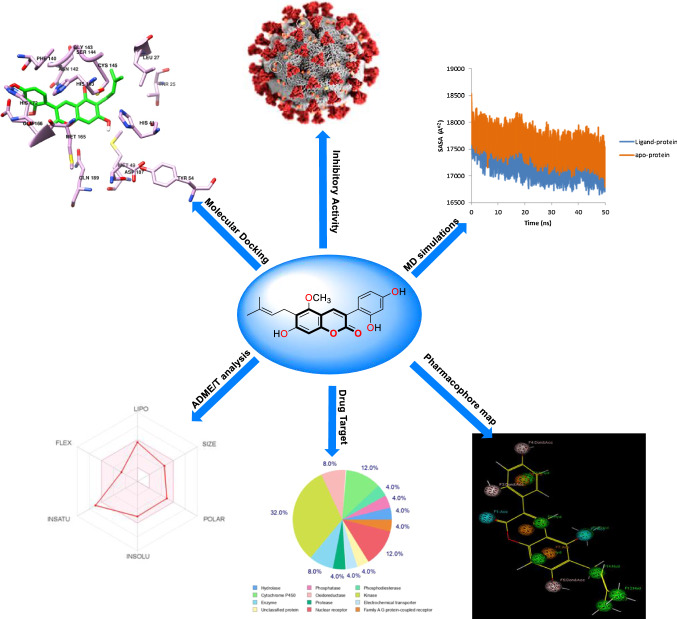
Abstract
Abstract
Coronavirus disease 2019 (COVID-19) is a pandemic viral disease caused by SARS-CoV-2 that generated serious damages for both the human population and the global economy. Therefore, it is currently considered as one of the most important global health problems of human societies and there is an urgent need for potent drugs or vaccines which can effectively combat this virus. The chymotrypsin-like protease (3CLpro) of SARS-CoV-2 plays a key role in the viral replication inside the host and thus is a promising drug target to design and develop effective antiviral drugs against SARS and other coronaviruses. This study evaluated some antiviral coumarin phytochemicals as potential inhibitors of coronaviruses 3CLpro by in silico approaches such as molecular docking, ADMET prediction, molecular dynamics simulation, and MM-PBSA binding energy calculation. Natural coumarin derivatives were docked to the 3CLpro of SARS-CoV-2 and for further investigation, docked to the 3CLpro of SARS-CoV and MERS-CoV. The docking scores of these natural compounds were compared with 3CLpro referenced inhibitors (ritonavir and lopinavir) and co-crystal inhibitor N3. Molecular docking studies suggested more than half of the coumarin phytochemicals had favorable interaction at the binding pocket of the coronaviruses 3CLpro and exhibited better binding affinities toward 3CLpro than ritonavir and lopinavir. Most antiviral phytochemicals interact strongly with one or both the catalytic dyad residues (His41 and Cys145) and the other key residues of SARS-CoV-2 main protease. Further, MD simulation and binding free energy calculations using MM-PBSA were carried out for three 3CLpro-coumarin complexes and 3CLpro-N3/lopinavir. The results confirmed that the 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate, and 3CLpro-inophyllum P complexes were highly stable, experience fewer conformation fluctuations and share a similar degree of compactness. Also, the pharmacokinetics and drug-likeness studies showed good results for the selected coumarin phytochemicals.Therefore, the coumarin phytochemicals could be used as antiviral agents in the treatment of COVID-19 after further studies.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-021-10230-6.
Keywords: SARS-CoV-2, COVID-19, Molecular docking, Molecular dynamics, Coumarin phytochemicals
Introduction
Coronaviruses (CoVs) are positive single-standard RNA viruses from the family Coronaviridae that can infect many hosts such as humans [1, 2]. The CoVs are placed within the subfamily orthocoronavirinae that categorized to four genera, Alpha-coronavirus, Beta-coronavirus, Gamma-coronavirus, and Delta-coronavirus [3, 4]. In the past two decades, two coronaviruses of the Beta-coronavirus family, including Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV), are zoonotic coronavirus that have caused global outbreaks and infected 1 million humans worldwide [5, 6]. The 2019 novel zoonotic coronavirus (SARS-CoV-2) was first identified and described in Wuhan city, the capital of Hubei province of China in the year 2019 that declared as COVID-19 by the World Health Organization (WHO) [7, 8] and was rapidly spread almost in all countries of the world [9] and became a global pandemic [10].
Humans can be infected by a virus through direct exposure to infected animals, direct contact human-to-infected humans with symptomatic or asymptomatic signs, and environmental contamination such as surfaces contaminated with droplets from coughing and sneezing of the symptomatic patients [11, 12]. This disease can show wide clinical features from asymptomatic state, mild, moderate to severe signs with multi-organ dysfunction in infected patients, such as respiratory symptoms like dry cough, running nose, dizziness, shortness of breath which might further advance to pneumonia, fever, headache, tiredness, conjunctivitis, loss of taste or smell, a rash on the skin, or discoloration of fingers or toes, aches, and pains, loss of speech or movement, malaise, and severe acute respiratory distress syndromes that causing death, also kidney failure with the fatal consequence, gastrointestinal disorders, including looseness of the bowels and diarrhea, sepsis and septic shock [12–14].
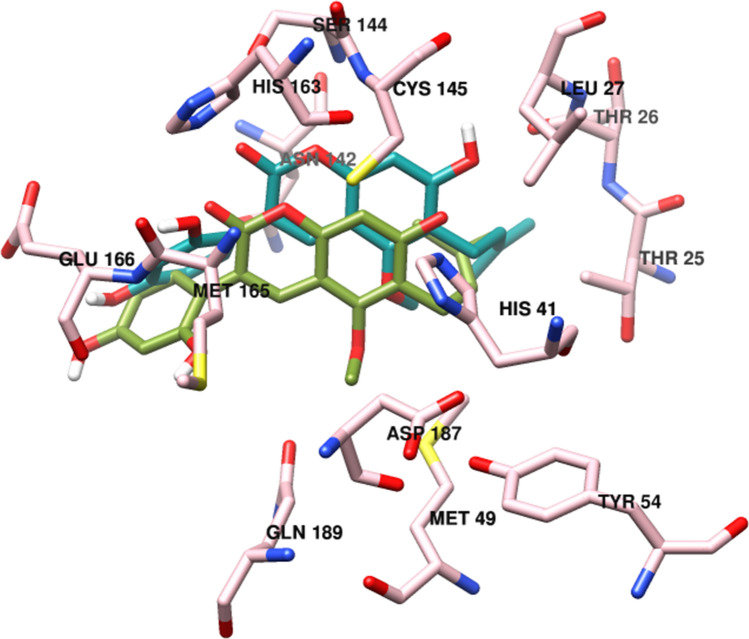
One of the attractive drug targets among coronavirus has been proposed to be 3-Chymotrypsin-like protease (3CLpro), called the main protease enzyme (Mpro), which contribute crucially to the essential maturation and replication and thus controls proteolytic cleavage as well as the processing of the large viral polyprotein orf1ab in combination with the papain-like protease (PLpro) [15]. 3CLpro protein selected as a candidate target for pan-CoV drug development, because it is critical for the viral replication or virulence, and highly conserved across all known CoVs and considered likely to be conserved in emerging CoVs. Moreover, the three-dimensional structure of the 3CLpro (with 306 amino acids, 6LU7) of SARS-CoV-2 like other coronaviruses such as MERS-CoV and SARS-CoV with 40 percent to 44 percent of the sequence homology involves 3 functional domains, including domain I (residues 8–101) and domain II (residues 102–184) consisting of 2-β barrel fold, which is similar to the chymotrypsin with a Cys-His catalytic dyad (Cyc145 and His41) situated in the cleft of domain I and II for SARS-CoV-2 catalytic activity, wherein Cys works as a nucleophile whereas His functions as a proton acceptor; and domain III (residues 201–306) also involves 5 α-helices linked to domain II through a long-loop area (residues 185 to 200) (Fig. S1) [16, 17]. The structure of 3CLpro complexed with a peptide-like inhibitor N3 and residues like His41, Phe140, Leu141, Asn142, Gly143, Cys145, His163, Met165, his172 and Gln189 show non-covalent interaction with N3 ligand. The ligand N3 forms hydrogen bonds (H-bond) with Gly143, Cys145, Glu166, and Gln189 residues in the binding pocket of this protease enzyme (Fig. S2) [18].
A key bicyclic heterocyclic is coumarin (2H-1-benzopyran-2-one) that is a natural secondary metabolite (SM) extracted from fungus, plants, bacteria, chemical synthesis, as well as essential oils, has been examined as one of the prominent structures to develop novel agents with higher specificity and affinity to different molecular targets showing antioxidant, anticancer, antiviral, anti-inflammatory and antileishmania activities [19–23]. Therefore, diverse families of plants like Umbelliferae, Clusiaceae, and Rutaceae have been used to isolate coumarins [19]. Moreover, natural compounds, synthetic and semi-synthetic drugs have been used against molecular targets of many viral proteins for inhibiting viral outbreak, which possess lower side effects and toxicity. Hence, they would be worthwhile candidates in the fight against diverse viruses like Covid-19 [24].
A lot of investigations referred to the inhibition impacts of diverse classes of natural coumarin phytochemicals (Fig. S3) on the functioning of viral proteins such as protease, integrase, reverse transcriptase as well as DNA polymerase, also, preventing viral entry against a wide range of human viruses such as hepatitis B and C, influenza, human immunodeficiency virus (HIV) and herpes simplex virus [19, 20, 25]. Coumarin compounds with similar structures including saxalin, psoralen, and bergapten have been known to prevent HIV replication [26]. Also, coumarins of mesoul and isomesoul have been reported to suppress HIV replication in jurkat T cell [27]. Kellerin, a sesquiterpene coumarin; rutamarin, a natural furanocoumarin; glycycoumarin, an aryl-coumarin, and osthole, a simple coumarin were reported to be anti-HSV and anti-HCV agents [28, 29]. Also, other studies have reported that some of the natural coumarins such as xanthotoxin, glycycoumarin, oxypeucedanin, pranferol and heraclenol have anti-HIV activity [24, 30].
In this study, we have investigated 50 natural coumarin phytochemicals isolated from plants to explore and identify the binding affinities and interactions of these phytochemicals against the coronavirus 3CLpro by molecular modeling approaches. The best compounds selected based on binding affinity were further investigated by molecular dynamics (MD) simulations and binding free energy calculations in which the selected compounds may be used as inhibitors against 3CLpro of SARS-CoV-2 and Covid-19 disease.
Material and methods
Protein structure
The 3CLpro cleavage sites on the polyproteins of coronaviruses are highly conserved, and their sequence and substrate specifications for coronaviruses of SARS-CoV-2, SARS-CoV, and MERS-CoV are identical [31]. This sequential similarity provides the insight for comparing SARS-CoV-2 with its previous counterparts leading to the identification of potent compounds to inhibit or control the replication of SARS- CoV-2. Therefore, the crystal structures of coronaviruses 3CLpro which were used in the docking analysis with sequence similarity were taken from the protein data bank (PDB) (http://www.rcsb.org) with the corresponding PDB identification codes [SARS-CoV-2 (6LU7), SARS-CoV (2DUC) and MERS-CoV (2YNA)]. 6LU7, 2DUC and 2YNA (PDB ID) were chosen as 3CLpro receptors because these have resolution values of 2.16, 1.70, and 1.50 Å, respectively. During the preparation process of the proteins using the Molecular Operation Environment (MOE) software, their water molecules and original ligands were removed, while polar hydrogen’s and Gasteiger charges were added to each protein. The protein structures were minimized by the energy minimization algorithm of MOE using the MMFF94X force field with the conjugate gradient method. Then, the protein structures were saved for molecular docking studies.
Ligand preparation
Bioactive coumarin phytochemicals (50 compounds) from different aromatic and medicinal plants and the 3CLpro reference inhibitors (ritonavir and lopinavir) were retrieved in SDF format from the PubChem database (www.pubchem.ncbi.nlm.nih.gov) and ChemSpider (www.chemspider.com). Then, all the ligands and inhibitors were converted into MOL2 format using Chemoffice Bio 3D Ultra (version 12.0, Cambridge Soft Corporation, Cambridge, 2010) and were optimized using AM1 semi-empirical method by Hyperchem software.
Molecular docking study
The molecular docking calculations were carried out using MOE software to predict the mode of interaction of the coumarin molecules and reference inhibitors with the active site of coronaviruses 3CLpro and to determine the binding affinities of these compounds with coronaviruses 3CLpro. Within MOE, the flexibility of ligands is considered while the proteins are considered as a rigid structure. Site finder [32–34] was used for the selection of the active site of the 3CLpro protein and the active site was defined with at least one atom within a distance cut off of 4.5 Å at ligand in the crystal structure of 3CLpro. The docking was done using the triangle matcher placement algorithm in combination with the London dG scoring function and force field as the refinement method. The best conformation of the ligands was further evaluated by the binding energies (s-score, kcal/mol), and interactions between the ligands and proteins were analyzed by the LigX module in MOE and UCSF chimera software.
Validation of docking
Docking protocol was validated by re-docking of the co-crystalized ligand (N3) into the 3CLpro structure (6LU7). As can be seen in Fig. S4, N3 molecule bound into similar positions of 3CLpro in comparison with its original crystallographic form and the docked structure had a RMSD of 1.669 Å after superimposing onto the native co-crystallized complex which indicates the validity of the method used.
In silico evaluation of physicochemical and pharmacokinetics properties
Various pharmacokinetic properties of the best-identified phytochemicals and the reference inhibitors with significant binding affinity for 3CLpro of SARA-CoV-2 were evaluated based on pharmacokinetics and physicochemical features such as drug-likeness rules (Lipinski [35], Veber [36], Egan [37], Ghose and Muegge [38]), lipophilicity (Log Po/w), water solubility, Log S, polar surface area (TPSA), number of rotatable bonds and medicinal chemistry (PAINS, Brenk, Lead likeness, synthetic accessibility) methods were analyzed using Swiss ADME and pkCSM-pharmacokinetics web tools. The canonical SMILES of the phytochemicals were copied from Chem Draw to calculate ADMET and drug likeness properties using default parameters. Also, the P450 site of metabolism (SOM), the PASS prediction, and the molecular target studies were calculated using RS-Web Predictor 1.0, PASS-Way2Drug server, and Swiss target prediction [39, 40].
Pharmacophore mapping
Pharmacophores constitute the main structural scaffold for a set of active compounds that is responsible for their biological activity and is frequently being used in drug design initiatives. Pharmacophore mapping is used to find important features with a set of functional groups for receptor binding. The pharmacophore protocol of MOE has been used to construct the most representative features of the glycycoumarin and 6LU7 active site, which are indicated as spheres that represent the pharmacophore of glycycoumarin and essential interaction points with the key residues on ligand binding of the 3CLpro.
Molecular dynamics simulation of 3CLpro of SARS-CoV-2
MD simulations are used for validation of molecular docking results and offer a reliable assessment of the potential stability of a protein–ligand complex [41]. In this study, MD simulations were carried out for the apo SARS-CoV-2 3CLpro; 3CLpro complexed with co-crystal inhibitor N3 and lopinavir as control and the complexes of 3CLpro with three selected coumarin phytochemicals obtained from molecular docking studies, which helped to get more insight into protein and docked complexes in biological conditions. The MD simulations were carried out using the GROningen MaChine for chemical simulations (GROMACS, version 5.1.2) with GROMOS96 43al force field [42–44] for 50 ns at the real physiological condition and aqueous solution at T = 310 K (37 °C) and P = 1 atm. The topology files of glycycoumarin, Inophyllum P, oxypeucedanin hydrate, N3, and lopinavir were obtained from the PRODRG server [45] while the topology file of 3CLpro protein was prepared by the GROMACS. The 3CLpro and 3CLpro-ligand systems were solvated and fully immersed in the dodecahedron box with simple point charge (SPC) waters and neutralized by adding four sodium ions and energy minimized using the steepest descent algorithm [1, 15, 18, 32] for 50,000 steps and energy tolerance of 500 kJ/mol in a periodic boundary condition and equilibrated to achieve the appropriate volume under NVT ensemble [46]. The particle-Mesh Ewald (PME) method was used with a Fourier grid spacing of 1 Å to calculate the long-range electrostatic interactions. The short-range Lennard–Jones and Coulomb interactions were calculated by a cutoff value of 14 Å, and the final MD production was carried out with a time step of 2 fs for 50 ns in the NPT ensemble [46, 47]. The root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), radius of gyration (Rg), solvent accessible surface area (SASA), and total intermolecular hydrogen bonds (H bonds) were calculated from MD simulations.
The binding free energy calculations
Molecular Mechanic/Poisson–Boltzmann surface Area (MM-PBSA) method [48] was used to calculate the binding free energy of the ligand–protein complex in the explicit solvent [49] using the g-mmpbsa script program [50]. The MD simulation trajectory of 50 ns was considered for the calculation of different components of the binding energy of the ligand and 3CLpro complex. The free energy of binding was calculated using the following equation:
Here, represent total binding free energy, while others show the free energy of ligand–protein complex, protein, and ligand, respectively.
the , and represent interactions among bonded, van der Waals, and electrostatic states. In contrast, the polar and nonpolar interactions to the solvation free energy are presented by the and , respectively, and -T∆S is the entropy contribution at temperature T.
Results and discussion
The main aim of this study was to identify potential coumarin phytochemicals as inhibitors for the 3CLpro protein of SARS-CoV-2. 3CLpro was selected because of its important role in viral replication. The molecular docking of all coumarin phytochemicals with 3CLpro was performed and the compounds that showed a strong binding affinity for 3CLpro were selected for further investigations. The ADMET properties of the top coumarin phytochemicals were analyzed and then, these compounds were evaluated through MD simulations and calculated free energy of binding for the compounds using MM-PBSA. This in silico study was undertaken to identify potential antiviral compound for COVID-19 (Fig. 1).
Fig. 1.
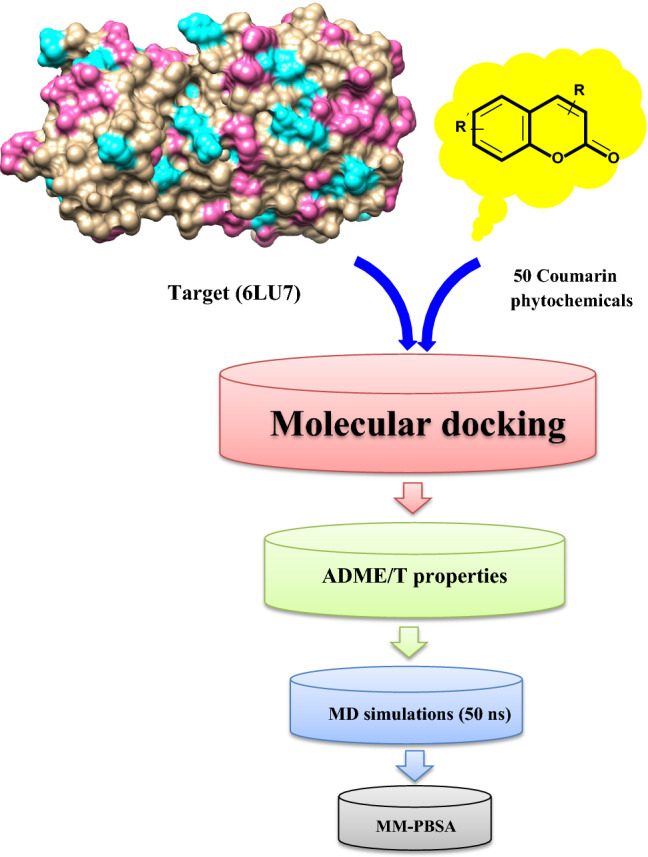
Schematic representation of various steps of the methodology to identify the phytochemical based coumarin inhibitors of 3CLpro
Molecular docking
The molecular docking approach to identify potential hits has become one of the most popular methods for structure-based computer-aided drug discovery (SB-CADD). In this study, the molecular docking is done using MOE software to find out the best candidates among the 50 coumarin phytochemicals based on their binding scores. The results of the binding affinities from the docking analysis of the coumarin phytochemicals to 3CLpro of SARS-CoV-2 ranged from -5.0 to -11.0 kcal/mol (Fig. 2).
Fig. 2.
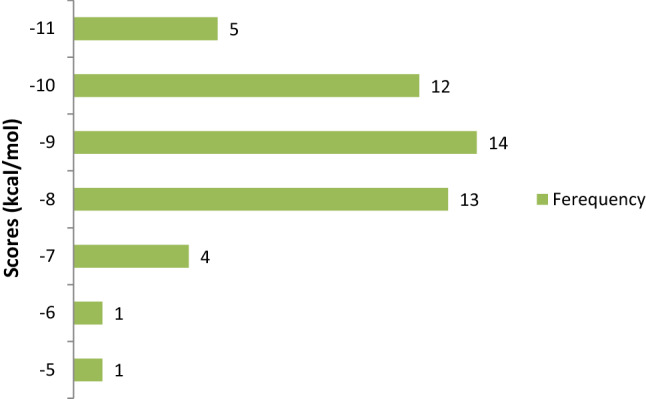
Frequency distribution of 50 coumarin phytochemicals over the range of docking scores
The coumarin phytochemicals with more negative binding score values had higher binding affinities for 3CLpro and were ranked higher. The binding energy scores of the coumarin phytochemicals to 3CLpro of SARS-CoV-2, along with their binding energy scores to 3CLpro of SARS-CoV and MERS-CoV, are shown in Table S1.
As shown by Table S1, N3, lopinavir, and ritonavir that are considered as the co-crystal inhibitor of 3CLpro and reference inhibitors exhibited a docking score of − 10.92, − 6.84 and − 10.89 kcal/mol for 3CLpro of SARS-CoV-2, whereas docking scores of N3, ritonavir, and lopinavir for 3CLpro of SARS-CoV equaled − 9.59 kcal/mol, − 8.10 kcal/mol and − 9.97 kcal/mol and for 3CLpro of MERS-CoV equaled − 9.30 kcal/mol, − 8.17 kcal/mol and − 9.37 kcal/mol. More than 50% of the coumarin phytochemicals showed docking score against SARS-coronavirus, which surpassed that of reference inhibitors.
The compounds glycycoumarin, Inophyllum P, oxypeucedanin hydrate, and mesuol were found to exhibit the best docking scores of − 11.89, − 11.43, − 11.76, and − 11.17 kcal/mol, respectively, among the coumarin phytochemicals against 3CLpro of SARS-CoV-2, while glycycoumarin had the highest binding affinity to that of SARS-CoV and MERS-CoV (Table 1). Therefore, glycycoumarin was the top docked compound to 3CLpro that interacted strongly with the target protein of the coronavirus. Analysis of the interactions of the best coumarin phytochemicals and reference inhibitors with amino acid residues of 3CLpro of coronaviruses (Table 1) showed that these compounds majorly interacted with the hotspot residues through hydrophobic interactions and with H-bonding under 4.0 Å (particularly with Cys145 and His41).
Table 1.
Interacting amino acid (aa) residues of 3CLpro of coronaviruses with the best coumarin phytochemicals
| Bioactive compound | Coronavirus | Interacted residues | aa residue involved in H-bonding (Bond Distance) |
|---|---|---|---|
| Ritonavir | SARS-CoV-2 | His 41, Cys145, Gly143, Met165, His164, Glu166, Asn142, Met49, Gln189, Thr26, Thr24, Thr25, Thr45, Ser46 | Glu166 (2.48), Thr25 (3.72) |
| Lopinavir | His 41, Cys145, Gly143, Met165, His164, Glu166, Asn142, Leu141, Phe140, Met49, Gln189, Asp187 | Gln189 (2.08) | |
| Glycycoumarin | His 41, Cys145, Ser144, Gly143, Met165, His164, Glu166, Asn142, Leu141, Phe140, Met49, Gln189, Asp187, Arg188, Tyr54 | Cys145 (2.46), Ser144 (1.91), Gln189 (2.15) | |
| Inophyllum P | His 41, Cys145, Ser144, Gly143, Met165, His164, Glu166, Asn142, Leu141, Phe140, Met49, Gln189, Leu27 | Cys145 (2.607), Ser144 (2.23), Leu141 (3.18) | |
| Mesuol | His 41, Cys145, Ser144, Gly143, Met165, His164, Glu166, Asn142, Leu141, Phe140, Met49, Gln189, Ser46 | Cys145 (2.608), Ser144 (3.77), Asn142 (1.27) | |
| Oxypeucedanin hydrate | His 41, Cys145, Ser144, Gly143, Met165, His164, Glu166, Asn142, Leu141, Phe140, Met49, Gln189, His163, Thr25, Thr26, Leu27 | Cys145 (3.04), Ser144 (2.21), His163 (2.86), His164 (3.04) | |
| Glycycoumarin | SARS-CoV | Gln189, Met165, His164, Cys145, His41, Arg188, Asp48, Cys44, Thr25, Glu47, Thr24, Thr45, Ala46 | His164 (2.41), Cys44 (2.51), Thr24 (2.30) |
| Inophyllum G2 | Gln189, Met165, His164, Cys145, His41, Arg188, Asp48, Cys44, Glu47, His163, Glu166, Met49, Asn142, Leu27, Thr26 | His164 (2.862), Glu166 (3.09), Asn142 (2.47) | |
| licopyranocoumarin | Phe140, Met165, His164, Cys145, His41, Ser144, Asp48, Cys44, Glu47, His163, Glu166, Met49, Asn142, Leu25, Thr24, Asp48, His172 | His163 (2.35), Thr25 (2.29, thr45 (3.72) | |
| Wedelolactone | Phe140, Met165, His164, Cys145, His41, Ser144, Asp48, Glu47, His163, Glu166, Met49, Asn142, His172, Leu141, Gly143 | Glu166 (2.07), Ser144 (1.72), Leu141 (2.02) | |
| Glycycoumarin | MERS-CoV | Met6, Asp294, Gln299, Asp295, Met298, Asn156, Glu157, Glu155 | Met6 (2.21), Asp295 (2.16), Asn156 (1.93) |
| licopyranocoumarin | Met6, Asp294, Gln299, Asp295, Met298, Asn156, Glu157, Glu155, Thr13, Lys5, Phe291, Ala8, Ser7, | Gln299 (1.790), Asn156 (3.58), Gly157 (3.21) | |
| Wedelolactone | Met6, Asp294, Gln299, Asp295, Met298, Asn156, Glu157, Glu155, Thr130, Phe291, Ala8, Phe115, Ser116, Thr154, | Glu155 (2.56), ser116 (2.48), Thr130 (3.22), Asp295 (2.84) | |
| Isomesuol | Met6, Asp294, Gln299, Asp295, Met298, Asn156, Glu157, Glu155, Ala8, Thr154, Ser116, Tyr153, Phe115, Phe291 | Gln299 (3.14), Ser114 (3.09) |
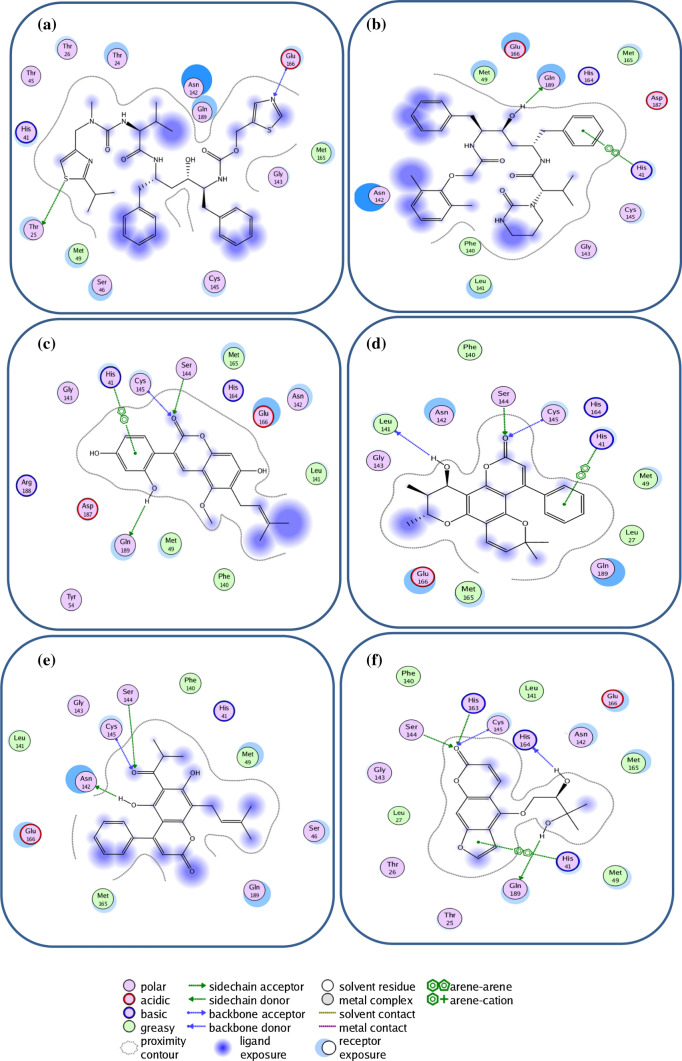
The results of the molecular docking of the best coumarin phytochemicals including glycycoumarin, Inophyllum P, mesuol, oxypeucedanin hydrate and reference inhibitors in the active site of SARS-CoV-2 3CLpro illustrated by their corresponding 2D interaction plots that the selected compounds interacted with either both (Cys145 and His41) or at least one catalytic dyad residue, detected by MOE (Fig. 3 and Fig. S5) [1, 18, 32, 43, 51]. The selected compounds and Ritonavir and lopinavir exhibit similar binding modes due to the parallel orientations of the ligands and their same key residues (Fig. 3), such as His41, Met49, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, Met165, His164, Glu166, Gln189, and Thr190. The results of ligand–protein binding interaction showed that ritonavir and lopinavir as reference inhibitors were docked into the active site and catalytic dyad (Cys145 and His41) of SARS-CoV-2. Ritonavir could form two hydrogen bonds with the side chain of Thr25 and the backbone of Glu166 (Fig. 3a), while lopinavir with a considerably higher binding energy (− 10.890 kcal/mol) than Ritonavir showed significant π-π stacking interaction with His41 of the catalytic dyad and form one hydrogen bond with the side chain of Gln189 (Table 1, Fig. 3b) and also, both of the inhibitors had hydrophobic interactions with surrendering residues. Glycycoumarin, a reported anti-viral agent known to inhibit hepatitis C virus (HCV) [29] and giant cell formation in HIV [30, 52], had significant binding to the catalytic dyad of SARS-Cov-2 that interacted with His41 by π-π stacking interaction and form hydrogen bonds with the backbone of Cys145 and also side chain of Ser144 and Gln189 along with hydrophobic interactions with the other residues such as Met49, Met165, Leu141 and Phe140 (Table 1, Fig. 3c). Inophyllum P, as an anti-viral used to inhibit HIV-1 reverse transcriptase [30], was stabilized through hydrogen bonding with the catalytic residue of Cys145, the backbone of Leu141 and the side chain of Ser144, π-π stacking interaction with the catalytic residue of His41, and hydrophobic interactions with the other surrounding residues (Fig. 3d). Mesuol, as simple coumarin that suppresses HIV-1 replication in Jurkat T cells [53], formed one hydrogen bond with the catalytic residue of Cys145 and two hydrogen bonds with the side chain of Asn142 and Ser144 and also, the other interactions with neighboring residues (Fig. 3e). Oxypeucedanin hydrate, a furanocoumarin reported to have anti-HIV effects [26, 54], stabilized the active site of 3CLpro of SARS-CoV-2 through hydrogen bonding interactions with the catalytic residue of the catalytic dyad (Cys145) and Ser144, His163, His164 and Gln189, π-π stacking interaction with the catalytic residue His41 and hydrophobic interactions with surrounding amino acid residues (Fig. 3f). The carbonyl oxygen of the coumarin ring mediated the three hydrogen bonds with the backbone of Cys145 and the side chain of Ser144 and His163. The side chain of Gln189 and backbone of his164 established hydrogen bonds with different hydroxyl groups.
Fig. 3.
2D view of the binding conformation of ligands in binding pocket of SARS-CoV-2 3CLpro, a Ritonavir, b Lopinavir, c Glycycoumarin, d Inophyllum P, e Mesuol, f Oxypeucedanin hydrate
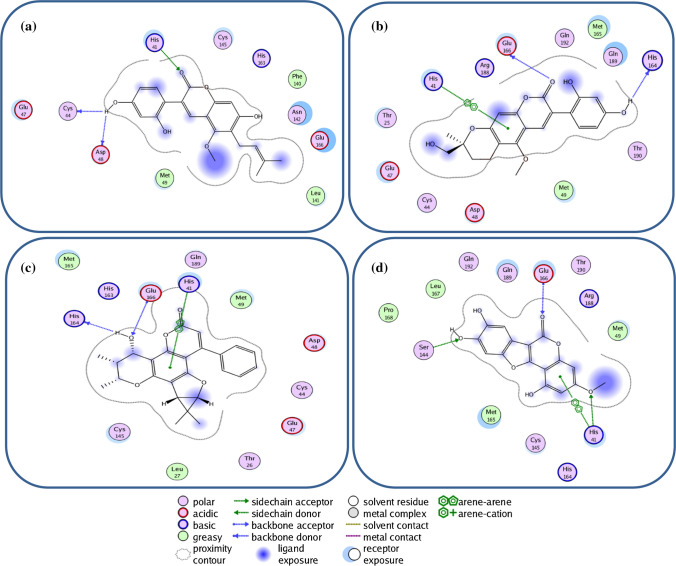
Like the ligand–protein binding interaction of glycycoumarin to SARS-CoV-2 3CLpro that targeted the Cys-His catalytic dyad (Cys145 and His41) along with the other binding residues, the docking analysis showed that the SARS-CoV 3CLpro interacted with the same ligand differently. Glycycoumarin interacted with His41 in catalytic dyad and Cys44 and Asp48 by hydrogen bonding interactions (Fig. 4a). For licopyranocoumarin, hydrogen bonding interactions with His164 and Glu166 were observed and it further interacted with His41 via π-π stacking interaction (Fig. 4b). The interaction of Inophyllum G2 with SARS-CoV3CLpro showed a hydrogen bonding to His164 and Glu166 while π-π stacking interaction was observed between His41 and ligand (Fig. 4c). His41, Ser144, and Glu166 residues of the protein interacted with wedelolactone via hydrogen bonds while His41 interacted via π-π stacking interaction to wedelolactone (Fig. 4d). These residues in the active site of SARS-CoV 3CLpro were also conserved for the Cys-His catalytic dyad binding hotspot.
Fig. 4.
2D representation of 3CLpro amino acid interactions of SARS-CoV with coumarin phytochemicals; a glycycoumarin, b licopyranocoumarin, c Inophyllum G2, d wedelolactone
Glycycoumarin, licopyranocoumarin, wedelolactone, and isomesuol interacted with MERS-CoV 3CLpro that is different from the other two coronaviruses. Hydrogen bonds were observed between glycycoumarin/isomesuol/wedelolactone and Asp295 while extra hydrogen bonds were also observed for glycycoumarin with Met6 and Asn156, wedelolactone with Glu155, Ser116 and Thr130, and isomesuol with Glu155 while licopyranocoumarin interacted via hydrogen bonding interactions to Gly157, Asn156 and Gln299. In addition to this, the other residues (Val4, Ala8, Thr154, Phe291, Asp294, and Met298) are involved in forming hydrophobic interactions with these compounds (Table1 and Fig. 5a-d).
Fig. 5.
2D representation of 3CLpro amino acid interactions of MERS-CoV with coumarin phytochemicals; a Glycycoumarin, b Licopyranocoumarin, c Wedelolactone and d Isomesuol
The results of this study showed that 8 compounds (glycycoumarin, Inophyllum P, mesuol, oxypeucedanin hydrate, glycycoumarin, Inophyllum G2, licopyranocoumarin, wedelolactone) with a considerable inhibitory tendency towards the SARS-coronavirus were identified from coumarin phytochemicals. All of the compounds such as glycycoumarin, Inophyllum P, mesuol, and oxypeucedanin hydrate were docked into the active site and were interacting with the catalytic dyad (Cys-His) of 3CLpro protein of the coronaviruses in a similar pattern as ritonavir and lopinavir (Fig. 3).
ADMET properties and drug likeness
In the next step, we employed pkCSM online-server (http://biosig.unimelb.edu.au/pkcsm/prediction) for predicting the ADMET features of the selected compounds and applied the Lipinski’s rule of five to screen the coumarin phytochemicals and filtered the orally bioavailable compounds. After that, we assessed the synthetic availability of the coumarin derivatives by the Swiss ADME online-tool (http://www.swissadme.ch/index.php). Its scale demonstrated a score of 1 for a relatively simple synthetic route, whereas a score closer to 10 had a high structural complexity, and was therefore difficult to synthesize.
According to the studies [55, 56], water solubility has been considered to be crucial to approximate the absorption of the medicines in the body, which has been given in the log (mol/L) ( insoluble < -10 < poorly soluble < -6 < moderately < -4 < soluble < -2 < very soluble < 0 < highly soluble). The intestinal absorption of all the selected compounds (88.20–97.44%) revealed an acceptable absorption feature. Furthermore, the blood/brain partition coefficient (log BB) of the top compounds indicated a lower opportunity for crossing the blood–brain barrier (BBB). Results have also shown that steady-state volume of distribution (VDss, log L/kg) value of glycycoumarin has been < − 0.15, which reflects more distribution of the above compound in the plasma rather than in the tissues whereas Inophyllum P, oxypeucedanin hydrate, and mesuol showed more distribution in the tissues (Table 3). For metabolism, two compounds mesuol and Inophyllum P were predicted as the substrate for the CYP450 3A4 subtype, also these compounds might be metabolized by CYP2D6. At the same time, the selected compounds could not inhibit the CYP450 2D6 subtype; however, compounds like mesuol, Inophyllum P and glycycoumarin might inhibit 2C6, 2C19, as well as CYP450 3A4 subtypes and all the selected compounds, could inhibit CYP450 1A2 subtype. Based on the prediction of the total clearance, hepatic and renal tissue can be used to clear such phytochemicals. The expected toxicity represents the fact that each compound did not show any skin sensitization and has been not detrimental to the liver. Moreover, Ames test has been used to reveal the anticipated toxicity, reflecting that oxypeucedanin hydrate has been not mutagenic. Additionally, the synthetic accessibility score of the selected phytochemicals equaled 3.55–5.12. With regard to Tables 2 and 3, bioavailability and ADMET (Rule of Five, Veber, Ghose, Muegge, & Egan) are in the reasonable ranges for the selected phytochemicals in solubility and lipophilicity. According to each parameter, it is possible to employ coumarin phytochemicals as the antiviral agents to treat COVID-19.
Table 3.
In Silico ADME/T prediction of the top binding coumarin phytochemicals
| Compound | Absorption | Distribution | Metabolism | Excretion | Toxicity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water solubility | Intestinal absorption (human) | Blood brain barrier Permeability | VDss (human) | CYP | Total Clearance | AMES toxicity | |||||||
| 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | |||||||
| substrate | inhibitor | ||||||||||||
| (logmol/L) | Numeric (%Absorbed) | (log BB) | (log (L/kg) | Categorical | (Yes/No) | Numeric (log ml/min/kg) | (Yes/No) | ||||||
| glycycoumarin | -4.08 | 88.20 | -1.14 | -0.28 | No | No | Yes | Yes | Yes | No | Yes | 0.78 | Yes |
| Inophyllum P | -5.08 | 96.48 | -0.43 | 0.32 | No | Yes | Yes | Yes | Yes | No | Yes | 0.56 | Yes |
| Mesuol | -5.41 | 96.68 | -0.10 | -0.05 | No | Yes | Yes | Yes | Yes | No | Yes | 0.80 | Yes |
| Oxypeucedanin hydrate | -3.09 | 97.44 | -0.66 | 0.33 | No | No | Yes | No | No | No | No | 0.75 | No |
Table 2.
Drug likeness properties of the top binding coumarin phytochemicals
| Drug Likeness Properties | Glycycoumarin | Inophyllum P | Mesuol | Oxypeucedanin hydrate |
|---|---|---|---|---|
| Molecular weight (g/mol)a | 368.38 | 404.46 | 392.44 | 304.29 |
| Consensus Log Po/wb | 3.54 | 4.14 | 4.56 | 1.79 |
| Log Sc | − 5.06 | − 5.28 | − 5.87 | − 2.69 |
| Num. H-bond acceptord | 6 | 5 | 5 | 6 |
| Num. H-bond donore | 3 | 1 | 2 | 2 |
| No of Rotatable Bondsf | 4 | 1 | 5 | 4 |
| Molar Refractivity | 104.20 | 116.97 | 115.49 | 80.34 |
| Lipinski | Yes | Yes | Yes | Yes |
| Ghose | Yes | Yes | Yes | Yes |
| Veber | Yes | Yes | Yes | Yes |
| Egan | Yes | Yes | Yes | Yes |
| Muegge | Yes | Yes | No | Yes |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 |
| TPSA (A2)g | 100.13 | 68.90 | 87.74 | 93.04 |
| Synthetic accessibility (SA) | 3.55 | 5.12 | 3.88 | 3.79 |
| Solubility (mol/l) | 1.79e-06 | 5.20e06 | 1.36e06 | 2.05e03 |
| PAINSh | 0 alert | 0 alert | 0 alert | 0 alert |
| Brenkj | 2 alert | 1 alert | 2 alert | 1 alert |
| Leadlikeness | No | No | No | Yes |
The general recommended ranges are as follows:
aMolecular weight, < 500
bPredictedoctanol/water partition coefficient, − 0.4 to + 5.6
cPredicted aqueous solubility, < -5.0
dNumber of hydrogen bond acceptors, < 10
eNumber of hydrogen bond donor, < 5
fRotatable bonds, < 10
gPolar surface area, < 140 A°2
hPan-Assay Interference
jStructural Alert
PASS prediction for antiviral activity
Studies have also considered PASS as a popular tool employed in nearly every drug industry with regard to the analyses of the structure–activity relationship [57]. It gives the prediction score for biological activities on the ratio of probability to be active (Pa) and probability to be inactive (Pi). A higher Pa means the biological activity is having more probability for a compound. Moreover, researchers identified the biological activity spectra of top coumarin chemicals which are deposited [58] in the PASS database. Table 4 presents the prediction results of ten biological activities for the top scoring coumarins. Notably, the present research results indicated the major utility of the PASS plan to predict the biological activities of a coumarin-phytochemical based on the respective coumarin structure that has been shown by an average prediction coefficient equal to 0.85 (Pa ranging from 0.243 to 0.937 when Pa > Pi) for the selected 4 compounds. Besides, the results revealed a number of biological activities of the selected compounds amongst the ten experimented activities. However, as shown in Table 5, for several instances PASS was unable to provide predictions for some of the biological activities.
Table 4.
The PASS prediction results of the biological activities of the coumarin phytochemicals series
| No | Biological activities | Glycycoumarin | Inophyllum P | Mesuol | Oxypeucedanin hydrate | ||||
|---|---|---|---|---|---|---|---|---|---|
| Pa | Pi | Pa | Pi | Pa | Pi | Pa | Pi | ||
| 1 | Membrane integrity agonist | 0.937 | 0.002 | – | – | 0.683 | 0.058 | 0.720 | 0.051 |
| 2 | HMOX1 expression enhancer | 0.635 | 0.015 | – | – | 0.265 | 0.115 | 0.355 | 0.072 |
| 3 | Chlordeconereductase inhibitor | 0.803 | 0.02 | – | – | 0.243 | 0.193 | 0.344 | 0.132 |
| 4 | HIF1A expression inhibitor | 0.936 | 0.004 | 0.867 | 0.008 | 0.348 | 0.135 | 0.512 | 0.053 |
| 5 | Histidine kinase inhibitor | 0.709 | 0.008 | 0.765 | 0.005 | 0.467 | 0.035 | 0.498 | 0.029 |
| 6 | Aldehyde oxidase inhibitor | 0.807 | 0.009 | – | – | – | – | 0.654 | 0.023 |
| 7 | Antimutagenic | 0.877 | 0.003 | – | – | – | – | 0.735 | 0.005 |
| 8 | Mucomembranous protector | 0.648 | 0.081 | – | – | 0.307 | 0.234 | – | – |
| 9 | TP53 expression enhancer | 0.901 | 0.005 | 0.484 | 0.098 | 0.616 | 0.045 | 0.580 | 0.056 |
| 10 | Chemopreventive | 0.807 | 0.004 | 0.665 | 0.008 | 0.615 | 0.009 | 0.596 | 0.010 |
Pa: prediction of activity spectra for substances; Pi: probable inactivity
Table 5.
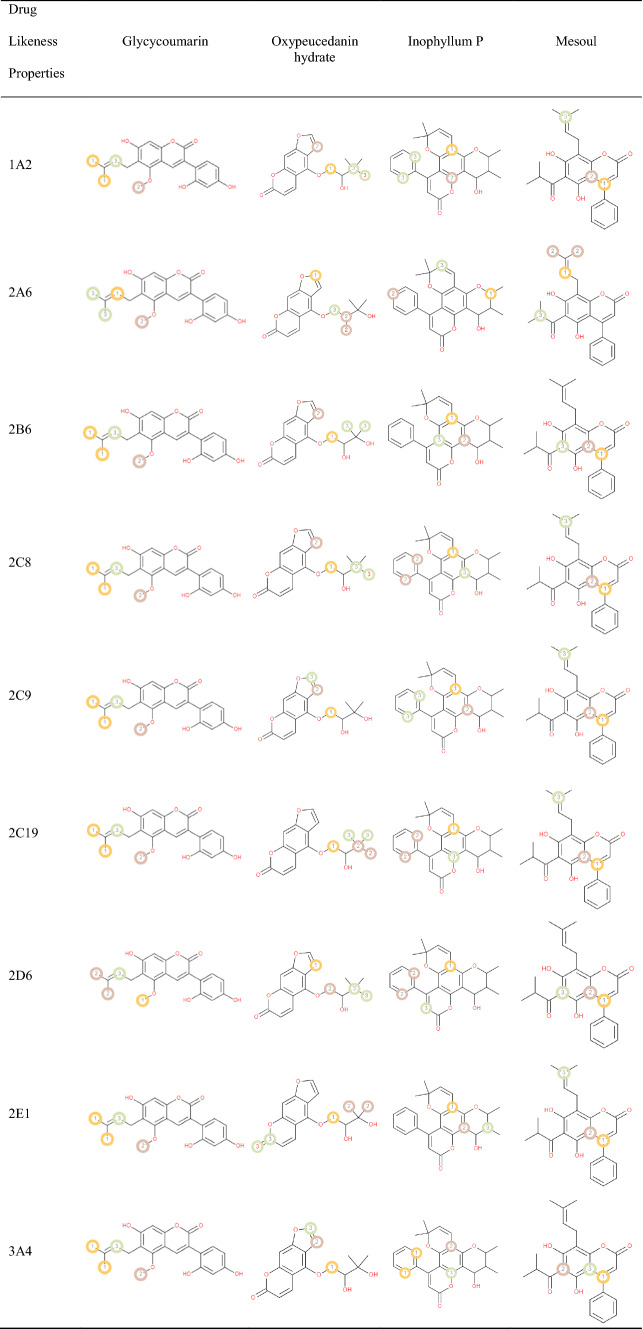
List of the P450 sites of metabolism prediction study of the Glycycoumarin, Oxypeucedanin hydrate Inophyllum P and Mesoul molecules
Table 5 reports probable sites of metabolism (SOM) through CYPs 1A2, 2B6, 2A6, 2C8, 2C19, 2E1, 3A4 and 2D6 of glycycoumarin, oxypeucedanin hydrate, Inophyllum P and mesoul. Probable sites of a chemical compound, wherein the metabolism by the isoforms of CYP450 enzymes happen, are marked by circles on the molecule's chemical structure [37]. According to P450 SOM prediction, glycycoumarin possessed 4 sites for CYP 450 2A6, CYP 450 1A2, CYP 450 2C19, CYP 450 2B6, CYP 450 2D6, CYP 450 2C8, CYP 450 3A4 and CYP 450 2E1. Oxypeucedanin hydrate had 5 sites of metabolism (SOMs) for the CYP 450 2C19 enzyme, 4 sites for CYP 450 1A2, 2A6, 2B6, 2C8, and 2D6, 3 sites for CYP 450 2C9, 2E1, and 3A4. The P450 SOM predictions for Inophyllum P showed 4 sites of SOMs for the CYP 450 1A2, 2C8, 2C9, 2C19, 2D6, and 3A4 and 3 sites for CYP 450 2A6, 2B6, and 2E1. Mesoul had 4 sites of SOMs for the CYP 450 2A6, and 3 sites for the CYP 450 1A2, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4.
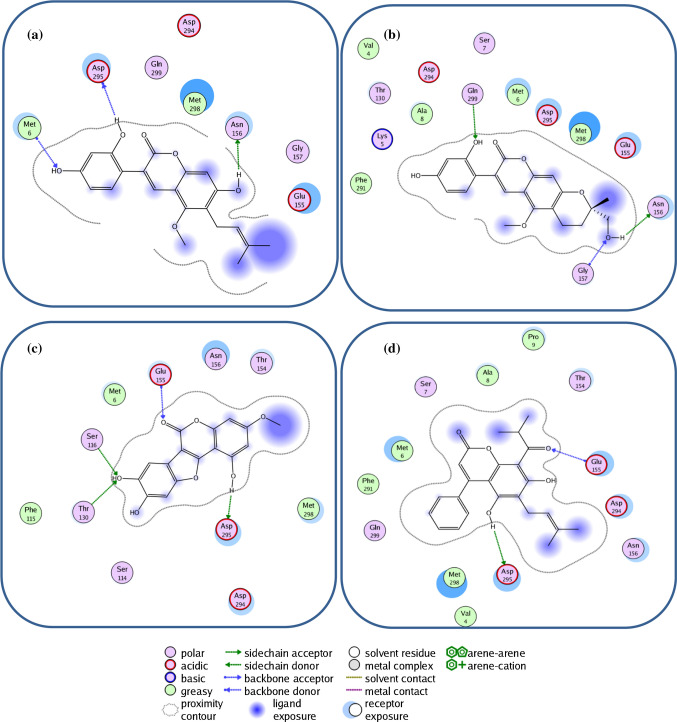
Target prediction
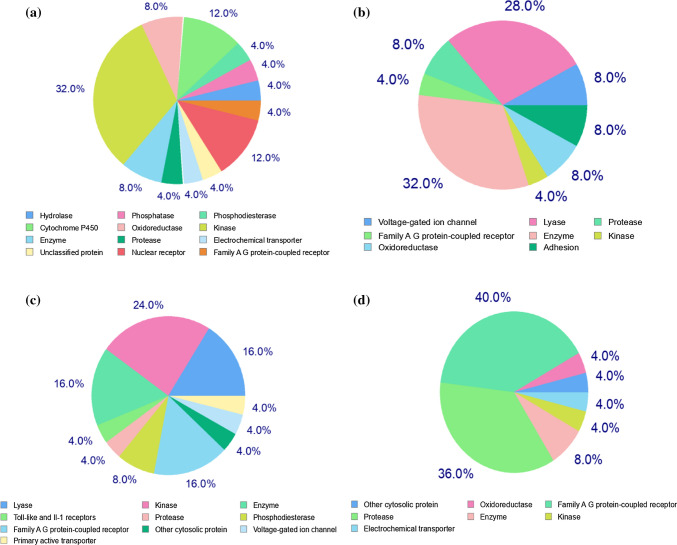
Molecular target studies are important to find the phenotypic side effects or potential cross reactivity caused by the action of glycycoumarin, oxypeucedanin hydrate, Inophyllum P, and mesuol as the best coumarin compounds. Figure 6 shows the % bioactivity of the top coumarins with respect to selected protein targets viz. enzymes, kinases, oxidoreductases, proteases, Family A G protein-coupled receptor, phosphodiesterases, and lyases as a pie-chart. Analysis revealed that proteases, Family A G protein-coupled receptors, and kinases were the main predicted targets for all the proposed compounds. The pie chart of the glycycoumarin predicted 32% of Kinase, 4% of Protease, 4% of Phosphatase, 4% of Hydrolase, 12% of Cytochrome P450, 8% of Enzyme (including: Estradiol 17-betadehydrogenase2, Dynamin-1, N-acylsphingosineamidohydrolase, Phospholipase A2, Aldo–keto reductase family 1 member B10), 8% of Oxidoreductase, 12% of Nuclear receptors, 4% of Electrochemical transporters, 4% of Phosphodiesterase, 4% of Unclassified protein and 4% of Family A G protein-coupled receptor. The possible sites of the target which the glycycoumarin may bind were Kinase, Cytochrome P450 and Nuclear receptors which stimulate the drug reaction accordingly. Also, this analysis presents an explanation for the use of glycycoumarin, oxypeucedanin hydrate, Inophyllum P, and Mesuol as SARS-CoV-2 main protease inhibitors.
Fig. 6.
Top-25 of target predicted for a Glycycoumarin, b Oxypeucedanin hydrate, c Inophyllum P and d Mesuol
Pharmacophore mapping
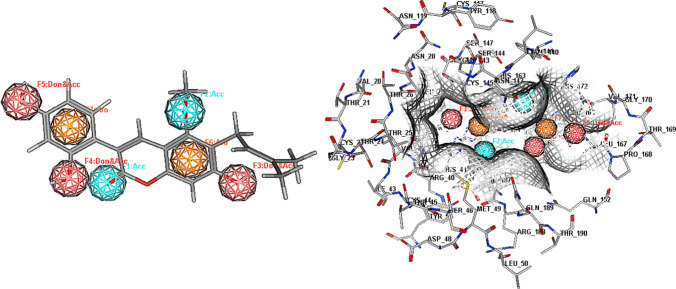
We implemented pharmacophore mapping for glycycoumarin that including two hydrogen-bond acceptor features, three hydrogen-bond donor and acceptor features and two aromatic ring features. Moreover, it created an acceptable number of significant contacts with 3CLpro pharmacophore (Fig. 7) and the features of the glycycoumarin compound were found to directly correspond to some key amino acids including His41, Gly143, Cys145, Asn142, Ser144, Glu166, Gln189, and His164, which play a critical role in 3CLpro inhibition activity. As shown in Fig. 7, the hydroxyl groups of the glycycoumarin that formed multiple direct hydrogen bond interactions with Asn142, His164 and Glu166 mapped the F3-F5 features. The methoxy group of the glycycoumarin showing a hydrogen bond interaction with Gln189 overlaid the F2 feature, while the carbonyl group that enabled considerable interactions with Cys145 and Ser144 mapped the F1 feature. Moreover, the benzene rings of the glycycoumarin that formed hydrophobic interactions with His41 and Phe140 mapped the F6-F7 features.
Fig. 7.
Pharmacophore Mapping of glycycoumarin in the binding site of 3CLpro. Cyan color-hydrogen bond acceptor, orange color-aromatic, dark pink color- hydrogen bond acceptor and donor
Molecular dynamics simulation study
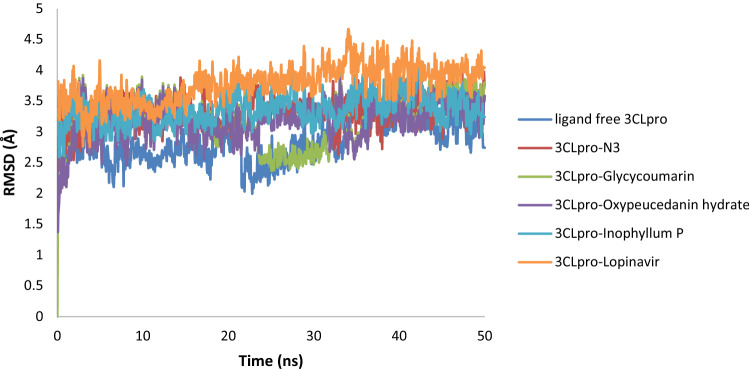
Molecular dynamics (MD) simulation is an imperative method to explore the conformational stability of virtual complexes and the contribution of key amino acid residues in ligand binding. The MD simulations for 3CLpro-glycycoumarin, 3CLpro-oxypeucedaninhydrate, and 3CLpro-Inophyllum P complexes along with that of three other systems (ligand free 3CLpro, 3CLpro-N3, and 3CLpro-lopinavir) were done for 50 ns to analyze the stability of these docked phytochemical compounds and evaluate the possible binding modes of the ligands. As depicted in Fig. 8, the backbone RMSD value of ligand free 3CLpro increased gradually until ~ 3.32 Å (0–5 ns), and then the RMSD value from 5 to 34 ns maintained a constant value (~ 2.77–2.88 Å). The value increased from 34 to 43 ns (~ 3.88–3.86 Å) and then decreased and reached 3.40 Å and remained almost the same till the end of the MD simulation. The RMSD value of the 3CLpro-N3 complex was ~ 3.22 Å at 22.50 ns, which rose to ~ 3.42 Å at 23.50 ns and persisted at the same value till 50 ns. The RMSD value for 3CLpro-lopinavir was found to remain almost constant (~ 3.84–4.04 Å) from 15 to 50 ns with some marginal fluctuations. The RMSD value of the 3CLpro-glycycoumarin complex increased from ~ 3.22 Å (at 2 ns) up to ~ 3.54 Å (at 22.50 ns). Then, within the next 10 ns, the value was decreased (~ 2.62 Å) and then, increased gradually until 3.65 Å and remained almost constant till the end of the MD run with some marginal fluctuations. For the 3CLpro-oxypeucedaninhydrate, the RMSD value increased gradually and reached to ~ 3.66 Å at 15 ns. Then, the RMSD value slightly decreased and persisted at ~ 3.20 Å from 18.30 ns till the end of the MD run. For 3CLpro-Inophyllum P, the RMSD value was found to remain almost constant (~ 3.28–3.46 Å) from 5.0 ns to 50.0 ns with some marginal fluctuations. The average RMSD values for ligand free 3CLpro, 3CLpro-N3 and 3CLpro-lopinavir systems were found to be 2.89 Å, 3.33 Å, and 3.78 Å, respectively, whereas the average RMSD values of 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes were 3.12 Å, 3.18 Å, and 3.37 Å, respectively. RMSD results showed that these three complexes were stable and the stability of 3CLpro-glycycoumarin was highest among all of them. Also, 3CLpro-coumarin complexes were more stable than the 3CLpro-lopinavir complex. In RMSD of 3CLpro with N3, lopinavir, glycycoumarin, oxypeucedanin hydrate, and Inophyllum P, the change was slightly more than ligand free 3CLpro that seemed that binding of the ligand with 3CLpro increased the conformational flexibility of 3CLpro.
Fig. 8.
RMSD plots of ligand free 3CLpro, 3CLpro-N3, 3CLpro-lopinavir, 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate, and 3CLpro-Inophyllum P complexes of SARS-CoV-2
The RMSF could be used to understand fluctuations as well as the flexibility of each residue in the different regions of simulated proteins. The average values of RMSF for ligand free 3CLpro, 3CLpro-N3, and 3CLpro-lopinavir were 1.85 Å, 1.80 Å, and 1.81 Å, respectively (Fig. 9). In ligand free 3CLpro system, most of the amino acid residues within the domain I and II of this system had RMSF fluctuation below 3.0 Å and only, residues 72, 155, and 168–170 showed higher fluctuations (up to 3.0 Å). The RMSF plot in Fig. 9 showed that 3CLpro-N3 and 3CLpro-lopinavir suffer more conformational fluctuations in domain III. The RMSF plot of the 3CLpro-N3 complex showed that very few amino acid residues within domain I, II, and III (residues 155, 215–217, 221–223, 274, and 278) have an RMSF value of more than 3.0 Å. The RMSF plot of 3CLpro-lopinavir showed more or less similar conformational fluctuations that of the ligand free 3CLpro system. The fluctuations for many amino acid residues of domain I and II were reduced upon the binding of lopinavir to 3Clpro. Also, the RMSF plot in Fig. 9 showed that 3CLpro-N3 and 3CLpro-lopinavir complexes suffer more conformational fluctuations in domain III. Analysis of RMSF plots showed that 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes had similar trends of dynamic fluctuation and RMSF distributions with average values of 1.78 Å, 1.78 Å and, 1.79 Å. These values indicated that 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes showed lower conformational fluctuation as compared to ligand free 3CLpro and 3CLpro-N3/lopinavir complexes. The fluctuation of various specific amino acid residues (Thr25, Thr26, His41, and Met49 in domain I; Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164, and Glu166 of domain II and Gln189 and Thr190 of domain III) in 3CLpro-coumarin complexes was less than that in ligand free 3CLpro and 3CLpro-N3/lopinavir complexes indicating that these residues within the binding pocket of 3CLpro protein interacted with these three coumarins while the loop regions and the C-terminal and N-terminal of the protein were largely fluctuating in all systems. His41 and Cys145 residues of the catalytic dyad in SARS-CoV-2 3CLpro showed RMSF of 1.48 Å and 1.30 Å, respectively. The His41 residue showed RMSF of 1.40 Å and 1.46 Å for 3CLpro-N3/lopinavir complexes, respectively, whereas in 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes His41 exhibited the RMSF of 1.28 Å, 1.31 Å, and 1.36 Å, respectively. The Cys145 residue exhibited the RMSF of 1.22 Å, 1.27 Å, 0.94 Å, 1.10 Å, and 1.19 Å for N3, lopinavir, glycycoumarin, oxypeucedanin hydrate, and Inophyllum P-protein systems, respectively, indicating the stability of the target protein with smaller conformational changes and the lower fluctuations in binding residues of catalytic dyad for all three 3CLpro-coumarin complexes than that of the 3CLpro-N3/lopinavir complex.
Fig. 9.
RMSF plot of ligand free 3CLpro and the 3CLpro-ligand complexes of SARS-CoV-2
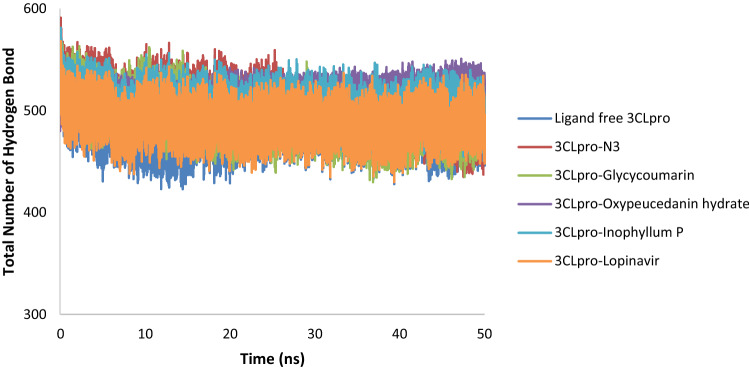
RMSD and RMSF analysis revealed that the SARS-CoV-2 3CLpro-glycycoumarin docking complex was highly stable during 50 ns simulations. Hydrogen bonding plays a significant role in determining the stability of a ligand–protein complex. The average numbers of intermolecular hydrogen bonds for ligand free 3CLpro, 3CLpro-N3, and 3CLpro-lopinavir were 481, 503, and 489, respectively, whereas the average numbers of intermolecular hydrogen bonds for 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate, and 3CLpro-Inophyllum P complexes were found to be 494, 516, and 502, respectively (Fig. 10). The highest number of hydrogen bonds was observed for 3CLpro-oxypeucedanin hydrate, while the lowest number of hydrogen bonds was observed in ligand free over the 50 ns simulations. 3CLpro-ligand complexes possessed a greater number of hydrogen bonds compared to ligand free 3CLpro which these hydrogen bonds stabilized the protein–ligand complexes during simulation.
Fig. 10.
Total number of H-bond count throughout the simulation for ligand free 3CLpro and the 3CLpro-ligand complexes of SARS-CoV-2
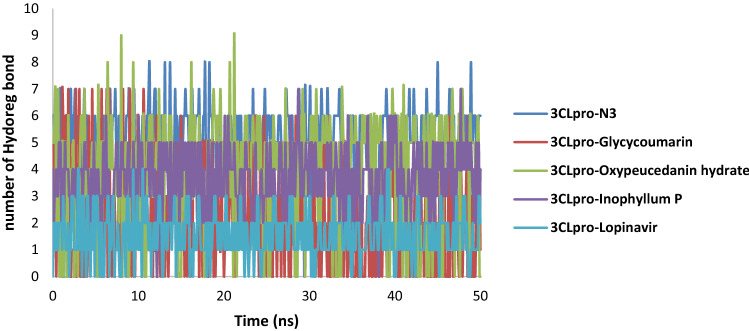
Analysis of the main protease-ligand complexes revealed most of the compounds form Hydrogen bonds with the amino acid residues of the binding pocket (Fig. 11). In the 3CLpro-N3 complex, the majority of conformations formed up to 3 hydrogen bonds during the MD simulation and a small number of conformations exhibited less than 1 and greater than 6 hydrogen bonds. For the 3CLpro-lopinavir complex, lopinavir formed 1 to 2 hydrogen bonds with residues of the binding pocket. In 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes, the number of hydrogen bonds formed was between 2 and 5 in the whole simulation while 3CLpro-glycycoumarin complex showed changes in bonding. More hydrogen bonds (> 5) were between 0 and 13 ns, after 13 ns the hydrogen bonds decreased to less than 5, and the last 15 ns, the hydrogen bond was between 2 and 4. This might suggest that there was a conformational change around glycycoumarin in the binding site during simulation. Overall, the results showed that all three 3CLpro-coumarin complexes were highly stable.
Fig. 11.
Number of intermolecular hydrogen bonds between 3CLpro of SARS-CoV-2 and N3, glycycoumarin, oxypeucedanin hydrate, Inophyllum P and lopinavir
The radius of gyration (Rg) parameter is used to describe the compactness and rigidity of the ligand–protein complex during MD simulation, in which less compactness (more unfolded) depicts a higher Rg value with conformational entropy, while low Rg values explain strong compactness and higher structural stiffness (more folded). As shown in Fig. 12, average Rg values of 3CLpro-N3 complex (21.13 Å) and 3CLpro-lopinavir complex (21.18 Å) were found to be in a similar range with ligand free 3CLpro (21.14 Å). The average Rg value for 3CLpro-glycycoumarin (21.03 Å), 3CLpro-oxypeucedanin hydrate (21.09 Å), and 3Clpro-Inophyllum P (21.13 Å) systems was slightly lower than that of the other three systems (ligand free 3CLpro, 3CLpro-N3, and 3CLpro-lopinavir). In an argument with the above observation, these molecules did not induce structural changes and were relatively more rigid than the N3, lopinavir, and ligand free 3CLpro and all three 3CLpro-coumarin complexes were compact throughout the simulation, indicating that the complexes were well converged.
Fig. 12.
Radius of gyration (Rg) plot ligand free 3CLpro and the 3CLpro-ligand complexes of SARS-CoV-2
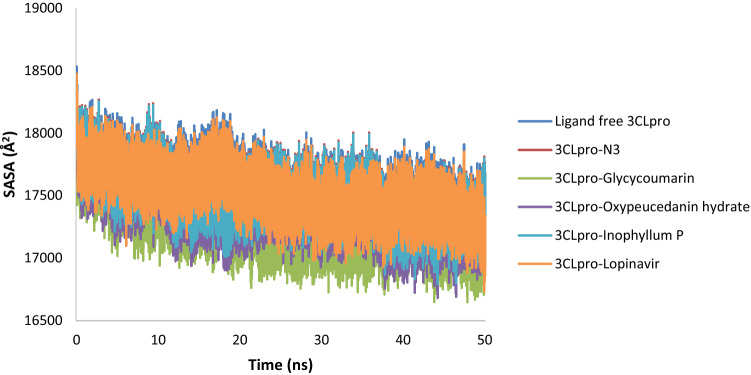
Solvent Accessible Surface Area (SASA) value indicates the degree of expansion of protein volume in each system over the simulation time. The average SASA values of the 3CLpro-N3 complex (~ 17,501.49 Å2) and the 3CLpro-lopinavir complex (~ 17,578.51 Å2) were higher than the three 3CLpro-coumarin complexes suggesting an expansion of 3CLpro during the interaction with N3 and lopinavir. The average SASA values of 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate, and 3CLpro-Inophyllum P complexes were 17,264.84 Å2, 17,377.37 Å2, and 17,487.35 Å2, respectively. These values indicated that all three 3CLpro-coumarin complexes were slightly lower than that of the ligand free 3CLpro (17,578.51 Å2) and 3Clpro-N3/lopinavir (Fig. 13), suggesting that the binding of glycycoumarin, oxypeucedanin hydrate, and Inophyllum P potentially could reduce 3CLpro protein expansion.
Fig. 13.
Solvent accessible surface area (SASA) plot ligand free 3CLpro and the 3CLpro-ligand complexes of SARS-CoV-2
MM-PBSA binding free energy calculation
The binding free energy of the ligand–protein complex was performed to revalidate the ligand affinity to the target receptor for the ligand–protein complex predicted by the molecular docking studies. The MM-PBSA free energy values of the 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate and 3CLpro-Inophyllum P complexes, as well as 3CLpro-N3 and 3CLpro-lopinavir, were calculated from 50 ns trajectories corresponding to every 5 ns time interval. Each energy term, including van der Waals energy, electrostatic energy, polar solvation energy, solvent accessible surface area (SASA) energy, and total binding free energy of the systems was given in Table 6. The calculated ΔG binding energy values of 3CLpro-N3 and 3CLpro-lopinavir complexes were found to be -56.25 kJ/mol and -40.94 kJ/mol (Table 6). On the contrary, the binding free energy values of 3CLpro-glycycoumarin, 3CLpro-oxypeucedanin hydrate, and 3CLpro-Inophyllum P complexes were − 60.31 kJ/mol, -58.86 kJ/mol and − 57.75 kJ/mol and these negative values of ΔG binding energy indicated that the selected compounds favorably interact with the target protein of 3CLpro. Among all the 3CLpro-coumarin complexes, the 3CLpro-glycycoumarin complex exhibited the highest binding free energy, while the 3CLpro-Inophyllum P complex showed the lowest binding free energy. According to the results of Table 6, the major favorable contributors were van der Waals (ΔEvdW) and electrostatic (ΔEelec) interactions and SASA energy while the polar component of solvation (ΔG polar) contributed unfavorably to the binding of glycycoumarin, oxypeucedanin hydrate, and Inophyllum P to 3CLpro and the selected coumarin phytochemicals may inhibit the SARS-CoV-2 main protease.
Table 6.
Binding free energy for glycycoumarin, oxypeucedanin hydrate and Inophyllum P and 3CLpro of SARS-CoV-2 calculated by MM-PBSA analysis
| Complex | Van der Waal energy (ΔEvdW) (Kj/mol) | Electrostatic energy (ΔEelec) (Kj/mol) | Polar solvation energy (ΔG polar) (Kj/mol) | SASA energy (Kj/mol) | Binding energy (Kj/mol) |
|---|---|---|---|---|---|
| 3CLpro-N3 | − 51.67 | − 45.39 | 54.58 | − 13.77 | − 56.25 |
| 3CLpro-Lopinavir | − 33.24 | − 38.83 | 43.76 | − 12.63 | − 40.94 |
| 3CLpro-glycycoumarin | − 44.16 | − 58.58 | 58.59 | − 16.15 | − 60.31 |
| 3CLpro-Oxypeucedanin hydrate | − 48.97 | − 53.79 | 63.13 | − 19.23 | − 58.86 |
| 3CLpro-Inophyllum P | − 53.48 | − 45.78 | 56.49 | − 14.97 | − 57.75 |
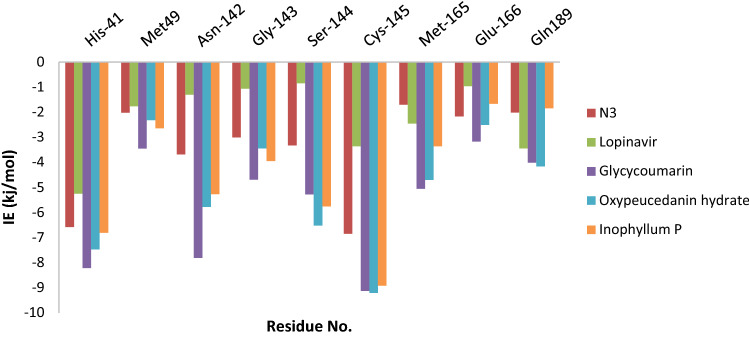
For determining the key residues involved in the ligand activities as well as understanding the interactions of the ligand with the 3CLpro protein residues, total binding free energy decomposed into the contribution energy of diverse residues at the active site of 3CLpro protein with all the five ligands has been computed and showed that the most contributive residues were Met49, His41, Gly143, Asn142, Cys145, Ser144, Glu166, Gln189, and Met165. Figure 14 depicts the respective energy contribution. These findings agree with, and mutually support, previously reported results of the main interacting residues within the 3CLpro active site that are deemed critical for effective ligand binding [59, 60].
Fig. 14.
binding free energies of the residues which have considerable interactions with N3, Lopinavir, glycycoumarin, Oxypeucedanin hydrate and Inophyllum P
The results indicated that catalytic dyad (His41 and Cys145) in the 3CLpro-coumarin complexes had a major energy contribution in binding affinity of 3CLpro compared to that of the 3CLpro-N3/lopinavir complexes. From Fig. 14, it was found that interactions of Cys 145 (− 9.18, − 9.21 and − 8.92 kJ/mol) by hydrogen bonds with glycycoumarin, oxypeucedanin hydrate, and Inophyllum P compounds had the biggest energy contribution to these complexes; and Asn142 (− 7.81, − 5.77 and − 5.27 kJ/mol) was also found to be one of the most important residues for activity by hydrogen bonds with glycycoumarin, oxypeucedanin hydrate, and Inophyllum P compounds, respectively. Whereas the binding free energy values of Cys145 and Asn142 residues in the 3CLpro-N3 complex were − 6.85 kJ/mol and − 3.68 kJ/mol and for 3CLpro-lopinavir complex were found to be − 3.36 kJ/mol and − 1.30 kJ/mol, respectively. Ser144 residue made a significant contribution with glycycoumarin, oxypeucedanin hydrate, and Inophyllum P through hydrogen bond with − 5.28 kJ/mol, − 6.52 kJ/mol and − 5.76 kJ/mol values of the binding free energy. Also, Gly143, Glu166, and Gln189 contributed significantly to the interaction energy with glycycoumarin, oxypeucedanin hydrate, and Inophyllum P by hydrogen bond formation with these compounds; hence the contributions of Gly143, Glu166 and Gln189 were advantageous for affinity binding. His41, Met49, and Met165 could have hydrophobic interactions as well as π − π stacking with the selected coumarin phytochemicals and make significant positive contributions to the binding of ligands with 3CLpro. In addition to the catalytic dyad, key residues of Asn142, Gly143, Glu166, and Gln189 favorably contribute to the binding affinity and verify the reliability of the molecular docking results.
According to the simulation results, the initial docked structure and the ultimate structure of the 3CLpro-glycycoumarin complex had been in a similar binding pocket and showed that ligand–protein conformation was stable after the simulation and docking results of the glycycoumarin with 3CLpro was reliable. Figure 15 presents the structure superposition of the complex following simulation. With regard to the 3D conformation results of glycycoumarin, the interaction of a majority of the residues (Met 49, His41, Leu141, Phe140, Ser144, Asn142, Cys145, Gly143, Met165, His163, Gln189, and Glu166) and glycycoumarin in the initial docked and ultimate 3CLpro-glycycoumarin complex after 50 ns simulations did not change. However, glycycoumarin form hydrogen bonds with the residues Cys145, Gln189 and Ser144 as well as novel hydrogen bonds with Thr25, Glu166 and Asn142. Notably, the substituted coumarin area of the glycycoumarin could create hydrophobic interactions with the Phe140, Met41, Met165, and Leu141 residues. Hence, such a binding interaction would be beneficial to the coumarin compounds' stability in the binding pocket of 3CLpro protein. Also, for investigation of the system condition during simulation, the 3CLpro-glycycoumarin structure was extracted from trajectories for every 10 ns (Fig. S6). These snapshots proved the fixed orientation of glycycoumarin at the active site of the 3CLpro throughout the simulation.
Fig. 15.
Superposition of molecular docking result and MD structure of compound glycycoumarin with 3CLpro after 50 ns simulations. The residues of active site (pink), docked glycycoumarin (dark cyan) and MD glycycoumarin (olive drab)
Due to their natural origin, coumarin phytochemical enjoys several benefits such as potentially less side effects, lower toxicity, and issues regarding the administration of a majority of the synthetic and semi-synthetic medicines. According to the present research, glycycoumarin, oxypeucedanin hydrate, and Inophyllum P compounds in addition to being of natural origin, drug-likely and particularly, having antiviral properties, also displayed comparable binding energy values with that of N3 and lopinavir. Therefore, further experimental investigations are suggested to explore probable preclinical and clinical efficiency of the phytochemicals like glycycoumarin oxypeucedanin hydrate and Inophyllum P to inhibit protease protein and treat COVID-19.
Future perspective
The potential for the emergence of novel CoVs and the mutating nature of CoVs in the future, make the development of broad spectrum of the antivirals necessary. As future perspectives, research should aim at the development of protease inhibitor antiviral compounds, which play a crucial role in the fusion of the virus to the host cell membrane, suppressing the entry of the virus. Also, based on of these studies, future research should be conducted on the application of already existing antiviral drugs, and plant-based traditional medicines on SARS-CoV-2 infected patients to find out if the expected benefits can be seen in the treatment process. For this purpose, randomized controlled trials should be carried out to obtain more accurate results. In addition, production of new vaccines and virus neutralizing antibodies to target the proposed viral molecular structures should be considered.
Conclusion
3-Chymotrypsin-like main protease (3CLpro) is an attractive target for the inhibition of the viral replication cycle and the treatment of SARS-CoV-2 infections. The aim of this study was to investigate the antiviral potential of a set of coumarin phytochemical compounds against coronavirus 3CLpro using in silico approaches. These inhibitors could inhibit the 3CLpro with a highly conserved inhibitory effect to both SARS-CoV2 and SARS-CoV. Among the studied 50 coumarin phytochemicals, glycycoumarin, Inophyllum P, mesuol and oxypeucedanin hydrate displayed the highest binding affinity with the best negative energy scores and interacted with one or both of the catalytic residues (His41 and Cys145) of 3CLpro through hydrophilic and hydrophobic bonding. MD results revealed that glycycoumarin, oxypeucedanin hydrate and Inophyllum P compounds are stable within the active site of 3CLpro of SARS-CoV-2 with significant binding free energies of − 60.31 kJ/mol, − 58.86 kJ/mol, and − 57.75 kJ/mol and also, the pharmacokinetics and ADMET evaluation indicate their efficiency as drug molecules. Based on these findings, most of the coumarin phytochemicals can be used to design effective antiviral drugs against SARS-CoV-2.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Clinical Biochemistry Research Center, Basic Health Sciences Institute; Shahrekord University of Medical Sciences for financial support of this research. The study was supported financially by the grant (No. 3364) provided by Shahrekord University of Medical Sciences and also was approved by the Ethics Committee of Shahrekord University of Medical Sciences (IR.SKUMS.REC.1399.117).
Abbreviations
- COVID-19
Coronavirus disease 2019 pneumonia
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- MERS-CoV
Middle East respiratory syndrome coronavirus
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- WHO
World Health Organization
- 3CLpro
3-Chymotrypsin-like protease
- Mpro
Main protease enzyme
- PLpro
Papain-like protease
- HIV
Human immunodeficiency virus
- MD
Molecular dynamics
- PDB
Protein data bank
- GROMACS
GROningen MaChine for chemical simulations
- RMSD
Root-mean-square deviation
- RMSF
Root-mean-square fluctuation
- Rg
Radius of gyration
- SASA
Solvent accessible surface area
- SPC
Simple point charge
- MM-PBSA
Molecular Mechanic/Poisson–Boltzmann surface Area
Declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any study with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chatterjee S, Maity A, Chowdhury S, Islam MA, Muttinini RK, Sen D. In silico analysis and identification of promising hits against novel coronavirus 3C-like main protease enzyme. J Biomol Struct Dyn. 2020;2020:1–14. doi: 10.1080/07391102.2020.1787228. [DOI] [PubMed] [Google Scholar]
- 2.Parvez M, Alam S, Karim M, Hasan M, Jaman J, Karim Z, Tahsin T, Hasan M, Hosen MJ. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int J Biol Macromole. 2020;163:1787–1797. doi: 10.1016/j.ijbiomac.2020.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-Ldrissi M, Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz DA, Graham AL. Potential maternal and infant outcomes from (Wuhan) coronavirus 2019-nCoV infecting pregnant women: lessons from SARS, MERS, and other human coronavirus infections. Viruses. 2020;12(2):194. doi: 10.3390/v12020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui DS. Epidemic and emerging coronaviruses (severe acute respiratory syndrome and Middle East respiratory syndrome) Clin Chest Med. 2017;38(1):71–86. doi: 10.1016/j.ccm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallingal A, Thachan Kundil V, Ayyolath A, Karlapudi AP, Muringayil Joseph T. Molecular modeling study of tectoquinone and acteoside from Tectona grandis linn: a new SARS-CoV-2 main protease inhibitor against COVID-19. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1832580. [DOI] [PubMed] [Google Scholar]
- 8.Kodchakorn V, Poovorawan Y, Suwannakarn K, Kongtawelert P. Molecular modelling investigation for drugs and nutraceuticals against protease of SARS-CoV-2. J Mol Graph Model. 2020 doi: 10.1016/j.jmgm.2020.107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Rohaimi AH, Al Otaibi F. Novel SARS-CoV-2 outbreak and COVID19 disease; a systemic review on the global pandemic. Genes Dis. 2020 doi: 10.1016/j.gendis.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, Megawati D, Hayati Z, Wagner AL, Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Pub Health. 2020 doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 13.Lechien JR, Chiesa-Estomba CM, Place S, Van Laethem Y, Cabaraux P, Mat Q, Huet K, Plzak J, Horoi M, Hans S. Clinical and epidemiological characteristics of 1420 European patients with mild-to-moderate coronavirus disease 2019. J Intern Med. 2020 doi: 10.1111/joim.13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam R, Parves MR, Paul AS, Uddin N, Rahman MS, Mamun AA, Hossain MN, Ali MA, Halim MA. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan A, Ali SS, Khan MT, Saleem S, Ali A, Suleman M, Babar Z, Shafiq A, Khan M, Wei D-Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro) J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1779128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Identification of alkaloids from Justicia adhatoda as potent SARS CoV-2 main protease inhibitors: An in silico perspective. J Mol Struct. 2020 doi: 10.1016/j.molstruc.2020.129489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra S, Pandey A, Manvati S. Coumarin: an emerging antiviral agent. Heliyon. 2020;6(1):e03217. doi: 10.1016/j.heliyon.2020.e03217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan MZ, Osman H, Ali MA, Ahsan MJ. Therapeutic potential of coumarins as antiviral agents. Eur J Med Chem. 2016;123:236–255. doi: 10.1016/j.ejmech.2016.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, Zhe-Ling F, Yi-Tao W, Li-Gen L. Anticancer carbazole alkaloids and coumarins from Clausena plants: a review. Chin J Nat Med. 2017;15(12):881–888. doi: 10.1016/S1875-5364(18)30003-7. [DOI] [PubMed] [Google Scholar]
- 22.Gonçalves GA, Spillere AR, Das Neves GM, Kagami LP, Von Poser GL, Canto RFS, Eifler-Lima VL. Natural and synthetic coumarins as antileishmanial agents: a review. Eur. J. Med. Chem. 2020;203:112514. doi: 10.1016/j.ejmech.2020.112514. [DOI] [PubMed] [Google Scholar]
- 23.Witaicenis A, Seito LN, da Silveira Chagas A, de Almeida Junior LD, Luchini AC, Rodrigues-Orsi P, Cestari SH, Di Stasi LC. Antioxidant and intestinal anti-inflammatory effects of plant-derived coumarin derivatives. Phytomedicine. 2014;21(3):240–246. doi: 10.1016/j.phymed.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Zhou P, Takaishi Y, Duan H, Chen B, Honda G, Itoh M, Takeda Y, Kodzhimatov OK, Lee K-H. Coumarins and bicoumarin from Ferula sumbul: anti-HIV activity and inhibition of cytokine release. Phytochemistry. 2000;53(6):689–697. doi: 10.1016/S0031-9422(99)00554-3. [DOI] [PubMed] [Google Scholar]
- 25.Venugopala KN, Rashmi V, Odhav B. Review on natural coumarin lead compounds for their pharmacological activity. Biomed Res Int. 2013 doi: 10.1155/2013/963248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shikishima Y, Takaishi Y, Honda G, Ito M, Takeda Y, Kodzhimatov OK, Ashurmetov O, LEE KH. Chemical constituents of Prangos tschimganica; structure elucidation and absolute configuration of coumarin and furanocoumarin derivatives with anti-HIV activity. Chem. Pharm. Bull. 2001;49(7):877–880. doi: 10.1248/cpb.49.877. [DOI] [PubMed] [Google Scholar]
- 27.Márquez N, Sancho R, Bedoya LM, Alcamí J, López-Pérez JL, San Feliciano A, Fiebich BL, Muñoz E. a natural occurring 4-phenylcoumarin, inhibits HIV-1 replication by targeting the NF-κB pathway. Antivir. Res. 2005;66(2–3):137–145. doi: 10.1016/j.antiviral.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Xu B, Wang L, González-Molleda L, Wang Y, Xu J, Yuan Y. Antiviral activity of (+)-rutamarin against Kaposi's sarcoma-associated herpesvirus by inhibition of the catalytic activity of human topoisomerase II. Antimicrob Agents Chemother. 2014;58(1):563–573. doi: 10.1128/AAC.01259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adianti M, Aoki C, Komoto M, Deng L, Shoji I, Wahyuni TS, Lusida MI, Fuchino H, Kawahara N, Hotta H. Anti-hepatitis C virus compounds obtained from Glycyrrhiza uralensis and other Glycyrrhiza species. Microbiol Immunol. 2014;58(3):180–187. doi: 10.1111/1348-0421.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez RW. Antiviral activity of compounds isolated from plants. Pharm Biol. 2003;41(2):107–157. doi: 10.1076/phbi.41.2.107.14240. [DOI] [Google Scholar]
- 31.Chen Y, Yiu CPB, Wong K-Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: Virtual screening reveals velpatasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Said MA, Albohy A, m.A. abdelrahman, H.S. Ibrahim, Important of glutamine 189 flexibility in SARS-CoV-2 main protease: lesson learned from in silico virtual screening of ChEMBL database and molecular dynamicd. Eur. J. Pharm. Sci. 2021;160:105744. doi: 10.1016/j.ejps.2021.105744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaki AA, Al-Karmalawy AA, El-Amierc YA, Ashour A. Molecular docking reveals the potential of Cleome amblyocarpa isolated compounds to inhibit COVID-19 virus main protease. New J Chem. 2020;44:16752–16758. doi: 10.1039/D0NJ03611K. [DOI] [Google Scholar]
- 34.Shah S, Chaple D, Arora S, Yende S, Mehta C, Nayak U. Prospecting for cressa cretica to treat COVID-19 via in silico molecular docking models of the SARSCoV-2. J Biomol Struct Dy. 2020 doi: 10.1080/07391102.2021.1872419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 36.Veber DF, Johnson SR, Cheng H-Y, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 37.Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 38.Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem. 2001;44(12):1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]
- 39.Enmozhi SK, Raja K, Sebastine I, Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasanth D, Murahari M, Chandramohan V, Panda SP, Atmakuri LR, Guntupalli C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 42.Ibrahim MAA, Abdelrahman AHM, Hussien T, Badr EAA, Mohamed TA, El-Seedi HR, Pare PW, Efferth T, Hegazy MEF. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitor. Comput Biol Med. 2020 doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar V, Dhanjal JK, Kaul SC, Wadhwa R, Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schüttelkopf AW, Van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Cryst D: Biol Crystal. 2004;60(8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 45.Pant S, Singh M, Ravichandiran V, Murty U, Srivastava HK. Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elfiky AA, Azzam EB. Novel Guanosine Derivatives against MERS CoV polymerase: an in silico perspective. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umesh D, Kundu C, Selvaraj SK, Singh V.K. D. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1763202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aldeghi M, Bodkin MJ, Knapp S, Biggin PC. Statistical analysis on the performance of Molecular Mechanics Poisson-Boltzmann Surface Area versus absolute binding free energy calculations: bromodomains as a case study. J Chem Inf Model. 2017;57(9):2203–2221. doi: 10.1021/acs.jcim.7b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. Kumari, R. Kumar, O.S.D.D. Consortium, A. Lynn, g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations, J. Chem. Inf. Model. 54(7) (2014) 1951-1962. https://doi.org/10.1021/ci500020m [DOI] [PubMed]
- 51.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Sayed KA. Natural products as antiviral agents studies in natural products chemistry. Amsterdam: Elsevier; 2000. [Google Scholar]
- 53.Reutrakul V, Leewanich P, Tuchinda P, Pohmakotr M, Jaipetch T, Sophasan S, Santisuk T. Cytotoxic coumarins from Mammea harmandii. Planta Med. 2003;69(11):1048–1051. doi: 10.1055/s-2003-45154. [DOI] [PubMed] [Google Scholar]
- 54.Shikishima Y, Takaishi Y, Honda G, Ito M, Takfda Y, Kodzhimotov O, Ashurmetov O, Lee K. Chemical constituents of prangos tschimganica; structure elucidation and absolute configuration of coumarin and furano coumarin derivatives with anti-HIV activity. Chem Pharm Bull. 2001;49:877–880. doi: 10.1248/cpb.49.877. [DOI] [PubMed] [Google Scholar]
- 55.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Nour MY, Ibrahim MM, Elsaman T. Ellagic acid, kaempferol, and quercetin from acacia nilotica: promising combined drug with multiple mechanisms of action. Curr Pharmacol Rep. 2019;5:255–280. doi: 10.1007/s40495-019-00181-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chinnasamy P, Arumugam R. In silico prediction of anticarcinogenic bioactivities of traditional anti-inflammatory plants used by tribal healers in Sathyamangalam wildlife Sanctuary, India. Egypt J Basic Appl Sci. 2018;5(4):265–279. doi: 10.1016/j.ejbas.2018.10.002. [DOI] [Google Scholar]
- 58.Sethi P, Bansal Y, Bansal G. Synthesis and PASS-assisted evaluation of coumarin-benzimidazole derivatives as potential anti-inflammatory and anthelmintic agents. Med Chem Res. 2018;27:61–71. doi: 10.1007/s00044-017-2036-1. [DOI] [Google Scholar]
- 59.Al-SharI NA. Tackling COVID-19: identification of potential main protease inhibitors via structural analysis, virtual screening, molecular docking and MM-PBSA calculations. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1800514. [DOI] [PubMed] [Google Scholar]
- 60.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J Chem Inf Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.