CoV‐2‐RNA positivity in COVID‐19‐suspected patients in Paris's area, France, of 16.2% on November 17, 2020 (Santé publique France 2020; https://www.santepubliquefrance.fr/
).
The C t values of N gene detection by the reference Liferiver rRT‐PCR were used to classify nasopharyngeal samples according to their level of SARS‐CoV‐2 RNA load; C t of 20 and 33 were taken as thresholds of very high and high SARS‐CoV‐2 RNA load, respectively, as previously stated. 14 , 15
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
4. DISCUSSION
We herein evaluated the practicability and analytical performances of VitaPCR™ SARS‐CoV‐2 Assay and Instrument, a rapid point‐of‐care NAAT, by reference to multiplex rRT‐PCR for SARS‐CoV‐2 RNA detection. Overall, the usability of the VitaPCR™ SARS‐CoV‐2 Instrument was high with most items getting mean notes over four on a five‐point Likert's scale. The satisfaction questionnaire indicated high appreciation of the VitaPCR™ NAAT for its easy storage condition of reagents, the short duration of analysis in only 20 min, the little place occupied by the VitaPCR™ Instrument and the possibility of point‐of‐care use of the VitaPCR™ Instrument, although the platform showed less developed and practical computing and that it allowed only one sample to be analyzed at a time. In the event of significant viral load (i.e., N gene C t values < 33 by reference rRT‐PCR), the VitaPCR™ NAAT showed high sensitivity (≥95%) and specificity (≥99%) for SARS‐CoV‐2 RNA detection, with excellent concordance, reliability, and accuracy of the VitaPCR™ platform with the reference multiplex rRT‐PCR, and PPVs and NPVs above 94%. However, in the event of low viral load, the analytical performances were the only medium. Taken together, these observations demonstrate that the VitaPCR™ NAAT harbored excellent practicability and analytical performances in case of significant contagiousness, which makes it suitable to be used as point‐of‐care NAAT in various hospital and nonhospital settings where rapid molecular diagnosis of SARS‐CoV‐2 is necessary.
Our observations confirm the previous analytical evaluation of the VitaPCR™ platform for SARS‐CoV‐2 RNA molecular detection by Fournier and colleagues, 16 reporting similar high sensitivity (99.3%), specificity (94.7%), PPV (88.6%), and NPV (99.7%) of the VitaPCR™ SARS‐CoV‐2 Assay and Instrument. In addition, our findings indicate high usability and satisfaction concerning the VitaPCR™ platform and also demonstrate that the VitaPCR™ platform may preferentially be used in patients with significant viral load.
The practicability of VitaPCR™ SARS‐CoV‐2 Assay and Instrument was assessed by usability and satisfaction questionnaires completed by trained health care professionals. Overall, the assay with its dedicated platform was easy to use and received excellent appreciation for practical routine use. All reagents appeared very accessible contained in the kit. The assay required only approximately 2–5 min of hands‐on time. The transfer of the sample to the reagent tube and the intuitiveness of the computer interface were considered potential weaknesses. The very fast time of the result with the VitaPCR™ platform and the possibility of point‐of‐care use was particularly appreciated. Taken together, these observations allow easy deployment of the VitaPCR™ platform in facilities where licensed technologists may not be available or where technologists are unfamiliar with high‐complexity PCR assays. Placement of the VitaPCR™ platform in areas such as emergency departments may be a solution to the replacement of insufficiently sensitive antigen tests for COVID diagnosis.
Our results clearly show that the analytical performances of the VitaPCR™ SARS‐CoV‐2 Assay and Instrument were much better in the event of a high viral load, that is, in the case of significant viral load. These observations demonstrate the interest in the VitaPCR™ SARS‐CoV‐2 Assay and Instrument as a rapid rule‐in test for COVID‐19 with samples at a high viral load, in symptomatic patients for example, and caution its use as a singular rule‐out test especially in the setting of samples with low viral loads. The SARS‐CoV‐2 RNA positive subpopulation of our clinical samples collection was characterized by a wide range of C t‐values with medium and low C t‐values dominating. This allowed the calculation of sensitivity and specificity values with higher relevance for clinical practice. The C t‐dependent evaluation showed very good sensitivity for highly and moderately SARS‐CoV‐2 positive samples (C t ≤ 33). In contrast, the sensitivity of the assay with specimens containing only limited viral load was medium. Thus, the VitaPCR™ SARS‐CoV‐2 Assay and Instrument may have limited suitability for the determination of the SARS‐CoV‐2 infection status of patients with low viral excretion. COVID‐19 infection would not be detected in patients in the early or late phase of the infection typically associated with a low viral load. Otherwise and remarkably, the high performances of the VitaPCR™ SARS‐Cov‐2 Assay were obtained with no requirement for prior RNA extraction. These performances are reminiscent to those obtained with other NAATs for SARS‐CoV‐2 RNA qualitative detection, 17 , 18 , 19 , 20 , 21 although there are sometimes significant analytical differences between the analyzers according to the populations tested and the quality and processing of sampling. 17 , 18 , 22 In addition, in our series, we have stratified the nasopharyngeal samples according to the level of viral load, indirectly evaluated by the value of the C t of the N gene according to the reference rRT‐PCR. Indeed, there is a trend to a natural gradual decrease of the SARS‐CoV‐2 RNA load in the nasopharyngeal samples overtime during the course of infection, at the origin of varying levels of contagiousness. 23 Our results clearly show that the analytical performances of the VitaPCR™ platform were much better in the event of a high viral load, that is, in the case of significant viral load. These observations demonstrate the interest in the VitaPCR™ SARS‐CoV‐2 Assay as a rapid rule‐in test for COVID‐19 with samples at high viral load, in symptomatic patients for example, and caution its use as a singular rule‐out test especially in the setting of samples with lower viral loads.
In the last decade, multiplex molecular detection for the simultaneous detection of respiratory pathogens has been developed with high analytical performances. 24 However, the high cost of hospital molecular platforms, the complexity of the equipment required, the need for trained professionals and the delay of 2–4 h to obtain the results with frequent unavailability on weekends and during the night impaired the use of conventional NAATs for respiratory viral infections such as influenza or SARS‐CoV‐2 diagnosis even in hospital settings. Finally, the implementation of point‐of‐care molecular testing for SARS‐CoV‐2 RNA could deliver benefits to patient care admitted to hospital with COVID‐19‐related respiratory illness in terms of more rapid confirmation of coronavirus infection, a shorter length of stay, more rapid isolation practices of patients tested positive, and shorter turnaround time, as previously shown for influenza as compared to laboratory testing. 25
The limitations of our study include a relatively small sample size, inability to control for sampling variability, and lack of an additional comparator method to discern the discrepancies between VitaPCR™ and multiplex rRT‐PCR results. Another possible limitation is the freezing of nasopharyngeal samples before their use with the VitaPCR™ platform. Nevertheless, this factor might not have significantly influenced the results, as the samples were handled using the dedicated swab of the kit, and further tested in VitaPCR™ Instrument within the recommendations stipulated in the package insert.
Finally, our observations confirm high usability and good appreciation of VitaPCR™ platform for its short duration of analysis (20 min), the little place occupied, and the possibility of point‐of‐care use, although only a single sample analysis is considered as a limiting factor. Furthermore, the analytical performances of the VitaPCR™ platform were high and close to those of reference rRT‐PCR in the event of significant viral load. In conclusion, the VitaPCR™ SARS‐CoV‐2 Assay is an accurate rapid point‐of‐care NAAT, suitable for clinical practice for the rapid diagnosis of COVID‐19, especially in patients with COVID‐19‐suspected symptoms.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTION
Frédéric Fitoussi, Raphaël Dupont, and Laurent Bélec conceived and designed the research; Laurent Bélec carried out the practicability study; Frédéric Fitoussi and Raphaël Dupont performed the experiments; Serge Tonen‐Wolyec performed statistical analyses; all authors analyzed the results and drafted the manuscript.
ACKNOWLEDGMENTS
The authors are grateful to Biosynex, for providing for the tests for the study. Dr. Serge Tonen‐Wolyec was the recipient of the ERASMUS program between the University of Kisangani, and the University of Liège.
Fitoussi F, Dupont R, Tonen‐Wolyec S, Bélec L. Performances of the VitaPCR™ SARS‐CoV‐2 Assay during the second wave of the COVID‐19 epidemic in France. J Med Virol. 2021;93:4351–4357. 10.1002/jmv.26950
REFERENCES
- 1. World Health Organization . Coronavirus disease (COVID‐19) outbreak. https://www.who.int. Accessed December 18, 2020.
- 2. Worldometer. https://www.worldometers.info/coronavirus/. Accessed March 21, 2021.
- 3. Péré H, Podglajen I, Wack M, et al. Nasal swab sampling for SARS‐CoV‐2: a convenient alternative in times of nasopharyngeal swab shortage. J Clin Microbiol. 2020;58(6):e00721‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581:465‐469. [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization . Laboratory testing for 2019 novel coronavirus (2019‐nCoV) in suspected human cases. Interim guidance; 2020. https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117. Accessed March 21, 2021.
- 6. Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID‐19: the disease and tools for detection. ACS Nano. 2020;14(4):3822‐3835. [DOI] [PubMed] [Google Scholar]
- 7. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Likert R. A technique for the measurement of attitudes. Arch Psychol. 1932;140:1‐55. [Google Scholar]
- 9. Newcombe RG. Two‐sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857‐872. [DOI] [PubMed] [Google Scholar]
- 10. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37‐46. [Google Scholar]
- 11. Landlis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159‐174. [PubMed] [Google Scholar]
- 12. Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32‐35. [DOI] [PubMed] [Google Scholar]
- 13. Journal Officiel de la République Française . Ordonnance n° 2010‐49 du 13 janvier 2010 relative à la biologie médicale. https://www.legifrance.gouv.fr/jorf/id/JORFTEXT000021683301/. Accessed October 09, 2020.
- 14. Société Française de Microbiologie (SFM) . Avis du 25 septembre de la Société Française de Microbiologie (SFM) relatif à l′interprétation de la valeur de Ct (estimation de la charge virale) obtenue en cas de RT‐PCR SARS‐CoV‐2 positive sur les prélèvements cliniques réalisés à des fins diagnostiques ou de dépistage. Version 1 _ 25/09/2020; 2020. https://www.sfm-microbiologie.org/wp-content/uploads/2020/09/Avis-SFM-valeur-Ct-excre%CC%81tion-virale-Version-Finale-25092020.pdf. Accessed March 21, 2021.
- 15. Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS‐CoV‐2 in infected patients. Clin Infect Dis. 2020;71(15):793‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fournier PE, Zandotti C, Ninove L, et al. Contribution of VitaPCR SARS‐CoV‐2 to the emergency diagnosis of COVID‐19. J Clin Virol. 2020;133:104682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Basu A, Zinger T, Inglima K, et al. Performance of Abbott ID Now COVID‐19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City Academic Institution. J Clin Microbiol. 2020;58(8):e01136‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dinnes J, Deeks JJ, Cochrane COVID‐19 Diagnostic Test Accuracy Group , et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldenberger D, Leuzinger K, Sogaard KK, et al. Brief validation of the novel GeneXpert Xpress SARS‐CoV‐2 PCR assay. J Virol Methods. 2020;284:113925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Österdahl MF, Lee KA, Lochlainn MN, et al. Detecting SARS‐CoV‐2 at point of care: preliminary data comparing loop‐mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis. 2020;20(1):783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolters F, van de Bovenkamp J, van den Bosch B, et al. Multi‐center evaluation of cepheid xpert® xpress SARS‐CoV‐2 point‐of‐care test during the SARS‐CoV‐2 pandemic. J Clin Virol. 2020;128:104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lephart PR, Bachman MA, LeBar W, et al. Comparative study of four SARS‐CoV‐2 Nucleic Acid Amplification Test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. Diagn Microbiol Infect Dis. 2021;99(1):115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Concato C, Piccioni L, Ranno S, et al. Comparison of the Allplex™ Respiratory Panel Assays and the automated Fast Track Diagnostics Respiratory pathogens 21 assay for the diagnosis of pediatric respiratory viral infections. Arch Virol. 2020;165(5):1191‐1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berry L, Lansbury L, Gale L, Carroll AM, Lim WS. Point of care testing of Influenza A/B and RSV in an adult respiratory assessment unit is associated with improvement in isolation practices and reduction in hospital length of stay. J Med Microbiol. 2020;69(5):697‐704. [DOI] [PMC free article] [PubMed] [Google Scholar]


