Abstract
Wildlife migrations provide important ecosystem services, but they are declining. Within the Greater Yellowstone Ecosystem (GYE), some elk Cervus canadensis herds are losing migratory tendencies, which may increase spatiotemporal overlap between elk and livestock (domestic bison Bison bison and cattle Bos taurus), potentially exacerbating pathogen transmission risk.
We combined disease, movement, demographic and environmental data from eight elk herds in the GYE to examine the differential risk of brucellosis transmission (through aborted foetuses) from migrant and resident elk to livestock.
For both migrants and residents, we found that transmission risk from elk to livestock occurred almost exclusively on private ranchlands as opposed to state or federal grazing allotments. Weather variability affected the estimated distribution of spillover risk from migrant elk to livestock, with a 7%–12% increase in migrant abortions on private ranchlands during years with heavier snowfall. In contrast, weather variability did not affect spillover risk from resident elk.
Migrant elk were responsible for the majority (68%) of disease spillover risk to livestock because they occurred in greater numbers than resident elk. On a per‐capita basis, however, our analyses suggested that resident elk disproportionately contributed to spillover risk. In five of seven herds, we estimated that the per‐capita spillover risk was greater from residents than from migrants. Averaged across herds, an individual resident elk was 23% more likely than an individual migrant elk to abort on private ranchlands.
Our results demonstrate links between migration behaviour, spillover risk and environmental variability, and highlight the utility of integrating models of pathogen transmission and host movement to generate new insights about the role of migration in disease spillover risk. Furthermore, they add to the accumulating body of evidence across taxa that suggests that migrants and residents should be considered separately during investigations of wildlife disease ecology. Finally, our findings have applied implications for elk and brucellosis in the GYE. They suggest that managers should prioritize actions that maintain spatial separation of elk and livestock on private ranchlands during years when snowpack persists into the risk period.
Keywords: Brucellosis Brucella abortus, cross‐species pathogen spillover, elk Cervus canadensis, human–wildlife conflict, partial migration, resource selection function, wildlife disease
The authors examined the influence of elk migration in the risk of disease spillover. Their findings demonstrate links between migration, spillover risk, and environmental variability, highlighting the utility of integrating disease transmission and movement models to generate novel insights about the influence of migration in spillover risk.

1. INTRODUCTION
Traditionally, epidemiological models have considered the temporal dynamics of pathogen transmission while frequently overlooking the role of movement in host–pathogen interactions (Diekmann et al., 2012; Dougherty et al., 2018). Host movements are often an essential component of transmission dynamics, however, especially for diseases with highly mobile hosts and long transmission periods (Dougherty et al., 2018; Plowright et al., 2017; White et al., 2018; Zidon et al., 2017). Seasonal migrations are a form of movement whereby animals take advantage of cyclical fluctuations in resources, escape predation and insect harassment, find mates, and avoid seasonably uninhabitable landscapes (Alerstam et al., 2003; Avgar et al., 2014; Dingle & Drake, 2007). These movements likely influence pathogen transmission both within and across host species, but these influences are seldom quantified (Altizer et al., 2011; Plowright et al., 2017; Teitelbaum et al., 2018).
Across taxa, many populations display a characteristic form of within‐population variation in migration behaviour, with migrants moving seasonally between distinct ranges, and residents remaining in the same area throughout the year (Chapman et al., 2011). This within‐population variation in individual behaviour is known as partial migration and offers unique opportunities to evaluate the role of migration in pathogen transmission dynamics by examining differences in transmission potential between migrants and residents. In many systems, residents exhibit higher disease prevalence than migrants (Akbar et al., 2012; Hall et al., 2014; Poulin et al., 2012). This may be because seasonal migration allows migrants to escape from infected habitats or because the energetic demands of migration disproportionately kill infected animals (Altizer et al., 2011; Bradley & Altizer, 2005; Mysterud et al., 2016). Here we examine the role of partial migration on the risk of pathogen transmission from elk Cervus canadensis to cattle Bos taurus and domestic bison Bison bison (hereafter livestock; Table 1), which we refer to as spillover risk.
TABLE 1.
Glossary of terms
| Term | Definition |
|---|---|
| Abortion risk | The daily relative risk of brucellosis‐induced abortion events in elk, Rxt (Equation 4) |
| Grazing land | Private ranchlands with ≥0.4 hectares of grazing area and state and federal grazing allotments when livestock were potentially present |
| Livestock | Domestic bison and cattle |
| Per‐capita spillover risk | The average spillover risk per adult female elk |
| Risk period | The period when most brucellosis‐induced abortions in elk occur, 15 February–30 June |
| Spillover risk | Abortion risk on grazing land |
Partial migration behaviour is common in elk populations in the Rocky Mountain West (Cole et al., 2015; Eggeman et al., 2016; Hebblewhite & Merrill, 2011; Jones et al., 2014; Middleton et al., 2013). In some of these populations, the number of migrant elk is decreasing and the number of resident elk is increasing, perhaps in response to changes in land use, predation risk, habitat conditions or climate (Cole et al., 2015; Hebblewhite & Merrill, 2011; Middleton et al., 2013). In the Greater Yellowstone Ecosystem (GYE), migration behaviour in elk herds has the potential to affect disease spillover risk.
Brucellosis is a zoonotic disease caused by the bacterium Brucella abortus, which induces, and is transmitted by, reproductive failures (abortions or nonviable births) in cattle, bison and elk (Cheville et al., 1998). Transmission occurs when individuals have direct contact with B. abortus bacteria in infected foetuses, placentas or birthing fluids (Cheville et al., 1998). Depending upon conditions, the bacteria can remain viable on tissue, soil or vegetation for several weeks, although scavengers typically remove aborted foetuses prior to loss of viability (Aune et al., 2012; Cook et al., 2004). In elk, almost all brucellosis‐induced abortions occur between February and June, with a peak from March through May (Cross et al., 2015). Although brucellosis was nearly eradicated from the United States, it still persists in the elk and bison populations of the GYE (National Academies of Sciences Engineering and Medicine, 2017; Ragan, 2002). Elk are responsible for the rare, but increasing, number of livestock infections in Idaho, Montana and Wyoming (Brennan et al., 2017; Kamath et al., 2016; Rhyan et al., 2013). These spillover events are of considerable concern for livestock managers because of the costs of quarantine and trade restrictions (National Academies of Sciences Engineering and Medicine, 2017). In addition, brucellosis is expanding into new elk populations in the GYE (Cross et al., 2010; Kamath et al., 2016).
Spillover risk from elk to livestock involves complex interactions among brucellosis seroprevalence, demography and density, distribution, the timing of abortions in elk, and the distribution and density of livestock (National Academies of Sciences Engineering and Medicine, 2017). The timing of spring elk migration in the GYE is influenced by snow conditions and plant phenology, and coincides with the period of greatest transmission risk for brucellosis (Cross et al., 2015; Jones et al., 2014; Rickbeil et al., 2019; White et al., 2010). At the onset of the transmission risk period, migrant and resident elk occur together on lower elevation winter ranges that are often managed as private ranchlands (Rayl et al., 2019). As the transmission period progresses, however, migrant elk begin moving 10–100 s of kilometres to summer range on publicly owned lands at higher elevations, thereby decoupling their distribution from resident elk (Barker et al., 2019; Rickbeil et al., 2019; White et al., 2010).
We examined elk‐to‐livestock spillover risk in space and time, focusing on the role that migration, weather variability, disease prevalence and demography play in influencing this risk. We hypothesized that weather variability would affect the spillover risk from migrant elk because of its influence on plant phenology and the timing of migration. Consequently, we predicted that migrant elk would generate more spillover risk during years with heavier snowfall, whereas we expected risk from resident elk to be unaffected by variability in annual weather conditions. Furthermore, we hypothesized that elk migration would lower the per‐capita risk of pathogen spillover because we expected commingling risk with livestock to be reduced as elk migrated away from winter range. Our results offer insight into the role migration plays in the risk of disease spillover at the wildlife–livestock interface and have practical implications for the management of elk and brucellosis in the GYE.
2. MATERIALS AND METHODS
We did not have data quantifying contact rates of livestock with infected elk foetuses, nor data on how frequently that contact results in infection. Therefore, we relied on an approach that coupled spatiotemporal estimates of elk and livestock distribution with disease and demographic data to quantify spillover risk. To evaluate the role of migration and weather variability in the risk of brucellosis transmission from elk to livestock, we followed the same general approach of Merkle et al. (2018) and Rayl et al. (2019). We (a) identified migrant and resident groups from each elk herd, (b) estimated the occurrence of migrant and resident groups using resource selection functions (RSFs; Manly et al., 2002), (c) combined our RSF elk occurrence predictions with estimates of adult female elk abundance, seroprevalence, pregnancy rates and transmission timing to predict the daily relative risk of brucellosis‐induced abortion events, which is proportional to the number of brucellosis‐induced abortion events and (d) estimated the proportion of transmission risk from migrant and resident elk occurring on public and private lands and within private ranchland and federal and state livestock allotments during low, average and heavy snowfall years. Our analytical framework required us to synthesize disparate ecological, epidemiological and behavioural datasets collected at varying times between 2005 and 2017. In all the analyses, we selected the most recent years of data available because our intent was to provide a contemporary snapshot of pathogen transmission risk in our study system.
2.1. Study area
We studied elk from eight Greater Yellowstone Ecosystem herds (collectively, ~18,500 individuals) that wintered in southwest Montana, USA (Figure 1). The area is characterized by long, cold winters and short, dry summers. These herds occupied a matrix of private, Bureau of Land Management (BLM), U.S. Fish and Wildlife Service (USFWS), National Park Service (NPS), U.S. Forest Service (USFS) and state government lands (Figure 1). Elevations range from 1,200 to 3,900 m. Within our study area, livestock occurred either on private land or on public land grazing allotments. Public land grazing allotments are permitted or leased to livestock owners, and consist of areas of land designated for grazing a specific number and kind of livestock for a specified period of time.
FIGURE 1.
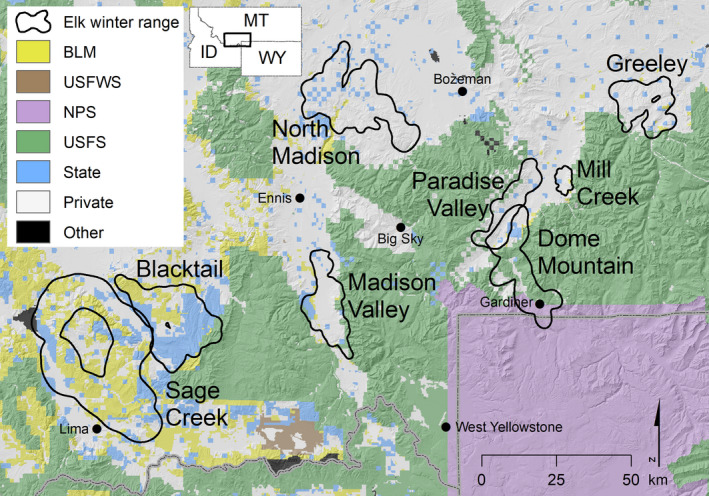
Winter ranges of the Madison Valley (3,993 elk), Dome Mountain (3,888 elk), North Madison (2,878 elk), Sage Creek (2,850 elk), Greeley (1,509 elk), Blacktail (1,357 elk), Paradise Valley (1,222 elk) and Mill Creek (786 elk) elk herds in the southwest Montana portion of the Greater Yellowstone Ecosystem, USA, with the matrix of Bureau of Land Management (BLM), U.S. Fish and Wildlife Service (USFWS), National Park Service (NPS), U.S. Forest Service (USFS), state government (State), private (Private) and other (Other) lands in the region. Number of elk estimated from survey data collected during winter 2016 or 2017. Shading depicts hillshade of elevation
2.2. Elk collaring and monitoring
We captured adult female elk ≥2 years of age by helicopter net‐gunning or chemical immobilization during January–March 2005–2015. We radio‐collared elk with Global Positioning System (GPS) collars programmed to acquire a location every 0.5, 1 or 2 hr (GPS 3300L, Lotek Wireless Inc.), and monitored each individual for 1–5 years. We tested hunter‐harvested and research‐captured adult female elk for exposure to B. abortus, and used the proportion of positive results from these tests during 2011–2017 to estimate herd seroprevalence. We followed Montana Department of Fish, Wildlife, and Parks biomedical protocols for free‐ranging cervidae in Montana during capture and handling procedures (Protocol FWP04‐2018). We classified 15 February to 30 June, the period when most abortions occur, as the transmission risk period (hereafter risk period; Table 1), and limited our analyses to this time period (Cross et al., 2015). Within the risk period, we defined winter (15 February–31 March; elk on winter range), spring (1 April–31 May; elk migrating to summer range) and summer (1 June–30 June; elk on summer range) seasons. We based our seasons upon elk movement and aggregation patterns, the typical timing of snow melt and green‐up, and to be consistent with seasons defined in our previous work (Rayl et al., 2018, 2019). Our risk period dataset included 223 elk monitored from February 2005–June 2015 (280 elk‐years, 1,475,613 locations; see Appendix S1 in Supporting Information).
2.3. Identifying migrants and residents
Although seasonal migration likely occurs along a continuum of movement tactics (Cagnacci et al., 2016; Dingle & Drake, 2007), our goal was to differentiate between the two dominant space use tactics that we hypothesized were most influential to spillover risk between elk and livestock. Therefore, we sought to discriminate between elk that remained inside or adjacent to winter range (residents) and elk that migrated away from winter range (migrants). To do so, we estimated the overlap of seasonal kernels to classify individual elk‐years as migrant or resident (Barker et al., 2019; Fieberg & Kochanny, 2005). Unlike prior studies, which have typically calculated the overlap between individual seasonal home ranges to differentiate between migrants and residents, we calculated the overlap between each herd's winter range and individual elk‐year post‐migration home ranges. We delineated a winter range for each herd using that herd's GPS locations during the winter season (excluding migratory portions of individual datasets when elk initiated migration during winter or did not return from summer range until winter) to create 99% contours of bivariate normal kernels with the reference bandwidth (Worton, 1989). We used individual elk‐year GPS locations from August to create 95% contours of bivariate normal kernels with the reference bandwidth as an estimate of post‐migration home ranges. For 13 individual elk‐years without GPS location data in August, we used July GPS locations to estimate post‐migration home ranges. We classified individual elk‐years as migrant when their post‐migration home range did not overlap winter range, and as resident when they did. Because we did not monitor individual elk for an equal number of years, we used the proportion of individuals classified as migrants during their first year of monitoring to estimate the proportion of migrants in each herd.
2.4. Resource selection function development
We used RSFs to describe the spatiotemporal relationship between the relative probability of female elk occurrence and landscape attributes. We fit RSFs separately for migrants and residents from each herd in winter, spring and summer because we expected resource selection to vary seasonally, among herds, and between migrants and residents. Because our objective was to identify fine‐scale spatiotemporal overlap of elk abortion events with areas of potential livestock presence, we used third‐order RSFs (selection of patches within individual home ranges) to characterize habitat selection (Meyer & Thuiller, 2006). We estimated RSFs by comparing habitat characteristics of observed locations with an equal number of available locations. We randomly sampled available locations from within a 99% contour of a bivariate normal kernel generated with the reference bandwidth for each individual‐year in each season (Worton, 1989). We randomly assigned available locations to a specific day drawn with replacement from the distribution of days of the corresponding elk‐year‐season observed locations. For all migrant and resident groups with >1 individual‐year (14 of 15 groups), we using a generalized linear mixed model (GLMM) with a binomial distribution, logit link and individual‐year as the random intercept to estimate the RSF. The random intercept accounted for unbalanced sample sizes among individual‐years, and the non‐independence of GPS locations (Gillies et al., 2006). The GLMM‐derived RSFs for migrants and residents from each herd in each season took the form:
| (1) |
where w(x) represented the RSF scores, βu was the selection coefficient for explanatory variable xu for the ith observation and the jth individual‐year, and γ 0 j was the random intercept for the jth individual‐year. For Madison Valley residents (n = 1 individual‐year), we used a generalized linear model (GLM) with a binomial distribution and logit link to estimate the RSF. The GLM‐derived RSF took the form:
| (2) |
In our RSFs, we included variables previously demonstrated to be important predictors of elk occurrence during the risk period (Merkle et al., 2018; Proffitt et al., 2011; Rayl et al., 2019). We incorporated a suite of topographic variables (elevation, slope, terrain position index [calculated as the difference between the elevation of a cell and the mean elevation of its nearest 80 surrounding cells], and solar radiation during the risk period; 30‐m resolution, U.S. Geological Survey National Elevation Dataset), distance to motorized roads (pseudothreshold [natural logarithm transformed distance + 1] functional form; 30‐m resolution, U.S. Department of Commerce, Bureau of the Census), landcover type (consolidated into four categories: forest, shrub, agriculture, grass [reference category]; 30‐m resolution, 2011 National Land Cover Database), snow cover (indicating presence of snow; 500‐m spatial and 8‐day temporal resolution, MODIS data; Hall et al., 2002), overall productivity or biomass of a pixel each year calculated as the annual integrated Normalized Difference Vegetation Index (NDVI, 250‐m resolution, MODIS data; Pettorelli et al., 2005) and the daily NDVI value of a pixel (250‐m resolution; scaled between 0 and 1). We assigned daily values of snow cover to each pixel using the pixel value from the 8‐day snow cover interval that encompassed that day. To derive daily NDVI values, we followed the methods of Bischof et al. (2012) and Merkle et al. (2016) to construct a smoothed and scaled NDVI time series for each pixel (see section 2 in Merkle et al. (2016) for details).
Prior to building seasonal RSFs, we evaluated whether a linear or quadratic functional form for elevation, slope, solar radiation and daily NDVI was better supported. For each migrant and resident group from each herd in each season, we built univariate GLMMs or GLMs for the functional forms of each variable, and determined the form with the most support among all tactic‐specific groups (i.e. all migrants or residents) using Akaike Information Criterion for small sample sizes (AICc; Burnham & Anderson, 2002). We evaluated collinearity between pairs of covariates before building seasonal RSFs. When we detected collinearity (Pearson's correlation coefficient ≥0.7), we built GLMMs or GLMs for each covariate, and excluded the covariate with less AICc support. We also assessed our seasonal RSF models (without quadratic terms) for multicollinearity using the variance inflation factor (VIF), and detected no issues (VIFs for all variables ≤4.43; Dormann et al., 2013; Graham, 2003). We derived maximum‐likelihood estimates for GLMMs using adaptive Gauss–Hermite approximation with 5 integration points (Bolker et al., 2009). To evaluate the predictive ability of our seasonal RSF models, we used 10 repetitions of fivefold cross‐validation with 10 bins of equal size, calculating the average Spearman rank correlation (r s) between the withheld data and the ranked bins (Boyce et al., 2002).
2.5. Predicting abortion and spillover risk
We built our RSFs using NDVI and snow cover data corresponding to the time period when individual elk were monitored, which allowed us to quantify the relationship between elk occurrence, vegetation phenology and snowfall. We then identified representative low, average and heavy snowfall years that occurred during our study period (Merkle et al., 2018; see Appendix S2). To evaluate the influence of weather variability on brucellosis transmission risk, we predicted each migrant and resident group's distribution using NDVI and snow cover datasets from these representative snowfall years (see below for details).
The distribution of ungulates is a function of, not only environmental factors but also cognitive factors associated with sociality, spatial fidelity, memory and learning (Jesmer et al., 2018; Merkle et al., 2017; Wolf et al., 2009). As a result, elk herds in the GYE show strong fidelity to seasonal ranges and migration routes and individual herd ranges tend to be concentrated within larger areas of suitable habitat (Boyce et al., 2003; Kauffman et al., 2018; Rayl et al., 2019; White et al., 2010). Therefore, we developed a new technique using a sliding window approach to limit the spatiotemporal extent over which we mapped each migrant or resident group's RSF predicted values to areas likely to have been occupied by that group on that day. First, we resampled all covariate grids from their original resolution to 250 m by calculating the mean pixel value that fell within the extent of the output 250‐m pixel. Then, for each of the three weather scenarios, we estimated the predicted relative probability of group use u(x, t) per 250‐m pixel x, per time step t (in days), as:
| (3) |
where i = refers to pixels 1 through n for time step t, wxt is the daily predicted RSF value of the relative probability of use by elk for a 250‐m pixel x and Kxt is the daily predicted value of elk availability (0 or 1) for pixel x. In Equation 3, Kxt limits the spatiotemporal extent of the predicted relative probility of use to areas likely to have been occupied by that group on that day. We employed a sliding window approach to estimate Kxt. For every time step t, we generated a 99% contour of a bivariate normal kernel with the reference bandwidth using elk‐group locations from t − 15 days to t + 15 days. We assigned pixels within the contour a Kxt value of 1 and pixels outside the contour a Kxt value of 0. During the first 15 days of the risk period, we estimated the Kxt kernel using elk‐group locations from the first 31 days of the risk period because some groups lacked location data prior to the start of the risk period (because of capture timing). The denominator in Equation 3 ensures that . equals 1, thereby allowing us to compare the daily predicted relative probability of use among groups. We also estimated the density experienced by migrant and resident groups during the risk period to explore how elk density changed from February through June, and the consequent implications for brucellosis transmission (see Appendix S3).
For each weather scenario, we calculated the daily relative risk of abortion events Rxt (hereafter abortion risk; Table 1) per 250 m pixel x, per time step t (in days), as:
| (4) |
where u(x, t) is the daily predicted relative probability of group use for pixel x from Equation 2, Fgh is the estimated number of female elk from group g and herd h, Sh is the average brucellosis seroprevalence estimated for herd h, y is a mean pregnancy rate of 90% (K. Proffitt, unpubl. data) and pt is the predicted daily probability of aborting given an individual is seropositive and pregnant (Cross et al., 2015; see Appendix S3). Equation 4 calculates a relative estimate, which is proportional to the number of abortion events, and can be directly compared among groups because the denominator in Equation 3 ensured that equals 1. We used samples from hunter‐harvested and research‐captured adult female elk to estimate herd seroprevalence (see Montana Fish Wildlife and Parks, 2015 for details on how serostatus was determined). We did not account for uncertainty in our estimates of Rxt because of computational limitations associated with deriving error estimates for u(x, t) on a cell‐by‐cell basis, and because accurate methods to do so for Fgh across the region were not available at the time of this analysis. While unaccounted for uncertainty associated with Fgh, Sh, y and pt may bias estimates of Rxt high or low, these biases are likely to affect each pixel and migrant group similarly. Therefore, general conclusions and results are likely to be invariant across the range of variability associated with each parameter.
We combined Rxt estimates with landownership data to estimate the daily and cumulative abortion risk from each migrant and resident group occurring on private, BLM, USFWS, NPS, USFS, and state government lands across the three weather scenarios. We did not consider the distribution of livestock in these calculations. We then calculated these same metrics for areas with potential livestock grazing to quantify the potential for elk‐to‐livestock spillover risk on the landscape. We defined areas of potential livestock grazing as private ranchlands in Montana with ≥0.4 hectares of grazing area (http://svc.mt.gov/msl/mtcadastral; Proffitt et al., 2011), and federal (USFS, BLM) and state (Wildlife Management Area) grazing allotments in Montana when livestock were potentially present on the allotments during the risk period (hereafter grazing land; Table 1; see Appendix S4). We used turnout dates from BLM and USFS grazing records from 2014 (Wells et al., 2019) and state grazing records from 2017 to determine when livestock were present on federal and state allotments. We defined spillover risk as abortion risk on private ranchlands during the risk period or on allotments with turnout dates during the risk period (Table 1). Therefore, abortion risk on livestock allotments outside of turnout dates did not contribute to our estimate of spillover risk. Our comparisons of spillover risk between migratory tactics and among herds and weather scenarios relied on the assumption that livestock contact with infected foetuses and the risk of infection were positively correlated with abortion risk.
We combined our estimates of the number of adult female elk in each group and spillover risk to calculate the average spillover risk per adult female elk (hereafter per‐capita spillover risk; Table 1). Collared elk from three migrant groups spent a portion of the risk period on BLM, USFS and private lands in Idaho where we did not have data on public and private grazing. To account for this in our estimate of per‐capita spillover risk, we reduced the estimated number of female elk from these groups by the daily predicted probability of group use that occurred in Idaho for each weather scenario. We conducted all analyses in program R version 3.3.1, using lme4 to fit GLMMs (R Development Core Team, 2016).
3. RESULTS
From our sample of 223 elk (280 elk‐years), we identified 152 migrants and 71 residents. We identified one fully resident herd, and seven herds with both migrant and resident individuals (Figure 2). We collected multiple years of data from 37 elk in five herds. Seven of these 37 elk switched migratory tactics between years, including 6 of 18 elk from the Paradise Valley herd and one of six from the Blacktail herd. As migrants departed from winter range, the density of elk declined for both migrant and resident groups (Figure 3).
FIGURE 2.
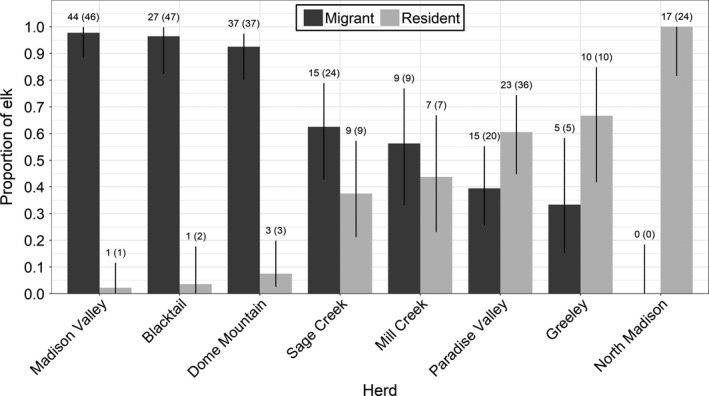
The estimated proportion of migrant and resident individuals in eight Greater Yellowstone Ecosystem elk herds. The number of collared elk (number of individual elk‐years) in each group is given at the top of the 95% binomial confidence intervals (black lines)
FIGURE 3.
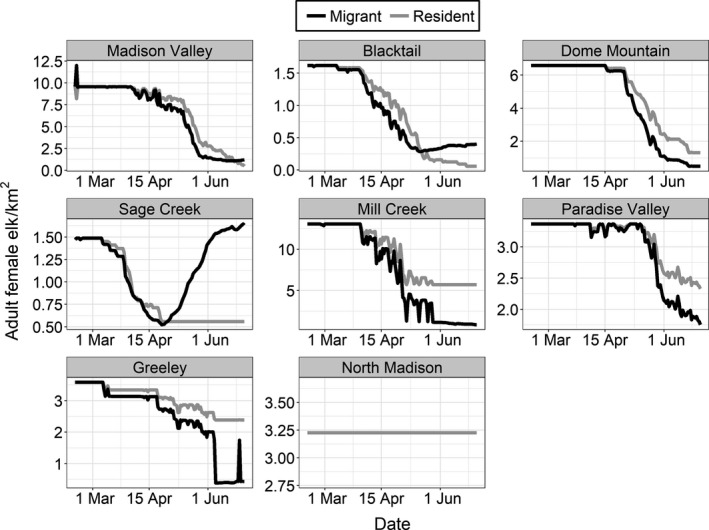
Daily density estimates experienced by migrant and resident female elk from eight herds (arranged from most to least migratory) in the Greater Yellowstone Ecosystem during the risk period (15 February–30 June). Density estimates were derived from survey data collected during winter 2016 or 2017
Patterns of resource selection varied among herds and groups, but there was no clear distinction between selection patterns of migrant and resident elk groups within herds or among winter (15 February–31 March), spring (1 April–31 May) or summer (1 June–30 June) seasons (see Appendix S5: Figure S1). In general, both migrant and resident elk selected areas at moderate elevation and slope, with higher terrain position index (i.e. on ridges) and intermediate to high solar radiation. Furthermore, they typically avoided forested landcover, motorized roads and snow cover, showed variable responses to shrub landcover and selected agricultural landcover, intermediate values of daily NDVI (i.e. surrogate for phenology stage), and higher annual integrated NDVI (i.e. surrogate for biomass). The average r s of our migrant and resident RSFs from 10 repetitions of our fivefold cross‐validation procedure was 0.98 (range = 0.93–1.00) in winter, 0.99 (range = 0.95–1.00) in spring and 0.97 (range = 0.87–1.00) in summer, indicating strong predictive ability (see Appendix S5: Table S1).
Seroprevalence did not differ among migrants and residents in our sample of collared elk, but sample sizes were relatively small for some herds (see Appendix S3: Figure S1). Estimated brucellosis seroprevalence ranged from a high of 53% (95% CI = 36%–70%) for the Mill Creek herd to a low of 2% (95% CI = 1%–7%) for the Greeley herd (see Appendix S3: Table S2). We estimated that the Madison Valley herd accounted for 41% of the abortion risk each year (i.e. 100 × [Madison Valley abortion risk/abortion risk from all herds] = 41%), whereas the Greeley herd accounted for <1%. These widely varying estimates among herds resulted from differences in brucellosis seroprevalence and the estimated number of female elk surveyed during the most recent elk trend counts (see Appendices S3 and S6).
Across weather scenarios, we calculated that 50%–56% of migrant and 77%–78% of resident abortion risk was on private lands (e.g. 100 × [migrant abortion risk on private lands/migrant abortion risk on all lands] = 50%–56%; Figure 4). During the average snowfall year, we estimated that approximately 50% of migrant abortion risk was on private lands, 25% on USFS lands, 13% on NPS lands, 8% on state government lands, 4% on BLM lands and 0% on USFWS lands. In that same year, we estimated that approximately 78% of resident abortion risk was on private lands, 8% on state government lands, 5% on both USFS and BLM lands, 4% on NPS lands, and 0% on USFWS lands. In contrast, we estimated that 98%–99% of spillover risk was on private ranchlands for both migrants and residents (e.g. 100 × [migrant spillover risk on private ranchlands/migrant spillover risk on grazing land] = 98%–99%). Spillover risk on private ranchlands represented 74%–75% of the total abortion risk from residents, and 49%–54% for migrants, depending on weather scenario. Migrant elk were responsible for 68% of spillover risk because they occurred in greater numbers than resident elk (Figure 5a, see Appendix S6: Table S1).
FIGURE 4.
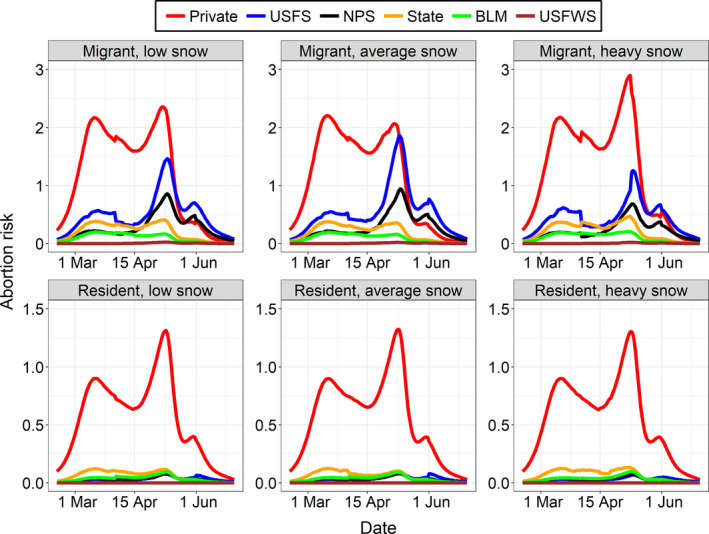
Abortion risk occurring on private (Private), U.S. Forest Service (USFS), National Park Service (NPS), state government (State), Bureau of Land Management (BLM), and U.S. Fish and Wildlife Service (USFWS) lands for migrant and resident female elk from eight Greater Yellowstone Ecosystem herds during representative low, average and heavy snowfall years that occurred during the study period
FIGURE 5.
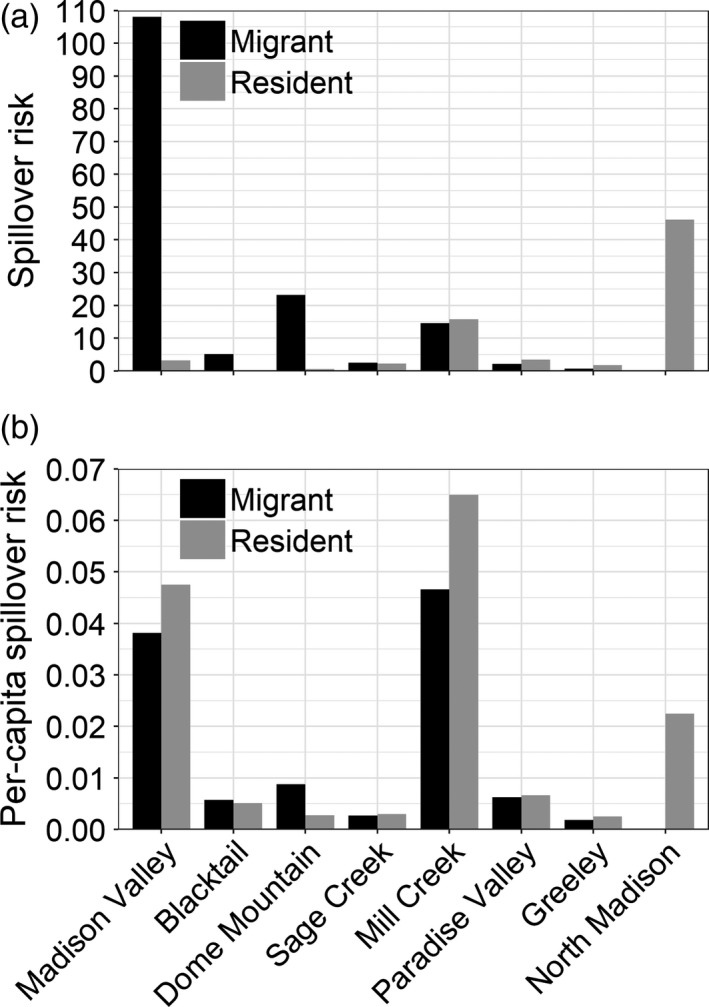
(a) Estimated cumulative spillover risk for migrant and resident female elk occurring on private ranchlands during the risk period (15 February–30 June) for eight Greater Yellowstone Ecosystem herds and (b) estimated cumulative per‐capita spillover risk for migrant and resident female elk occurring on private ranchlands during the risk period. Values in both panels were averaged across weather scenarios
We found support for our hypothesis that weather variability would affect spillover risk for migrant, but not resident elk. We estimated that the distribution of spillover risk for migrants was somewhat sensitive to changes in snow cover, with 7%–12% more risk occurring on private ranchlands during heavy snowfall years than during low or average snowfall years. Conversely, we estimated that the distribution of spillover risk on private ranchlands for residents changed <1% during low, average and heavy snowfall years. We also found support for our hypothesis that elk migration lowered per‐capita spillover risk. Although migrants were responsible for 68% of spillover risk because of their greater numbers, for five of seven herds with both migrants and residents, per‐capita spillover risk on private ranchlands was greater for residents than for migrants (Figure 5b). Averaged across weather scenarios and herds, we estimated that per‐capita spillover risk on private ranchlands was 23% greater for residents than for migrants.
4. DISCUSSION
We combined ecological, epidemiological and behavioural data from >200 elk in eight GYE herds with livestock distribution data to examine the influence of partial migration on abortion and spillover risk. By incorporating these datasets, we revealed that most spillover risk from elk to livestock was on private ranchlands in early spring. Furthermore, we found that migrant elk generated the majority of spillover risk because they were more abundant than resident elk. On a per‐capita basis, however, we estimated that migrant elk were 23% less risky to livestock than resident elk because they migrated off of private ranchlands during the risk period.
Our synthetic approach is uncommon in disease ecology because of the challenges associated with merging data streams collected at varying spatial and temporal resolutions and scales (Dougherty et al., 2018; Plowright et al., 2017; Rayl et al., 2019). Other studies have integrated host movement or density data to estimate pathogen transmission risk or exposure rates within a host species (e.g. Borg et al., 2017; Proffitt et al., 2015; Russell et al., 2015). Indeed, there have been a number of previous studies that have examined the potential for brucellosis spillover risk from wildlife to livestock, but none to date that have examined the role of host migration in spillover risk (Kilpatrick et al., 2009; Merkle et al., 2018; Proffitt et al., 2011; Rayl et al., 2019). Our analysis accounted for many of the major components of elk‐to‐livestock brucellosis spillover risk, including host movement, distribution, density, prevalence and transmission timing, and suggested important links between migratory behaviour and the risk of disease spillover. The approach we developed is applicable to other disease systems such as avian influenza and bighorn Ovis canadensis and domestic sheep (O. aries) pneumonia, although the challenge is to have all the necessary datasets align in space, time and resolution (Cross et al., 2019; Rayl et al., 2019).
As we had hypothesized, our findings provided evidence that weather variability affected spillover risk from elk to livestock in the GYE. Previous work in this system has documented delayed elk migration in spring following winters with heavier snowfall and later vegetation green‐up (Rickbeil et al., 2019; White et al., 2010). Our results correspond with these findings and demonstrate the cascading effects this environmental variability can have on spillover risk. As predicted, we found that heavier snowfall did not influence spillover risk from residents, but that it had a modest influence on the distribution of spillover risk from migrants. During heavy snow years, we estimated that 7%–12% more migrant spillover risk was on private ranchlands, likely because snow cover delayed departure from winter range. Under future climate change scenarios, decreased snowpack and advanced snowmelt are expected in the Rocky Mountains (Gergel et al., 2017). These changes may induce earlier departure from winter range, thereby reducing spillover risk from migrant elk on private ranchlands (Rickbeil et al., 2019; White et al., 2010).
In support of our second hypothesis, we found evidence that the spring migration of elk alleviated per‐capita spillover risk. As migrants moved from lower‐elevation winter range to higher‐elevation summer range they moved away from private ranchlands where spillover risk was highly concentrated. Importantly, however, and in contrast to differences in per‐capita spillover risk, we found that migrant elk generated the majority of spillover risk in our system because of their greater abundance. It has been hypothesized that the number of migrants and residents in partially migratory populations is maintained by density‐dependent demography or behavioural switching between movement tactics (Kaitala et al., 1993; Lundberg, 2013). In partially migratory ungulate populations, migrants frequently outnumber residents, and this is what we observed among the elk in our study (Fryxell et al., 1988; Fryxell & Sinclair, 1988; Sawyer et al., 2016; Figure 2). Recent studies in the Rocky Mountains, however, have demonstrated that the benefits of elk migration relative to residency may be declining as a result of altered landscapes, climate regimes or predator guilds (Barker et al., 2019; Hebblewhite & Merrill, 2011; Middleton et al., 2013). For example, Barker et al. (2019) found that resident elk had access to higher quality forage than migrant elk because of the availability of irrigated agriculture in valley bottoms where residents resided year‐round. If the fitness benefits of migration in our system decrease, this may affect the demographic balance of migrants and residents, and therefore, also influence the risk of disease spillover. How these potential demographic changes may alter the risk of disease spillover in the future remains unknown, however. If resident elk were to increase in number while elk carrying capacity remains static, this will likely translate into increased risk because of the greater spillover potential associated with resident behaviour. On the other hand, if elk herds decline in size as a result of a decreasing number of migrants, the risk of disease spillover will likely decline as well.
An ancillary benefit of migration may be a reduction in disease or parasite exposure for hosts (Altizer et al., 2000; Folstad et al., 1991; Piersma, 1997). In temperate environments, cervids frequently occur at higher densities during winter, which may enhance the risk of pathogen transmission (Conner et al., 2008). In our study area, the migration of elk off of winter range lowered the conspecific density experienced by both migrant and resident groups during the risk period (Figure 3). This decline in density may reduce elk‐to‐elk transmission risk of brucellosis, as well as other density‐dependent diseases. By the end of the risk period, migrant groups typically occurred at lower densities than resident groups. It is important to note, however, that our density estimates did not account for commingling of migrant elk from different herds during migration and on summer range, and thus are likely underestimates. Similar observations of higher conspecific density of resident elk compared to migrant elk have been observed elsewhere in Montana (Barker et al., 2019). Whether or not these changes in density result in differences in pathogen transmission within migrant and resident elk groups requires further investigation. In our sample of collared elk, seroprevalence did not differ among migrants and residents, consistent with other analyses, but sample sizes were relatively small for some herds (see Appendix S3: Figure S1; Yang et al., 2019).
Our work clearly illustrates the value of including spatiotemporal variability for both reservoir and host populations during examinations of spillover risk. There was a striking contrast between our estimates of abortion risk, which did not consider the spatiotemporal distribution of livestock, and our estimates of spillover risk, which did. If we relied only on our estimates of abortion risk, we would have erroneously concluded that 44%–50% of transmission risk for migrant elk and 22%–23% for resident elk was on state and federal lands (Figure 4). In contrast, when we incorporated our spatiotemporal estimates of livestock distribution, we found almost no spillover risk (<2%) outside of private ranchlands for both migrant and resident elk. This suggests that the current timing of livestock stocking on state and federal allotments is effective at preventing commingling of elk and livestock during the risk period, at least for the elk herds in the Montana brucellosis designated surveillance area.
Importantly, we were unable to account for several sources of spatial and temporal variability in our analyses. Most significantly, we did not have detailed data on the spatiotemporal distribution of livestock on private ranchlands. As a result, we most likely overestimated risk on this grazing type because we assumed that livestock were always present. Similarly, we did not have information about the space use of livestock within individual allotments, which likely changes annually. Additionally, we did not have data quantifying contact rates of livestock with infected elk foetuses, how frequently that contact results in infection or the environmental persistence of B. abortus. Aune et al. (2012) found that B. abortus can remain viable on foetal tissues and soil or vegetation for 21–81 days depending on month, temperature and exposure to sunlight. We expect that aborted foetuses will not remain on the landscape for that long in our study area because they will likely be removed by scavengers much more quickly (Cook et al., 2004). Further research is needed to estimate foetal scavenging rates for our study area. Finally, as in Merkle et al. (2018) and Rayl et al. (2019), we did not include a temporal transmission component within elk herds, and therefore, could not predict disease dynamics across consecutive years. Further research is needed to collect finer‐resolution data on the distribution of livestock, and to incorporate temporal models of pathogen transmission into future predictions of risk.
Our estimates of abortion and spillover risk should be viewed somewhat cautiously, as we were unable to include estimates of variance in our predictions. Instead, as in Merkle et al. (2018) and Rayl et al. (2019), we assumed that the number of female elk, seroprevalence, the proportion of migrants and residents, abortion timing, pregnancy rates, potential livestock distribution, and space‐use predictions were all known without error. Incorporating uncertainty from seroprevalence, the proportion of migrants and residents, abortion timing and the number of female elk into our analyses would be relatively straightforward, but computationally demanding. Such an effort would rigorously propagate error through only a portion of Equation 4, as we do not currently have fine‐resolution spatiotemporal data on livestock distribution. Additionally, it would be challenging to quantify uncertainty in the predicted probability of elk use across space and time given existing computing capacity. Because this unaccounted‐for uncertainty is likely to affect individuals within herds similarly; however, it would be unlikely to alter inferences about within‐herd differences between migrant and resident spillover risk (i.e. Figure 5b). It could, though, affect conclusions of among‐herd comparisons (i.e., Figure 5a). In the future, as computational capacity increases, it would be useful to quantify this error. Doing so would generate information that could be used to identify optimal data collection and surveillance strategies to minimize uncertainty in risk predictions.
Although animal movements likely impact disease dynamics, it is uncommon and difficult to synthesize host movements with disease ecology (Altizer et al., 2011; Dougherty et al., 2018; White et al., 2018; but see Guber et al., 2016; Merkle et al., 2018; Rayl et al., 2019). These unified approaches are necessary, however, to properly understand the effects that complex movement behaviours, such as migration, may have on host–pathogen dynamics. In this work, we used an integrated modelling framework to enumerate spillover risk in space and time, and found significant links between migration behaviour, the potential for pathogen transmission and environmental variability. Further research is needed to determine how density‐dependent demography, behavioural switching between movement tactics and environmental change may influence these links, and therefore, the distribution of spillover risk.
AUTHORS' CONTRIBUTIONS
N.D.R., J.A.M., K.M.P., E.S.A. and P.C.C. conceived the study; K.M.P., E.S.A., J.D.J. and J.A.G. collected and prepared the data; N.D.R. performed the analyses; N.D.R. wrote the manuscript, and all authors contributed to revisions and gave final approval for publication.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
Funding was provided by Montana Fish, Wildlife and Parks (FWP) through an agreement with Montana Department of Livestock and the Animal and Plant Health Inspection Service of the U.S. Department of Agriculture. Additional funding was provided by the U.S. Geological Survey. We thank the many FWP staff for their efforts in helping with landowner contacts, field operations and continued support of this project. We thank pilots N. Cadwell, B. Malo, M. Shelton, M. Stott and R. Swisher for their work in capturing elk. We thank F. Thompson and the Bureau of Land Management (BLM) for compiling the BLM livestock allotment data. We thank S. Wells, L. McNew, D. Tyers and the U.S. Forest Service (USFS) forest supervisors of the Greater Yellowstone Ecosystem for assembling the USFS livestock allotment data. We thank J. Cunningham, K. Loveless and D. Waltee for their work collecting elk count data. We thank A. Brennan, K. Szcodronski and K. Manlove for helpful discussions about this work. We thank Q. Kujala for helpful comments that improved this manuscript and for securing the funding to conduct this work. We thank the Editor‐in‐Chief, Associate Editor, M. Kauffman and three anonymous reviewers for valuable comments that improved this manuscript. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Rayl ND, Merkle JA, Proffitt KM, et al. Elk migration influences the risk of disease spillover in the Greater Yellowstone Ecosystem. J Anim Ecol. 2021;90:1264–1275. 10.1111/1365-2656.13452
Handling Editor Bethany Hoye
DATA AVAILABILITY STATEMENT
Data available from the Dryad Digital Repository https://doi.org/10.5061/dryad.34tmpg4j2 (Rayl et al. 2020).
REFERENCES
- Akbar, H. , Pinçon, C. , Aliouat‐Denis, C.‐M. , Derouiche, S. , Taylor, M.‐L. , Pottier, M. , Carreto‐Binaghi, L.‐H. , González‐González, A. E. , Courpon, A. , Barriel, V. , Guillot, J. , Chabé, M. , Suarez‐Alvarez, R. O. , Aliouat, E. M. , Dei‐Cas, E. , & Demanche, C. (2012). Characterizing pneumocystis in the lungs of bats: Understanding pneumocystis evolution and the spread of pneumocystis organisms in mammal populations. Applied and Environmental Microbiology, 78(22), 8122–8136. 10.1128/AEM.01791-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alerstam, T. , Hedenström, A. , & Åkesson, S. (2003). Long‐distance migration: Evolution and determinants. Oikos, 103(2), 247–260. 10.1034/j.1600-0706.2003.12559.x [DOI] [Google Scholar]
- Altizer, S. , Bartel, R. , & Han, B. A. (2011). Animal migration and infectious disease risk. Science, 331(6015), 296–302. 10.1126/science.1194694 [DOI] [PubMed] [Google Scholar]
- Altizer, S. M. , Oberhauser, K. S. , & Brower, L. P. (2000). Associations between host migration and the prevalence of a protozoan parasite in natural populations of adult monarch butterflies. Ecological Entomology, 25(2), 125–139. 10.1046/j.1365-2311.2000.00246.x [DOI] [Google Scholar]
- Aune, K. , Rhyan, J. C. , Russell, R. , Roffe, T. J. , & Corso, B. (2012). Environmental persistence of Brucella abortus in the Greater Yellowstone Area. Journal of Wildlife Management, 76(2), 253–261. 10.1002/jwmg.274 [DOI] [Google Scholar]
- Avgar, T. , Street, G. , & Fryxell, J. M. (2014). On the adaptive benefits of mammal migration. Canadian Journal of Zoology, 92(6), 481–490. 10.1139/cjz-2013-0076 [DOI] [Google Scholar]
- Barker, K. J. , Mitchell, M. S. , Proffitt, K. M. , & DeVoe, J. D. (2019). Land management alters traditional nutritional benefits of migration for elk. Journal of Wildlife Management, 83(1), 167–174. 10.1002/jwmg.21564 [DOI] [Google Scholar]
- Bischof, R. , Egil Loe, L. , Meisingset, E. L. , Zimmermann, B. , Van Moorter, B. , & Mysterud, A. (2012). A migratory northern ungulate in the pursuit of spring: Jumping or surfing the green wave? American Naturalist, 180, 407–424. [DOI] [PubMed] [Google Scholar]
- Bolker, B. M. , Brooks, M. E. , Clark, C. J. , Geange, S. W. , Poulsen, J. R. , Stevens, M. H. H. , & White, J. S. S. (2009). Generalized linear mixed models: A practical guide for ecology and evolution. Trends in Ecology & Evolution, 24(3), 127–135. 10.1016/j.tree.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Borg, N. J. , Mitchell, M. S. , Lukacs, P. M. , Mack, C. M. , Waits, L. P. , & Krausman, P. R. (2017). Behavioral connectivity among bighorn sheep suggests potential for disease spread. Journal of Wildlife Management, 81(1), 38–45. 10.1002/jwmg.21169 [DOI] [Google Scholar]
- Boyce, M. S. , Mao, J. S. , Merrill, E. H. , Fortin, D. , Turner, M. G. , Fryxell, J. , & Turchin, P. (2003). Scale and heterogeneity in habitat selection by elk in Yellowstone National Park. Ecoscience, 10(4), 421–431. 10.1080/11956860.2003.11682790 [DOI] [Google Scholar]
- Boyce, M. S. , Vernier, P. R. , Nielsen, S. E. , & Schmiegelow, F. K. A. (2002). Evaluating resource selection functions. Ecological Modelling, 157, 281–300. 10.1016/S0304-3800(02)00200-4 [DOI] [Google Scholar]
- Bradley, C. A. , & Altizer, S. (2005). Parasites hinder monarch butterfly flight: Implications for disease spread in migratory hosts. Ecology Letters, 8(3), 290–300. 10.1111/j.1461-0248.2005.00722.x [DOI] [Google Scholar]
- Brennan, A. , Cross, P. C. , Portacci, K. , Scurlock, B. M. , & Edwards, W. H. (2017). Shifting brucellosis risk in livestock coincides with spreading seroprevalence in elk. PLoS ONE, 12(6), 1–16. 10.1371/journal.pone.0178780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham, K. P. , & Anderson, D. R. (2002). Model selection and multimodel inference: A practical information‐theoretic approach. Springer. [Google Scholar]
- Cagnacci, F. , Focardi, S. , Ghisla, A. , van Moorter, B. , Merrill, E. H. , Gurarie, E. , Heurich, M. , Mysterud, A. , Linnell, J. , Panzacchi, M. , May, R. , Nygård, T. , Rolandsen, C. , & Hebblewhite, M. (2016). How many routes lead to migration? Comparison of methods to assess and characterize migratory movements. Journal of Animal Ecology, 85(1), 54–68. 10.1111/1365-2656.12449 [DOI] [PubMed] [Google Scholar]
- Chapman, B. B. , Brönmark, C. , Nilsson, J. Å. , & Hansson, L. A. (2011). Partial migration: An introduction. Oikos, 120(12), 1761–1763. 10.1111/j.1600-0706.2011.20070.x [DOI] [Google Scholar]
- Cheville, N. F. , McCullough, D. R. , & Paulson, L. R. (1998). Brucellosis in the Greater Yellowstone Area. National Academy Press. [Google Scholar]
- Cole, E. K. , Foley, A. M. , Warren, J. M. , Smith, B. L. , Dewey, S. R. , Brimeyer, D. G. , Fairbanks, W. S. , Sawyer, H. , & Cross, P. C. (2015). Changing migratory patterns in the Jackson elk herd. Journal of Wildlife Management, 79(6), 877–886. 10.1002/jwmg.917 [DOI] [Google Scholar]
- Conner, M. M. , Ebinger, M. R. , Blanchong, J. A. , & Cross, P. C. (2008). Infectious disease in cervids of North America: Data, models, and management challenges. Annals of the New York Academy of Sciences, 1134, 146–172. 10.1196/annals.1439.005 [DOI] [PubMed] [Google Scholar]
- Cook, W. E. , Williams, E. S. , & Dubay, S. A. (2004). Disappearance of bovine fetuses in northwestern Wyoming. Wildlife Society Bulletin, 32(1), 254–259. 10.2193/0091-7648(2004)32[254:dobfin]2.0.co;2 [DOI] [Google Scholar]
- Cross, P. C. , Cole, E. K. , Dobson, A. P. , Edwards, W. H. , Hamlin, K. L. , Luikart, G. , Middleton, A. D. , Scurlock, B. M. , & White, P. J. (2010). Probable causes of increasing brucellosis in free‐ranging elk of the Greater Yellowstone Ecosystem. Ecological Applications, 20(1), 278–288. 10.1890/08-2062.1 [DOI] [PubMed] [Google Scholar]
- Cross, P. C. , Maichak, E. J. , Rogerson, J. D. , Irvine, K. M. , Jones, J. D. , Heisey, D. M. , Edwards, W. H. , & Scurlock, B. M. (2015). Estimating the phenology of elk brucellosis transmission with hierarchical models of cause‐specific and baseline hazards. Journal of Wildlife Management, 79(5), 739–748. 10.1002/jwmg.883 [DOI] [Google Scholar]
- Cross, P. C. , Prosser, D. J. , Ramey, A. M. , Hanks, E. M. , & Pepin, K. M. (2019). Confronting models with data: The challenges of estimating disease spillover. Philosophical Transactions of the Royal Society B: Biological Sciences, 374, 20180435. 10.1098/rstb.2018.0435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann, O. , Heesterbeek, H. , & Britton, T. (2012). Mathematical tools for understanding infectious disease dynamics (Vol. 7). Princeton University Press. [Google Scholar]
- Dingle, H. , & Drake, V. A. (2007). What is migration? BioScience, 57(2), 113–121. 10.1641/B570206 [DOI] [Google Scholar]
- Dormann, C. F. , Elith, J. , Bacher, S. , Buchmann, C. , Carl, G. , Carré, G. , Marquéz, J. R. G. , Gruber, B. , Lafourcade, B. , Leitão, P. J. , Münkemüller, T. , McClean, C. , Osborne, P. E. , Reineking, B. , Schröder, B. , Skidmore, A. K. , Zurell, D. , & Lautenbach, S. (2013). Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography, 36(1), 27–46. 10.1111/j.1600-0587.2012.07348.x [DOI] [Google Scholar]
- Dougherty, E. R. , Seidel, D. P. , Carlson, C. J. , Spiegel, O. , & Getz, W. M. (2018). Going through the motions: Incorporating movement analyses into disease research. Ecology Letters, 21(4), 588–604. 10.1111/ele.12917 [DOI] [PubMed] [Google Scholar]
- Eggeman, S. L. , Hebblewhite, M. , Bohm, H. , Whittington, J. , & Merrill, E. H. (2016). Behavioural flexibility in migratory behaviour in a long‐lived large herbivore. Journal of Animal Ecology, 85(3), 785–797. 10.1111/1365-2656.12495 [DOI] [PubMed] [Google Scholar]
- Fieberg, J. , & Kochanny, C. O. (2005). Quantifying home‐range overlap: The importance of the utilization distribution. Journal of Wildlife Management, 69(4), 1346–1359. 10.2193/0022-541x(2005)69[1346:qhotio]2.0.co;2 [DOI] [Google Scholar]
- Folstad, I. , Nilssen, A. C. , Halvorsen, O. , & Andersen, J. (1991). Parasite avoidance: The cause of post‐calving migrations in Rangifer? Canadian Journal of Zoology, 69(9), 2423–2429. 10.1139/z91-340 [DOI] [Google Scholar]
- Fryxell, J. M. , Greever, J. , & Sinclair, A. R. E. (1988). Why are migratory ungulates so abundant? The American Naturalist, 131(6), 781–798. 10.1086/284822 [DOI] [Google Scholar]
- Fryxell, J. M. , & Sinclair, A. R. E. (1988). Causes and consequences of migration by large herbivores. Trends in Ecology & Evolution, 3(9), 237–241. 10.1016/0169-5347(88)90166-8 [DOI] [PubMed] [Google Scholar]
- Gergel, D. R. , Nijssen, B. , Abatzoglou, J. T. , Lettenmaier, D. P. , & Stumbaugh, M. R. (2017). Effects of climate change on snowpack and fire potential in the western USA. Climatic Change, 141(2), 287–299. 10.1007/s10584-017-1899-y [DOI] [Google Scholar]
- Gillies, C. S. , Hebblewhite, M. , Nielsen, S. E. , Krawchuk, M. A. , Aldridge, C. L. , Frair, J. L. , Saher, D. J. , Stevens, C. E. , & Jerde, C. L. (2006). Application of random effects to the study of resource selection by animals. Journal of Animal Ecology, 75(4), 887–898. 10.1111/j.1365-2656.2006.01106.x [DOI] [PubMed] [Google Scholar]
- Graham, M. H. (2003). Confronting multicollinearity in ecological multiple regression. Ecology, 84(11), 2809–2815. 10.1890/02-3114 [DOI] [Google Scholar]
- Guber, A. K. , Williams, D. M. , Dechen Quinn, A. C. , Tamrakar, S. B. , Porter, W. F. , & Rose, J. B. (2016). Model of pathogen transmission between livestock and white‐tailed deer in fragmented agricultural and forest landscapes. Environmental Modelling and Software, 80, 185–200. 10.1016/j.envsoft.2016.02.024 [DOI] [Google Scholar]
- Hall, D. K. , Riggs, G. A. , Salomonson, V. V. , Digirolamo, N. E. , & Bayr, K. J. (2002). MODIS snow‐cover products. Remote Sensing of Environment, 83, 181–194. 10.1016/S0034-4257(02)00095-0 [DOI] [Google Scholar]
- Hall, R. J. , Altizer, S. , & Bartel, R. A. (2014). Greater migratory propensity in hosts lowers pathogen transmission and impacts. Journal of Animal Ecology, 83(5), 1068–1077. 10.1111/1365-2656.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebblewhite, M. , & Merrill, E. H. (2011). Demographic balancing of migrant and resident elk in a partially migratory population through forage‐predation tradeoffs. Oikos, 120(12), 1860–1870. 10.1111/j.1600-0706.2011.19436.x [DOI] [Google Scholar]
- Jesmer, B. R. , Merkle, J. A. , Goheen, J. R. , Aikens, E. O. , Beck, J. L. , Courtemanch, A. B. , Hurley, M. A. , McWhirter, D. E. , Miyasaki, H. M. , Monteith, K. L. , & Kauffman, M. J. (2018). Is ungulate migration culturally transmitted? Evidence of social learning from translocated animals. Science, 361, 1023–1025. 10.1007/978-3-030-24436-1_17 [DOI] [PubMed] [Google Scholar]
- Jones, J. D. , Kauffman, M. J. , Monteith, K. L. , Scurlock, B. M. , Albeke, S. E. , & Cross, P. C. (2014). Supplemental feeding alters migration of a temperate ungulate. Ecological Applications, 24(7), 1769–1779. 10.1890/13-2092.1 [DOI] [PubMed] [Google Scholar]
- Kaitala, A. , Kaitala, V. , & Lundberg, P. (1993). A theory of partial migration. The American Naturalist, 142(1), 59–81. 10.1086/285529 [DOI] [Google Scholar]
- Kamath, P. L. , Foster, J. T. , Drees, K. P. , Luikart, G. , Quance, C. , Anderson, N. J. , Clarke, P. R. , Cole, E. K. , Drew, M. L. , Edwards, W. H. , Rhyan, J. C. , Treanor, J. J. , Wallen, R. L. , White, P. J. , Robbe‐Austerman, S. , & Cross, P. C. (2016). Genomics reveals historic and contemporary transmission dynamics of a bacterial disease among wildlife and livestock. Nature Communications, 7, 11448. 10.1038/ncomms11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, M. J. , Meacham, J. E. , Sawyer, H. , Steingisser, A. Y. , Rudd, W. J. , & Ostlind, E. (2018). Wild migrations: Atlas of Wyoming's ungulates. Oregon State University Press. [Google Scholar]
- Kilpatrick, A. M. , Gillin, C. M. , & Daszak, P. (2009). Wildlife‐livestock conflict: The risk of pathogen transmission from bison to cattle outside Yellowstone National Park. Journal of Applied Ecology, 46(2), 476–485. 10.1111/j.1365-2664.2008.01602.x [DOI] [Google Scholar]
- Lundberg, P. (2013). On the evolutionary stability of partial migration. Journal of Theoretical Biology, 321, 36–39. 10.1016/j.jtbi.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Manly, B. F. J. , McDonald, L. L. , Thomas, D. L. , McDonald, T. L. , & Erickson, W. P. (2002). Resource selection by animals: Statistical design and analysis for field studies. 10.2307/5247 [DOI] [Google Scholar]
- Merkle, J. A. , Cross, P. C. , Scurlock, B. M. , Cole, E. K. , Courtemanch, A. B. , Dewey, S. R. , & Kauffman, M. J. (2018). Linking spring phenology with mechanistic models of host movement to predict disease transmission risk. Journal of Applied Ecology, 55(2), 810–819. 10.1111/1365-2664.13022 [DOI] [Google Scholar]
- Merkle, J. A. , Monteith, K. L. , Aikens, E. O. , Hayes, M. M. , Hersey, K. R. , Middleton, A. D. , Oates, B. A. , Sawyer, H. , Scurlock, B. M. , & Kauffman, M. J. (2016). Large herbivores surf waves of green‐up during spring. Proceedings of the Royal Society B: Biological Sciences, 283(1833), 20160456. 10.1098/rspb.2016.0456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle, J. A. , Potts, J. R. , & Fortin, D. (2017). Energy benefits and emergent space use patterns of an empirically parameterized model of memory‐based patch selection. Oikos, 126(2), 185–195. 10.1111/oik.03356 [DOI] [Google Scholar]
- Meyer, C. B. , & Thuiller, W. (2006). Accuracy of resource selection functions across spatial scales. Diversity and Distributions, 12(3), 288–297. 10.1111/j.1366-9516.2006.00241.x [DOI] [Google Scholar]
- Middleton, A. D. , Kauffman, M. J. , McWhirter, D. E. , Cook, J. G. , Cook, R. C. , Nelson, A. A. , Jimenez, M. D. , & Klaver, R. W. (2013). Animal migration amid shifting patterns of phenology and predation: Lessons from a Yellowstone elk herd. Ecology, 94(6), 1245–1256. 10.1890/11-2298.1 [DOI] [PubMed] [Google Scholar]
- Montana Fish Wildlife and Parks . (2015). Targeted elk brucellosis surveillance project 2011–2015 comprehensive report. Montana Fish, Wildlife and Parks. [Google Scholar]
- Mysterud, A. , Qviller, L. , Meisingset, E. L. , & Viljugrein, H. (2016). Parasite load and seasonal migration in red deer. Oecologia, 180(2), 401–407. 10.1007/s00442-015-3465-5 [DOI] [PubMed] [Google Scholar]
- National Academies of Sciences Engineering and Medicine . (2017). Revisiting brucellosis in the Greater Yellowstone Area. 10.17226/24750 [DOI] [PubMed] [Google Scholar]
- Pettorelli, N. , Vik, J. O. , Mysterud, A. , Gaillard, J. M. , Tucker, C. J. , & Stenseth, N. C. (2005). Using the satellite‐derived NDVI to assess ecological responses to environmental change. Trends in Ecology & Evolution, 20(9), 503–510. 10.1016/j.tree.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Piersma, T. (1997). Do global patterns of habitat use and migration strategies co‐evolve with relative investments in immunocompetence due to spatial variation in parasite pressure? Oikos, 80(3), 623–631. 10.2307/3546640 [DOI] [Google Scholar]
- Plowright, R. K. , Parrish, C. R. , McCallum, H. , Hudson, P. J. , Ko, A. I. , Graham, A. L. , & Lloyd‐Smith, J. O. (2017). Pathways to zoonotic spillover. Nature Reviews Microbiology, 15(8), 502–510. 10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin, R. , Closs, G. P. , Lill, A. W. T. , Hicks, A. S. , Herrmann, K. K. , & Kelly, D. W. (2012). Migration as an escape from parasitism in New Zealand galaxiid fishes. Oecologia, 169(4), 955–963. 10.1007/s00442-012-2251-x [DOI] [PubMed] [Google Scholar]
- Proffitt, K. M. , Anderson, N. , Lukacs, P. , Riordan, M. M. , Gude, J. A. , & Shamhart, J. (2015). Effects of elk density on elk aggregation patterns and exposure to brucellosis. Journal of Wildlife Management, 79(3), 373–383. 10.1002/jwmg.860 [DOI] [Google Scholar]
- Proffitt, K. M. , Gude, J. A. , Hamlin, K. L. , Garrott, R. A. , Cunningham, J. A. , & Grigg, J. L. (2011). Elk distribution and spatial overlap with livestock during the brucellosis transmission risk period. Journal of Applied Ecology, 48(2), 471–478. 10.1111/j.1365-2664.2010.01928.x [DOI] [Google Scholar]
- R Development Core Team . (2016). R: A language and environment for statistical computing. R Development Core Team. [Google Scholar]
- Ragan, V. E. (2002). The Animal and Plant Health Inspection Service (APHIS) brucellosis eradication program in the United States. Veterinary Microbiology, 90(1–4), 11–18. 10.1016/S0378-1135(02)00240-7 [DOI] [PubMed] [Google Scholar]
- Rayl, N. D. , Merkle, J. A. , Proffitt, K. M. , Almberg, E. S. , Jones, J. D. , Gude, J. A. , & Cross, P. C. (2020). Data from: Elk migration influences the risk of disease spillover in the Greater Yellowstone Ecosystem. Dryad Digital Repository, 10.5061/dryad.34tmpg4j2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayl, N. D. , Proffitt, K. M. , Almberg, E. S. , Jones, J. D. , Merkle, J. A. , Gude, J. A. , & Cross, P. C. (2018). Estimating the risk of elk‐to‐livestock brucellosis transmission in Montana. Montana Fish, Wildlife and Parks. [Google Scholar]
- Rayl, N. D. , Proffitt, K. M. , Almberg, E. S. , Jones, J. D. , Merkle, J. A. , Gude, J. A. , & Cross, P. C. (2019). Modeling elk‐to‐livestock transmission risk to predict hotspots of brucellosis spillover. Journal of Wildlife Management, 83(4), 817–829. 10.1002/jwmg.21645 [DOI] [Google Scholar]
- Rhyan, J. C. , Nol, P. , Quance, C. , Gertonson, A. , Belfrage, J. , Harris, L. , Straka, K. , & Robbe‐Austerman, S. (2013). Transmission of brucellosis from elk to cattle and bison, Greater Yellowstone Area, USA, 2002–2012. Emerging Infectious Diseases, 19(12), 1992–1995. 10.3201/eid1912.130167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickbeil, G. J. M. , Merkle, J. A. , Anderson, G. , Atwood, M. P. , Beckmann, J. P. , Cole, E. K. , Courtemanch, A. B. , Dewey, S. , Gustine, D. D. , Kauffman, M. J. , McWhirter, D. E. , Mong, T. , Proffitt, K. , White, P. J. , & Middleton, A. D. (2019). Plasticity in elk migration timing is a response to changing environmental conditions. Global Change Biology, 25(7), 2368–2381. 10.1111/gcb.14629 [DOI] [PubMed] [Google Scholar]
- Russell, R. E. , Gude, J. A. , Anderson, N. J. , & Ramsey, J. M. (2015). Identifying priority chronic wasting disease surveillance areas for mule deer in Montana. Journal of Wildlife Management, 79(6), 989–997. 10.1002/jwmg.914 [DOI] [Google Scholar]
- Sawyer, H. , Middleton, A. D. , Hayes, M. M. , Kauffman, M. J. , & Monteith, K. L. (2016). The extra mile: Ungulate migration distance alters the use of seasonal range and exposure to anthropogenic risk. Ecosphere, 7(10), e01534. 10.1002/ecs2.1534 [DOI] [Google Scholar]
- Teitelbaum, C. S. , Huang, S. , Hall, R. J. , & Altizer, S. (2018). Migratory behaviour predicts greater parasite diversity in ungulates. Proceedings of the Royal Society B: Biological Sciences, 285(1875). 10.1098/rspb.2018.0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, S. L. , McNew, L. B. , Tyers, D. B. , Van Manen, F. T. , & Thompson, D. J. (2019). Grizzly bear depredation on grazing allotments in the Yellowstone Ecosystem. Journal of Wildlife Management, 83(3), 556–566. 10.1002/jwmg.21618 [DOI] [Google Scholar]
- White, L. A. , Forester, J. D. , & Craft, M. E. (2018). Dynamic, spatial models of parasite transmission in wildlife: Their structure, applications and remaining challenges. Journal of Animal Ecology, 87(3), 559–580. 10.1111/1365-2656.12761 [DOI] [PubMed] [Google Scholar]
- White, P. J. , Proffitt, K. M. , Mech, L. D. , Evans, S. B. , Cunningham, J. A. , & Hamlin, K. L. (2010). Migration of northern Yellowstone elk: Implications of spatial structuring. Journal of Mammalogy, 91(4), 827–837. 10.1644/08-MAMM-A-252.1.Key [DOI] [Google Scholar]
- Wolf, M. , Frair, J. , Merrill, E. , & Turchin, P. (2009). The attraction of the known: The importance of spatial familiarity in habitat selection in wapiti Cervus elaphus . Ecography, 32(3), 401–410. 10.1111/j.1600-0587.2008.05626.x [DOI] [Google Scholar]
- Worton, B. J. (1989). Kernel methods for estimating the utilization distribution in home‐range studies. Ecology, 70(1), 164–168. 10.2307/1938423 [DOI] [Google Scholar]
- Yang, A. , Gomez, J. P. , Haase, C. G. , Proffitt, K. M. , & Blackburn, J. K. (2019). Effects of brucellosis serologic status on physiology and behavior of Rocky Mountain elk (Cervus canadensis Nelsoni) in southwestern Montana, USA. Journal of Wildlife Diseases, 55(2), 304–315. 10.7589/2018-01-011 [DOI] [PubMed] [Google Scholar]
- Zidon, R. , Garti, S. , Getz, W. M. , & Saltz, D. (2017). Zebra migration strategies and anthrax in Etosha National Park. Namibia. Ecosphere, 8(8), 10.1002/ecs2.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Data available from the Dryad Digital Repository https://doi.org/10.5061/dryad.34tmpg4j2 (Rayl et al. 2020).


