Abstract
The goals of PhenX (consensus measures for Phenotypes and eXposures) are to promote the use of standard measurement protocols and to help investigators identify opportunities for collaborative research and cross‐study analysis, thus increasing the impact of individual studies. The PhenX Toolkit (https://www.phenxtoolkit.org/) offers high‐quality, well‐established measurement protocols to assess phenotypes and exposures in studies with human participants. The Toolkit contains protocols representing 29 research domains and 6 specialty collections of protocols that add depth to the Toolkit in specific research areas (e.g., COVID‐19, Social Determinants of Health [SDoH], Blood Sciences Research [BSR], Mental Health Research [MHR], Tobacco Regulatory Research [TRR], and Substance Abuse and Addiction [SAA]). Protocols are recommended for inclusion in the PhenX Toolkit by Working Groups of domain experts using a consensus process that includes input from the scientific community. For each PhenX protocol, the Toolkit provides a detailed description, the rationale for inclusion, and supporting documentation. Users can browse protocols in the Toolkit, search the Toolkit using keywords, or use Browse Protocols Tree to identify protocols of interest. The PhenX Toolkit provides data dictionaries compatible with the database of Genotypes and Phenotypes (dbGaP), Research Electronic Data Capture (REDCap) data submission compatibility, and data collection worksheets to help investigators incorporate PhenX protocols into their study design. The PhenX Toolkit provides resources to help users identify published studies that used PhenX protocols. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol: Using the PhenX Toolkit to support or extend study design
Keywords: data collection protocols, environmental exposures, genome‐wide association studies (GWAS), phenotypes, standard measures
INTRODUCTION
The PhenX Toolkit (https://www.phenxtoolkit.org/) is a catalog of recommended measurement protocols intended to help standardize data collection. In the PhenX Toolkit, a “protocol” is the methodology used to collect the data to address a specific concept or measure. PhenX protocols address a wide range of research domains and are suitable for use in genomic, biomedical, clinical, epidemiologic, and translational research studies (Stover, Harlan, Hammond, Hendershot, & Hamilton, 2010). The Toolkit provides detailed protocols for data collection as well as tools to help investigators incorporate these protocols into their research studies. Use of PhenX protocols facilitates collaboration and cross‐study analyses, potentially increasing the scientific impact of individual studies (Hamilton et al., 2011). The PhenX Toolkit is a freely available resource that is intentionally created through a consensus process. PhenX is driven by the scientific community, including oversight by a Steering Committee, recommendations of Working Group (WG) experts, and feedback from the broader scientific community (Maiese et al., 2013). The PhenX Steering Committee established the criteria for WGs to consider as they decide which protocols to recommend for inclusion in the Toolkit (Table 1). PhenX WGs use these guidelines as they review and select well‐established, broadly validated protocols. The PhenX Toolkit currently includes 29 research domains (Table 2) and 6 specialty collections (Table 3) that together include 874 protocols. Research domains add breadth to the Toolkit, whereas specialty collections add depth to the Toolkit in specific areas of research (Figs. 1 and 2). Several National Institutes of Health (NIH) institutes have issued guide notices announcing the availability of PhenX protocols for specific areas of research. Social Determinants of Health (SDoH) and COVID‐19 specialty collections have recently been added to the Toolkit.
Table 1.
Criteria for Selecting Measurement Protocols for the PhenX Toolkit a
|
|
|
|
|
|
|
|
|
| Additional criteria |
|
|
|
|
|
Note: The PhenX Steering Committee defined criteria to guide the selection of the measurement protocols to be included in the PhenX Toolkit.
Table 2.
The 29 PhenX Research Domains a
| Alcohol, tobacco and other substances (19) |
| Anthropometrics (31) |
| Cancer (12) |
| Cancer outcomes and survivorship (15) |
| Cardiovascular (14) |
| Demographics (16) |
| Diabetes (16) |
| Environmental exposures (15) |
| Gastrointestinal (15) |
| Genomic medicine implementation (15) |
| Geriatrics (16) |
| Infectious diseases and immunity (17) |
| Neurology (33) |
| Nutrition and dietary supplements (15) |
| Obesity (14) |
| Ocular (18) |
| Oral health (15) |
| Pediatric development (18) |
| Physical activity and physical fitness (19) |
| Pregnancy (21) |
| Psychiatric (31) |
| Psychosocial (25) |
| Rare genetic conditions (20) |
| Reproductive health (24) |
| Respiratory (21) |
| Skin, bone, muscle, and joint (10) |
| Smoking cessation, harm reduction, and biomarkers (11) |
| Social environments (19) |
| Speech, language, and hearing (23) |
Note: The number in parentheses is the number of protocols in the domain.
Table 3.
PhenX Specialty Collections a
| Blood sciences research |
|
|
|
| COVID‐19 |
|
|
|
|
|
|
| Mental health research |
|
|
|
|
|
| Social determinants of health |
|
|
|
| Substance abuse and addiction |
|
|
|
|
|
|
| Tobacco regulatory research |
|
|
|
|
|
Note: The number in parentheses is the number of protocols in the collection.
Figure 1.
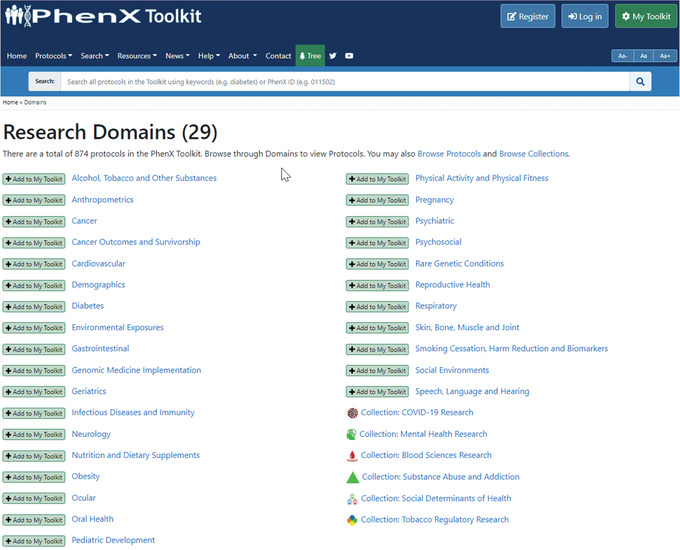
PhenX research domains and collections.
Figure 2.
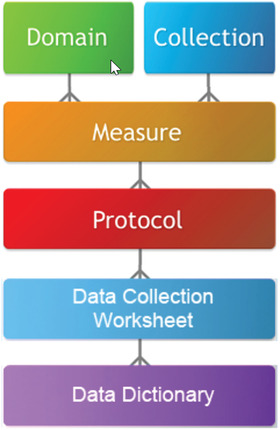
A PhenX domain consists of quantitative and qualitative measures with a unifying theme, whereas collections consist of measures with a shared characteristic, target population, or topic. Measures included in a collection may cut across multiple research domains.
When designing a new study or expanding an existing study, investigators may visit the Toolkit to search, browse, review, and select PhenX protocols to include in their study. Using PhenX protocols can help investigators ensure that their study data will be compatible with data from other studies that implement these standard data collection methods (Phillips et al., 2017). Knowing that PhenX protocols are recommended by experts, investigators can be confident that they are adding high‐quality protocols to their studies even when those protocols are from research topics outside their realm of expertise.
The Basic Protocol provides step‐by‐step guidance on using the PhenX Toolkit to identify and incorporate standard phenotype and environmental exposure protocols into a study (Hamilton et al., 2011). The Basic Protocol walks through browsing and searching the Toolkit for measurement protocols, reviewing text of and additional information about identified protocols, and using Toolkit features and resources to download protocols for inclusion in a study.
USING THE PhenX TOOLKIT TO SUPPORT OR EXTEND STUDY DESIGN
The PhenX Toolkit is a web‐based resource that makes it easy for researchers to identify standard data collection measurement protocols to include in their studies. The value of standard measures is widely recognized (Bennett et al., 2011) because they facilitate cross‐study analysis, allow studies to be combined to increase statistical power, and allow comparisons between studies to facilitate validation of results. Without standard protocols, investigators must harmonize data collected using different but conceptually related methodologies, which can be a time‐consuming and difficult process.
Materials
An up‐to‐date Web browser, such as Google Chrome, Microsoft Edge, Safari, and Firefox, is required for this protocol
Example scenario
An investigator with expertise in diabetes is designing a new research study. She has several diabetes protocols in her study design and would like to consider expanding the scope of her study to include protocols related to cardiovascular health, environmental health, and SDoH. She is unsure of the recommended standard data to collect and collection methods to follow for a few of these measures and visits the PhenX Toolkit to identify recommended protocols for phenotypes and exposures that could be included in her new study. She navigates the Toolkit to consider protocols that extend beyond her area of expertise to add to the study design. Additionally, the investigator knows that data collected using PhenX protocols can be readily compared with or combined with data from other studies that have also used the same PhenX protocols.
The investigator follows these steps to explore and identify protocols in the PhenX Toolkit:
-
1Navigate to the PhenX Toolkit website at https://www.phenxtoolkit.org/.
- The investigator notes several ways to access information from the home page, including a search bar and buttons such as “Research Domains” and “Browse Protocol Tree” (Fig. 3).
To start, the investigator decides to use the search bar on the home page.
Figure 3.

The PhenX Toolkit home page: https://www.phenxtoolkit.org/.
-
2Search the Toolkit for “diabetes” using the search bar.
- The search bar allows users to type in words, phrases, or topics of interest. For this study, the investigator is going to search the term “diabetes.”
- This search returns 27 protocols. (Fig. 4 displays the search results.)
- The investigator already plans to include a few bioassays in her study and is curious to see what bioassays are included in the Toolkit. Using the filters on the left under “Refine by:”, she selects “Bioassay” under “Data Collection Mode” (Fig. 5).
- The investigator sees “Oral Glucose Tolerance Test” listed among the search results. She has already included this test in her study design. Clicking that link displays a description of the protocol, details of specimen collection, specific instructions for administration, and related protocols. Essential Protocols collect information necessary for data interpretation, and are suggested in the left‐hand panel. Related Protocols are other protocols in the Toolkit that may be of interest. Related Protocols are suggestions only and are not required for interpretation of a protocol. Tabs on this page can be accessed to obtain additional material, including information on administration, details, source, variables, and measure. After reviewing this information, the investigator notes that this protocol is the same one she has already included in her new study design.
- Upon further review of each tab, the investigator sees on the “Details” tab (Fig. 6) that links are provided to other research standards such as caDSR Common Data Elements (CDE), Logical Observation Identifiers Names and Codes (LOINC), and Human Phenotype Ontology. These additional resources and links to other research standards will be nice to review because she regularly uses the Oral Glucose Tolerance Test protocol. She also notes the quick access to directly download a Data Collection Worksheet (DCW) or Data Dictionary (DD), which are available through the “Download” drop‐down menu above the “Protocol” panel (Fig. 7).
Figure 4.
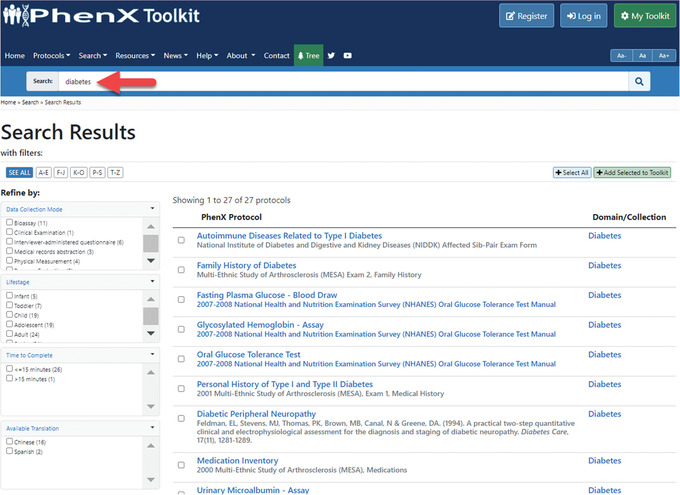
Search bar results for diabetes protocols in the PhenX Toolkit.
Figure 5.
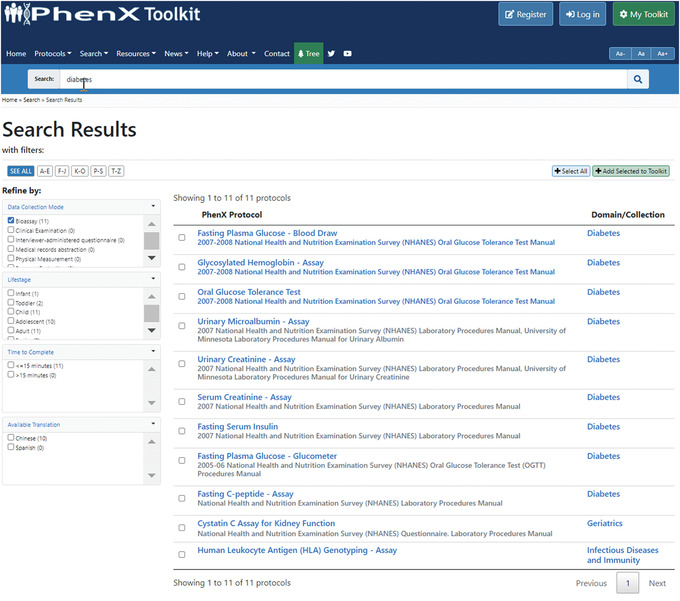
Filtered search results enable quick discovery of relevant protocols.
Figure 6.
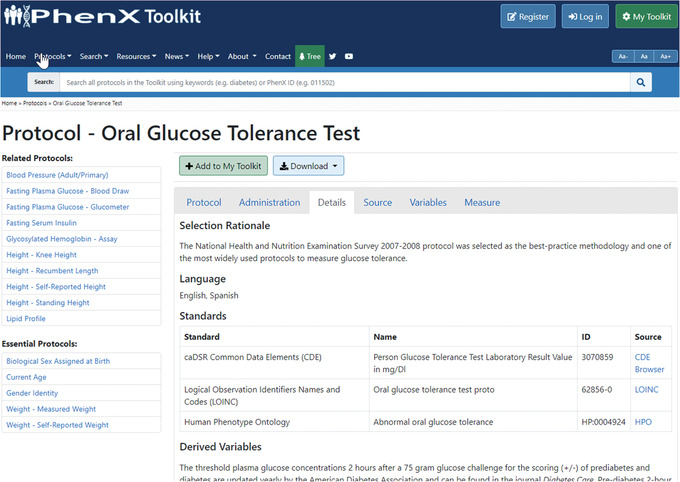
The “Details” panel on the “Protocol” page.
Figure 7.
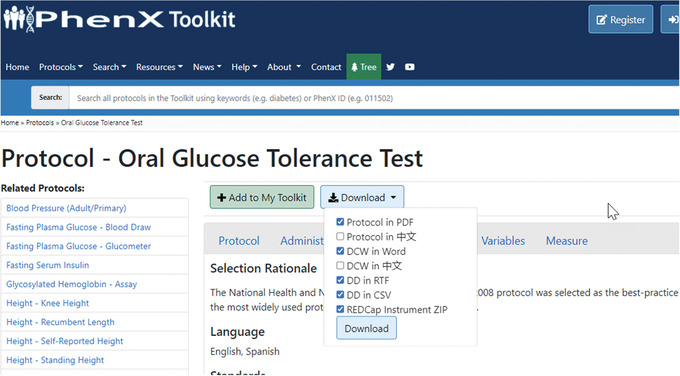
Protocol resources can be downloaded from the “Protocol” panel.
-
3Select protocols via “Add to My Toolkit.”
- Because the investigator already uses the Oral Glucose Tolerance Test, she decides to add the protocol to My Toolkit, so she can review additional resources displayed in the “Details” panel (Fig. 6). After she adds the protocol to My Toolkit, she can save it to access and download later (Fig. 7). She will also have quick access to directly download a Data Collection Worksheet (DCW) or Data Dictionary (DD), which are available at the top of the “Details” panel.
- More information and guidance on My Toolkit can be found at https://www.youtube.com/watch?v=Gz_4LLaQmr0.
-
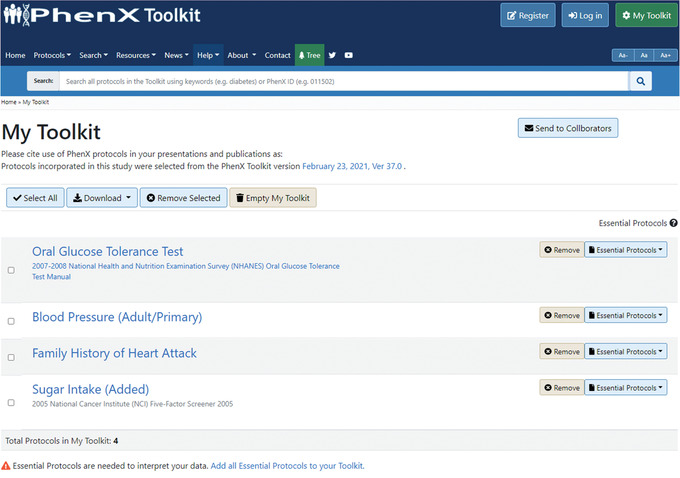
4Use My Toolkit and review Essential Protocols.
-
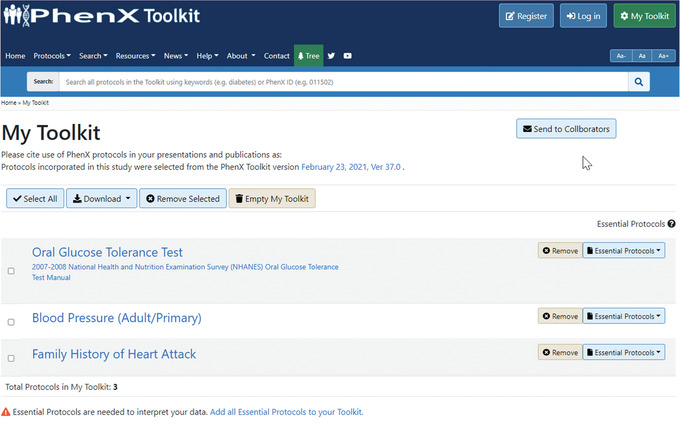
In My Toolkit, the investigator may see that there are some Essential Protocols associated with the protocols in My Toolkit. An alert indicates that Essential Protocols are needed to interpret the data. In My Toolkit, each of the Essential Protocols can be reviewed and added to My Toolkit. For example, the Current Age protocol is essential to interpreting the Oral Glucose Tolerance Test protocol (Fig. 8).Alternatively, users can quickly add Essential Protocols by selecting “Add all Essential Protocols to your Toolkit” (Fig. 9).Clicking the Essential Protocol name will bring the user to the “Details” panel for that protocol. After adding the suggested Essential Protocols to My Toolkit, the investigator logs off to resume later. The protocols she has selected are saved in My Toolkit.
- Satisfied with identifying the Oral Glucose Tolerance Test, the investigator goes back to the PhenX Toolkit home page and this time decides to click “Research Domains” (Fig. 10). She notices the Cardiovascular Domain (Fig. 11) and wonders if the PhenX Toolkit includes cardiovascular protocols that might be appropriate to consider for her new study. When she clicks “Cardiovascular,” the investigator now sees the 14 protocols that make up the Cardiovascular domain (Fig. 12).
- After scanning the 14 protocols in the Cardiovascular domain, she notices protocols that she would like to review further and clicks the protocol names to view additional details for each. She adds “Blood Pressure (Adult/Primary)” and “Family History of Heart Attack” to My Toolkit (Fig. 13).
-
How to become a TK Registered User: To access the ability to save protocols and return to them at a later date, the user can register and create a profile. First, click the “Add to My Toolkit” button to save one or more protocols for convenience. (Fig. 14). After registering, users can access features such as the ability to name and save multiple Toolkits and share them with collaborators. Registered users can also opt to receive the PhenX Toolkit Newsletter to learn about new content releases and access the Link Your Study feature.Exploring the Toolkit, the investigator finds additional standard protocols relevant to her study design. The following steps detail how to use Smart Search, Advanced Search, and the Protocol Tree Browser.
-
Figure 8.
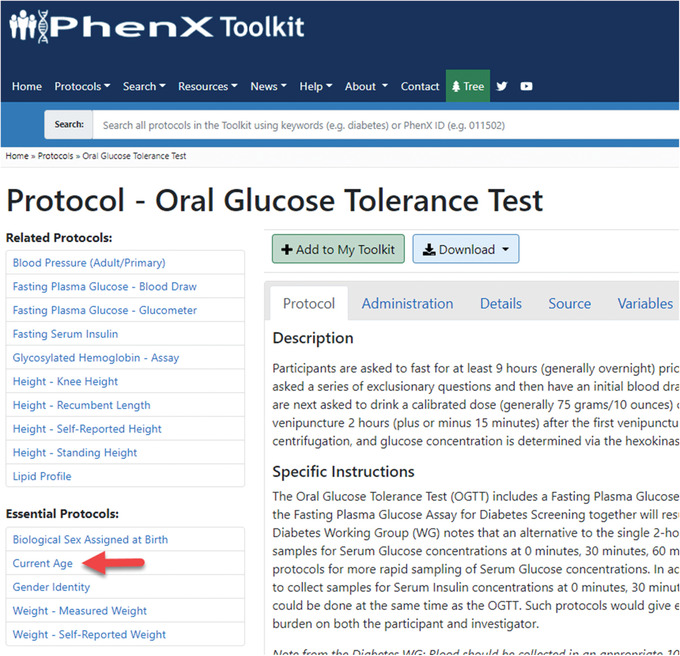
The Current Age protocol is essential to interpreting the Oral Glucose Tolerance Test protocol.
Figure 9.
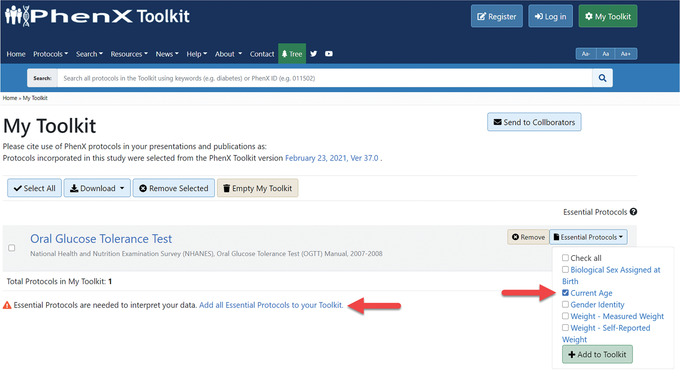
The My Toolkit page allows the investigator to save protocols of interest to download or browse later. Essential Protocols can be added individually or together by selecting the active link next to the alert at the bottom of My Toolkit.
Figure 10.

Browsing research domains from the home page.
Figure 11.
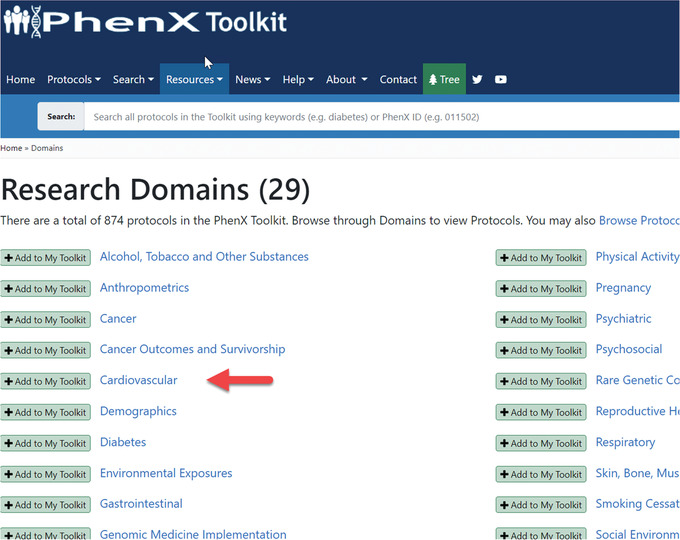
Browsing the research domains page.
Figure 12.
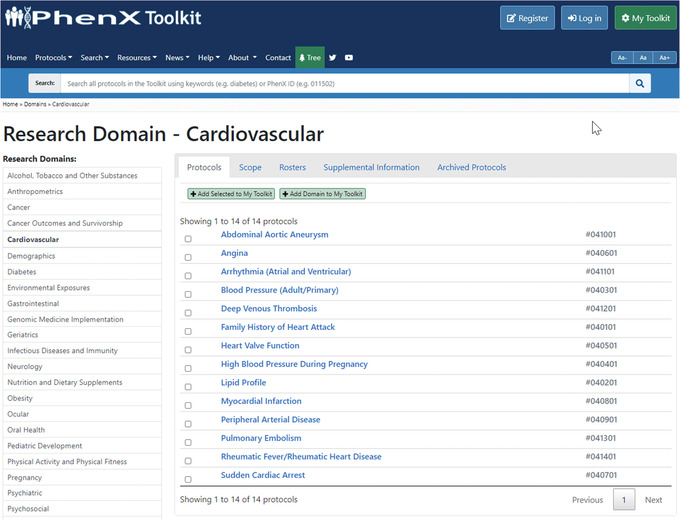
Browsing protocols in the Cardiovascular Domain.
Figure 13.
Adding Cardiovascular Domain protocols to My Toolkit.
Figure 14.
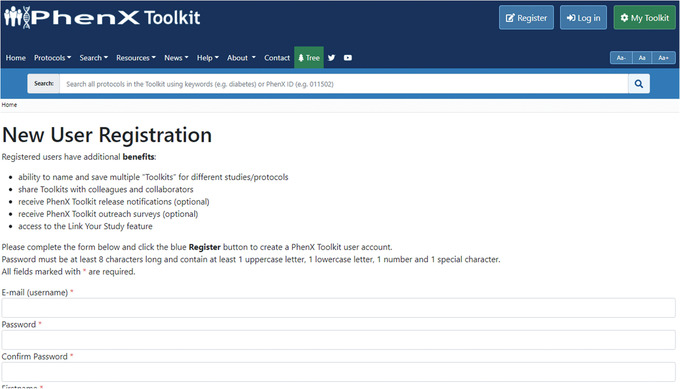
Registered users can save protocol selections, share with collaborators and access studies linking PhenX protocols.
-
5Use Smart Search to find all protocols from a specific source (e.g., National Health Interview Survey [NHIS]).
- The Smart Search query bar at the top of the page allows a user to search all protocols in the Toolkit for those with matching metadata, including protocol source, specific instructions for protocol administration, and descriptions.
- Because the investigator is interested in identifying protocols assessing social determinants of health and nutrition, she decides to search the Toolkit for a large national study, which she thinks might contain these types of protocols. She chooses NHIS. The investigator types the search term “NHIS” into the Smart Search bar at the top of the PhenX Toolkit webpage. The search returns 50 relevant results (Fig. 15).
- The investigator selects the “Sugar Intake (Added)” protocol, which looks like a good match to include in her new diabetes study. She adds this protocol to My Toolkit (Fig. 16).
Figure 15.
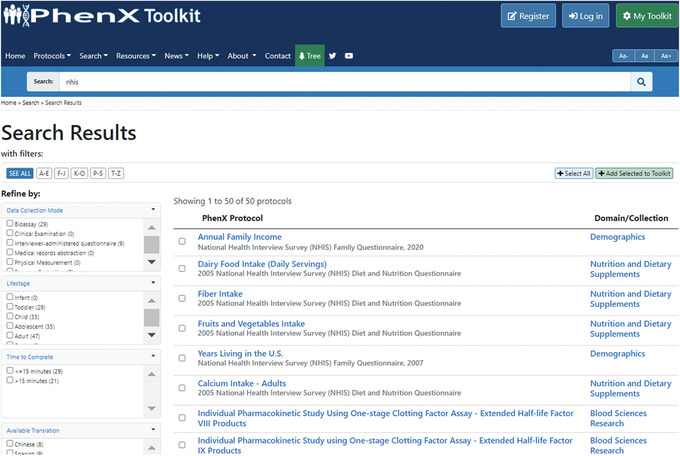
Using the Smart Search query for National Health Interview Survey (NHIS) protocols.
Figure 16.
Adding an NHIS protocol (Sugar Intake) to My Toolkit.
-
6
Advanced Search under the “Search” drop‐down menu on the PhenX Toolkit home page is where search operations can be fine‐tuned (Fig. 17). The investigator decides to search here for a protocol that measures healthcare access, and has a couple of options. Smart Search combs through protocol metadata and key fields, so it has high specificity in its function. Smart Search is also the default operation of the search bar at the top of each page. Another search option is Text Search only, found on the Advanced Search page. The advanced Text Search looks for word matches in the entire protocol text rather than just key metadata fields. A full Text Search of any relevant search term can be used to review more broadly relevant results and any Supplemental Information, which are listed at the end of the results. The investigator types “healthcare access” into Advanced Search and clicks the Text Search button (Fig. 18). The results are displayed, and she sees “Access to Healthcare Services.” After reviewing the protocol, she decides to add it to My Toolkit.
More information and guidance on searching the Toolkit can be found at https://www.youtube.com/watch?v=yAycTLYdFAM.
Figure 17.

Navigating to Advanced Search.
Figure 18.
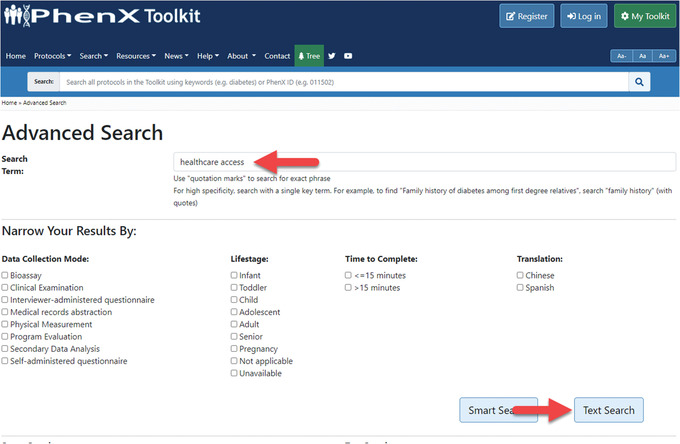
Executing a full Text Search.
-
7One protocol the investigator would still like to identify is a measure related to Environmental Health, which might affect diabetes. She goes back to the PhenX Toolkit home page and clicks the “Tree” button (Fig. 19). The PhenX Toolkit Protocol Tree Browser groups all protocols in the Toolkit by MeSH (Medical Subject Headings) terms (National Library of Medicine [NLM]; see Internet Resources). It is used for indexing, cataloging, and searching of biomedical and health‐related information. MeSH includes the subject headings appearing in MEDLINE/PubMed, the NLM Catalog, and other NLM databases.
- The user visits the Cross‐Domain Concept Tree Browser main page (Fig. 20).
- She clicks to open the ”Environmental Health” branch, followed by “Air Pollution,” then clicks the Air Quality Index protocol. Information about the protocol is then displayed, so she can easily review the protocol there or go to the protocol page (Fig. 20). She decides to add the Air Quality Index protocol to My Toolkit using the button on the top right of the screen.
- More information and guidance on using the Browse Protocol Tree can be found at https://www.youtube.com/watch?v=JSL7nqEF6AY.
Following these steps, the investigator identified several standard protocols for inclusion in her study design. The investigator can download protocol documents in real time or save them in My Toolkit, where she can download them individually or as a set. If the investigator is logged in as a registered user, her selections are saved, and she can return later to review and download the protocol documents and even share the protocols with collaborators directly from within the Toolkit.
Figure 19.
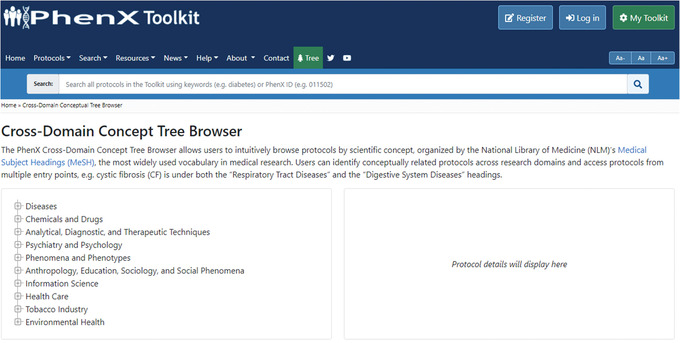
The Protocol Tree Browser.
Figure 20.
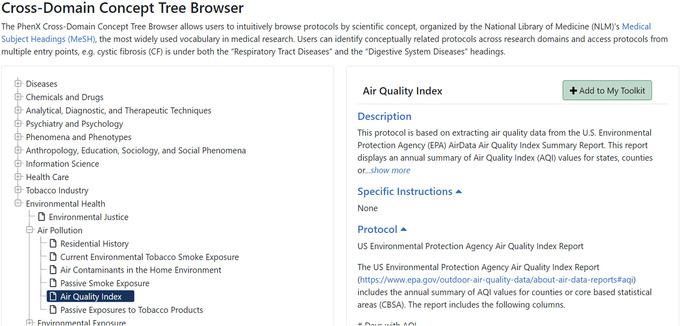
The protocol preview from the Protocol Tree Browser page.
COMMENTARY
Background Information
The features and parameters we describe next detail Toolkit resources designed to assist users in implementing standard data collection using these protocols. We also discuss how to appropriately cite use of the Toolkit and of protocols provided by the Toolkit. The standard procedure outlined in each protocol in the PhenX Toolkit ensures consistent data collection across different studies. Data need to be collected using the same methodology in order for the data to be truly comparable (Fig. 21). The Toolkit provides several resources to assist with these research implementation phases. Using standard protocols allows for comparisons among studies to validate results and allows results to be combined across studies to increase statistical power in downstream analyses. This ultimately increases the scientific impact of individual studies.
Figure 21.
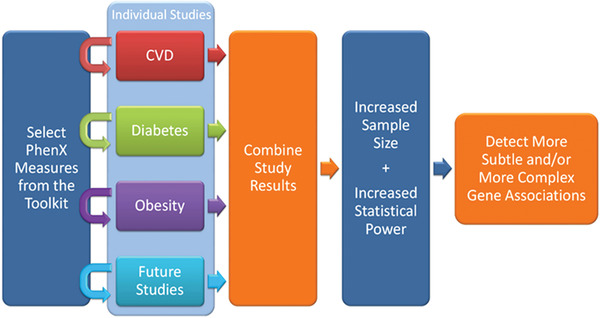
Using standard measures allows datasets across different domains to be combined and/or compared, increasing the scientific impact of individual studies in genome‐wide association studies.
Critical Parameters
Investigators should consider several factors when using the PhenX Toolkit. Users should understand the concept of Essential protocols and identify how to download and use Data Collection Worksheets and Data Dictionaries.
Users who would like to see some real‐world examples of studies citing the PhenX Toolkit or review funding opportunity announcements (FOAs) recommending use of PhenX measures can access this information from the drop‐down menu under the “Resources” tab on the navigation bar. From the “Resources” tab, the “Publications & Presentations” page shows publications for research studies that have used PhenX Toolkit protocols in their study design.
Toolkit Features
Data collection worksheet (DCW)
Toolkit users can download protocol details including Data Collection Worksheet (DCWs) directly from the “Details” panel on the protocol page or from My Toolkit. The DCW is provided as a Microsoft Word document to facilitate easy integration into existing study documentation. Investigators can generate basic or full reports for selected protocols and download a DCW (Fig. 22). Here the user can see details related to data collection for saved protocols.
Figure 22.
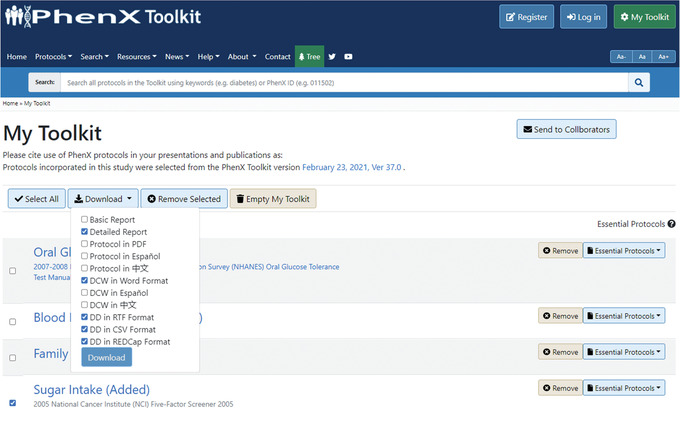
Downloading a basic or full report and Data Collection Worksheet from My Toolkit.
The Toolkit offers a DCW (Fig. 23) that helps investigators integrate PhenX protocols into their existing research. The DCW captures each data item required and collected in the measurement protocol, which helps ensure that investigators collect all necessary information during data collection.
Figure 23.

The Data Collection Worksheet is available in Word and PDF formats.
PhenX Data Dictionaries
PhenX Data Dictionaries (DDs) describe the variables associated with the selected protocols. These data dictionaries are available in three formats: (1) the RTF format captures skip patterns commonly used in epidemiologic studies (Fig. 24); (2) the CSV format is compatible with the database of Genotypes and Phenotypes (dbGaP) data submission packet (Fig. 25); and (3) the REDCap ZIP file format is suitable for upload to studies being implemented in REDCap (Fig. 26). The REDCap data dictionary can be imported as a standalone REDCap project or added to an existing project for data collection (Fig. 26).
Figure 24.
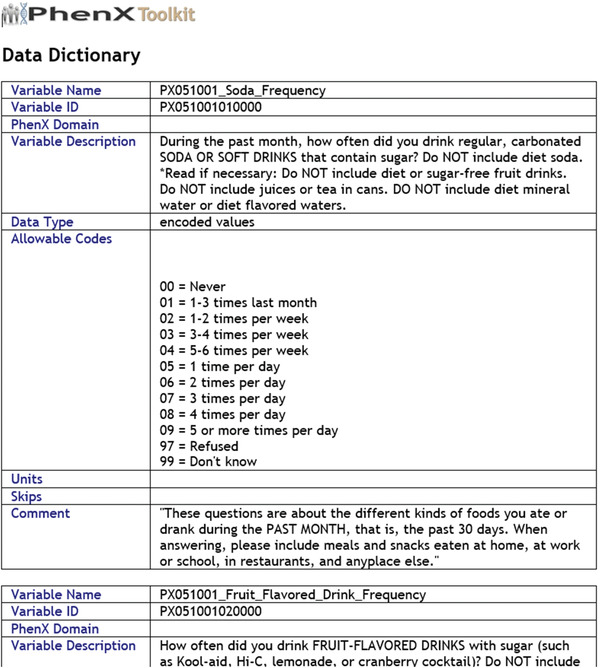
The PhenX Data Dictionary in Word format.
Figure 25.

The PhenX Data Dictionary in .csv format.
Figure 26.
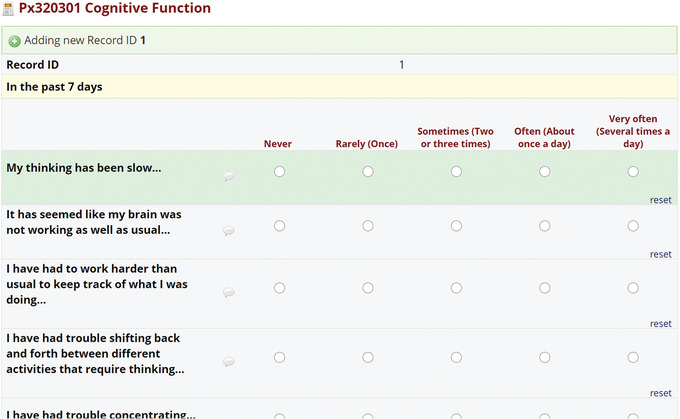
REDCap data dictionaries can be imported as new projects and can be used immediately for data collection.
Link Your Study (LYS)
The LYS feature collects basic study information from registered users who are using PhenX protocols in their research. The LYS feature helps users find other investigators employing the same PhenX protocols in their studies to facilitate opportunities for cross‐study analysis.
Publications Citing PhenX
To view papers citing the PhenX Toolkit, navigate to the “Resources” menu from the home page and select “Publications & Presentations,” then select “Publications Citing PhenX” (Fig. 27). Information on citing the PhenX Toolkit can be found at https://www.phenxtoolkit.org/help/citation.
Figure 27.
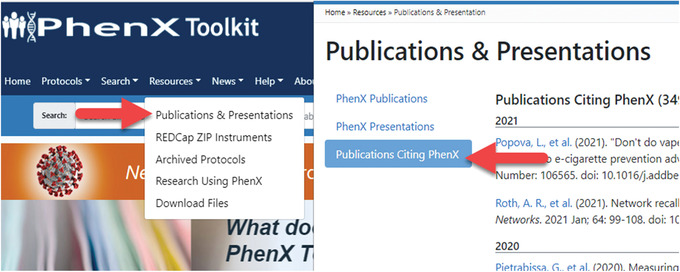
The “Publications Citing PhenX” page can be found under the “Resources” menu.
PhenX Citation
How to cite PhenX
Instructions for citing PhenX in publications are available on the home page (Fig. 28): https://www.phenxtoolkit.org/help/citation.
Figure 28.

Instructions on citing PhenX can be found from the home page.
How to cite the PhenX Toolkit
Hamilton, C. M., Strader, L. C., Pratt, J. G., Maiese, D., Hendershot, T., Kwok, R. K., Hammond, J. A., Huggins, W., Jackman, D., Pan, H., Nettles, D. S., Beaty, T. H., Farrer, L. A., Kraft, P., Marazita, M. L., Ordovas, J. M., Pato, C. N., Spitz, M. R., Wagener, D., … Haines, J. (2011). The PhenX Toolkit: Get the most from your measures. American Journal of Epidemiology, 174(3), 253–260. doi: 10.1093/aje/kwr193.
How to cite use of PhenX measures
“Measures incorporated in this study were selected from the PhenX Toolkit version February 23, 2021, Ver 37.0.” The date and version number should reflect the version from which the user downloaded the protocols for use.
Troubleshooting
The PhenX Toolkit provides recommended standard data collection protocols which are suitable for a variety of study designs. Because not all measurement protocols are suitable for all study designs, it is the investigator's responsibility to select appropriate protocols for inclusion in their study. The Toolkit home page includes a button to the “Toolkit Guidance” page (Fig. 29), where investigators can find links to references for conducting research with human subjects including Certificate of Confidentiality, the NIH Genomic Data Sharing Policy, A Code of Ethics for Public Health, the National Human Genome Research Institute (NHGRI) informed consent resource, and references pertaining to study design. The Toolkit Guidance guidelines are included in every downloaded PhenX report.
Figure 29.

The Toolkit Guidance is provided in basic and full reports.
Requirements Table
Each protocol has varying needs for equipment and expertise for administration (Fig. 30). By reviewing the Equipment Needs and Requirements (under the “Administration” tab), investigators can identify potential barriers or needs they will potentially encounter when administering a protocol. The “Administration” tab also describes personnel and training required for the protocol. Reviewing this information can help investigators determine their ability to follow the protocol and any needs they may encounter along the way. Knowing these needs up front can help investigators be best prepared for conducting a study.
Figure 30.
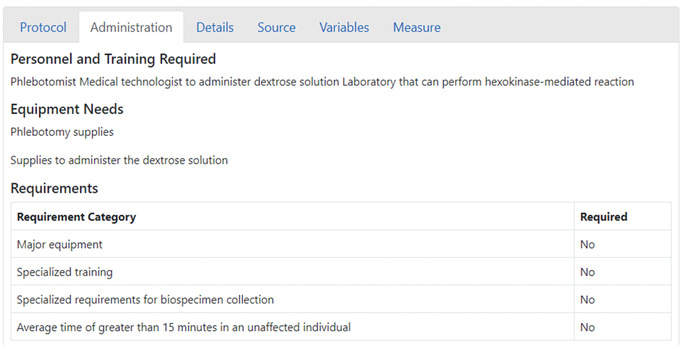
Requirements for the Oral Glucose Tolerance Test protocol.
Understanding Results
By including protocols from the PhenX Toolkit in new or existing studies, investigators will be able to combine or compare data from other studies incorporating the same PhenX protocols. This facilitates cross‐study analysis and increases the statistical power to identify genetic associations with complex diseases and traits, gene‐gene interactions, and gene‐environment interactions.
Promoting and incorporating standard measures ultimately increases the impact of individual studies. To date, 394 FOAs have been issued from NIH and the Department of Defense that encourage the use of PhenX protocols. Widespread adoption of PhenX protocols will augment the impact of biomedical research studies and ultimately improve the health and well‐being of the population.
Time Considerations
Brevity of protocol administration is considered when protocols are selected for inclusion in the Toolkit. Protocols that take more than 15 minutes, on average, for an unaffected individual are noted in the Toolkit. Protocols that meet the other requirements for inclusion and require the least burden (time is considered, among other factors) on the investigator and participant are ideal choices for inclusion in the Toolkit (Fig. 30).
Author Contributions
Lisa Cox: writing original draft. Stephen Hwang: writing original draft. Jonathan Haines: writing review and editing. Erin Ramos: writing review and editing. Catherine McCarty: writing review and editing. Mary Marazita: writing review and editing. Michelle Engle: writing original draft. Tabitha Hendershot: writing original draft. Huaqin (Helen) Pan: writing review and editing. Carol Hamilton: writing review and editing.
Conflict of Interest Statement
Authors have no financial or personal relationship between themselves and others that might bias their work.
Acknowledgments
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health, Award Number U41HG007050, with co‐funding from the National Institute on Drug Abuse (NIDA), the National Heart, Lung, and Blood Institute (NHLBI), the Office of the Director (OD), the Office of Behavioral and Social Sciences Research (OBSSR) of the National Institutes of Health (NIH), and Administrative Supplement Award Number 3U41HG007050‐08S1 funded by the Office of the Director (OD), the Office of Behavioral and Social Sciences Research (OBSSR), the National Institute On Minority Health And Health Disparities (NIMHD), and the National Cancer Institute (NCI) of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The authors would like to acknowledge the guidance of the PhenX Steering Committee, contributions of all Working Groups, Expert Review Panels, and research panel members, and the efforts of the overall RTI/NHGRI/NIH PhenX project team. Project participants are acknowledged at https://www.phenxtoolkit.org/about/teams.
Cox, L. A. , Hwang, S. , Haines, J. , Ramos, E. M. , McCarty, C. A. , Marazita, M. L. , Engle, M. L. , Hendershot, T. , Pan, H. , & Hamilton, C. M. (2021). Using the PhenX Toolkit to select standard measurement protocols for your research study. Current Protocols, 1, e149. doi: 10.1002/cpz1.149
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Literature Cited
- Bennett, S. N. , Caporaso, N. , Fitzpatrick, A. L. , Agrawal, A. , Barnes, K. , Boyd, H. A. , … Williams, K. (2011). Phenotype harmonization and cross‐study collaboration in GWAS consortia: The GENEVA experience. Genetic Epidemiology, 35(3), 159–173. doi: 10.1002/gepi.20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, C. M. , Strader, L. C. , Pratt, J. G. , Maiese, D. , Hendershot, T. , Kwok, R. K. , … Haines, J. (2011). The PhenX Toolkit: Get the most from your measures. American Journal of Epidemiology, 174(3), 253–260. doi: 10.1093/aje/kwr193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiese, D. R. , Hendershot, T. P. , Strader, L. C. , Wagener, D. K. , Hammond, J. A. , Huggins, W. , … Hamilton, C. M. (2013). PhenX—establishing a consensus process to select common measures for collaborative research. Research Triangle Park, NC: RTI Press. [PubMed] [Google Scholar]
- Phillips, M. , Grant, T. , Giampietro, P. , Bodurtha, J. , Valdez, R. , Maiese, D. R. , … Hamilton, C. M. (2017). PhenX measures for phenotyping rare genetic conditions. Genetics in Medicine, 19(7), 834–837. doi: 10.1038/gim.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, P. J. , Harlan, W. R. , Hammond, J. A. , Hendershot, T. , & Hamilton, C. M. (2010). PhenX: A toolkit for interdisciplinary genetics research. Current Opinion in Lipidology, 21(2), 136–140. doi: 10.1097/MOL.0b013e3283377395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Key References
This 2015 Current Protocols in Human Genetics article provides information on new content and features of the PhenX Toolkit.
This 2011 article is the first PhenX publication in Current Protocols in Human Genetics. It provides significant background on the history of PhenX and the PhenX Toolkit and serves as a point of reference for how the Toolkit has grown and evolved over time.
- Hendershot, T. , Pan, H. , Haines, J. , Harlan, W. R. , Marazita, M. L. , McCarty, C. A. , … Hamilton, C. M. (2015). Using the PhenX Toolkit to add standard measures to a study. Current Protocols in Human Genetics, 86, 1.21.1–1.21.17. doi: 10.1002/0471142905.hg0121s86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot, T. , Pan, H. , Haines, J. , Harlan, W. , Junkins, H. , Ramos, E. , & Hamilton, C. M. (2011). Using the PhenX Toolkit to add standard measures to your study. Current Protocols in Human Genetics, 71(1), 1.21.1–1.21.18. doi: 10.1002/0471142905.hg0121s71 [DOI] [PubMed] [Google Scholar]
Internet Resources
Phenxtoolkit.org is the primary website for the PhenX Toolkit. As described in this article, the Toolkit provides detailed protocols for collecting data and tools to help investigators incorporate these protocols into their studies. Using protocols from the PhenX Toolkit facilitates cross‐study analysis, potentially increasing the scientific impact of individual studies.
The PhenX web portal provides general information about the PhenX project and the consensus process used by PhenX to develop the contents of the Toolkit. The web portal also serves as a host for project communications such as the quarterly newsletter and secure portal sites for individual Working Groups.
The dbGaP is a National Center for Biotechnology Information–funded public repository for individual‐level phenotype, genotype, exposure, and sequence data, and the associations between them. Each PhenX protocol has a data dictionary that is compatible with the dbGaP data submission packet.
In addition to dbGaP compatibility, the PhenX Data Dictionaries are also available as REDCap .zip files that can be immediately integrated into studies implementing REDCap for data capture. Over 1.6 million users in 141 countries currently use REDCap software, and over 13,000 journal articles cite REDCap usage.
The NHGRI‐European Bioinformatics Institute GWAS Catalog is a publicly available repository of GWAS and their results and includes visualizations of variant‐trait associations mapped onto chromosomal positions on the human genome. The potential for GWAS to relate phenotypes to specific genetic variation is greatly increased when data can be combined or compared across multiple studies. The PhenX Toolkit facilitates replication and validation across studies by promoting standard measurement protocols, ultimately increasing the statistical power for identifying and replicating variants associated with complex diseases and with gene‐gene and gene‐environment interactions.
National Library of Medicine.
- PhenXToolkit: https://www.phenxtoolkit.org/
- PhenX web portal: https://www.phenx.org/
- Database of Genotypes and Phenotypes (dbGaP): https://www.ncbi.nlm.nih.gov/gap/
- Research Electronic Data Capture (REDCap): https://www.project‐redcap.org/
- Genome‐Wide Association Studies (GWAS) Catalog: https://www.ebi.ac.uk/gwas/
- https://www.nlm.nih.gov/mesh/meshhome.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


