Abstract
Key points
We report a novel method for the transient expression of SARS‐CoV‐2 envelope (E) protein in intracellular organelles and the plasma membrane of mammalian cells and Xenopus oocytes.
Intracellular expression of SARS‐CoV‐2 E protein increases intra‐Golgi pH.
By targeting the SARS‐CoV‐2 E protein to the plasma membrane, we show that it forms a cation channel, viroporin, that is modulated by changes of pH.
This method for studying the activity of viroporins may facilitate screening for new antiviral drugs to identify novel treatments for COVID‐19.
Abstract
The envelope (E) protein of coronaviruses such as SARS‐CoV‐1 is proposed to form an ion channel or viroporin that participates in viral propagation and pathogenesis. Here we developed a technique to study the E protein of SARS‐CoV‐2 in mammalian cells by directed targeting using a carboxyl‐terminal fluorescent protein tag, mKate2. The wild‐type SARS‐CoV‐2 E protein can be trafficked to intracellular organelles, notably the endoplasmic reticulum–Golgi intermediate complex, where its expression increases pH inside the organelle. We also succeeded in targeting SARS‐CoV‐2 E to the plasma membrane, which enabled biophysical analysis using whole‐cell patch clamp recording in a mammalian cell line, HEK 293 cells, and two‐electrode voltage clamp electrophysiology in Xenopus oocytes. The results suggest that the E protein forms an ion channel that is permeable to monovalent cations such as Na+, Cs+ and K+. The E current is nearly time‐ and voltage‐independent when E protein is expressed in mammalian cells, and is modulated by changes of pH. At pH 6.0 and 7.4, the E protein current is activated, whereas at pH 8.0 and 9.0, the amplitude of E protein current is reduced, and in oocytes the inward E current fades at pH 9 in a time‐ and voltage‐dependent manner. Using this directed targeting method and electrophysiological recordings, potential inhibitors of the E protein can be screened and subsequently investigated for antiviral activity against SARS‐CoV‐2 in vitro and possible efficacy in treating COVID‐19.
Keywords: envelope protein, ion channel, membrane targeting, SARS‐CoV‐2
Key points
We report a novel method for the transient expression of SARS‐CoV‐2 envelope (E) protein in intracellular organelles and the plasma membrane of mammalian cells and Xenopus oocytes.
Intracellular expression of SARS‐CoV‐2 E protein increases intra‐Golgi pH.
By targeting the SARS‐CoV‐2 E protein to the plasma membrane, we show that it forms a cation channel, viroporin, that is modulated by changes of pH.
This method for studying the activity of viroporins may facilitate screening for new antiviral drugs to identify novel treatments for COVID‐19.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) emerged in 2019, when an outbreak of presumed viral pneumonia was documented in Wuhan, China (Zhou et al. 2020 b). Following the onset of the epidemic in Wuhan in December 2019, the infectious agent responsible was isolated and identified as a betacoronavirus (Zhu et al. 2020). This novel coronavirus, termed SARS‐CoV‐2, is closely related to SARS‐CoV‐1 and MERS‐CoV (Andersen et al. 2020), and to a number of coronaviruses circulating in bats in Yunnan (Zhu et al. 2020), and especially to those associated with an area where zoonotic transmission to humans is strongly suspected, based on serological surveillance (Wang et al. 2018). Extensive spread of the virus was facilitated by international travel (Bedford et al. 2020; Gonzalez‐Reiche et al. 2020), and early seeding (Bedford et al. 2020; Worobey et al. 2020; Stadlbauer et al. 2021) combined with efficient human‐to‐human transfer (Shang et al. 2020), asymptomatic transmission (Arons et al. 2020; Moghadas et al. 2020) and slow public health measures in many countries (Pei et al. 2020) resulted in a global pandemic that has now infected and killed millions (World Health Organization, 2021). The infectious disease now referred to as COVID‐19 is complex and can be severe in highly susceptible individuals with known risk factors (e.g. advanced age, obesity, diabetes or hypertension) leading to high mortality rates (Cummings et al. 2020; Williamson et al. 2020; Zhou et al. 2020 a). Severe lower respiratory symptoms are common in these patients, such as pneumonia and acute respiratory distress syndrome (Mehta et al. 2020; Zhang et al. 2020), as well as findings more typical of systemic infection and sepsis, such as renal failure and disseminated intravascular coagulopathy (do Espírito Santo et al. 2020; Giannis et al. 2020; Klok et al. 2020).
The pathophysiology of infection with coronaviruses is proposed to be dependent on a group of accessory proteins, such as 3a (Castaño‐Rodriguez et al. 2018; Yue et al. 2018; Kern et al. 2020) and the envelope (E) protein (Nieto‐Torres et al. 2014) of SARS‐CoV‐1. These proteins are suspected of being ‘viroporins’ (Nieva et al. 2012; Kern et al. 2020), i.e. viral proteins that can form ion channels, and may play a role in the pathogenesis of SARS‐CoV‐2 (McClenaghan et al. 2020; Schoeman & Fielding, 2020).
The function of viroporins is not yet fully understood and some aspects of the biophysics are still controversial (McClenaghan et al. 2020). There is some evidence that these viroporins participate in viral propagation and pathogenesis (Nieva et al. 2012). Viroporins such as the influenza A virus M2 protein are permeable to cations, including H+ (Pinto et al. 1992; Schnell & Chou, 2008), and are believed to function in the process of intracellular acidification, whereby protons (H+) are transported from the lumen of the Golgi apparatus into the cytoplasm. This results in an increase in pH inside these organelles that is apparently required for the release of virions in influenza (Helenius, 1992), for appropriate assembly of hepatitis C virus (Wozniak et al. 2010), and for virus assembly and production in coronaviruses (Nieto‐Torres et al. 2015 b; Schoeman & Fielding, 2019).
The E protein of the coronavirus SARS‐CoV‐2 is a small protein of 75 amino acid residues that shows 95% sequence identity with that of SARS‐CoV‐1 (Bianchi et al. 2020). There is a consensus view that the E protein of SARS‐CoV‐1 plays an important role not only in viral replication but also in pathogenicity (Schoeman & Fielding, 2019). SARS‐CoV‐1 E, which has been shown to be important for optimal virus packaging and export (Nieto‐Torres et al. 2014), demonstrates several properties that are consistent with ion channel activity (Wilson et al. 2004; Torres et al. 2007). Computational models and biochemical analysis of the SARS‐CoV‐1 E protein have suggested that it may form as a 45 kDa pentamer of the 9 kDa E protein (Pervushin et al. 2009), and electrophysiological recordings in artificial planar bilayer membranes have been used to investigate E protein function (Torres et al. 2006; Nieto‐Torres et al. 2015 b). Recent studies in bacteria suggested an ion channel function for SARS‐CoV‐2 E (Singh Tomar & Arkin, 2020) and bilayer recordings suggest that it forms a cation channel (Xia et al. 2020). Topological analysis of the E protein suggests that the amino (N)‐terminus resides within intracellular organelles and the carboxyl (C)‐terminus in the cytoplasm (Duart et al., 2020), with a single transmembrane segment, the structure of which has been solved using NMR spectroscopy (Mandala et al. 2020). However, with regard to this point, detailed study of the E protein in mammalian cell membranes has not been possible.
Here we report a novel strategy for the expression of the SARS‐CoV‐2 E protein in mammalian cells that facilitates the characterization of its ion channel activity. Using a fluorescent carboxyl‐terminal tag (mKate2), we demonstrate that the E protein can be trafficked to the endoplasmic reticulum–Golgi intermediate complex (ER–GIC), resulting in increased pH within the organelle. We also succeeded for the first time in targeting the E protein to the plasma membrane for biophysical analysis, and were able to show that plasma membrane expression of the SARS‐CoV‐2 E protein in HEK 293 cells and Xenopus oocytes is associated with a current that is carried by cations and regulated by pH.
Methods
Cell culture
Human embryonic kidney (HEK) 293S cells (ATCC, cat. no. CRL‐3022) and NIH 3T3 cells (ATCC, cat. no. CRL‐1658) were cultured in Dulbecco's modified Eagle's medium (DMEM)–nutrient mixture F‐12 (Thermo Fisher Scientific, Waltham, MA, USA, cat no. 11320033) and DMEM (Thermo Fisher Scientific cat. no. 10313021), respectively. Both media were supplemented with GlutaMax‐I and penicillin–streptomycin and 10% fetal bovine serum (not heat‐inactivated, HyClone, cat. no. SH30071.03, Thermo Fisher Scientific) under normoxia (20% O2, 5% CO2, at 37°C). HEK 293S cells were subcultured once a week by gently pipetting and plated at a density of 105 cells ml−1. The culture medium was replaced every 3–4 days. NIH 3T3 cells were passaged using trypsin–EDTA (0.25%, Thermo Fisher Scientific, cat. no. 25200‐056) every other day. For electrophysiological recordings, cells were plated on 12 mm glass coverslips (Neuvitro, Camas, WA, USA).
Molecular biology constructs and transfection
Plasmid DNA constructs were generated using standard methods with restriction enzymes (New England BioLabs, Ipswich, MA, USA), DNA ligase (MightyMix, TaKaRa, Mountain View, CA, USA) and polymerase chain reaction (PCR) with Phusion polymerase (Thermo Fisher Scientific). Construct inserts for these experiments were synthesized (Integrated DNA Technologies, Coralville, IA, USA) and subcloned into pcDNA3 vector (Thermo Fisher Scientific). Schematic representations of the constructs are shown in Fig. 1. The initial construct encoding the E protein targeted to the intracellular site (SARS2‐E‐mKate2 in Fig. 1) was termed ‘WT’ and the construct encoding the E protein targeted to the plasma membrane (SARS2‐E‐Ala6‐ΔPBM‐PM‐mKate2 in Fig. 1) was termed ‘PM’. The PM construct without the fluorescent carboxyl‐terminal fluorescent tag (mKate2) was termed ‘untagged‐PM.’ Mock transfections were performed by using pcDNA3 vector with mKate2‐Nuclear Localization Signal (NLS) as the insert. The day before transfection, HEK 293S cells were plated at a cell density of 3 × 105 cells ml−1. When HEK 293S cells reached between 60 and 80% confluence, cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, cat. no. 11668027); 0.8 μg of the plasmid was mixed into 50 μl of serum‐free OptiMEM, and 2 μl of Lipofectamine 2000 added to another 50 μl of serum‐free OptiMEM. Plasmid and Lipofectamine were mixed and incubated 30 min at room temperature, and then added to the plate. After 4 h, the medium was changed to fresh HEK 293S culture medium and cells were then replated at the desired density for imaging and electrophysiological experiments. Cells were incubated for 18–36 h in a 5% CO2 incubator before recordings were initiated. Transfection efficiency was estimated by counting cells that show mKate2 fluorescence and was typically moderate, between 20 and 35% for physiological recordings, in order to minimize lipofection toxicity.
Figure 1. Targeting SARS‐CoV‐2 E protein to the plasma membrane of NIH 3T3 cells.
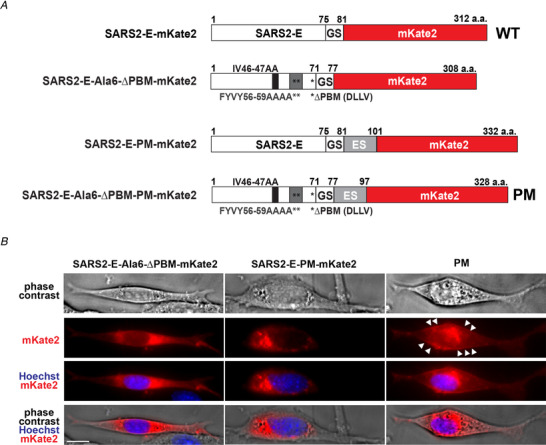
A, diagram of the SARS‐CoV‐2 E (SARS2‐E) constructs used for targeting experiments in NIH 3T3 cells. Numbers indicate the position of the amino acid (a.a.) in the sequence. B, representative phase contrast and fluorescence images of NIH 3T3 cells transfected with the constructs shown in A. Scale bar, 10 μm. The initial SARS2‐E‐mKate2 construct consisted of the cDNA encoding SARS‐CoV‐2 E protein, fused to a carboxyl (C)‐terminal fluorescent tag mKate2, separated by an intermediate spacer (GS) using glycine‐serine repeat (GSGSGS) and was termed WT for subsequent experiments (A, first construct). The construct SARS2‐E‐Ala6‐ΔPBM‐mKate2 (A, second construct) was made by mutating the PDZ‐binding motif (PBM) by replacement of six residues, 46–47 (A, described above the constructs) and 56–59 (A, indicated with double asterisks and described below the constructs) with alanine (Ala6) and deleting the C‐terminal ER retention signal DLLV (ΔPBM, indicated with single asterisk in the constructs). This construct resulted in expression of the E protein in organelles (B). A third construct (A) SARS2‐E‐PM‐mKate2 was made by insertion of a consensus Golgi export signal (ES) of Kir2.1 (KSRITSEGEYIPLDQIDINV) after the spacer, and again this construct resulted in expression of the E protein in organelles (B). The final construct SARS2‐E‐Ala6‐ΔPBM‐PM‐mKate2 (A) was made by inserting the Golgi export signal into the second construct, and this construct directed expression of the E protein to the plasma membrane (B, white arrowheads) as well as intracellular structures (B). We termed this final construct PM. [Color figure can be viewed at wileyonlinelibrary.com]
Channel subunit cRNA preparation and Xenopus laevis oocyte injection
Complementary RNA (cRNA) transcripts encoding WT and PM channel constructs were generated by in vitro transcription with the mMessage mMachine kit (Thermo Fisher Scientific) from linearized cDNA containing the channel coding regions in the pXOOM vector, in which Xenopus laevis β‐globin 5′‐ and 3′‐untranslated regions flank the insert. For the experiments in oocytes we used the untagged‐PM construct, the mKate2‐tagged PM construct (PM) and the mKate2‐tagged WT construct (WT). The cRNAs (30–60 ng) were injected into defolliculated stage V and VI Xenopus laevis oocytes obtained from a commercial supplier (Xenoocyte, Dexter, MI, US). The oocytes were stored for 3 days at 16°C in ND96 oocyte storage solution containing penicillin and streptomycin, with daily washing, prior to two‐electrode voltage‐clamp recording.
Immunofluorescence in HEK 293S cells
HEK 293S cells were transfected with mock, WT or PM constructs and plated on 12 mm glass coverslips (Neuvitro). Between 18 and 24 h after transfection, HEK 293S cells were washed in phosphate‐buffered saline (PBS), fixed for 15 min in cold methanol and permeabilized in 0.3% Triton X‐100 in PBS for 5 min. Non‐specific binding was blocked with 5% bovine serum albumin (BSA) for 60 min and the cells were then incubated with primary antibody mouse anti‐ERGIC‐53 (1:100, Enzo Life Sciences, Farmingdale, NY, USA) in 1.5% BSA overnight at 4 °C. The cells were rinsed three times with PBS for 5 min each and incubated with Alexa Fluor 488‐conjugated anti‐mouse antibodies (1:500, Thermo Fisher Scientific) in 1.5% BSA for 1 h at room temperature. The cells were rinsed three times with PBS for 5 min each, counterstained with 10 μm 4,6‐diamidino‐2‐phenylindole (DAPI) and mounted on coverslips with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA). Images were acquired on a confocal microscope (A1, Nikon Instruments), and then processed with ImageJ software (NIH).
Imaging experiments and pH measurements
For the imaging experiments in Figs 1 and 3, NIH 3T3 cells were plated at a cell density of 2.5 × 105 cells ml−1. The following day, the cells were transfected using Lipofectamine 3000 reagents (Thermo Fisher Scientific, cat. no. L3000001); 4 μg of plasmid was mixed into 125 μl of serum‐free OptiMEM with 5 μl of P3000 reagent. This was then added to another 125 μl of serum‐free OptiMEM containing 7.5 μl of Lipofectamine 3000. Plasmid/P3000‐lipofectamine complex was incubated for 15 min at room temperature, and then added to the plate. For the pH imaging experiments in Fig. 3, the medium was replaced 20–24 h after transfection, and 1 μm of Lysosensor Green DND‐189 (Thermo Fisher Scientific, cat. no. L7535) was added. The cells were incubated for 30 min in a 37 °C, 5% CO2 incubator. The medium was replaced one final time prior to imaging. The live cell imaging was conducted on a customized/automated fluorescence microscope (Ti‐U, Nikon) using an environmental chamber (Tokai‐Hit Co., Shizuoka‐ken, Japan) and the culture medium to maintain normal cell culture conditions (37 °C, 5% CO2, 20% O2). NIH 3T3 cells were used because of stronger adherence to glass surfaces and a larger cytoplasmic volume, which make these cells more suitable for longer‐term intracellular imaging, than HEK 293S cells. Transfection efficiency was estimated by counting cells that showed mKate2 fluorescence, and was typically between 25 and 35%. Fluorescence quantification and analysis was performed with ImageJ software and Prism 7 (GraphPad Software Inc., La Jolla, CA, USA). Representative images were also gathered on a Leica DMi8 microscope (Leica Microsystems, Wetzlar, Germany).
Figure 3. SARS‐CoV‐2 E protein expression decreases DND‐189 fluorescence in NIH 3T3 cells.
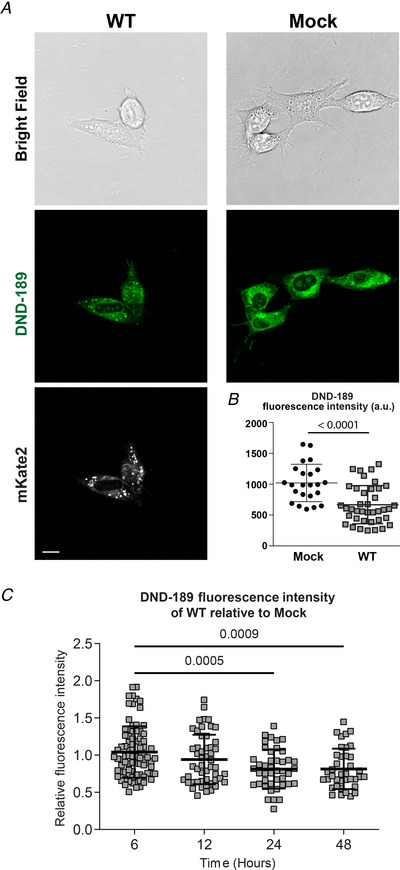
A, representative cell images of NIH 3T3 cells transfected with pcDNA3 vector (Mock) or WT. Top row, bright‐field images; second row, LysoSensor DND‐189 images (green fluorescence); third row, mKate2 fluorescence to confirm E protein expression. Scale bar, 10 μm. B, quantification of fluorescence intensity in mock (n = 23) and WT (n = 39) transfected cells showing a decrease in mean cell fluorescence intensity 24 h following WT expression (1021 ± 304 vs. 668 ± 308 a.u. for mock and WT, respectively), which is consistent with an increase in luminal pH within organelles. Fluorescence was quantified using ImageJ analysis software. Student's t‐test, P < 0.0001. C, the time course of changes in pH monitored via fluorescence intensity at 6, 12, 24 and 48 h post‐transfection (n = 83, 51, 43 and 38, respectively) showing a decreasing relative fluorescence intensity (r.f.u., relative to mock) over time, indicating an increase in organellar pH. One‐way ANOVA F (3,211) = 7.133, P = 0.0001, Bonferroni's post hoc test when compared to 6 h (1.04 ± 0.34 r.f.u.): 12 h (0.94 ± 0.33 r.f.u., P = 0.2022), 24 h (0.81 ± 0.26 r.f.u., P = 0.0005), 48 h (0.81 ± 0.27 r.f.u., P = 0.0009). In B and C, data were collected and pooled from three separate transfections for each condition. Data are presented as means ± SD. [Color figure can be viewed at wileyonlinelibrary.com]
Whole‐cell voltage‐clamp recording
Recordings in HEK 293S cells were performed between 18 and 36 h after transfection. A coverslip containing the transfected cells was submerged in a chamber and continuously perfused at a flow rate of 1–2 ml min−1 with Hepes‐buffered experimental saline solution at room temperature (22–25°C). HEK 293S cells were visualized under an upright light microscope (BX51WI, Olympus, Tokyo, Japan) coupled to a camera (C8484, Hamamatsu Photonics, Hamamatsu, Japan). Transfected cells were identified by the mKate2 fluorescence (illuminated with a mercury lamp or with LED at 525 nm). Pipettes were pulled from borosilicate glass (World Precision Instruments, Sarasota, FL, USA, TW150F‐4) with a resistance of 2–4 MΩ when filled with one of the following ‘intracellular’ pipette solutions: potassium gluconate or CsCl. Potassium gluconate‐based pipette solution contained (in mm): 130 potassium gluconate, 5 KCl, 4 NaCl, 10 Hepes, 0.2 EGTA, 4 ATP‐Mg, 0.3 GTP‐Mg and 10 phosphocreatine. CsCl pipette solutions contained (in mm): 140 CsCl, 1 MgCl2, 10 Hepes, 0.5 EGTA, 2 ATP‐Mg and 0.2 GTP‐Na. The CsCl internal solution is a calcium‐free solution ([Ca2+]i < 10 nm). All pipette solutions were adjusted to pH 7.25, and osmolality to 285 mOsm kg−1. The standard extracellular solution was (in mm): 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 10 Hepes and 10 glucose. For the permeability experiments, our standard saline solution was replaced with the following ‘external’ solutions: N‐methyl D‐glucamine+ (NMDG) solution contained (in mm): 140 NMDG, 1 MgCl2, 2 CaCl2, 10 Hepes and 10 glucose; and KCl external solution contained (in mm): 145 KCl, 1 CaCl2, 10 Hepes and 10 glucose. The pH of the external solutions was adjusted to 7.4 and the osmolality to 305 mOsm kg−1 with sucrose. Low pH 6.0 solution was buffered with MES (10 mm) and pH 8.0 solution with Hepes (10 mm).
Data were collected with a Multiclamp 700B amplifier (Molecular Devices, San Jose, CA, USA) and Clampex 10.6 (Molecular Devices). Data were collected at 10 kHz and low‐pass filtered at 1 kHz. A priori exclusion criteria were adopted and established at the beginning of the project in order to minimize data acquisition from multiple cell assemblies and extended syncytia, and these criteria stipulated that recordings in which C m > 50 pF (C m > 55 pF for WT), R m < 200 MΩ, or magnitude of holding current I h > 50 pA were not included for analysis. Series resistance (R s) was monitored throughout the experiments and recordings were discarded if R s changed >30% during an experiment or if R s > 20 MΩ. Recordings were performed using the whole‐cell voltage‐clamp variation of the patch clamp technique (Hamill et al. 1981) from individual transfected cells emitting the red fluorescence signal from mKate2. After attaining the stable whole‐cell configuration, the cells were held at −40 mV and current–voltage (I–V) relationships spanning the range from −100 mV to +100 mV were generated by applying rectangular voltage commands of 200 ms duration in 5 mV increments and 600 ms intervals between voltage steps. The I–V relationships were calculated from the steady‐state values of the current response using Clampfit 10.6 (Molecular Devices). The E currents were calculated after subtracting currents from mock‐transfected cells using Prism 8.0 (GraphPad). Capacitance was measured online using Clampex 10.6 (Molecular Devices) with an update rate at 100 Hz. The reversal potential (E rev) was measured for each ion substitution experiment following the mock subtraction procedure. The relative permeability ratios for cations were calculated from observed E rev values using an equation derived from the Goldman–Hodgkin–Katz current equation (Hille, 2001). Membrane voltages reported are uncorrected for a liquid junction potential of 4.6 mV (Na+ external solution), 0.7 mV (K+ external solution) and 0.6 mV (NMDG external solution), calculated using pClamp 10.6 software (Molecular Devices).
Two‐electrode voltage‐clamp in Xenopus oocytes
We performed two‐electrode voltage‐clamp recording at room temperature with an OC‐725C amplifier (Warner Instruments, Hamden, CT, USA) and pClamp10 software. Oocytes were placed in a small‐volume oocyte bath (Warner Instruments). Bath solution (in mm: 96 NaCl, 4 KCl, 1 MgCl2, 1 CaCl2, 10 Hepes; pH as stated) was introduced via gravity perfusion; pipettes were of 1–3 MΩ resistance when filled with 3 m KCl. Recordings were performed with voltage pulses between −100 mV and +70 mV at 10 mV increments for the prepulse from a holding potential of 0 mV and then holding at −30 mV for the tail pulse before returning to 0 mV. Clampfit (Molecular Devices) and Origin (OriginLab Corp., Northampton, MA, USA) were used to analyse data and produce figures.
Statistics
All statistical analyses were performed using Prism 7.0 or 8.0. All summary data are presented as means ± sd. Additional detailed statistical information is included in the figure legends.
Results
Expression of SARS‐CoV‐2 WT and PM targeting constructs in HEK 293S cells
We initiated our study by expressing the E protein of SARS‐CoV‐2 in two common mammalian cell lines, NIH 3T3 and HEK 293S cells, knowing that other renal epithelial cells (e.g. Vero E6) express high levels of angiotensin‐converting enzyme 2 (ACE2) (Li et al. 2003) and hence are efficiently infected by the SARS‐CoV‐2 virus (Zhou et al. 2020 b), making renal epithelial cells an important secondary target of the virus that is relevant to the pathogenesis of COVID‐19 (Regla‐Nava et al. 2013).
We first expressed the original construct of the SARS‐CoV‐2 E protein in NIH 3T3 cells and then engineered constructs designed to target the protein to the plasma membrane to allow us to characterize its electrophysiological properties. Initially, the cDNA encoding SARS‐CoV‐2 E protein was fused to a C‐terminal tag consisting of the modified red fluorescent protein mKate2 (Shcherbo et al. 2009) and we termed this construct ‘WT’ (Fig. 1 A and B). We then engineered this construct to directly express SARS‐CoV‐2 E proteins at the plasma membrane of mammalian cells. Deletion of the C‐terminal ER retention signal DLLV, which was first identified in the E protein of avian coronavirus (Lim & Liu, 2001) and in SARS‐CoV‐1 (Cohen et al. 2011), combined with the insertion of a consensus Golgi export signal from the mammalian ion channel Kir2.1 (Hofherr et al. 2005) inserted between E and mKate2 (Fig. 1 A and B), allowed us to detect mKate2 fluorescence in the plasma membrane in NIH 3T3 cells (Fig. 1 B). We designated this construct ‘PM’ for the subsequent electrophysiological and imaging experiments.
The subcellular distribution of the E protein was then characterized in HEK 293S cells with a confocal microscope (Fig. 2). Mock transfections were performed using a nuclear localization signal (NLS, PKKKRKV) fused to mKate2. The red fluorescence was confined to the nucleus, as expected (Fig. 2 A). Using immunofluorescence with an antibody against ERGIC‐53, we were able to show that the red fluorescence signal associated with expression of the WT construct is confined to intracellular organelles situated adjacent to the nucleus, and was largely co‐localized with the marker of the ER and the ER–GIC (Fig. 2 A and B). The PM construct allowed us to clearly detect mKate2 fluorescence in the plasma membrane of HEK 293S cells (Fig. 2 A).
Figure 2. Expression of SARS‐CoV‐2 WT and PM targeting constructs in HEK 293S cells.
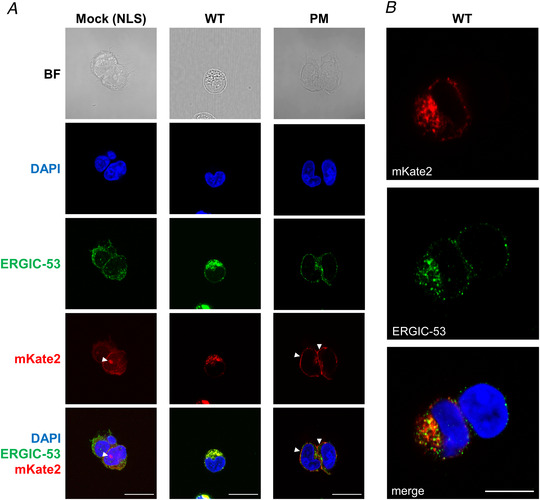
A, representative confocal images of HEK 293S cells transfected using three constructs: pcDNA3 vector‐mKate2‐NLS (mock), SARS2‐E‐mKate2 (WT) and SARS2‐E‐Ala6‐ΔPBM‐PM‐mKate2 (PM). We were able to study the subcellular distribution of the E protein in HEK 293S cells by visualizing the red fluorescence signal of mKate2. Immunolabelling with anti‐ERGIC‐53 shows that expression of the WT construct is largely confined to intracellular organelles adjacent to the nucleus, presumably the ER and the ER–Golgi intermediate compartment (anti‐ERGIC‐53). Deletion of the ER retention signal first identified in the SARS‐CoV‐1 E protein combined with the insertion of a consensus Golgi export signal from the mammalian ion channel Kir2.1, allowed us to detect the PM construct in the plasma membrane. Panels show bright field (BF) images, nuclei stained for DAPI, mouse anti‐ERGIC‐53, and the C‐terminal red fluorescent tag, mKate2. These images show nuclear expression of NLS (mock, white arrowheads), organelle expression of E protein (WT) and expression of E protein at the plasma membrane (PM, white arrowheads). The bottom row shows merged images for DAPI, ERGIC‐53 and mKate2. Scale bar, 20 μm. B, a representative image of HEK 293S cells transfected with WT construct, which shows at higher resolution the co‐localization of SARS‐CoV‐2 E protein (mKate2) with the marker for ERGIC‐53 surrounding the nuclei (DAPI staining). Scale bar, 10 μm. [Color figure can be viewed at wileyonlinelibrary.com]
SARS‐CoV‐2 E protein expression decreases DND‐189 fluorescence in NIH 3T3 cells
To investigate whether intracellular expression of SARS‐CoV‐2 E can alter the internal pH of organelles, NIH 3T3 cells were transfected with the WT E protein construct and imaged using the fluorescent pH‐sensitive reporter DND‐189 (Fig. 3 A). Our results show that E protein expression decreases the fluorescence of regions of interest that correspond to intracellular organelles in these cells (1021 ± 304 fluorescence arbitrary units (a.u.) vs. 668 ± 308 a.u. for mock and WT, respectively) (Fig. 3 B). To examine this effect of E protein expression on internal pH in greater detail, we conducted a time course experiment over 48 h post‐transfection, recording fluorescence at 6, 12, 24 and 48 h. These results show a decrease in fluorescence intensity from the DND‐189 reporter relative to mock‐transfected cells, which was statistically significant at 24 and 48 h (both 19% below mock, Fig. 3 C). Because the fluorescence of DND‐189 increases at lower pH, and declines at higher pH (pK a ≈ 5.2), these results suggest that, as SARS‐CoV‐2 E expression in mammalian cells progresses, there is a time‐dependent increase of internal pH of intracellular organelles such as lysosomes and ER–GIC.
Membrane currents from cells expressing SARS‐CoV‐2 E WT or PM constructs
To characterize the electrophysiological activity of the E protein, we used the whole‐cell configuration of the patch clamp technique to measure the membrane currents of mock, WT and PM constructs expressed in HEK 293S cells (Fig. 4 A). The I–V relationships were obtained using a Cs+‐based internal solution, and the results revealed no significant differences between the currents recorded in WT‐ and mock‐transfected cells (Fig. 4 B and C). A mock‐subtraction analysis confirmed that there is no current associated with expression of the WT E protein construct (Fig. 4 B and D), consistent with a lack of expression of the WT E protein in the plasma membrane. When the PM construct was expressed in HEK 293S cells, however, we observed a larger increase in total membrane conductance compared to that seen with the WT construct (Fig. 4 B and D). Membrane currents were larger in cells transfected with PM construct than in mock‐transfected cells (Fig. 4 C). The subtraction analysis confirmed that there is an additional current associated with expression of the PM construct (Fig. 4 D), consistent with presence of the PM version of the E protein in the plasma membrane. The E current was ∼±40 pA at ±100 mV (400 pS whole cell conductance), nearly voltage‐independent, and the E rev was −8.8 mV, 95% CI (−10.3, −7.4 mV), indicating that Na+ and Cs+ are nearly equally permeant through the pore of the channel.
Figure 4. Membrane currents from HEK 293S cells expressing SARS‐CoV‐2 E WT or PM constructs.
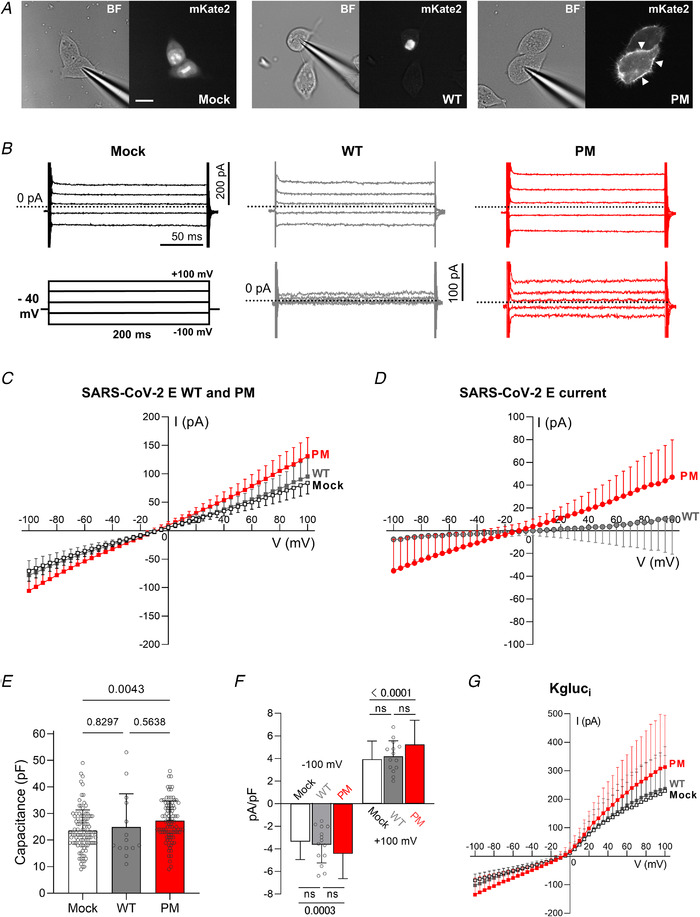
A, representative images of HEK 293S cells transfected with mock (NLS‐mKate2 into pcDNA3), WT and PM SARS‐CoV‐2 E protein constructs. Left panels, bright field images; right panels, mKate2 fluorescence. Scale bar, 20 μm. B, representative current records from HEK 293S cells expressing pcDNA3 vector (mock), WT and PM in top panels, and ‘mock subtracted’ traces of WT and PM in the bottom panels. Scale bars: 200 pA (vertical), 50 ms (horizontal). ‘Mock subtracted’ records are shown below. Scale bar, 100 pA. Dotted lines indicate zero current level. C, averaged I–V curves of mock (open squares, n = 102 from 24 transfections), PM (n = 95 from 22 transfections) and WT (n = 14 from 3 transfections) transfected cells. There was no difference between WT and mock (two‐way ANOVA, main difference between constructs, F (2,208) = 13.69, P < 0.0001, with Tukey's post hoc test, P = 0.7036 for WT vs. mock). When PM was expressed in HEK 293S cells, we observed higher total membrane conductance than with WT or mock (two‐way ANOVA, F (2,208) = 13.69, P < 0.0001, Tukey's post hoc: P < 0.0001 for PM vs. WT, and P < 0.0001 for PM vs. mock). D, subtracting mock‐transfected cell data from WT‐transfected data reveals no current. A larger voltage‐independent current is recorded in cells transfected with PM, and is revealed by digital subtraction (‘mock subtraction’) (two‐way ANOVA, main difference between constructs, F (1,107) = 3.334, P = 0.0742, and Bonferroni's post hoc test: P < 0.05 for V < −70 mV and >45 mV, P < 0.0001 at −100 mV and +100 mV). E, capacitance of PM (n = 95), mock (n = 102) and WT (n = 14) HEK 293S transfected cells (one‐way ANOVA, F (2,208) = 5.192, P = 0.0063, and Tukey's post hoc test: Mock vs. PM, P = 0.0043; Mock vs. WT, P = 0.8297; and PM vs. WT, P = 0.5638). F, current amplitudes in B were normalized to yield current density (pA/pF). Compared to mock‐transfected cells, the normalized current density of PM was greater at −100 mV (one‐way ANOVA, F (2,208) = 8.03, P = 0.0004, Tukey's post hoc test, Mock: −3.37 ± 1.58 pA pF−1 vs. PM: −4.44 ± 2.20 pA pF−1, P = 0.0003) and at +100 mV (one‐way ANOVA, F (2,208) = 12.35, P < 0.0001, Tukey's post hoc test, Mock: 3.93 ± 1.61 pA pF−1 vs. PM: 5.24 ± 2.15 pA pF−1, P < 0.0001). Current densities for WT were −3.64 ± 1.62 pA pF−1 at −100 mV (P = 0.3005, WT vs. PM) and 4.18 ± 1.38 pA pF−1 at +100 mV (P = 0.1227, WT vs. PM). No differences were found between Mock and WT (Tukey's post hoc test, at −100 mV: P = 0.8714, and at +100 mV: P = 0.8796). For visualization purposes, ns: not significant. G, using the whole‐cell configuration of the patch‐clamp technique with potassium gluconate in the pipette (Kgluci), cells transfected with the pcDNA3 vector only (mock, open squares, n = 13), WT (n = 7) and PM (n = 12) all exhibit a voltage‐dependent outward current. I–V relationships with potassium gluconate reveal a larger current in PM compared to mock‐transfected HEK 293S cells (mixed‐model two‐way ANOVA, main difference between constructs, F (2,29) = 0.3026, P = 0.011, and Tukey's post hoc test, PM vs. mock, P < 0.05 at V > +80 mV). The values plotted in the graphs are expressed as means ± SD. Currents in B–D and G were elicited by 200 ms commands from V h = −40 mV in 5 mV steps (from −100 mV to + 100 mV). [Color figure can be viewed at wileyonlinelibrary.com]
We compared cell capacitance between cell populations transfected with the various constructs to control for differences in cell size (Fig. 4 E). The average membrane capacitance of HEK 293S cells for the different constructs was 23.6 ± 7.7 pF for mock, 24.9 ± 12.5 pF for WT and 27.3 ± 7.5 pF for PM (Fig. 4 E). The modest increase in the capacitance of cells transfected with PM might be due to the expression of the E protein in the membrane. We normalized for capacitance and calculated the current density at −100 and +100 mV (Fig. 4 F). Cells that expressed the PM construct had higher inward (−4.44 ± 2.20 pA pF−1) and outward (5.24 ± 2.15 pA pF−1) current density when compared to mock (−100 mV: −3.37 ± 1.58 pA pF−1, P <0.0001; +100 mV: 3.93 ± 1.61 pA pF−1, P = 0.0003) (Fig. 4 F). These results show that the normalized current density contributed by the E protein is approximately ±1 pA pF−1. When using a high K+ pipette solution (potassium gluconate), we also observed a higher current amplitude with PM than with WT and mock (Fig. 4 G). A time‐ and voltage‐dependent outward current component was also present in mock‐transfected cells, and this is mainly because of an endogenous K+ current that has been widely reported in the literature (Jiang et al. 2002). This was effectively abolished in our experiments by replacing K+ with Cs+ in the pipette solution.
The E protein is modulated by pH and is permeable to small monovalent cations
We then explored the pH dependence of E protein ion channel activity using electrophysiological recordings. HEK 293S cells were transfected with PM and mock constructs, and recordings were performed 18–36 h after transfection. In these experiments, we used Cs+‐based internal solution, and currents from mock‐transfected cells were ‘mock subtracted’ to obtain the E current. Lowering extracellular pH (corresponding to greater luminal acidity in vivo) modified the voltage dependence of the E protein current so that the E current, which was nearly linear at pH 7.4, showed inward rectification at pH 6.0 (Fig. 5 A and C). This is consistent with easier movement of ions from the extracellular solution, which corresponds to the luminal compartment, to the cytoplasmic compartment. At pH 8.0, the E current was reduced across a wide range of holding potentials (V h < −85 mV and V h > +60 mV) (Fig. 5 B and D).
Figure 5. The E protein is modulated by pH and is permeable to small cations.
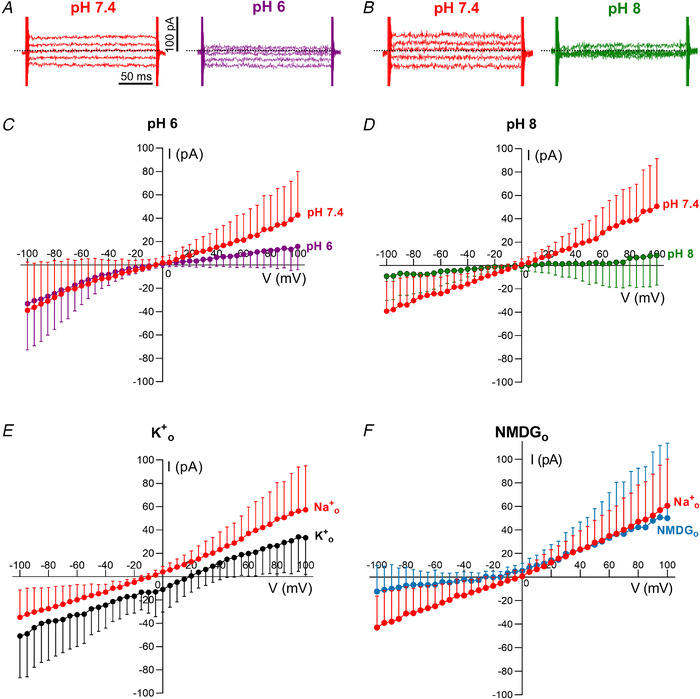
A and B, representative examples of mock‐subtracted traces in standard Na+ extracellular solution at pH 7.4 (A and B) and after exchange with a similar external solution at pH 6 (A) or pH 8 (B). Dotted lines indicate the zero current level. Scale bars: 100 pA (vertical), 50 ms (horizontal). C–F, averaged mock‐subtracted current–voltage relationships for whole‐cell current in PM‐transfected cells in response to changes of pH and ion substitution in the external solution. Currents were elicited by 200 ms commands from V h = −40 mV in 5 mV steps (from −100 mV to +100 mV). C, the mock‐subtracted SARS‐CoV‐2 E current is linear at pH 7.4 but shows inward rectification when the external solution is exchanged for one at pH 6 (Mock = 11, PM = 12, from 5–6 transfections; two‐way ANOVA, main effect of pH: F (1,11) = 4.079, P = 0.0667, Bonferroni's post hoc test, P < 0.05 at V >75 mV, P < 0.0001 at +100 mV). D, SARS‐CoV‐2 E current measured at pH 7.4 is reduced when exposed to an external solution at pH 8 (Mock = 9, PM = 8, from 4–5 transfections; two‐way ANOVA, main effect of pH: F (1,7) = 6.628, P = 0.0368, Bonferroni's post hoc test, P < 0.05 at V h < −85 mV (P = 0.0017 at −100 mV) and V h > +60 mV (P < 0.0001 at +100 mV)). E, the SARS‐CoV‐2 E protein is permeable to K+. The standard external solution containing Na+ was replaced by a high K+ solution and the E rev shifted from −8.5 mV (95% CI: −11.7, −5.5 mV) in Na+ o to 18.7 mV (17.2, 20.3 mV) in high K+ o (Mock = 10, PM = 11, from 4–5 transfections; two‐way ANOVA, main effect of K+, F (1,10) = 20.85, P = 0.0010). The relative permeability ratio P K: P Na was estimated as 2.9 (2.8, 3.2). F, SARS‐CoV‐2 E protein is not permeable to NMDG (Mock = 11, PM = 10, from 4–5 transfections; two‐way ANOVA, main effect of NMDG: F (1,9) = 14.26, P = 0.0044, Bonferroni's post hoc test, P < 0.05 at V h < −85 mV (P = 0.0032 at −100 mV)). The relative permeability ratio of P Na: P Cs was estimated as 0.9 (0.7, 1.0). The values plotted in the graphs are expressed as means ± sd. [Color figure can be viewed at wileyonlinelibrary.com]
By expressing the SARS‐CoV‐2 E protein in the plasma membrane, we were able to record the currents and to alter either the internal or external solutions to investigate the ion selectivity of the proposed ion channels composed of E proteins. Replacement of external Na+ with K+ revealed that both of these monovalent cations are able to pass through the ion channel formed by the E protein (Fig. 5 E) with the reversal potential being shifted from −8.5 mV in external Na+ to +18.7 mV in external K+. These data indicate that K+ is approximately threefold more permeant than either Cs+ or Na+ within the E channel pore. Replacement of external Na+ with N‐methyl‐D‐glucamine+ (NMDG) resulted in a loss of inward current (Fig. 5 F), suggesting that this bulky cation is unable to permeate through the pore, and also that Cl– ions do not easily permeate the ion channel. The small negative shift in the reversal potential indicates that Cs+ is slightly more permeant than Na+ (P Na: P Cs estimated as 0.9). Anion permeability was undetectable above the level of r.m.s. noise (∼4 pA), yielding an estimate for cation/anion permeability ratio >10.
PM but not WT produces robust currents in Xenopus oocytes, decreased at high pH
The small amplitude of the SARS‐CoV‐2 E protein current in HEK 293S cells may make it difficult to use this mammalian cell‐based recording platform for drug screening. Therefore, to improve the robustness and throughput using our engineered E protein constructs, we initiated additional electrophysiological experiments, expressing the E protein in Xenopus oocytes. Using two‐electrode voltage clamp recording of currents in Xenopus laevis oocytes injected with cRNA encoding WT or untagged‐PM, we discovered that untagged‐PM generates robust currents (>±10 μA) at pH 7.5 that were linear across the voltage range studied (−100 to 70 mV) (Fig. 6 A), and the current amplitude increased with the quantity of PM cRNA injected (Fig. 6 C). Although the untagged version of the PM construct was used for the majority of oocyte experiments, we found that the presence or absence of the mKate2‐epitope tag did not alter the E current amplitude (Fig. 6 C). In contrast, cRNA generated from the WT construct did not generate significant currents (Fig. 6 A and C). Expression of the untagged‐PM construct depolarized the resting V m to −14.5 ± 5.8 mV, versus −21.4 ± 4.0 mV for WT‐injected oocytes and −19.1 ± 5.1 mV for non‐injected oocytes (n = 20–25; P = 0.0064, non‐injected vs. PM) (Fig. 6 B). The PM current was modified when the bath pH was changed (Fig. 7 C and D) (one‐way ANOVA comparing normalized currents of pH 6.0, 7.5 and 9.0 at −100 mV, P = 0.0189, and at +50 mV, P = 0.0077). The mean peak current observed at pH 6.0 (Fig. 7 A, C and D) was larger than that at pH 9.0 (Fig. 7 B, C and D), whereas non‐injected oocytes did not generate significant currents at pH 6.0 (Fig. 7 C). Unlike the HEK 293 cell experiments, we did not see marked inward rectification at pH 6 in oocytes, but we did observe a time‐dependent decline in current amplitude at hyperpolarized potentials. This was variable between oocytes and in each oocyte was more prominent at pH 9.0 than at pH 6.0 (Fig. 7 A and B). Such a current decline might perhaps arise due to the collapse of electrochemical gradients, or it may reflect a voltage‐dependent blockade of the channel by an extracellular cation at a site that is only revealed at higher extracellular pH. Overall, the oocyte‐based electrophysiological recordings allowed us to observe E currents that seem broadly similar in their biophysical properties to those recorded from the mammalian cells, but with a much smaller relative contribution from endogenous currents compared with recordings in mammalian cells, and a much larger current amplitude, resulting in a greatly improved signal‐to‐noise ratio.
Figure 6. SARS‐CoV‐2 E PM expresses robust currents when expressed in Xenopus oocytes.
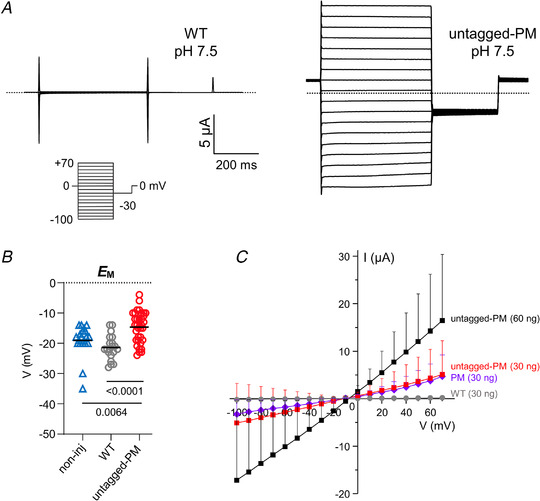
A, exemplar current traces for Xenopus oocytes expressing WT (left panel) and untagged‐PM (right panel) as indicated, at pH 7.5 (30 ng cRNA). Voltage protocol and scale bars are shown in the inset. Dashed lines indicate zero current level. B, scatter plot of the unclamped membrane potential (E M) for oocytes expressing untagged‐PM (n = 35), WT (n = 20) or non‐injected (non‐inj) oocytes (triangles, n = 20) (statistical analysis by one‐way ANOVA (P = 0.0002) and Tukey's post hoc test, untagged‐PM vs. WT, P < 0.0001; untagged‐PM vs. non‐inj, P = 0.0064; WT vs. non‐inj, P = 0.3290). C, mean peak current versus voltage for oocytes after injection of 30 ng (mKate2‐tag) PM cRNA (diamonds, n = 5), 30 ng (squares, n = 17) or 60 ng ( squares, n = 8) untagged‐PM cRNA, or after injection of 30 ng WT cRNA (circles, n = 15). The mean peak current at +50 mV was equivalent between oocytes expressing tagged (3.1 ± 2.8 μA, n = 5) and untagged (3.6 ± 5.1 μA, n = 17) versions of the PM construct (Student's t‐test, P = 0.8336). cRNA encoding WT, PM and untagged‐PM constructs was generated from cDNA in the pXOOM vector. Two batches of oocytes were used for WT and untagged‐PM constructs and one batch for the PM construct. [Color figure can be viewed at wileyonlinelibrary.com]
Figure 7. The E protein is modulated by pH when expressed in Xenopus oocytes.
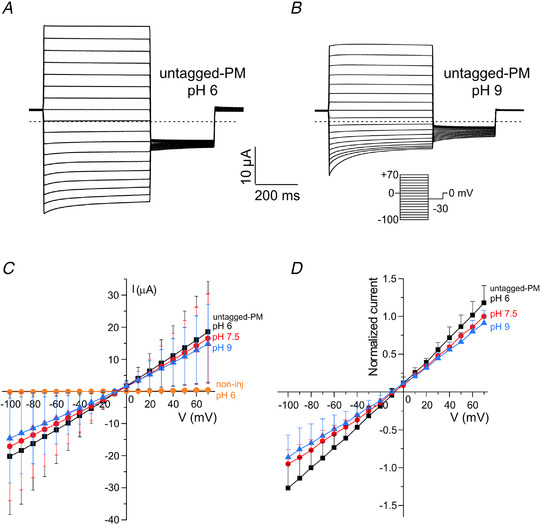
A and B, exemplar current traces for untagged‐PM expressed (60 ng cRNA) in a single Xenopus oocyte at bath pH 6.0 (A) and pH 9.0 (B). Dashed lines indicate zero current level. Voltage protocol and scale bars inset. C, mean peak current versus voltage for oocytes after injection of 60 ng untagged‐PM cRNA, each studied at bath pH of 6 (squares), 7.5 (circles) and 9.0 (triangles) (n = 8), or non‐injected at pH 6 (squares, n = 20). D, mean data after normalization for each oocyte to the peak current of PM at +70 mV, pH 7.5. Statistical analysis by one‐way ANOVA and Tukey's post hoc test comparing normalized currents at −100 mV (main difference between pH solutions, P = 0.0189; pH 6.0 (−1.27 ± 0.32) vs. pH 7.5 (−0.95 ± 0.19), P = 0.0764; pH 6.0 vs. pH 9.0 (−0.86 ± 0.30), P = 0.0200; pH 7.5 vs. pH 9.0, P = 0.7980) and +50 mV (P = 0.0077; pH 6 (0.86 ± 0.15) vs. pH 7.5 (0.74 ± 0.04), P = 0.1150; pH 6 vs. pH 9 (0.66 ± 0.12), P = 0.0059; pH 7.5 vs. pH 9.0, P = 0.360). Data are presented as means ± sd. Two batches of oocytes were used for the experiment. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
The E protein of SARS‐CoV‐1 is essential for virus propagation (DeDiego et al. 2007) and has been implicated in its pathogenicity (Nieto‐Torres et al. 2014, 2015 b), although the mechanisms remain unclear. To characterize the function of the E protein of SARS‐CoV‐2 for this study, we generated two innovative constructs for the expression of the E protein in mammalian cells. This system of transient expression enabled us to study its dual functions: the ion channel activity of SARS‐CoV‐2 E when it is expressed in the plasma membrane, and its effect on proton homeostasis when expressed in intracellular organelles. Several groups have shown that epitope‐tagged E protein of SARS‐CoV‐1 localizes in the ER–GIC (Nieto‐Torres et al. 2011; Cohen et al. 2011). Our results in HEK 293S cells with the red fluorescent tag mKate2 and the ER–GIC marker ERGIC‐53 show a perinuclear localization of WT SARS‐CoV‐2 E protein that is consistent with ER–GIC targeting. Although an effect of mKate2 on localization may not be totally eliminated, previous studies in SARS‐CoV‐1 have shown that subcellular localization of the E protein is similar for tagged and untagged forms (Nieto‐Torres et al. 2011; Cohen et al. 2011). In addition, our results in Xenopus oocytes show large currents of similar amplitude for tagged and untagged versions of the PM constructs, suggesting that the presence of the mKate2 tag has no major deleterious effects on the function of the E protein. Therefore, the mKate2 tag is a useful reporter to investigate the localization and function of E proteins.
One important role of coronaviral viroporins is in regulation of the pH within the lumen of intracellular organelles such as the Golgi apparatus. For example, expression of the E protein of infectious bronchitis virus (IBV) (Westerbeck & Machamer, 2019) increased intra‐organelle pH, inducing neutralization of Golgi pH, resulting in a protective effect that allows the IBV spike protein to be sequestered from proteolysis, and promotes virus assembly and release (Westerbeck & Machamer, 2019). The viroporin p7 of hepatitis C virus (HCV) also induces alkalinization within the Golgi that is required for HCV production and propagation (Wozniak et al. 2010). Alterations in the ionic environment of the ER and Golgi by SARS‐CoV‐1 E are proposed to activate the cell stress response and apoptosis pathways (DeDiego et al. 2011). Our pH imaging results show that SARS‐CoV‐2 E protein expression decreases the fluorescence of regions that correspond to intracellular organelles such as lysosome, ER–GIC and Golgi apparatus, and this is consistent with an increase in the pH of these compartments, as reported for the E protein of IBV (Westerbeck & Machamer, 2019). The time course of the pH change that we observed in NIH 3T3 cells is also consistent with previous work on the kinetics of coronavirus E protein expression (Nieto‐Torres et al. 2011; Venkatagopalan et al. 2015). These findings suggest that SARS‐CoV‐2 E protein has a similar pathophysiological effect on intracellular proton homeostasis as other viroporins, which may result in facilitated viral assembly and enhanced spike protein stability in infected cells. Although an interaction between E and the spike protein remains to be demonstrated in coronaviruses (Schoeman & Fielding, 2019), expression of both E and membrane (M) proteins seems to be required for virus‐like particle formation (Huang et al. 2004; Schoeman & Fielding, 2019), including in SARS‐CoV‐2 (Boson et al. 2021). This interaction appears to take place between the cytoplasmic tails of M and E proteins (Corse & Machamer, 2003), and may enhance the activity of the E protein when expressed in the Golgi apparatus. The alteration of proton homeostasis may be a common consequence of coronaviral viroporin expression in infected mammalian cells and essential for viral function.
Viroporins form ion channels, and the main function of these is presumably to alter ionic gradients across organelle membranes (Nieva et al. 2012; Scott & Griffin, 2015). This feature may be crucial for not only promoting viral assembly and propagation but also facilitating pathogenesis by disturbing cellular homeostasis (Nieto‐Torres et al. 2015 a; Farag et al. 2020). Our PM construct is designed to expose the N‐terminus of the E protein at the exterior of the cell, with the C‐terminus being intracellular. This design allowed us to alter external pH and ions in our electrophysiological experiments as a model of the physiological conditions within the Golgi apparatus, while the cytoplasmic conditions of pH and ions remain the same (represented by the pipette solution in the whole‐cell recording configuration). Our results suggest that the E protein has ion channel activity that can be regulated by protons. A recent study in planar lipid bilayers also showed the E protein of SARS‐CoV‐2 to be permeable to both Na+ and K+ and to show sensitivity to pH changes (Xia et al. 2020). The selectivity for monovalent cations such as K+ and Na+ exhibited by SARS‐CoV‐2 E is similar to reports for the E protein of SARS‐CoV‐1 (Wilson et al. 2004, 2006) and p7 of HCV (Premkumar et al. 2004). However, the planar lipid bilayer platform using purified lipids and recombinant protein may be lacking in endogenous mammalian factors that can contribute to E protein function. As findings from the bilayer platform might not be pathophysiologically relevant, a simple biological role of the SARS‐CoV‐2 E protein can be proposed, tested and validated based on our platforms using these constructs in live mammalian cells and oocytes.
Under physiological conditions, the relevant value of the membrane potential for the E current is −80 mV (the potential of the Golgi lumen being +80 mV relative to the cytoplasm) (Matamala et al. 2019), and we can conclude that when the E protein is present in the Golgi apparatus membrane, the low pH within the lumen would promote the activation of the channel, which is permeable to smaller cations, eventually lowering cytoplasmic K+ in the infected cell. Alkaline pH, on the other hand, would inhibit the flow of cations across the organelle membrane. In this context, it is interesting to note that droplets suspended within exhaled air are somewhat alkaline, at approximately pH 8 (Vaughan et al. 2003). This implies that, when intact virus particles are ejected from the human airway in respiratory droplets, the more alkaline conditions would result in the E protein channel present in the virus envelope being mainly closed, so that the exhaled virion would then be essentially sealed off from the external environment, perhaps prolonging viral survival. Other viroporins, such as the M2 protein of influenza A virus, can function as a proton channel (Pinto et al. 1992) with a high proton selectivity over monovalent cations (Chizhmakov et al. 1996). It is possible that the E protein itself is also permeable to H+, and our pH imaging results are consistent with this. However, direct measurements of such very small (∼5 pA) currents are at the limit of resolution of our present recording techniques in HEK 293S cells.
While the experiments in HEK 293S cells enabled us to characterize electrophysiological properties of SARS‐CoV‐2 E protein, the small currents measured in this system would make screening of blockers against the E protein difficult and arduous. We therefore expressed the PM construct in Xenopus laevis oocytes, and we observed that currents of 10–20 μA were generated at −100 mV. Increasing extracellular pH reduced the amplitude of the E current, consistent with the pH dependence observed in HEK 293S cells, and WT did not generate any observable current in oocytes. We did observe some differences between E currents recorded in oocytes versus mammalian cells. Such findings are not unprecedented for channels expressed using these two different expression systems, as previously reported for other channels such as KCNQ1–KCNE3 (Abbott, 2016). One likely explanation for the voltage‐dependent fade or ‘inactivation’ seen in the present study in oocytes is the possible voltage‐dependent block of the channel by an external cation, perhaps due to the deprotonation of a binding site at pH 9. Alternatively, the current decay might reflect the collapse in electrochemical gradients that is sometimes seen in oocytes when recording extremely large currents, although such effects should be independent of pH. Taken together, our results suggest that the oocyte‐based recording platform using these engineered constructs might prove applicable for drug discovery studies aimed at identifying channel blockers or inhibitors of SARS‐CoV‐2 E protein.
The potential for the coronavirus E proteins to serve as targets for therapeutic intervention seems clear. The E protein has been proposed to play an important role in pathogenesis of SARS‐CoV (Nieto‐Torres et al. 2014; Jimenez‐Guardeño et al. 2014) and SARS‐CoV‐2 (Schoeman & Fielding, 2020), and the history of the M2 channel blockers as antiviral drugs for the treatment of influenza A (Dolin et al. 1982) suggests that this approach could be applied to SARS‐CoV‐2. Several computational modelling studies (Gupta et al. 2020; Chernyshev, 2020), as well as experimental studies in bacteria (Singh Tomar & Arkin, 2020) and lipid bilayers (Xia et al. 2020), have highlighted the potential for identifying new and existing drugs that act by this mechanism. The inhibitor 5‐(N,N‐ hexamethylene)‐amiloride (HMA) is known to be effective against SARS‐CoV‐1 E (Pervushin et al. 2009), and amantadine was also shown to inhibit SARS‐CoV‐1 E (Torres et al. 2007) as well as M2 (Pinto et al. 1992) and p7 (Griffin et al. 2003). HMA had stronger affinity than amantadine in docking studies with SARS‐CoV‐2 E protein (Mandala et al. 2020), whereas both compounds showed weak inhibitory activity in lipid bilayer experiments and showed cytotoxicity at these high concentrations (Xia et al. 2020). Other potential inhibitors for SARS‐CoV‐2 E include compounds derived from berbamine, which reduced the secretion of cytokines (Xia et al. 2020), and memantine (Singh Tomar & Arkin, 2020). Our E‐current recording platform will be useful in the evaluation of these and other drug candidates, including existing drugs that can be repurposed. The need for new viroporins has become more acute in recent months, with the emergence of novel viral variants that seem likely to become dominant (Washington et al. 2021) as well as others that have the potential to escape from the acquired immunity generated by vaccination or as a consequence of prior SARS‐CoV‐2 infection (Cele et al. 2021; Wibmer et al. 2021).
The method described here for the expression of the SARS‐CoV‐2 E protein in the plasma membrane of mammalian and amphibian cells is widely applicable, with minor modifications, to other coronaviruses, and more broadly to the study of any viroporins. The application of this technique should greatly facilitate screening and validation of novel compounds that block the ion channel formed by the E protein of SARS‐CoV‐2, some of which might prove to have antiviral activity and may find clinical utility in the treatment of COVID‐19.
Additional information
Competing interests
The authors declare no competing financial interests.
Author contributions
N.L.H., M.Y. and D.C.‐G. conceived the study and designed the experiments. M.Y. and R.B. designed and prepared the constructs, R.B., D.C.‐G. and M.Y. performed the imaging experiments, and D.C.‐G. and G.W.A. performed the electrophysiology experiments. All of the authors contributed to data analysis and interpretation. N.L.H., D.C.‐G. and M.Y. wrote the paper, all authors edited the paper. All authors have read and approved the final version of this manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This work was supported by Columbia Stem Cell Initiative (to M.Y.), the Department of Anesthesiology at CUIMC (to N.L.H.) and Columbia University Irving Medical Centre Dean's Office Fund (to M.Y. and N.L.H.). G.W.A. was supported by NIH, National Institute of General Medical Sciences (GM130377).
Supporting information
Statistical Summary Document
Acknowledgements
The authors thank M. Shelanski, E. Passegue, C.W. Emala and A. Brambrink for logistical support of our laboratories during the pandemic, K. Yoshida, C. Abate‐Shen, Y. Huang, D.D. Ho and A. De Silva for helpful discussion and support, and M.A. Wall and W. Baird for providing masks during the COVID‐19 epidemic in New York City.
Biographies
David Cabrera Garcia earned his PhD in Functional and Molecular Biology from the University of Oviedo, Spain, studying the toxicological and electrophysiological effects of marine toxins on neurons. He is currently a postdoctoral researcher at Columbia University where he investigates the effects of acute alcohol on the electrophysiology of neuronal populations in the frontal cortex. His research interests include GABAergic interneurons, neural circuits and ion channel physiology.

Ramsey Bekdash is a PhD candidate in the Molecular Pharmacology and Therapeutics Graduate Program at the Columbia University Medical Center. His work pertains to metabolite biosensor development and drug repurposing studies for cardiac arrhythmias in Dr Masayuki Yazawa's laboratory. He completed his undergraduate studies at University of Maryland, where he earned a dual degree in General Biology and Chemistry. His research interests include synthetic biology, biosensor development, and optogenetics.

Edited by: Ian Forsythe & Peying Fong
D. Cabrera‐Garcia and R. Bekdash are joint first authors.
Linked articles: This article is highlighted in a Perspectives article by Fong. To read this article, visit https://doi.org/10.1113/JP281601.
This is an Editor's Choice article from the 1 June 2021 issue.
Contributor Information
Masayuki Yazawa, Email: my2387@cumc.columbia.edu.
Neil L. Harrison, Email: nh2298@cumc.columbia.edu.
Data availability statement
Any supplementary information, chemical compound information, and source data are available from the corresponding author (M.Y.) upon reasonable request.
References
- Abbott GW (2016). KCNE1 and KCNE3: The yin and yang of voltage‐gated K+ channel regulation. Gene 576, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC & Garry RF (2020). The proximal origin of SARS‐CoV‐2. Nat Med 26, 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom‐Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA; Public Health–Seattle and King County and CDC COVID‐19 Investigation Team . (2020). Presymptomatic SARS‐CoV‐2 infections and transmission in a skilled nursing facility. N Engl J Med 382, 2081–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T, Greninger AL, Roychoudhury P, Starita LM, Famulare M, Huang ML, Nalla A, Pepper G, Reinhardt A, Xie H, Shrestha L, Nguyen TN, Adler A, Brandstetter E, Cho S, Giroux D, Han PD, Fay K, Frazar CD, Ilcisin M, Lacombe K, Lee J, Kiavand A, Richardson M, Sibley TR, Truong M, Wolf CR, Nickerson DA, Rieder MJ, Englund JA; Seattle Flu Study Investigators , Hadfield J, Hodcroft EB, Huddleston J, Moncla LH, Müller NF, Neher RA, Deng X, Gu W, Federman S, Chiu C, Duchin JS, Gautom R, Melly G, Hiatt B, Dykema P, Lindquist S, Queen K, Tao Y, Uehara A, Tong S, MacCannell D, Armstrong GL, Baird GS, Chu HY, Shendure J & Jerome KR (2020). Cryptic transmission of SARS‐CoV‐2 in Washington state. Science 370, 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M, Benvenuto D, Giovanetti M, Angeletti S, Ciccozzi M & Pascarella S (2020). Sars‐CoV‐2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed Res Int 2020, 4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset F‐L, Lavillette D & Denolly S (2021). The SARS‐CoV‐2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus‐like particles. J Biol Chem 296, 100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño‐Rodriguez C, Honrubia JM, Gutiérrez‐Álvarez J, DeDiego ML, Nieto‐Torres JL, Jimenez‐Guardeño JM, Regla‐Nava JA, Fernandez‐Delgado R, Verdia‐Báguena C, Queralt‐Martín M, Kochan G, Perlman S, Aguilella VM, Sola I & Enjuanes L (2018). Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. MBio 9, e02325–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S, Gazy I, Jackson L, Hwa S‐H, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa M‐YS, NGS‐SA, COMMIT‐KZN Team , Lessells RJ, de Oliveira T & Sigal A (2021). Escape of SARS‐CoV‐2 501Y.V2 variants from neutralization by convalescent plasma. medRxiv, 10.1101/2021.01.26.21250224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyshev A (2020). Pharmaceutical targeting the envelope protein of SARS‐CoV‐2: The screening for inhibitors in approved drugs. ChemRxiv, 10.26434/chemrxiv.12286421.v1. [DOI] [Google Scholar]
- Chizhmakov IV, Geraghty FM, Ogden DC, Hayhurst A, Antoniou M & Hay AJ (1996). Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol 494, 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JR, Lin LD & Machamer CE (2011). Identification of a golgi complex‐targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J Virol 85, 5794–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E & Machamer CE (2003). The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, Aaron JG, Claassen J, Rabbani LRE, Hastie J, Hochman BR, Salazar‐Schicchi J, Yip NH, Brodie D & O'Donnell MR (2020). Epidemiology, clinical course, and outcomes of critically ill adults with COVID‐19 in New York City: A prospective cohort study. Lancet 395, 1763–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego ML, Alvarez E, Almazan F, Rejas MT, Lamirande E, Roberts A, Shieh W‐J, Zaki SR, Subbarao K & Enjuanes L (2007). A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego ML, Nieto‐Torres JL, Jiménez‐Guardeño JM, Regla‐Nava JA, Álvarez E, Oliveros JC, Zhao J, Fett C, Perlman S & Enjuanes L (2011). Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog 7, e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Espírito Santo DA, Lemos ACB & Miranda CH (2020). In vivo demonstration of microvascular thrombosis in severe COVID‐19. J Thromb Thrombolysis 50, 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolin R, Reichman RC, Madore HP, Maynard R, Linton PN & Webber‐Jones J (1982). A controlled trial of amantadine and rimantadine in the prophylaxis of influenza A infection. N Engl J Med 307, 580–584. [DOI] [PubMed] [Google Scholar]
- Duart G, García‐Murria MJ, Grau B, Acosta‐Cáceres JM, Martínez‐Gil L & Mingarro I (2020). SARS‐CoV‐2 envelope protein topology in eukaryotic membranes. Open Biol 10, 200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag NS, Breitinger U, Breitinger HG & El Azizi MA (2020). Viroporins and inflammasomes: a key to understand virus‐induced inflammation. Int J Biochem Cell Biol 122, 105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannis D, Ziogas IA & Gianni P (2020). Coagulation disorders in coronavirus infected patients: COVID‐19, SARS‐CoV‐1, MERS‐CoV and lessons from the past. J Clin Virol 127, 104362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Reiche AS, Hernandez MM, Sullivan MJ, Ciferri B, Alshammary H, Obla A, Fabre S, Kleiner G, Polanco J, Khan Z, Alburquerque B, van de Guchte A, Dutta J, Francoeur N, Melo BS, Oussenko I, Deikus G, Soto J, Sridhar SH, Wang YC, Twyman K, Kasarskis A, Altman DR, Smith M, Sebra R, Aberg J, Krammer F, García‐Sastre A, Luksza M, Patel G, Paniz‐Mondolfi A, Gitman M, Sordillo EM, Simon V & van Bakel H (2020). Introductions and early spread of SARS‐CoV‐2 in the New York City area. Science 369, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin SDC, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MPG & Rowlands DJ (2003). The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett 535, 34–38. [DOI] [PubMed] [Google Scholar]
- Gupta MK, Vemula S, Donde R, Gouda G, Behera L & Vadde R (2020). In‐silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn (in press), doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B & Sigworth FJ (1981). Improved patch‐clamp techniques for high‐resolution current recording from cells and cell‐free membrane patches. Pflugers Archl 391, 85–100. [DOI] [PubMed] [Google Scholar]
- Helenius A (1992). Unpacking the incoming influenza virus. Cell 69, 577–578. [DOI] [PubMed] [Google Scholar]
- Hille B (2001). Ionic Channels of Excitable Membranes, 3rd edn. Sinauer Associates Inc., Sunderland, MA. [Google Scholar]
- Hofherr A, Fakler B & Klöcker N (2005). Selective Golgi export of Kir2.1 controls the stoichiometry of functional Kir2.x channel heteromers. J Cell Sci 118, 1935–1943. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yang Z, Kong W & Nabel GJ (2004). Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: Implications for assembly and vaccine production. J Virol 78, 12557–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Sun X, Cao K & Wang R (2002). Endogenous Kv channels in human embryonic kidney (HEK‐293) cells. Mol Cell Biochem 238, 69–79. [DOI] [PubMed] [Google Scholar]
- Jimenez‐Guardeño JM, Nieto‐Torres JL, DeDiego ML, Regla‐Nava JA, Fernandez‐Delgado R, Castaño‐Rodriguez C & Enjuanes L (2014). The PDZ‐binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog 10, e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern DM, Sorum B, Hoel CM, Sridharan S, Remis JP, Toso DB & Brohawn SG (2020). Cryo‐EM structure of the SARS‐CoV‐2 3a ion channel in lipid nanodiscs. bioRxiv, 10.1101/2020.06.17.156554. [DOI] [Google Scholar]
- Klok FA, Kruip MJHA, van der Meer NJM, Arbous MS, Gommers DAMPJ, Kant KM, Kaptein FHJ, van Paassen J, Stals MAM, Huisman MV & Endeman H (2020). Incidence of thrombotic complications in critically ill ICU patients with COVID‐19. Thromb Res 191, 145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasllieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greeneugh TC, Choe H & Farzan M (2003). Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KP & Liu DX (2001). The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre‐Golgi compartments and physical interaction between the envelope and membrane proteins. J Biol Chem 276, 17515–17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A & Hong M (2020). Structure and drug binding of the SARS‐CoV‐2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol 27, 1202–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamala E, Castillo C, Vivar JP & Brauchi S (2019). Imaging the electrical activity of organelles in living cells. bioRxiv, 10.1101/578765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClenaghan C, Hanson A, Lee S‐J & Nichols CG (2020). Coronavirus proteins as ion channels: Current and potential research. Front Immunol 11, 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS & Manson JJ (2020). COVID‐19: Consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadas SM, Fitzpatrick MC, Sah P, Pandey A, Shoukat A, Singer BH & Galvani AP (2020). The implications of silent transmission for the control of COVID‐19 outbreaks. Proc Natl Acad Sci U S A 117, 17513–17515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto‐Torres JL, DeDiego ML, Álvarez E, Jiménez‐Guardeño JM, Regla‐Nava JA, Llorente M, Kremer L, Shuo S & Enjuanes L (2011). Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 415, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto‐Torres JL, DeDiego ML, Verdiá‐Báguena C, Jimenez‐Guardeño JM, Regla‐Nava JA, Fernandez‐Delgado R, Castaño‐Rodriguez C, Alcaraz A, Torres J, Aguilella VM & Enjuanes L (2014). Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 10, e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto‐Torres JL, Verdiá‐Báguena C, Castaño‐Rodriguez C, Aguilella V & Enjuanes L (2015a). Relevance of viroporin ion channel activity on viral replication and pathogenesis. Viruses 7, 3552–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto‐Torres JL, Verdiá‐Báguena C, Jimenez‐Guardeño JM, Regla‐Nava JA, Castaño‐Rodriguez C, Fernandez‐Delgado R, Torres J, Aguilella VM & Enjuanes L (2015b). Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 485, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Madan V & Carrasco L (2012). Viroporins: Structure and biological functions. Nat Rev Microbiol 10, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S, Kandula S & Shaman J (2020). Differential effects of intervention timing on COVID‐19 spread in the United States. medRxiv, 10.1101/2020.05.15.20103655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Tan E, Parthasarathy K, Lin X, Jiang FL, Yu D, Vararattanavech A, Tuck WS, Ding XL, Torres J, Soong TW, Liu DX & Torres J (2009). Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog 5, e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ & Lamb RA (1992). Influenza virus M2 protein has ion channel activity. Cell 69, 517–528. [DOI] [PubMed] [Google Scholar]
- Premkumar A, Wilson L, Ewart GD & Gage PW (2004). Cation‐selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett 557, 99–103. [DOI] [PubMed] [Google Scholar]
- Regla‐Nava JA, Jimenez‐Guardeño JM, Nieto‐Torres JL, Gallagher TM, Enjuanes L & DeDiego ML (2013). The replication of a mouse adapted SARS‐CoV in a mouse cell line stably expressing the murine SARS‐CoV receptor mACE2 efficiently induces the expression of proinflammatory cytokines. J Virol Methods 193, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR & Chou JJ (2008). Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D & Fielding BC (2019). Coronavirus envelope protein: Current knowledge. Virol J 16, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D & Fielding BC (2020). Is there a link between the pathogenic human coronavirus envelope protein and immunopathology? A review of the literature. Front Microbiol 11, 2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C & Griffin S (2015). Viroporins: Structure, function and potential as antiviral targets. J Gen Virol 96, 2000–2027. [DOI] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A & Li F (2020). Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A 117, 11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova G V., Solovieva EA, Chepurnykh T V., Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW & Chudakov DM (2009). Far‐red fluorescent tags for protein imaging in living tissues. Biochem J 418, 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Tomar PP & Arkin IT (2020). SARS‐CoV‐2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem Biophys Res Commun 530, 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D, Tan J, Jiang K, Hernandez MM, Fabre S, Amanat F, Teo C, Arunkumar GA, McMahon M, Capuano C, Twyman K, Jhang J, Nowak MD, Simon V, Sordillo EM, van Bakel H & Krammer F (2021). Repeated cross‐sectional sero‐monitoring of SARS‐CoV‐2 in New York City. Nature 590, 146–150. [DOI] [PubMed] [Google Scholar]
- Torres J, Maheswari U, Parthasarathy K, Ng L, Liu DX & Gong X (2007). Conductance and amantadine binding of a pore formed by a lysine‐flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci 16, 2065–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Parthasarathy K, Lin X, Saravanan R, Kukol A & Liu DX (2006). Model of a putative pore: the pentameric α‐helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys J 91, 938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan J, Ngamtrakulpanit L, Pajewski TN, Turner R, Nguyen TA, Smith A, Urban P, Hom S, Gaston B & Hunt J (2003). Exhaled breath condensate pH is a robust and reproducible assay of airway acidity. Eur Respir J 22, 889–894. [DOI] [PubMed] [Google Scholar]
- Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA & Hogue BG (2015). Coronavirus envelope (E) protein remains at the site of assembly. Virology 478, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Li S‐Y, Yang X‐L, Huang H‐M, Zhang Y‐J, Guo H, Luo C‐M, Miller M, Zhu G, Chmura AA, Hagan E, Zhou J‐H, Zhang Y‐Z, Wang L‐F, Daszak P & Shi Z‐L (2018). Serological evidence of bat SARS‐related coronavirus infection in humans, China. Virol Sin 33, 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Barrett KMS, Larsen BB, Anderson C, White S, Cassens T, Jacobs S, Levan G, Nguyen J, Ramirez JM, Rivera‐Garcia C, Sandoval E, Wang X, Wong D, Spencer E, Robles‐Sikisaka R, Kurzban E, Hughes LD, Deng X, Wang C, Servellita V, Valentine H, De Hoff P, Seaver P, Sathe S, Gietzen K, Sickler B, Antico J, Hoon K, Liu J, Harding A, Bakhtar O, Basler T, Austin B, Isaksson M, Febbo PG, Becker D, Laurent M, McDonald E, Yeo GW, Knight R, Laurent LC, de Feo E, Worobey M, Chiu C, Suchard MA, Lu JT, Lee W& Andersen KG (2021). Genomic epidemiology identifies emergence and rapid transmission of SARS‐CoV‐2 B.1.1.7 in the United States. medRxiv, 10.1101/2021.02.06.21251159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerbeck JW & Machamer CE (2019). The infectious bronchitis coronavirus envelope protein alters golgi pH to protect the spike protein and promote the release of infectious virus. J Virol 93, e00015‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Lambson BE, Vermeulen M, van den Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN & Moore PL (2021). SARS‐CoV‐2 501Y.V2 escapes neutralization by South African COVID‐19 donor plasma. bioRxiv, 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans SJW, Smeeth L & Goldacre B (2020). Factors associated with COVID‐19‐related death using OpenSAFELY. Nature 584, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Gage P & Ewart G (2006). Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Mckinlay C, Gage P & Ewart G (2004). SARS coronavirus E protein forms cation‐selective ion channels. Virology 330, 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2021). Weekly epidemiological update – 12 January 2021. https://www.who.int/publications/m/item/weekly‐epidemiological‐update%14;12‐january‐2021.
- Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, Rambaut A, Suchard MA, Wertheim JO & Lemey P (2020). The emergence of SARS‐CoV‐2 in Europe and North America. Science 370, 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM & Weinman SA (2010). Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog 6, e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Shen X, He Y, Pan X, Wang Y, Yang F, Fang S, Wu Y, Zuo X, Xie Z, Jiang X, Chi H, Meng Q, Zhou H, Zhou Y, Cheng X, Chen T, Xin X, Jiang H, Xiao G, Zhao Q, Zhang L‐K, Shen J, Li J & Gao Z (2020). SARS‐CoV‐2 envelope protein causes acute respiratory distress syndrome (ARDS)‐like pathological damage and constitutes an antiviral target. bioRxiv, 10.1101/2020.06.27.174953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Nabar NR, Shi CS, Kamenyeva O, Xiao X, Hwang IY, Wang M & Kehrl JH (2018). SARS‐coronavirus open reading frame‐3a drives multimodal necrotic cell death. Cell Death Dis 9, 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhou P, Wei Y, Yue H, Wang Y, Hu M, Zhang S, Cao T, Yang C, Li M, Guo G, Chen X, Chen Y, Lei M, Liu H, Zhao J, Peng P, Wang C‐Y & Du R (2020). Histopathologic changes and SARS‐CoV‐2 immunostaining in the lung of a patient with COVID‐19. Ann Intern Med 172, 629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H & Cao B (2020a). Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet 395, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF & Shi ZL (2020b). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF & Tan W (2020). A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical Summary Document
Data Availability Statement
Any supplementary information, chemical compound information, and source data are available from the corresponding author (M.Y.) upon reasonable request.


