Abstract
Dendritic cells (DCs) are key regulators of the immune system that shape T cell responses. Regulation of T cell induction by DCs may occur via the intracellular enzyme indoleamine 2,3‐dioxygenase 1 (IDO), which catalyzes conversion of the essential amino acid tryptophan into kynurenine. Here, we examined the role of IDO in human peripheral blood plasmacytoid DCs (pDCs), and type 1 and type 2 conventional DCs (cDC1s and cDC2s). Our data demonstrate that under homeostatic conditions, IDO is selectively expressed by cDC1s. IFN‐γ or TLR ligation further increases IDO expression in cDC1s and induces modest expression of the enzyme in cDC2s, but not pDCs. IDO expressed by conventional DCs is functionally active as measured by kynurenine production. Furthermore, IDO activity in TLR‐stimulated cDC1s and cDC2s inhibits T cell proliferation in settings were DC‐T cell cell‐cell contact does not play a role. Selective inhibition of IDO1 with epacadostat, an inhibitor currently tested in clinical trials, rescued T cell proliferation without affecting DC maturation status or their ability to cross‐present soluble antigen. Our findings provide new insights into the functional specialization of human blood DC subsets and suggest a possible synergistic enhancement of therapeutic efficacy by combining DC‐based cancer vaccines with IDO inhibition.
Keywords: cDC1, cDC2, plasmacytoid dendritic cells, IDO, epacadostat
Among human blood dendritic cell subsets, cDC1s uniquely express biologically active IDO at steady‐state, whereas cDC2s express IDO upon IFN‐γ‐ or TLR agonist‐mediated signaling. The enzyme converts tryptophan into kynurenine, thereby limiting T cell proliferation. Selective inhibition of IDO by epacadostat impedes IDO‐mediated immunosuppressive effects in conventional DCs.
Introduction
Dendritic cells (DCs) are professional antigen‐presenting cells that can both induce and regulate T cell priming. One of the mechanisms by which DCs can negatively regulate T cell responses is via the intracellular enzyme indoleamine 2,3‐dioxygenase 1 (IDO). IDO catalyzes breakdown of tryptophan into N‐formyl‐kynurenine, the first and rate‐limiting step in tryptophan catabolism. Tryptophan deprivation, sensed by ribosomal kinase GCN2, and tryptophan metabolites such as kynurenine, inhibit effector T cell proliferation and induce anergy or apoptosis [1, 2, 3]. In regulatory T cells (Tregs), however, GCN2 signaling leads to activation and induces a strong suppressive phenotype [4]. Furthermore, both IDO‐mediated GCN2 activation and binding of kynurenine to the aryl hydrocarbon receptor promote de novo Treg differentiation from naïve CD4+ T cells [5, 6].
IDO appears to play an important role in inducing tolerance and limiting excessive inflammation, for instance in the placenta [7]. In some settings, however, IDO activity might actually be detrimental to the host. Cancer cells and myeloid‐derived suppressor cells often express IDO to inhibit antitumor immunity and high IDO expression is associated with poor prognosis and shorter overall survival in several cancer types, including ovarian cancer [8], endometrial cancer [9], colorectal cancer [10], and melanoma [11]. Primary DCs are currently under investigation for their use in cancer vaccines, which aim at boosting the antitumor‐specific T cell response. Because IDO activity inhibits T cell function, expression of IDO by human DC subsets may have important implications for the efficacy of DC‐based immunotherapy.
Multiple types of inflammatory signals may induce IDO expression in DCs. IDO1 gene promoters contain several interferon (IFN) response elements, allowing transcription of IDO upon signaling by type I IFNs, and more effectively by type II IFN (IFN‐γ) [12, 13]. Other inducers of IDO include ligation of CD40 and Toll‐like receptors (TLRs), and reverse signaling by B7 molecules upon ligation by CTLA‐4 [14, 15, 16, 17]. Importantly, IDO expression at the protein level does not necessarily correlate with biological activity of the enzyme. For instance, a combination of different signals was shown to be required to achieve full enzymatic activity in human monocyte‐derived DCs (moDCs) [14, 18].
Most studies on IDO expression by DCs have been performed with murine DCs and human moDCs. In contrast, less is known about the function of IDO in human primary DCs, which are subdivided into three major subsets: plasmacytoid DCs (pDCs) and type 1 and type 2 conventional DCs (cDC1s and cDC2s). Interestingly, RNA profiling revealed high IDO expression by human steady‐state and activated cDC1s [19, 20, 21, 22]. Our recent comparative proteomic analysis of steady‐state human DC subsets likewise revealed IDO protein to be uniquely expressed by cDC1s [23]. Here, we compared the expression and activity of IDO in immature and stimulated human peripheral blood DCs and studied the effects on T cell proliferation.
Results
Immature cDC1s selectively express high levels of functionally active IDO
High expression of IDO by human cDC1s compared to cDC2s and pDCs was first reported on RNA level by Crozat et al. [19], using a dataset generated with RNA microarray [24]. More recently, single‐cell RNA‐sequencing (RNA‐seq) analysis of steady‐state human blood DC subsets also demonstrated IDO to be uniquely associated with cDC1s [20, 21]. Furthermore, IDO1 gene expression was higher in cDC1s compared to cDC2s and pDCs in human tonsil [25], and thymus and spleen [26]. We confirm these findings by combining a bulk RNA‐seq dataset of blood cDC1s (Mathan et al. manuscript in preparation) and a bulk RNA‐seq dataset of blood cDC2s and pDCs [27], both generated by our group. Comparing the total prevalence of the genes that Villani et al. found to be specific for cDC1s [20] revealed that IDO1 is, together with XCR1 and CADM1, the most uniquely expressed gene by cDC1s, compared to cDC2s and pDCs (Fig. 1A). Moreover, IDO1 gene expression in cDC1s is increased upon stimulation with TLR3 or TLR7/8 agonists (Fig. 1B).
Figure 1.
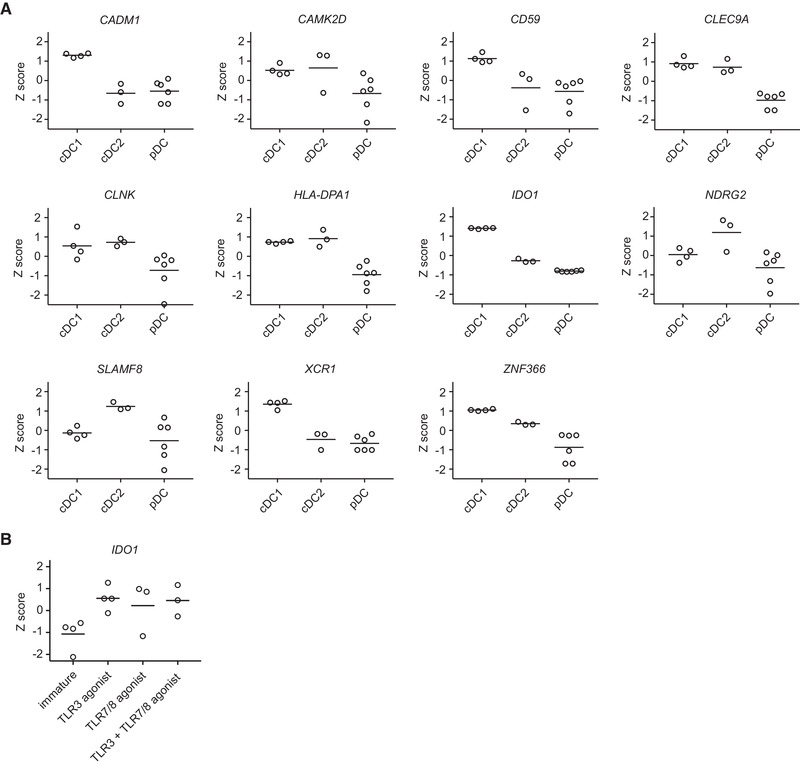
IDO1 is among the genes that best defines peripheral blood cDC1s under homeostatic conditions and its expression is increased upon stimulation with TLR agonists. Gene expression was analyzed from RNA‐seq datasets. Z scores were calculated using normalized and log transformed counts per million values. Means are depicted. (A) Expression of selected genes was compared between immature cDC1s, immature cDC2s, and immature (unstimulated or IL‐3‐treated) pDCs, of 4 (cDC1) or 3 (cDC2 and pDC) different donors. (B) IDO1 gene expression among cDC1s of three or four different donors cultured overnight in absence or presence of a clinical grade TLR3 agonist (Hiltonol) and/or TLR7/8 agonist (protamine‐RNA).
Comparative proteomic analysis in steady‐state human blood DC subsets revealed that also at the protein level, IDO was highly expressed by cDC1s, while the signal was below detection limit for cDC2s and pDCs (Fig. 2A) [23]. Analysis of immature blood DCs by flow cytometry confirmed significantly higher IDO expression by cDC1s compared to cDC2s and pDCs (Fig. 2B).
Figure 2.
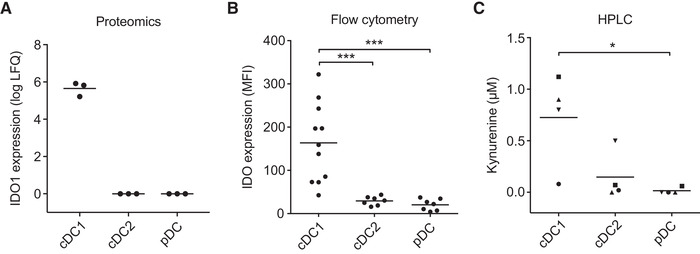
Immature cDC1s, but not cDC2s and pDCs, express functionally active IDO. (A) IDO1 protein expression among steady‐state human blood DCs of three different donors was analyzed from a proteomics dataset. Means are depicted. (B) IDO intracellular protein expression was analyzed by flow cytometry in DCs cultured overnight. The graph shows the geometric mean fluorescent intensity (MFI) of IDO expression subtracted by the MFI of the isotype control, with at least seven different donors per condition from 10 independent experiments. Means are depicted. Significance was determined by one‐way ANOVA with Bonferroni correction (*** P < 0.001). (C) IDO activity was analyzed by measuring l‐kynurenine in supernatants of 24‐h DC cultures by HPLC. Results are from six different donors from four independent experiments; in one experiment (squares), pooled supernatants from 3 different donors were used. Means are depicted and symbols correspond to measurements belonging to the same donors. Significance was determined by repeated measures one‐way ANOVA with Bonferroni correction (* P < 0.05).
We next analyzed IDO activity across DC subsets by measurement of L‐kynurenine levels in supernatants from 24‐hour DC cultures by high‐performance liquid chromatography (HPLC). cDC1s converted around 1 μM tryptophan into kynurenine, whereas the same number of pDCs and cDC2s produced less kynurenine (Fig. 2C). Taken together, immature human peripheral blood cDC1s express functionally active IDO at significantly higher levels than cDC2s and pDCs.
IDO is induced in cDC2s and upregulated in cDC1s by TLR ligation or IFN‐γ stimulation
Upon activation, DCs acquire the capacity to migrate to the draining lymph node and express co‐stimulatory molecules needed to activate T cells. Hence, DC‐based vaccines using mature DCs induce superior immunological responses compared to vaccines using immature DCs [28]. We therefore extended our analysis of IDO expression to TLR ligand‐matured DCs (Supporting Information Fig. 1). Furthermore, since IFN‐γ is a potent inducer of IDO and is highly produced by effector T cells, IFN‐γ stimulation is relevant in vivo and was therefore also included in the analysis. DC subsets isolated from peripheral blood were cultured overnight with the indicated stimuli and IDO protein expression was analyzed by flow cytometry (Fig. 3A and B), while IDO enzyme activity was determined by measurement of kynurenine production in supernatants by HPLC (Fig. 3C). Importantly, while immature cDC2s showed only minimal levels of IDO expression and activity, stimulation with IFN‐γ or the combination of TLR3‐agonist polyinosinic:polycytidylic acid (poly(I:C)) with TLR7/8 agonist resiquimod (R848) increased both protein expression and enzyme activity in this DC subset. cDC1s stimulated with IFN‐γ or the combination of poly(I:C) and R848 maintained and further increased IDO protein expression and activity, to levels higher than found in cDC2s (Supporting Information Fig. 2). High kynurenine production by IFN‐γ‐stimulated cDC1s was reduced upon addition of the IDO1‐specific inhibitor epacadostat (Supporting Information Fig. 3), indicating that kynurenine production is IDO‐dependent. In contrast, pDCs stimulated with IFN‐γ, R848 or TLR9‐agonist CpG oligodeoxynucleotide class C (CpG‐C) did not upregulate IDO expression or activity (Fig. 3B and C). Thus, TLR ligation or IFN‐γ stimulation induces IDO expression and activity in cDC1s and cDC2s, but not pDCs.
Figure 3.
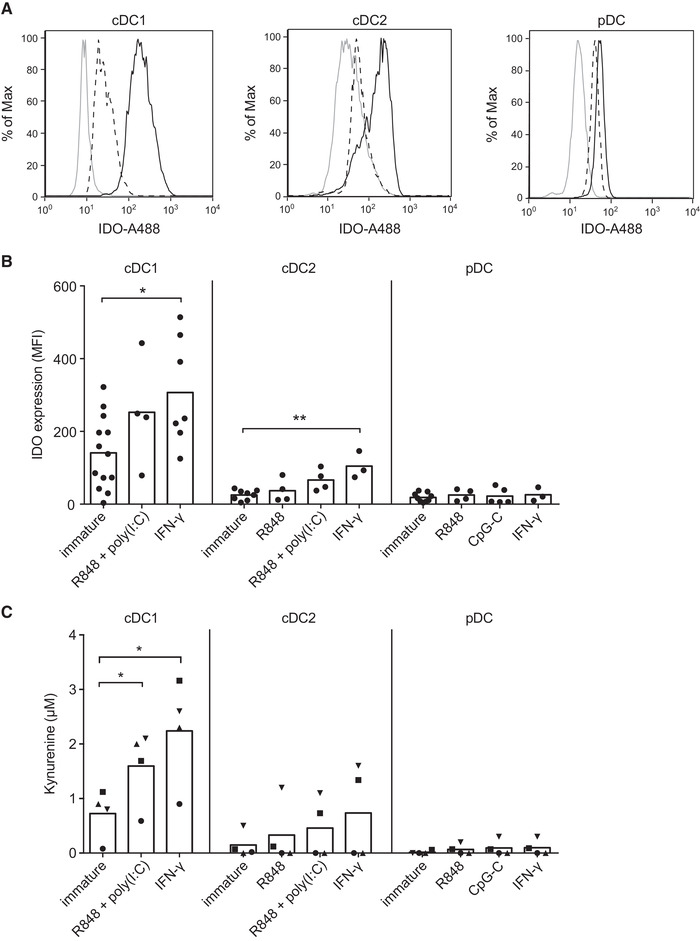
IDO protein expression and activity among stimulated blood DC subsets. (A) Flow cytometry histograms from a representative donor showing isotype control (solid grey line), IDO expression in absence of stimulus (dashed black line), and IDO expression for either IFN‐γ or CpG‐C stimulation (solid black line) for cDCs or pDCs, respectively. (B) The figure shows the geometric mean fluorescence intensity (MFI) of IDO expression subtracted by the MFI of the isotype control, with at least 3 different donors per condition from 10 independent experiments. Bars represent means. Significance was determined by one‐way ANOVA with Bonferroni correction, comparing stimulated DCs with immature control (* P < 0.05; ** P < 0.01). (C) IDO activity was analyzed by measuring l‐kynurenine in supernatants of 24‐h DC cultures by HPLC. The results are from six different donors from four independent experiments; in one experiment (squares), pooled supernatants from 3 different donors were used. Bars represent means and symbols correspond to measurements belonging to the same donors. Significance was determined by repeated measures one‐way ANOVA with Bonferroni correction, comparing stimulated DCs with their immature control (* P < 0.05).
Epacadostat‐treatment of cDC1s and cDC2s prevents IDO‐mediated inhibition of T cell proliferation
Human blood DC subsets express high levels of stimulatory molecules upon maturation and are known for their T cell stimulatory capacity [29]. However, IDO activity can inhibit T cell responses by depleting tryptophan and by increasing toxic kynurenine metabolites. To test whether IDO expressed by human blood DC subsets affects T cell proliferation, peripheral blood lymphocytes (PBLs) were stimulated with anti‐CD3/anti‐CD28 antibody‐coated beads in DC‐conditioned medium. T cell proliferation was inhibited by supernatant of cDC1s or cDC2s matured with poly(I:C) and R848 or stimulated with IFN‐γ (Fig. 4A); this inhibition could be partially rescued by specifically blocking IDO activity with epacadostat, with statistically significant differences for the conditions where cDCs were stimulated with TLR ligands. In contrast, treating pDCs with epacadostat did not affect T cell proliferation. These findings are in accordance with the IDO expression and activity data and underline the functional relevance of IDO activity by human cDCs on T cell proliferation.
Figure 4.
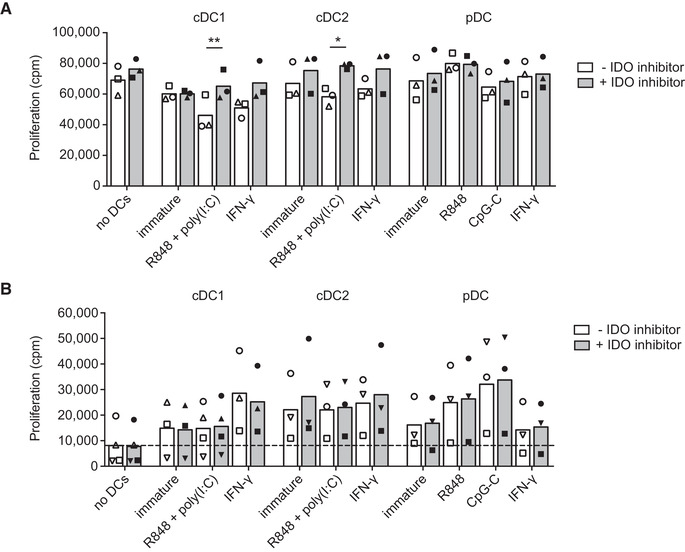
IDO expressed by cDCs inhibits T cell proliferation. Blood DCs were stimulated with indicated stimuli and/or epacadostat in medium containing 10 μM tryptophan. (A) PBLs were stimulated with anti‐CD3/CD28‐coated beads in supernatants of 48‐hour DC cultures and proliferation was measured after three days by tritiated thymidine incorporation. Mean proliferation in counts per minute (cpm) of three different donors from 3 independent experiments with technical triplicates (cDC2, pDC) or duplicates (cDC1) is shown. Symbols correspond to measurements belonging to the same donors. Significance was determined by two‐tailed paired t‐test comparing absence vs presence of IDO inhibitor (* P < 0.05; ** P < 0.01). (B) Allogeneic PBLs were cultured with overnight‐stimulated DCs and proliferation was measured after another three days by tritiated thymidine incorporation. Mean proliferation of at least three different donors from four independent experiments with technical triplicates (cDC2, pDC) or duplicates (cDC1) are shown. Symbols correspond to measurements belonging to the same donors. Significance was determined by two‐tailed paired t‐test, comparing absence versus presence of IDO inhibitor.
The effect of IDO on DC‐induced T cell proliferation was further studied in a mixed lymphocyte reaction (MLR). All three primary DC subsets induced allogenic T cell proliferation, but inhibition of IDO did not substantially affect T cell proliferation (Fig. 4B), in contrast to the assay with DC‐conditioned medium. Yet, addition of epacadostat to moDCs stimulated with LPS and IFN‐γ to express high levels of IDO [30] was able to increase allogeneic T cell proliferation (Supporting Information Fig. 4). This suggests that IDO activity in primary cDCs has only limited inhibitory effects in settings were these cells have contact‐mediated interactions with T cells. While poly(I:C) and R848 stimulation induces IDO activity (Fig. 3), this potent combination of TLR agonists is also known to strongly upregulate costimulatory molecules and cytokine production [29]. These findings might therefore reflect the complex interplay of inhibitory and stimulatory molecules expressed or secreted by DCs in T cell priming.
Epacadostat is currently under investigation in several clinical trials. Although its effects as a single treatment appear limited, several clinical studies suggest that IDO inhibition might potentiate the effects of other cancer immunotherapies [31]. To achieve a successful combination of epacadostat with DC‐based immunotherapy, the inhibitor should not negatively affect DC function. Cross‐presentation of exogenous antigens on MHC class I enables DCs to prime CD8+ T cells, which is crucial for induction of anti‐tumor immunity. In humans, all blood DC subsets are capable of cross‐presenting soluble antigens [32], although cDC1s are superior at cross‐presenting cellular antigens [33, 34, 35, 36]. We therefore tested the effect of epacadostat on the capacity of cDC1s, cDC2s and pDCs of HLA‐A2.1+ donors to cross‐present soluble gp100(272‐300) peptide to T cells transduced with a T cell receptor specific for the gp100(280‐288) peptide. Cross‐presentation of gp100(272‐300) peptide requires intracellular processing in order to release the HLA‐A2.1‐restricted gp100(280‐288) epitope. In line with our expectations, all DC subsets effectively cross‐presented soluble gp100(272‐300) (Fig. 5). IDO inhibition by epacadostat did not significantly change the ability of any of the three primary DC subsets, nor moDCs (Supporting Information Fig. 5), to cross‐present soluble antigen.
Figure 5.
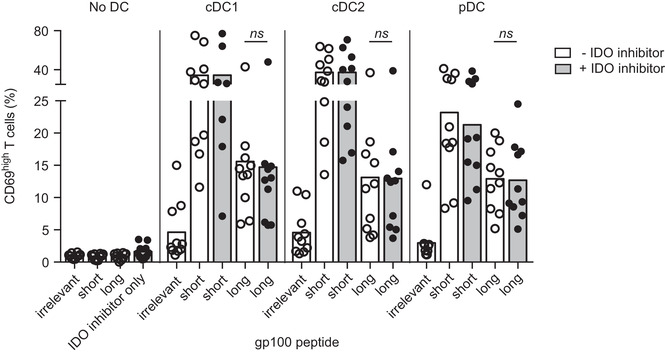
IDO inhibition by epacadostat does not affect the ability of DCs to cross‐present soluble antigen to T cells. Blood DCs were stimulated overnight with R848 (pDCs) or R848 and poly(I:C) (cDCs) in medium containing 1 μM tryptophan, with or without epacadostat, and loaded with gp100(154‐162) (irrelevant), gp100(280‐288) (short), or gp100(272‐300) (long) peptide. After overnight culture, T cells expressing the T cell receptor recognizing gp100(280‐288) were added at a 1:5 DC:T cell ratio for 24 h. CD69 expression by T cells was used as a readout for DC‐induced T cell stimulation and was measured by flow cytometry. Mean frequency of CD69high cells among live CD3+ T cells is shown, with at least seven different donors per condition from 11 independent experiments, with technical duplicates for cDC2 and pDC conditions. Significance was determined by two‐tailed paired t‐test, comparing absence versus presence of IDO inhibitor (ns, non‐significant).
The IDO inhibitor 1‐methyl‐tryptophan has been reported to partially inhibit the maturation of moDCs in response to certain TLR agonists [15]. We, therefore, tested whether epacadostat treatment affects the expression of membrane molecules on DC subsets that are normally upregulated upon maturation, as well as the production of IL‐12 by cDC subsets. Epacadostat treatment did not substantially change the expression of MHC‐I (HLA‐ABC), MHC‐II (HLA‐DR), CD40, CD80, CD83, CD86, CCR7, PD‐L1, or IL‐12 by any of the three DC subsets upon maturation by TLR3 and/or TLR7/8 agonists (Supporting Information Fig. 6A and B). Taken together, epacadostat enhances T cell proliferation induced by TCR cross‐linking in the presence of supernatant of TLR‐stimulated cDCs, while it does not interfere with DC maturation or their ability to cross‐present soluble antigen. These properties make this IDO inhibitor an interesting candidate to enhance the efficacy of DC‐based cancer immunotherapies.
Discussion
DCs can limit T cell responses via IDO activity. Although IDO is well‐characterized in murine DCs and human moDCs, much less is known about its activity and function in human primary DCs. Here, we compared the expression and activity of IDO across the three major human DC subsets found in peripheral blood. We describe a functional role for IDO in cDCs, in particular in cDC1s.
RNA expression profiling of steady‐state human DC subsets showed unique IDO1 expression by cDC1s [19, 20, 21]. We confirm that among immature DCs, IDO1 is one of the most uniquely expressed genes by cDC1s. Using flow cytometry, we demonstrate that IDO is also uniquely expressed at the protein level by immature cDC1s, which corroborates our previous study using proteomics [23]. We expand on this data by showing that IDO in immature cDC1s is biologically active, as it leads to the conversion of tryptophan into kynurenine. Upon stimulation with IFN‐γ or TLR agonists, its expression and activity is further increased and can limit T cell proliferation. Of note, human cDC1s have also been reported to specifically express the IDO1‐paralog gene IDO2 [19, 22], but its expression at the protein level, activity, and functional role have not been investigated here.
Human cDC1s are considered the equivalent of murine CD8α+ DCs [19, 33, 34, 36, 37, 38]. Likewise, murine CD8α+ DCs can mediate T cell suppression via IDO [39]. Interestingly, CD8α+ DCs constitutively express IDO, but the enzyme requires additional signaling through IFN‐γ or CTLA‐4 to become biologically active [40, 41]. We show that although signaling via IFN‐γ increases IDO expression and activity in human cDC1s, it is not needed per se for its enzymatic activity. Human cDC1s, like their murine counterpart, are excellent cross‐presenters and primers of T cells [37]. Expression of immunosuppressive IDO by this potent pro‐inflammatory immune cell seems paradoxical. Yet, cDC1s express the C‐type lectin receptor CLEC9A to efficiently induce immune responses towards dead cell antigens [33]. It is crucial that during homeostasis, apoptotic self‐antigens do not provoke an immune response, for this may lead to systemic autoimmunity. IDO has been reported to regulate tolerance to apoptotic antigens in mice [42]. Hence, preferential expression of IDO in cDC1s could function as a safeguard mechanism, ensuring default immunosuppressive function in absence of pathogen‐ or danger‐associated molecular patterns. In addition, upregulation of IDO by stimulated cDC1s might serve as a negative feedback mechanism to prevent excessive T cell stimulation. These hypotheses should be tested in future studies.
cDC2s do not express IDO when immature, but upregulate it upon exposure to TLR3 and TLR7/8 agonist or IFN‐γ. Production of kynurenine by human cDC2s has also been reported in response to LPS, E. coli, or prostaglandin E2 [43, 44]. In our assays, the supernatant from TLR‐stimulated cDC1s as well as cDC2s had a functional impact on T cell proliferation induced by TCR crosslinking. Inhibition of T cell proliferation was dependent on IDO and independent of cell‐cell contact, probably by depleting tryptophan and/or enriching T cell inhibitory breakdown products of tryptophan. Stimulatory cell‐cell contact between cDCs and T cells in an MLR seems to overpower IDO‐mediated inhibition, as blocking IDO did not boost T cell proliferation in this setting. Still, IDO activity in cDCs might become functionally relevant in the tumor microenvironment, where the enzyme's activity can contribute to other immunosuppressive signals.
Our data show that immature human pDCs exhibit no or very low IDO expression, which is in accordance with a study by Trabanelli et al. [44], as well as RNA expression profiling of steady‐state human DC subsets [19]. We also found very low expression and activity of IDO in pDCs stimulated with CpG‐C, R848, or IFN‐γ. Nevertheless, this does not rule out the possibility that human pDCs are able to express functional IDO in other settings. Indeed, IDO expression by human pDCs has been reported upon stimulation by HIV, CpG‐A or CpG‐B [45, 46]. Gerlini et al. identified IDO‐expressing pDCs in metastatic sentinel lymph nodes of melanoma patients [47]. pDCs from control lymph nodes and the blood of these patients did not express IDO, suggesting that expression of the enzyme by human pDCs can be induced by tumors.
By using the IDO1‐selective inhibitor epacadostat, we were able to increase T cell proliferation in conditioned medium of cDC1s or cDC2s. Epacadostat was demonstrated to be safe in phase I/II clinical trials, and is currently being tested in multiple phase III trials in combination with checkpoint blockade [31, 48]. Our results suggest that epacadostat could alternatively be used in combination with DC‐based cancer immunotherapies to increase inflammatory responses. Besides combining systemic epacadostat treatment with DC‐based cancer therapies, epacadostat might also be used during DC vaccine preparation. Treating conventional DCs ex vivo with epacadostat prior to injection into the patient could potentially enhance efficacy of immunostimulatory DC vaccines.
Materials and methods
RNA sequencing and proteomic analysis
RNA‐seq analysis was performed on a dataset of cDC2s and pDCs deposited at the Gene Expression Omnibus (accession number: GSE89442) [27] and a dataset of cDC1s (Mathan et al. manuscript in preparation). Data were analyzed using the R platform for statistical computing. Count values from both datasets were normalized and log‐transformed, before calculating Z scores using the edgeR package [49]. Proteomics analysis was performed on a dataset of cDC1s, cDC2s and pDCs deposited at the ProteomeXchange Consortium (dataset identifier: PXD004678) [23].
Cell isolation and culture
Human primary DCs were isolated from buffy coats or apheresis products (Sanquin, Amsterdam, The Netherlands) obtained from healthy volunteers after written informed consent and according to institutional guidelines. Peripheral blood mononuclear cells were obtained via ficoll density gradient centrifugation. DCs were isolated by FACS sorting after enrichment with Dynabeads Human DC enrichment kit (Invitrogen, Carlsbad, CA). The remaining cells were incubated with anti‐Lin1‐FITC, anti‐HLA‐DR‐PE‐Cy7 (both BD Biosciences, Franklin Lakes, NJ), anti‐CD1c‐BV421 (Biolegend, San Diego, CA), anti‐CD141‐APC and anti‐CD304‐PE (both Miltenyi Biotec, Bergisch Gladbach, Germany). The cells were sorted on a BD Aria II from lineage negative, HLA‐DRhigh cells into three different populations, based on the expression of CD141, CD1c or CD304 to obtain cDC1s, cDC2s, or pDCs, respectively. In some experiments as indicated, DC subsets were isolated from PBLs with magnetic microbeads by MACS sorting (Miltenyi Biotec). PBLs were prepared by depleting monocytes via anti‐CD14‐conjugated microbeads or adherence to plastic culture flasks. To generate immature moDCs, monocytes were cultured in X‐VIVO 15 medium supplemented with 2% human serum, 300 IU/mL IL‐4 and 450 IU/mL GM‐CSF (both ImmunoTools, Friesoythe, Germany) for 7 days. For mature moDCs, a maturation cocktail was added for the last day of the culture period.
Cells were cultured in either X‐VIVO 15 medium (Lonza, Basel, Switzerland) supplemented with 2% human serum (Sigma–Aldrich, Saint Louis, MO), hereinafter described as complete X‐VIVO medium, or in custom‐made tryptophan‐free RPMI 1640 (Gibco/Thermo Fisher Scientific, Waltham, MA) supplemented with 2 mM ultraglutamine (Lonza), 1% antibiotic‐antimycotic (Gibco), 10% human serum albumin (Sanquin) and the indicated concentration of l‐tryptophan (Merck, Darmstadt, Germany), hereinafter described as complete RPMI medium. DCs were incubated with stimuli as indicated: 4 μg/mL R848, 20 μg/mL poly(I:C), 5 μg/mL CpG‐C (all Enzo Life Sciences, Farmingdale, NY), 100 ng/mL LPS (Sigma–Aldrich), or 10 ng/mL IFN‐γ (Thermo Scientific). In the absence of activation stimulus, 10 ng/mL recombinant human IL‐3 (CellGenix) was added to pDC cultures as survival factor.
Flow cytometry
Flow cytometry experiments were performed according to the guidelines by Cossarizza et al. [50]. To analyze IDO expression, DCs were isolated by MACS sorting and cultured for 16 h in complete X‐VIVO medium with indicated stimuli. The next day, cells were stained extracellularly with anti‐CD141‐PE for cDC1s, anti‐CD1c‐PE for cDC2s, or anti‐CD304‐PE (all Miltenyi Biotec) for pDCs. Cells were subsequently fixed, permeabilized, and stained intracellularly with anti‐IDO‐A488 (clone #700838; R&D systems, Minneapolis, MN). Cells were measured on a BD FACS Calibur and data were analyzed by FlowJo software (see Supporting Information Fig. 7 for the gating strategy).
To analyze maturation marker expression upon treatment with epacadostat, DCs were isolated by FACS sorting and cultured at 1 × 104 DCs/well for 16 hours in complete RPMI medium with 1 or 10 μM l‐tryptophan, indicated stimuli and with or without 10 μM epacadostat (INCB024360; Selleck Chemicals, Houston, TX). After overnight culture, cells were labeled with fixable viability dye eFluor‐780 (eBioscience), anti‐CD80‐FITC (Biolegend), anti‐CD83‐APC, anti‐CD86‐PE‐Cy7, anti‐HLA‐ABC‐V450, anti‐HLA‐DR‐BV510 (all BD Biosciences), anti‐PD‐L1‐APC (Biolegend), CD40‐PE (Immunotech/Beckman Coulter, Brea, CA), and anti‐CD1b/c‐FITC (Diaclone, Besancon Cedex, France) or anti‐CD303‐FITC (Miltenyi Biotec). Expression of maturation markers on live DCs was determined on a BD FACSVerse and data were analyzed by FlowJo software.
Cytokine detection
Supernatants of DCs cultured at 1 × 104 DCs/200μL/well with indicated stimuli were analyzed with standard sandwich ELISAs detecting IL‐12p70 (Biolegend).
Kynurenine quantification
DCs were isolated by FACS sorting and 80 × 103 DCs were cultured at 0.5 × 106 cells/mL for 24 h in complete X‐VIVO medium and indicated stimuli. Conditioned media were analyzed for l‐kynurenine by HPLC. Samples were pretreated with perchloric acid to precipitate proteins. Samples were pretreated (5:1) with 20% formic acid. 200 μL of samples was mixed with 200 μL buffer (0.77 g/L NH4Ac, 100 mg/L EDTA‐Na2, 375 mg/L NaCl, 37.5 mL/L MeOH; pH 5.35). Pure kynurenine (Sigma‐Aldrich) was used for calibration. Samples were injected into an Acquity Ultra Performance Liquid Chromatography (UPLC) system (Waters, Milford, MA), consisting of an autosampler, injector, column (Acquity UPLC HSS T3 1.8 μm; 2.1 × 100 mm) and UV detector (Waters) at 360 nm. Chromatographic separation was performed at an elution rate of 0.3 mL/min buffer.
T cell proliferation assays
For the proliferation assay with conditioned medium, DCs were isolated by FACS sorting and 2.5‐3 × 104 DCs were cultured for 48 hours in 100 μL complete RPMI medium with 10 μM L‐tryptophan, indicated stimuli and with or without 10 μM epacadostat. DCs from three different donors were pooled at the same ratio for each subset to obtain a sufficient number of cells and to even out donor variation. After 48 h, 75 μL of this conditioned medium was added to 75 μL complete RPMI medium containing 1.5 × 105 PBLs and 3 × 104 anti‐CD3/CD28 beads (Gibco). After 3 days, proliferation was assessed by adding 1 μCi [0.037 MBq]/well of tritiated thymidine (MP Biomedicals, Irvine, CA) to the cells. Tritium incorporation over 16 h was measured with a scintillation counter. All experiments were performed in triplicate for cDC2s and pDCs and in duplicate for cDC1s.
For classical MLR assays, DCs were isolated by FACS sorting and 2.5‐4 × 104 DCs (always the same numbers across subsets) were cultured for 16 h in 100 μL complete RMPI medium with 10 μM L‐tryptophan, indicated stimuli and with or without 10 μM epacadostat. Then, 1.5 × 105 PBLs were added and incubated with the DCs for 3 days. Proliferation was assessed by tritiated thymidine incorporation as above. All experiments were performed in triplicate for cDC2s and pDCs and in duplicate for cDC1s.
Antigen cross‐presentation assay
Jurkat T cells transduced with CD8 and the α and β chains of a T cell receptor specific for the HLA‐A2.1‐restricted gp100(280‐288) peptide (JE6.1 fl296; described previously [51]) were cultured in RPMI supplemented with 2 mM ultraglutamine, 1% antibiotic‐antimycotic and 10% fetal bovine serum (Greiner Bio‐One, Kremsmünster, Austria). Positivity for the gp100‐specific TCR was monitored by flow cytometry with anti‐TCR‐V‐β14‐PE (ImmunoTools).
DCs were isolated from buffy coats or apheresis products of HLA‐A2.1+ donors by FACS sorting and cultured at 1 × 104 DCs/well in complete RPMI with 1 μM L‐tryptophan, indicated stimuli and with or without 10 μM epacadostat. In addition, DCs were loaded with 1 μM gp100(154‐162), 1 μM gp100(280‐288), or 50 μM gp100(272‐300) peptide (all JPT peptide technologies, Berlin, Germany). After 16 h of culture, 5 × 104 JE6.1 fl296 T cells were added directly to the DCs and cultured for another 24 h. The cells were labeled with anti‐CD3‐FITC (BD Biosciences), anti‐CD69‐APC (eBioscience) and fixable viability dye eFluor‐780 to determine the percentage of CD69high cells among live CD3+ T cells. All experiments were performed in singlicate for conditions with cDC1s and in duplicate for conditions with cDC2s and pDCs.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software version 7 (GraphPad Software Inc, San Diego, CA). Details on the performed statistical tests can be found in the figure legends. Statistical significance was defined as: ns, P > 0.05; * P < 0.05; ** P < 0.01; *** P < 0.001.
Author contributions
S.P.S., J.J.P.v.B., and G.F.‐G. contributed equally as first authors. S.P.S., J.J.P.v.B., G.F.‐G., J.W., M.C.v.d.N., R.v.S., M.M.V., and P.B.H.G. performed the experiments. S.P.S., J.J.P.v.B., G.F.‐G., and J.T. analyzed the data. S.P.S., J.J.P.v.B., G.F.‐G., J.W., S.I.B., J.T., C.G.F., I.J.M.d.V., and G.S. wrote the manuscript. I.J.M.d.V. and G.S. supervised the research.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202048580.
Abbreviations
- cDC1s
type 1 conventional dendritic cells
- cDC2s
type 2 conventional dendritic cells
- CpG‐C
CpG oligodeoxynucleotide class C
- DCs
dendritic cells
- HPLC
high‐performance liquid chromatography
- IDO
indoleamine 2,3‐dioxygenase 1
- IFN
interferon
- LPS
lipopolysaccharide
- MLR
mixed lymphocyte reaction
- moDCs
monocyte‐derived dendritic cells
- PBLs
peripheral blood lymphocytes
- pDCs
plasmacytoid dendritic cells
- Poly(I:C)
polyinosinic:polycytidylic acid
- R848
resiquimod
- RNA‐seq
RNA‐sequencing
- TLRs
Toll‐like receptors
- Tregs
regulatory T cells
Supporting information
Supporting Information
Acknowledgments
This work was supported by a Radboudumc Ph.D. grant and Vici grant 918.14.655 from the Netherlands Organization for Scientific Research (NWO). C.G.F. is recipient of the NWO Spinoza Award and ERC Adv Grant ARTimmune (834618). J.T. is recipient of Dutch Cancer Society grant 10620. We thank Rob Woestenenk (cell sorting facility) for technical support.
Data availability statement
The data that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE89442, and in ProteomeXchange Consortium at http://www.proteomexchange.org, reference number PXD004678. Additional data are available from the corresponding author upon request.
References
- 1. Munn, D.H. , Sharma, M.D. , Baban, B. , Harding, H.P. , Zhang, Y. , Ron, D. , and Mellor, A.L. , GCN2 kinase in T cells mediates proliferative arrest and anergy induction in response to indoleamine 2,3‐dioxygenase. Immunity. 2005. 22: 633–642. [DOI] [PubMed] [Google Scholar]
- 2. Terness, P. , Bauer, T.M. , Röse, L. , Dufter, C. , Watzlik, A. , Simon, H. , and Opelz, G. , Inhibition of allogeneic T cell proliferation by indoleamine 2,3‐dioxygenase‐expressing dendritic cells: mediation of suppression by tryptophan metabolites. J. Exp. Med. 2002. 196: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frumento, G. , Rotondo, R. , Tonetti, M. , Damonte, G. , Benatti, U. , and Ferrara, G.B. , Tryptophan‐derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3‐dioxygenase. J. Exp. Med. 2002. 196: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharma, M.D. , Baban, B. , Chandler, P. , Hou, D.‐Y. , Singh, N. , Yagita, H. , Azuma, M. , et al., Plasmacytoid dendritic cells from mouse tumor‐draining lymph nodes directly activate mature Tregs via indoleamine 2,3‐dioxygenase. J. Clin. Invest. 2007. 117: 2570–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fallarino, F. , Grohmann, U. , You, S. , McGrath, B.C. , Cavener, D.R. , Vacca, C. , Orabona, C. , et al., The combined effects of tryptophan starvation and tryptophan catabolites down‐regulate T cell receptor zeta‐chain and induce a regulatory phenotype in naive T cells. J. Immunol. 2006. 176: 6752–6761. [DOI] [PubMed] [Google Scholar]
- 6. Mezrich, J.D. , Fechner, J.H. , Zhang, X. , Johnson, B.P. , Burlingham, W.J. , and Bradfield, C.A. , An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010. 185: 3190–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sedlmayr, P. , Blaschitz, A. , and Stocker, R. , The Role of Placental Tryptophan Catabolism. Front. Immunol. 2014. 5: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okamoto, A. , Nikaido, T. , Ochiai, K. , Takakura, S. , Saito, M. , Aoki, Y. , Ishii, N. , et al., Indoleamine 2,3‐Dioxygenase Serves as a Marker of Poor Prognosis in Gene Expression Profiles of Serous Ovarian Cancer Cells. Clin. Cancer Res. 2005. 11: 6030–6039. [DOI] [PubMed] [Google Scholar]
- 9. Ino, K. , Yoshida, N. , Kajiyama, H. , Shibata, K. , Yamamoto, E. , Kidokoro, K. , Takahashi, N. , et al., Indoleamine 2,3‐dioxygenase is a novel prognostic indicator for endometrial cancer. Br. J. Cancer. 2006. 95: 1555–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brandacher, G. , Perathoner, A. , Ladurner, R. , Schneeberger, S. , Obrist, P. , Winkler, C. , Werner, E.R. , et al., Prognostic Value of Indoleamine 2,3‐Dioxygenase Expression in Colorectal Cancer: Effect on Tumor‐Infiltrating T Cells. Clin. Cancer Res. 2006. 12: 1144–1151. [DOI] [PubMed] [Google Scholar]
- 11. Brody, J.R. , Costantino, C.L. , Berger, A.C. , Sato, T. , Lisanti, M.P. , Yeo, C.J. , Emmons, R. V. , et al., Expression of indoleamine 2,3‐dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle. 2009. 8: 1930–1934. [DOI] [PubMed] [Google Scholar]
- 12. Dai, W. , and Gupta, S.L. , Regulation of indoleamine 2,3‐dioxygenase gene expression in human fibroblasts by interferon‐gamma. Upstream control region discriminates between interferon‐gamma and interferon‐alpha. J. Biol. Chem. 1990. 265: 19871–19877. [PubMed] [Google Scholar]
- 13. Hassanain, H.H. , Chon, S.Y. , and Gupta, S.L. , Differential regulation of human indoleamine 2,3‐dioxygenase gene expression by interferons‐gamma and ‐alpha. Analysis of the regulatory region of the gene and identification of an interferon‐gamma‐inducible DNA‐binding factor. J. Biol. Chem. 1993. 268: 5077–5084. [PubMed] [Google Scholar]
- 14. Hwu, P. , Du, M.X. , Lapointe, R. , Do, M. , Taylor, M.W. , and Young, H.A. , Indoleamine 2,3‐dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 2000. 164: 3596–3599. [DOI] [PubMed] [Google Scholar]
- 15. Agaugué, S. , Perrin‐Cocon, L. , Coutant, F. , André, P. , and Lotteau, V. , 1‐Methyl‐tryptophan can interfere with TLR signaling in dendritic cells independently of IDO activity. J. Immunol. 2006. 177: 2061–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Von Bubnoff, D. , Scheler, M. , Wilms, H. , Fimmers, R. , and Bieber, T. , Identification of IDO‐positive and IDO‐negative human dendritic cells after activation by various proinflammatory stimuli. J. Immunol. 2011. 186: 6701–6709. [DOI] [PubMed] [Google Scholar]
- 17. Grohmann, U. , Orabona, C. , Fallarino, F. , Vacca, C. , Calcinaro, F. , Falorni, A. , Candeloro, P. , et al., CTLA‐4‐Ig regulates tryptophan catabolism in vivo. Nat. Immunol. 2002. 3: 1097–1101. [DOI] [PubMed] [Google Scholar]
- 18. Braun, D. , Longman, R.S. , and Albert, M.L. , A two‐step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic‐cell maturation. Blood. 2005. 106: 2375–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crozat, K. , Guiton, R. , Guilliams, M. , Henri, S. , Baranek, T. , Schwartz‐Cornil, I. , Malissen, B. , et al., Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol. Rev. 2010. 234: 177–198. [DOI] [PubMed] [Google Scholar]
- 20. Villani, A.‐C. , Satija, R. , Reynolds, G. , Sarkizova, S. , Shekhar, K. , Fletcher, J. , Griesbeck, M. , et al., Single‐cell RNA‐seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017. 356: eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. See, P. , Dutertre, C.‐A. , Chen, J. , Günther, P. , McGovern, N. , Irac, S.E. , Gunawan, M. , et al., Mapping the human DC lineage through the integration of high‐dimensional techniques. Science. 2017. 356: eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balan, S. , Ollion, V. , Colletti, N. , Chelbi, R. , Montanana‐Sanchis, F. , Liu, H. , Vu Manh, T.‐P. , et al., Human XCR1 + Dendritic Cells Derived In Vitro from CD34 + Progenitors Closely Resemble Blood Dendritic Cells, Including Their Adjuvant Responsiveness, Contrary to Monocyte‐Derived Dendritic Cells . J. Immunol. 2014. 193: 1622–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Worah, K. , Mathan, T.S.M. , Vu Manh, T.P. , Keerthikumar, S. , Schreibelt, G. , Tel, J. , Duiveman‐de Boer, T. , et al., Proteomics of Human Dendritic Cell Subsets Reveals Subset‐Specific Surface Markers and Differential Inflammasome Function. Cell Rep. 2016. 16: 2953–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindstedt, M. , Lundberg, K. , and Borrebaeck, C.A.K. , Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 2005. 175: 4839–4846. [DOI] [PubMed] [Google Scholar]
- 25. Durand, M. , Walter, T. , Pirnay, T. , Naessens, T. , Gueguen, P. , Goudot, C. , Lameiras, S. , et al., Human lymphoid organ cDC2 and macrophages play complementary roles in T follicular helper responses. J. Exp. Med. 2019. 216: 1561–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heidkamp, G.F. , Sander, J. , Lehmann, C.H.K. , Heger, L. , Eissing, N. , Baranska, A. , Lühr, J.J. , et al., Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci. Immunol. 2016. [DOI] [PubMed] [Google Scholar]
- 27. Mathan, T.S.M. , Textor, J. , Sköld, A.E. , Reinieren‐Beeren, I. , van Oorschot, T. , Brüning, M. , Figdor, C.G. , et al., Harnessing RNA sequencing for global, unbiased evaluation of two new adjuvants for dendritic‐cell immunotherapy. Oncotarget. 2017. 8: 19879–19893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Vries, I.J.M. , Lesterhuis, W.J. , Scharenborg, N.M. , Engelen, L.P.H. , Ruiter, D.J. , Gerritsen, M.‐J.P. , Croockewit, S. , et al., Maturation of dendritic cells is a prerequisite for inducing immune responses in advanced melanoma patients. Clin. Cancer Res. 2003. 9: 5091–5100. [PubMed] [Google Scholar]
- 29. Sittig, S.P. , Bakdash, G. , Weiden, J. , Sköld, A.E. , Tel, J. , Figdor, C.G. , de Vries, I.J.M. , et al., A Comparative Study of the T Cell Stimulatory and Polarizing Capacity of Human Primary Blood Dendritic Cell Subsets. Mediators Inflamm. 2016. 2016: 3605643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jochems, C. , Fantini, M. , Fernando, R.I. , Kwilas, A.R. , Donahue, R.N. , Lepone, L.M. , Grenga, I. , et al., The IDO1 selective inhibitor epacadostat enhances dendritic cell immunogenicity and lytic ability of tumor antigen‐specific T cells. Oncotarget. 2016. 7: 37762–37772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Komiya, T. , and Huang, C.H. , Updates in the Clinical Development of Epacadostat and Other Indoleamine 2,3‐Dioxygenase 1 Inhibitors (IDO1) for Human Cancers. Front. Oncol. 2018. 8: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tel, J. , Schreibelt, G. , Sittig, S.P. , Mathan, T.S.M. , Buschow, S.I. , Cruz, L.J. , Lambeck, A.J.A. , et al., Human plasmacytoid dendritic cells efficiently cross‐present exogenous Ags to CD8+ T cells despite lower Ag uptake than myeloid dendritic cell subsets. Blood. 2013. 121: 459–467. [DOI] [PubMed] [Google Scholar]
- 33. Jongbloed, S.L. , Kassianos, A.J. , McDonald, K.J. , Clark, G.J. , Ju, X. , Angel, C.E. , Chen, C.‐J.J. , et al., Human CD141+ (BDCA‐3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross‐presents necrotic cell antigens. J. Exp. Med. 2010. 207: 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bachem, A. , Güttler, S. , Hartung, E. , Ebstein, F. , Schaefer, M. , Tannert, A. , Salama, A. , et al., Superior antigen cross‐presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010. 207: 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiang, M.‐C. , Tullett, K.M. , Lee, Y.S. , Idris, A. , Ding, Y. , McDonald, K.J. , Kassianos, A. , et al., Differential uptake and cross‐presentation of soluble and necrotic cell antigen by human DC subsets. Eur. J. Immunol. 2016. 46: 329–339. [DOI] [PubMed] [Google Scholar]
- 36. Crozat, K. , Guiton, R. , Contreras, V. , Feuillet, V. , Dutertre, C.‐A. , Ventre, E. , Manh, T.‐P.V. , et al., The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J. Exp. Med. 2010. 207: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poulin, L.F. , Salio, M. , Griessinger, E. , Anjos‐Afonso, F. , Craciun, L. , Chen, J.‐L. , Keller, A.M. , et al., Characterization of human DNGR‐1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010. 207: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robbins, S.H. , Walzer, T. , Dembélé, D. , Thibault, C. , Defays, A. , Bessou, G. , Xu, H. , et al., Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome‐wide expression profiling. Genome Biol. 2008. 9: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grohmann, U. , Fallarino, F. , Silla, S. , Bianchi, R. , Belladonna, M.L. , Vacca, C. , Micheletti, A. , et al., CD40 ligation ablates the tolerogenic potential of lymphoid dendritic cells. J. Immunol. 2001. 166: 277–283. [DOI] [PubMed] [Google Scholar]
- 40. Mellor, A.L. , Baban, B. , Chandler, P. , Marshall, B. , Jhaver, K. , Hansen, A. , Koni, P.A. , et al., Cutting edge: induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J. Immunol. 2003. 171: 1652–1655. [DOI] [PubMed] [Google Scholar]
- 41. Fallarino, F. , Vacca, C. , Orabona, C. , Belladonna, M.L. , Bianchi, R. , Marshall, B. , Keskin, D.B. , et al., Functional expression of indoleamine 2,3‐dioxygenase by murine CD8 alpha(+) dendritic cells. Int. Immunol. 2002. 14: 65–68. [DOI] [PubMed] [Google Scholar]
- 42. Ravishankar, B. , Liu, H. , Shinde, R. , Chandler, P. , Baban, B. , Tanaka, M. , Munn, D.H. , et al., Tolerance to apoptotic cells is regulated by indoleamine 2,3‐dioxygenase. Proc. Natl. Acad. Sci. U. S. A. 2012. 109: 3909–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kassianos, A.J. , Hardy, M.Y. , Ju, X. , Vijayan, D. , Ding, Y. , Vulink, A.J.E. , McDonald, K.J. , et al., Human CD1c (BDCA‐1)+ myeloid dendritic cells secrete IL‐10 and display an immuno‐regulatory phenotype and function in response to Escherichia coli. Eur. J. Immunol. 2012. 42: 1512–1522. [DOI] [PubMed] [Google Scholar]
- 44. Trabanelli, S. , Očadlíková, D. , Ciciarello, M. , Salvestrini, V. , Lecciso, M. , Jandus, C. , Metz, R. , et al., The SOCS3‐Independent Expression of IDO2 Supports the Homeostatic Generation of T Regulatory Cells by Human Dendritic Cells. J. Immunol. 2014. 192: 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boasso, A. , Herbeuval, J.‐P. , Hardy, A.W. , Anderson, S.A. , Dolan, M.J. , Fuchs, D. , and Shearer, G.M. , HIV inhibits CD4+ T‐cell proliferation by inducing indoleamine 2,3‐dioxygenase in plasmacytoid dendritic cells. Blood. 2007. 109: 3351–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen, W. , Liang, X. , Peterson, A.J. , Munn, D.H. , and Blazar, B.R. , The indoleamine 2,3‐dioxygenase pathway is essential for human plasmacytoid dendritic cell‐induced adaptive T regulatory cell generation. J. Immunol. 2008. 181: 5396–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerlini, G. , Di Gennaro, P. , Mariotti, G. , Urso, C. , Chiarugi, A. , Pimpinelli, N. , and Borgognoni, L. , Indoleamine 2,3‐dioxygenase+ cells correspond to the BDCA2+ plasmacytoid dendritic cells in human melanoma sentinel nodes. J. Invest. Dermatol. 2010. 130: 898–901. [DOI] [PubMed] [Google Scholar]
- 48. Ricciuti, B. , Leonardi, G.C. , Puccetti, P. , Fallarino, F. , Bianconi, V. , Sahebkar, A. , Baglivo, S. , et al., Targeting indoleamine‐2,3‐dioxygenase in cancer: Scientific rationale and clinical evidence. Pharmacol. Ther. 2019. 196: 105–116. [DOI] [PubMed] [Google Scholar]
- 49. Robinson, M.D. , McCarthy, D.J. , and Smyth, G.K. , edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010. 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cossarizza, A. , Chang, H.D. , Radbruch, A. , Acs, A. , Adam, D. , Adam‐Klages, S. , Agace, W.W. , et al., Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019. 49: 1457–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schaft, N. , Lankiewicz, B. , Gratama, J.W. , Bolhuis, R.L.H. , and Debets, R. , Flexible and sensitive method to functionally validate tumor‐specific receptors via activation of NFAT. J. Immunol. Methods. 2003. 280: 13–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are openly available in Gene Expression Omnibus at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE89442, and in ProteomeXchange Consortium at http://www.proteomexchange.org, reference number PXD004678. Additional data are available from the corresponding author upon request.


