Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), a novel β coronavirus, has led to very significant global healthcare challenges. Development of cellular immunity following infection, or indeed vaccination, will be pivotal in gaining pandemic control, and has been shown to be protective in animal studies and from vaccination studies. 1 However, the extent to which immunocompromised patients are able to mount a ‘normal’ immune response to SARS‐CoV‐2 remains unknown. Collaborative cohort analyses reporting clinical sequelae following SARS‐CoV‐2 infection in patients with either a classical ‘Philadelphia Chromosome’ negative myeloproliferative neoplasm (MPN) or chronic myeloid leukaemia (CML) have recently been published. 2 , 3 Outcomes were varied but, to date, older patients, those with myelofibrosis (MF) and those who had sudden discontinuation of the Janus kinase (JAK) inhibitor (JAKi) ruxolitinib had worse outcomes. Chronic MPN, in particular MF, are associated with a pro‐inflammatory state and inherent dysregulation of pivotal natural killer (NK) cell, regulatory T‐cell and effector T‐cell function. 4 These heterogeneous defects are likely modified by patient age, disease subtype and stage. Moreover, complex adaptive immune responses to significant viral challenges may potentially be attenuated by tyrosine kinase inhibitors (TKI), JAKi and other cytoreductive therapies. In the present study, we characterise memory T‐cell responses following natural infection with SARS‐CoV‐2 in seven patients with a chronic myeloid disorder compared to a control cohort of healthcare workers.
Patients with a World Health Organization (WHO)‐defined MPN and healthy controls (HC) working in an acute healthcare setting were recruited in accordance with the Regional Research and Ethics Review Board. Peripheral blood mononuclear cells (PBMCs) were isolated using standard density centrifugation techniques and cryopreserved. T‐cell functionality was assessed using intracellular cytokine staining after incubation with SARS‐CoV‐2‐specific peptides covering the immunogenic domains of the spike (S)‐, membrane (M)‐ and nucleocapsid (N) protein (Miltenyi Biotech, Bergisch Gladbach, Germany). Briefly, cells were thawed and then rested for 18 h at 37°C, 5% CO2. Specific peptides and anti‐cluster of differentiation 28 (CD28) (BD Bioscience San Jose, CA, USA) were added for 3 h, followed by Brefeldin‐A (BFA) for an additional 3 h. Unstimulated cells were utilised as negative controls and Cytostim (Miltenyi Biotech) was added separately as a positive control. Cells were stained with a viability dye, stained with antibodies directed against surface markers, and fixed and permeabilised (BD CytoFix/Cytoperm) before staining with antibodies directed against intracellular cytokines. 5 , 6 Directly conjugated monoclonal antibodies with the following specificities were used; CD3 BUV395 (clone SK37), CD4 PE (clone M‐T477), tumour necrosis factor α (TNFα) (clone MAB11), interferon γ (IFNγ) APC (clone B27), interleukin 2 (IL‐2) PerCP‐5·5 (clone MQ1‐17H12). Live/dead staining was performed using Zombie NIR® amine reactive fluorescent dye (Biolegend, San Diego, CA, USA). Gating on the lymphocyte population, single cells, live cells, CD3+ cells, CD4+ cells and CD4– (CD8+) was performed. Analysis was performed on a BD Fortessa cytometer and results processed using Flowjo, version 10·5. Statistical analysis was performed using GraphPad Prism, version 8 (GraphPad Software Inc., La Jolla, CA, USA).
The patient‐ and healthcare worker cohorts (control) characteristics’ are summarised in Tables S1 and S2. In brief, there were six healthcare workers (three male, median age of 35·5 years) and seven patients (three CML, one MF, one polycythaemia vera and two essential thrombocythaemia; two male; median age of 48 years) with confirmed or presumed clinical exposure to SARS‐CoV‐2. Three individuals with no known exposure, positive polymerase chain reaction (PCR) or serology were also included (negative group). The patients with CML were on TKI therapy (one each for imatinib, bosutinib and ponatinib), whereas all four patients with MPN were treatment naïve. The median (range) time from illness/positive test to sampling for cases was 31 (6–38) weeks and for healthcare workers was 25·5 (22–38) weeks.
The results are expressed as mean increases in cytokine expression in CD4+ and CD8+ cells compared to baseline unstimulated samples, with a positive result considered to be that with a >twofold increase from baseline cytokine expression. Three HC and two patients with MPN, lacking detectable anti‐SARS‐CoV‐2 antibodies, but with a clear history of symptoms or exposure, demonstrated a ‘positive’ T‐cell response. This included one HC who had PCR confirmed infection, but lacking detectable antibodies 3 months after testing. Both patients with discordant T‐cell and antibody responses were on TKI therapy for chronic phase CML. Patients with chronic MPN without detectable antibodies had a T‐cell response that was broadly equivalent in many areas to that seen in those with detectable anti‐SARS‐CoV‐2 antibodies. For example, the mean proportion of CD8+ T cells, expressing TNFα and IFNγ in response to exposure to the M protein, was 0·51% in seronegative patients compared with 0·44% in the antibody‐positive group.
Patients with MPN with a characteristic history or confirmed previous infection using PCR or antibody testing (‘MPN‐positive group’) had a robust T‐cell response after exposure to immunogenic SARS‐CoV‐2 peptides, comparable to that seen in the HC‐positive group (not statistically significant), contrasting with the negative group. For example, patients with MPN demonstrated a polyfunctional T‐cell response with mean dual expression of TNFα and IL‐2 in 0·23% of CD4+ cells, following exposure to the M protein, compared with 0·11% in HC‐positive group and 0·01% of the negative group. (Figs 1 and 2). For N and S protein exposure, patients with MPN demonstrated a mean TNFα and IL‐2+ expression of 0·29% and 0·19% of CD4+ cells respectively, compared with 0·13% and 0·18% in the HC‐positive group and 0·03% (both) in the negative group. For CD8+ T cells, the mean TNFα and IFNγ+ expression in the MPN‐positive group, HC‐positive group and negative group, respectively, was 0·46%, 0·38% and 0% for M protein exposure; 0·22%, 0·34% and 0·01% for N protein exposure and 0·3%, 0·41% and 0% for S protein exposure.
Fig 1.
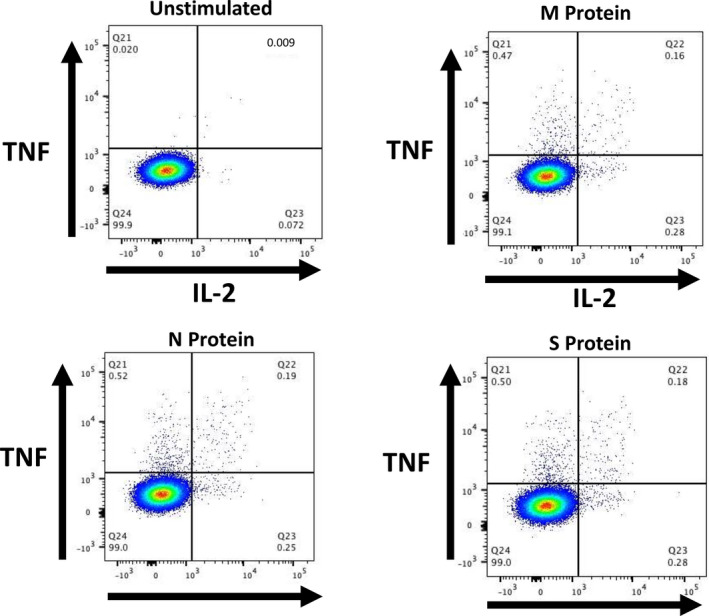
Illustrative composite flow cytometry plots from a patient (aged 65 years) with essential thrombocythaemia demonstrating CD4+ cells expressing tumour necrosis factor α (TNFα) (y axes) and interleukin 2 (IL‐2) (x axes). Top left, unstimulated cells; top right, M protein; bottom left, N protein; bottom right, S protein. [Colour figure can be viewed at wileyonlinelibrary.com]
Fig 2.
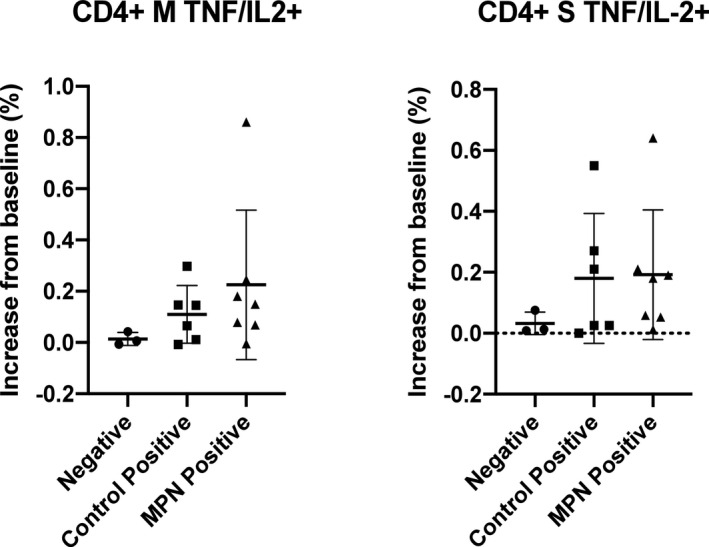
Comparative analyses between the myeloproliferative neoplasm (MPN)‐positive group and the healthy control‐positive group (both groups include those with characteristic history or confirmed previous infection using polymerase chain reaction or antibody testing) and the negative group (not meeting any of these criteria) for CD4+ cell expression of tumour necrosis factor α (TNFα) and interleukin 2 (IL‐2) in response to the M and S proteins.
Amongst patients with MPN a CD4+ T‐cell response was seen against each of the SARS‐CoV‐2 proteins investigated, which was broadly equivalent for cells expressing IL‐2 and TNFα; however, a modest increase was identified for IFNγ‐expressing cells directed against the M protein (CD4+ mean expression against the M protein 0·36%, N protein 0·13%, S protien 0·15%). For CD8+ T cells, expression of TNFα was similar for the N and S proteins, but slightly higher for the M protein (CD8+ mean expression 0·32%, 0·23% and 0·59% respectively). A polyfunctional response was observed in all patients with chronic MPN in either CD4+ or CD8+ T cells. Cytokine analysis in patients with MPN showed expression of TNFα, in response to each protein to be greater in CD4+ T cells compared with CD8+ (mean expression against M protein, 0·85% vs. 0·59%; N protein, 0·62% vs. 0·32%; S protein, 0·66% vs. 0·23%). In contrast, expression of IFNγ was greater in CD8+ T cells (mean expression against M protein, 0·81% vs. 0·36%; N protein, 0·38% vs. 0·13%; S protein, 0·24% vs. 0·14%). Of note, the patients with CML on TKI therapy had responses in keeping with HC, e.g. mean expression of TNFα/IL2+ in response to M protein was 0·19% (CML) versus 0·11% (HC positive).
The complex, frequently discordant, immune response following SARS‐CoV‐2 exposure is under extensive investigation at present. For the first time, we have demonstrated that patients with chronic MPN can mount functional memory T‐cell responses following natural SARS‐CoV‐2 exposure. For the CML cohort, there did not appear to be a significant attenuation of T‐cell responses compared to HC. This is potentially reassuring, given that TKIs possess potential suppressive effects due to ‘off target’ activity against Src kinases inhibiting T‐cell receptor signalling. 6 Similarly, the MPN cohort, albeit heterogeneous, did not demonstrate attenuated T‐cell responses compared to HC. However, of note, none were on active cytoreductive or JAKi therapy. Given the immunomodulatory effects of JAKi such as ruxolitinib on B‐cell, T‐cell, NK‐cell and dendritic‐cell function, delineation of immune responsiveness in SARS‐CoV‐2 infected patients with MPN on such agents is required. 4 , 7 , 8
T‐cell ‘fate’ following SARS‐CoV‐2 exposure appears dynamic, dependent on both host characteristics and indeed the severity of infection. 9 , 10 , 11 As suggested by Altmann et al., 12 responses to SARS‐CoV‐2 can in fact become ‘uncoupled’ with discordant B‐ and T‐cell priming and memory. Whether either CD4+ or CD8+ immune responsiveness can function as true correlates of protection remains unknown. However, the importance of a memory T‐cell response is intimated by evidence from the SARS‐CoV‐1 infection, whereby patients demonstrated relatively transient antibody responses with longer lasting T‐cell immunity. 13 Further understanding of such is paramount for correct timing of immunosuppressive drugs in the disease course, optimisation of assays to assess surrogate correlates of longer‐term ‘protection’ and underpinning of future vaccination strategies.
Barbui et al. 2 recently reported on 175 patients with MPN diagnosed with SARS‐CoV‐2 infection, demonstrating higher mortality compared to the non‐MPN population. Worse outcome was associated with older age, male sex, comorbidities, a diagnosis of MF, lymphopenia and respiratory support requirement. Higher risks were also identified for ruxolitinib discontinuation following infection. The CANDID CML study reported on 110 patients with SARS‐CoV‐2 infection, 70% of whom were on TKIs. 3 Univariate analysis revealed age >75 years, severe infection and use of imatinib (likely surrogate marker of older age) as adverse risk factors.
We appreciate that the present study is exploratory in nature, with a limited cohort size and comprises a range of chronic MPN conditions. We focussed solely on T‐cell functionality, did not include measurement of SARS‐CoV‐2‐neutralising antibodies or assessment for pre‐existing ‘cross‐reactive’ T cells. These data, however, represents an important ‘snapshot’ of SARS‐CoV‐2 mediated T‐cell modulation in chronic MPN albeit, as we lack longitudinal samples, it remains unclear if such responses are both durable and persistent. Nonetheless, despite these caveats, we have demonstrated that T cells from patients with chronic MPN are able to mount a memory response upon re‐exposure to key immunogenic proteins from SARS‐CoV‐2. Moreover, the time between infection and sampling was >6 months in five of the included patients with MPN, suggesting a durable T‐cell response is maintained in this group. We have previously demonstrated impaired B‐ and T‐cell responses to the annual influenza vaccine in patients with MPN compared to HC. 14 In addition, the need for repeat influenza A virus subtype H1N1 (H1N1)‐vaccination in CML (two doses) has been demonstrated to optimise responsiveness. 5 Hence, there is a requirement to urgently delineate comprehensive SARS‐CoV‐2 vaccine responses across these patient cohorts.
In conclusion, the present results are the first step in understanding adaptive immune responses in patients with chronic MPN following SARS‐CoV‐2 exposure. Comprehensive global assessment of the immunome within these patient groups is required paralleled with exploration of how pre‐existing immunity ‘imprinting’ affects cellular and humoral response to vaccination strategies.
Conflict of Interest
Patrick Harrington: research funding from Bristol Myers Squibb and Speaker fees from Incyte. Claire N. Harrison: Novartis; Speaker fees: Novartis, Jannsen, CTI, Celgene, Medscape; Advisory Board: Incyte, CTI, Sierra Oncology, Novartis, Celgene, Roche, AOP pharma, Geron, Astra Zenica. Shahram Kordasti: Celgene and Novartis research grant; Alexion speaker honorarium. Hugues de Lavallade: grants and speakers fees from Bristol Myers Squibb and Incyte; Speakers fees from Novartis and Pfizer. Donal P. McLornan: Speaker fees and advisory boards Novartis, Celgene and Jazz pharmaceuticals.
Supporting information
Table S1. Patients with MPN – clinical characteristics.
Table S2. Healthcare workers: characteristics of positive cases.
Figure S1. Comparative analyses between the MPN‐positive group, the healthy control‐positive group (and the negative group for CD8+ cell expression of TNFα and IFNγ in response to M and S proteins.
Figure S2. Comparative analyses of monofunctional versus polyfunctional T‐cell response in patients with chronic MPN in CD4+ T cells directed against S and N proteins.
Acknowledgements
Patrick Harrington designed the research study, performed the research, analysed the data and wrote the paper. Claire N. Harrison, Richard Dillon, Deepti H. Radia, Katayoun Rezvani, Kavita Raj, Claire Woodley, Natalia Curto‐Garcia, Jennifer O’Sullivan, Jamie Saunders, Shahram Kordasti and Sahra Ali assisted with patient recruitment and reviewed the paper. Hugues de Lavallade and Donal P. McLornan designed the research study and wrote the paper.
References
- 1. Bao L, Deng W, Gao H, Xiao C, Liu J, Xue J, et al. (2020). Lack of Reinfection in Rhesus Macaques Infected with SARS‐CoV‐2. bioRxiv 2020.03.13.990226. Available at: http://biorxiv.org/lookup/doi/10.1101/2020.03.13.990226 [Accessed January 14, 2021].
- 2. Barbui T, Vannucchi AM, Alvarez‐Larran A, Iurlo A, Masciulli A, Carobbio A, et al. High mortality rate in COVID‐19 patients with myeloproliferative neoplasms after abrupt withdrawal of ruxolitinib. Leukemia. 2021; 35:485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rea D, Mauro MJ, Cortes JE, Jiang Q, Pagnano KB, Ongondi M, et al. COVID‐19 in patients (pts) with chronic myeloid leukemia (CML): results from the International CML Foundation (iCMLf) CML and COVID‐19 (CANDID) Study. Blood. 2020;136:46–7. [Google Scholar]
- 4. McLornan DP, Khan AA, Harrison CN. Immunological consequences of JAK inhibition: friend or foe? Curr Hematol Malignancy Rep. 2015;10:370–9. [DOI] [PubMed] [Google Scholar]
- 5. de Lavallade H, Garland P, Sekine T, Hoschler K, Marin D, Stringaris K, et al. Repeated vaccination is required to optimize seroprotection against H1N1 in the immunocompromised host. Haematologica. 2011;96:307–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Lavallade H, Khoder A, Hart M, Sarvaria A, Sekine T, Alsuliman A, et al. Tyrosine kinase inhibitors impair B‐cell immune responses in CML through off‐target inhibition of kinases important for cell signaling. Blood. 2013;122:227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heine A, Held SAE, Daecke SN, Wallner S, Yajnanarayana SP, Kurts C, et al. The JAK‐inhibitor ruxolitinib impairs dendritic cell function in vitro and in vivo. Blood. 2013;122:1192–202. [DOI] [PubMed] [Google Scholar]
- 8. Keohane C, Kordasti S, Seidl T, Perez Abellan P, Thomas NSB, Harrison CN, et al. JAK inhibition induces silencing of T Helper cytokine secretion and a profound reduction in T regulatory cells. Br J Haematol. 2015;171:60–73. [DOI] [PubMed] [Google Scholar]
- 9. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID‐19: current state of the science. Immunity. 2020;52:910–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS‐CoV‐2 coronavirus in humans with COVID‐19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS‐CoV‐2 in UK convalescent individuals following COVID‐19. Nat Immunol. 2020;21:1336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Altmann DM, Boyton RJ. SARS‐CoV‐2 T cell immunity: specificity, function, durability, and role in protection. Science Immunol. 2020;5:eabd6160. [DOI] [PubMed] [Google Scholar]
- 13. Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus‐specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88:11034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alimam S, Ann Timms J, Harrison CN, Dillon R, Mare T, DeLavallade H, et al. Altered immune response to the annual influenza A vaccine in patients with myeloproliferative neoplasms. Br J Haematol. 2020. 10.1111/bjh.17096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patients with MPN – clinical characteristics.
Table S2. Healthcare workers: characteristics of positive cases.
Figure S1. Comparative analyses between the MPN‐positive group, the healthy control‐positive group (and the negative group for CD8+ cell expression of TNFα and IFNγ in response to M and S proteins.
Figure S2. Comparative analyses of monofunctional versus polyfunctional T‐cell response in patients with chronic MPN in CD4+ T cells directed against S and N proteins.


