Abstract
Sepsis and sepsis‐associated lung inflammation significantly contribute to the morbidity and mortality of critical illness. Here, we examined the hypothesis that neuronal guidance proteins could orchestrate inflammatory events during endotoxin‐induced lung injury. Through a targeted array, we identified netrin‐1 as the top upregulated neuronal guidance protein in macrophages treated with lipopolysaccharide (LPS). Furthermore, we found that netrin‐1 is highly enriched in infiltrating myeloid cells, particularly in macrophages during LPS‐induced lung injury. Transcriptional studies implicate hypoxia‐inducible factor HIF‐1α in the transcriptional induction of netrin‐1 during LPS treatment. Subsequently, the deletion of netrin‐1 in the myeloid compartment (Ntn1loxp/loxp LysM Cre) resulted in exaggerated mortality and lung inflammation. Surprisingly, further studies revealed enhanced natural killer cells (NK cells) infiltration in Ntn1loxp/loxp LysM Cre mice, and neutralization of NK cell chemoattractant chemokine (C‐C motif) ligand 2 (CCL2) reversed the exaggerated lung inflammation. Together, these studies provide functional insight into myeloid cell‐derived netrin‐1 in controlling lung inflammation through the modulation of CCL2‐dependent infiltration of NK cells.
Keywords: lung inflammation, netrin‐1, natural killer cells
Abbreviations
- A2BAR
adenosine 2B receptor
- BAL
bronchoalveolar lavage
- BALF
bronchoalveolar lavage fluid
- MΦ
macrophage
- NGP
neuronal guidance protein
- PMN
polymorphonuclear cells
1. INTRODUCTION
Persistent and uncontrolled inflammation is the hallmark of sepsis, and sepsis‐associated lung inflammation, which significantly contributes to morbidity and mortality of critical illness. 1 , 2 Inflammation by itself is an essential adaptive response to noxious stimuli such as infection and tissue injury. 3 , 4 , 5 , 6 , 7 However, in situations where an infectious burden is excessive or inflammation becomes dysregulated (ie, fails to terminate or resolve), inflammation can result in profound collateral tissue damage and systemic impact. 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 Indeed, endogenous mechanisms and immune‐modulators are programed to limit inflammation and restore tissue homeostasis. 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 Such mechanisms include negative feedback pathways that serve to dampen inflammation and active stimulation by pro‐resolution mediators that coordinate the termination of inflammation. 21 , 24 , 25 , 26 , 27 , 28 , 29 , 30 Myeloid cells, such as macrophages and neutrophils, are a crucial part of innate immunity and key drivers of the initiation and resolution of inflammation. For example, during lung inflammation, monocytes are recruited to the inflamed air space to differentiate into macrophages secreting cytokines that act locally to stimulate chemotaxis and activate neutrophils. 31 Therefore, understanding how myeloid cells contribute to the proper inflammatory responses are areas of intense investigation. 32 , 33 , 34 , 35 , 36 , 37 , 38
Neuronal guidance proteins (NGPs) were first characterized for their role in neurogenesis through their ability to act as chemoattractant and chemorepellent cues to guide axons to their target synapses. 39 , 40 , 41 , 42 However, increasing evidence has recognized NGPs as a class of immune‐modulators 43 , which can regulate inflammatory processes by limiting or promoting the migration of leukocytes during acute and chronic inflammatory conditions. 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 For example, netrin‐1, as one of the most investigated NGPs in immune modulation, is expressed by endothelial cells and plays a crucial role in inhibiting the migration of neutrophils to different chemoattractants. 55 In addition, several lines of evidence from mice with partial deletion of netrin‐1 (Ntn1+/−) indicated that netrin‐1 plays an important role in limiting the transmigration of leukocytes during models of acute lung injury, peritonitis, and colitis. 46 , 47 , 49 , 56 Additionally, exposure of hypoxia results in netrin‐1 induction in mucosal epithelial cells in a hypoxia‐inducible factor HIF‐1α‐dependent manner, and Ntn1+/− mice exhibit increased myeloperoxidase activity in the colon tissue upon hypoxia exposure. 47 Furthermore, recombinant netrin‐1 treatment inhibits chemokine (C‐C motif) ligand 2 (CCL2) and chemokine (C‐C motif) ligand 19‐driven macrophage migration in vitro. 57 Besides its role in leukocyte migration, netrin‐1 was shown to suppress inflammatory macrophage functions 58 , 59 and to promote resolution of inflammation by stimulating the production of specialized pro‐resolving mediators and tissue regeneration. 60 , 61 However, the functional role of myeloid cells‐derived netrin‐1 during lung inflammation has not been elucidated. Our studies demonstrated that, for the first time, myeloid cell‐specific expression of netrin‐1 confers lung protection through the modulation of CCL2‐dependent natural killer (NK) cell migration.
2. MATERIALS AND METHODS
2.1. Mice
Wild‐type (C57BL/6J), Ntn1loxp/loxp, 62 Hif1αloxp/loxp, 63 and LysM Cre 64 mice were purchased from Jackson Laboratory. Detailed information on the mice strains is listed in Supplementary Table 1. Mice were housed and bred in a pathogen‐free suite at the Center for Laboratory Animal Medicine and Care at the University of Texas Health Science Center at Houston (UTHealth). All experimental animal protocols were approved by the UTHealth Institutional Animal Care and Use Committee. Accounting for sex as a variable, for experiments using C57BL/6J or Ntn1loxp/loxp LysM Cre mice, experiments were performed with age‐ and weight‐matched equal numbers of male and female mice throughout all groups. In our experiments using Hif1αloxp/loxp LysM Cre mice, sex‐dependent differences in mice were not observed and we used age‐ and weight‐matched mice (Supplementary Figure 1).
2.2. Generation of Ntn1loxp/loxp LysM Cre+ and Hif1αloxp/loxp LysM Cre+ mice
To conditionally achieve myeloid cell‐specific deletion, Ntn1loxp/loxp and Hif1αloxp/loxp mice were crossbred with LysM Cre+ to generate Ntn1loxp/loxp LysM Cre and Hif1αloxp/loxp LysM Cre mice, respectively. Knockout in Ntn1loxp/loxp LysM Cre mice was confirmed by performing RT‐qPCR measuring knockout efficiency of the Ntn1 mRNA transcript levels in bone marrow and in bronchoalveolar lavage (BAL) cells of intratracheal LPS‐treated mice (Supplementary Figure 2A,B). Hif1αloxp/loxp LysM Cre mice have been previously genotyped and characterized. 65
2.3. Isolation of human polymorphonuclear cells (PMNs) and monocyte‐derived macrophages (hMDMs)
The protocol for the collection of human blood from healthy donors was approved by the Institutional Review Board at UTHealth and participant consent was obtained prior to the collection. Detailed information on the reagents is listed in the Supplementary Table 2. All centrifuge steps were performed at 4°C. In a 60 mL of syringe prefilled with 10 mL of citrate‐dextrose buffer (Sigma‐Aldrich), 50 mL of blood was obtained by venipuncture. Blood was then centrifuged at 400 ×g for 10 minutes. Plasma was transferred into two clean tubes and centrifuged again at 400 ×g for 10 minutes. The resulting cell pellets were added back to remaining blood and 20 mL of 3% dextran in normal saline was added to promote the sedimentation for 40 minutes. Supernatant was then transferred to new tubes and topped with HBSS (Thermo Fisher, Waltham, MA) and then, centrifuged at 400 ×g for 10 minutes. Samples were then treated with Red Blood Cell Lysis Solution (Miltenyi Biotec, US) and then, centrifuged at 400 ×g for 10 minutes. The resulting cell pellet was re‐suspended in 2.5 mL of gradient buffer (HBSS(−) +25 mM HEPES + 1 mM EDTA) and carefully layered on top of 10 mL of Ficoll‐Paque PLUS (GE Healthcare, Sweden) and then, centrifuged at 700 ×g for 30 minutes with no break. Interphase peripheral blood mononuclear cells (PBMCs) were carefully pipetted into two tubes and washed twice with cold HBSS (−)+25 mM HEPES+10% FCS. The remaining cell pellet, which consists of PMNs were also washed twice with cold HBSS+25 mM HEPES+10% FCS. PMNs were then cultured for experiments in DMEM+25 mM HEPES+20% FCS, 2 mM Gln, 1% of Antibiotic/Antimycotic solution. To obtain hMDMs, PBMCs were cultured for 7 days in macrophage differentiation media: RPMI 1640 (supplemented with 10% of heat inactivated fetal bovine serum and 10 ng/mL of recombinant human M‐CSF). Treatment with lipopolysaccharide (LPS, Sigma‐Aldrich) was performed using a concentration of 1 μg/mL.
2.4. RNA isolation and mRNA quantification by RT‐qPCR
Total RNA isolation was performed using a TRIzol Reagent extraction protocol according to the product manual (QIAzol Lysis Reagent, Qiagen). RNA concentration was measured using BioTek Cytation 5 with the Take3 plate (Winooski, VT, USA). Reverse transcription was performed with Thermo Fisher Applied Biosystems High Capacity Reverse Transcription Kit on a Bio‐Rad T100 thermal cycler (Hercules, CA, USA). Quantitative PCR was performed using TaqMan probes targeting netrin‐1 (Thermo Fisher, Human: Cat# 4331182 and Mouse: Cat# 4331182) and 18S for control (Thermo Fisher, cat# 4331182). PCR reactions were performed with TaqMan Universal PCR Master Mix (Thermo Fisher) on a Bio‐Rad real‐time PCR system.
2.5. Neuronal guidance peptide mRNA array
hMDMs were stimulated with LPS for 8 hours and then, collected for RNA isolation. Screening for Neuronal Guidance Peptide mRNA expression was performed using the Axon Guidance PrimerArray (cat.# PH006) and TB Green Premix Ex Taq II from Takara Bio Inc. (Shiga, Japan) according to the manufacture’s protocols.
2.6. Western blotting
Detailed information on the antibodies is listed in Supplementary Table 1. Protein was extracted using RIPA Buffer (Thermo Fisher, cat.# 89900) with protease and phosphatase inhibitor cocktails (New England Biolabs). Protein was quantified using BCA assay. After electrophoreses, membranes were blocked for 1 hour at room temperature in skim milk. Membranes were incubated in primary antibodies at 4°C overnight. Horseradish peroxidase‐conjugated secondary antibodies were used at 1:5000 dilution. Membranes were imaged on a Bio‐Rad ChemiDoc Touch Imaging System. ImageJ software (National Institutes of Health) was used for protein quantification.
2.7. Murine model of endotoxin‐induced lung injury
Lung injury was performed in mice aged 8‐10 weeks old. Anesthesia was achieved using intraperitoneal injection of 70 mg/kg of pentobarbital. Direct intratracheal intubation was achieved using the BioLite Intubation System (Braintree Scientific, Inc., Braintree, MA). After intubation, LPS (2 mg/mL of stock solution in phosphate‐buffered saline) was instilled into the lungs at a dose of 3.75 µg/g. Equivalent weight‐dosed volumes of phosphate‐buffered saline were used for vehicle controls. Mice were then monitored until the effects of anesthesia were no longer observed. Weights were measured daily and mice were euthanized when they experienced two consecutive days of 25% weight loss, were unable to eat or drink, or appeared grossly moribund. Mice were euthanized with a pentobarbital overdose (250 mg/kg) (Socumb, animal NDC Code: 11695‐4836‐5, Henry Schein Animal Health, Melville, NY) and subsequently exsanguinated by laceration of the inferior vena cava. Bronchoalveolar lavage (BAL) was collected with three 500 µL ice‐cold PBS washes and cells were counted using a hemocytometer. A 50 µL aliquot of BAL containing cells were centrifuged onto glass slides using a Rotofix 32A centrifuge (Hettich, Tuttlingen, Germany). Slides were stained with Hema3 Stat Pack (Fisher Scientific, Waltham MA) and counted for neutrophil percentage using light microscopy. The remaining BAL was spun at 300 ×g for 5 minutes and the supernatant (BAL Fluid) and cell pellets were flash‐frozen in liquid nitrogen, separately. Mouse lungs were perfused with pre‐chilled PBS via the right ventricle. Then, lungs were excised and snap‐frozen or inflated with 20 cm H2O hydrostatic pressure of 10% formalin (neutral buffered) through the tracheal cannula and then, submerged into the formalin.
2.8. Lung histology and lung injury scoring
Lungs were fixed for 24‐48 hours in formalin and processed using a Leica TP1020 Semi‐enclosed Benchtop Tissue Processor (Leica Camera, Wetzlar, Germany). Lung sections (5 micrometer thickness) were stained with hematoxylin & eosin and scored for levels of acute lung injury by a pathologist (blinded to the experimental information). Lungs were scored based on a previously described method with slight modification (no injury = 0; injury to 25% = 1; injury to 50% = 2; injury to 75% = 3; and diffused injury = 4). 66 Four injury categories were graded separately on each slide: (1) Atelectasis; (2) Cellularity; (3) alveolar wall thickening; (4) and perivascular edema. A total score was determined by adding up the individual scores.
2.9. Quantification of BAL fluid albumin, cytokine, and chemokine levels
Mouse BAL fluid albumin levels were measured to infer the levels of pulmonary edema using enzyme‐linked immunosorbent assays (ELISA) (Bethyl Laboratories). BAL fluid IL‐6, IL‐1β, and CCL2 levels were measured using ELISA. Detail information about the ELISA kits is included in Supplementary Table 2.
2.10. Immunohistochemistry
The lung slices were cut in 5 µm in thickness and mounted on glass slides. Antigen retrieval was performed with the citric acid‐based solution (Vector Laboratories) using a pressure cooker following deparaffinization and rehydration. The BAL cell slides were prepared with cytospin (Hettich ROTOFIX 32A centrifuge, Germany) and cells were fixed with freshly made 4% Paraformaldehyde (PFA) for 20 minutes (room temperature) followed by permeabilization (0.3% Triton X‐100, 20 minutes). Endogenous peroxidase activity was quenched (3% hydrogen peroxide solution, 5 minutes). After blocking with 2.5% normal goat serum, slices were incubated with primary antibodies (4°C, overnight). The following steps were performed with ABC‐HRP Kit (Vector Laboratories) and DAB (3,3′‐diaminobenzidine) according to the manual instruction followed by hematoxylin counterstaining. Negative control slides were incubated with the recombinant rabbit IgG (Abcam, Cat#ab172730) instead of primary antibody at the same concentration. A Leica DM2500 light microscope was used to evaluate the staining and pictures were taken with a Leica DMC5400 digital camera. Detailed information on the antibodies is listed in Supplementary Table 3.
2.11. Flow cytometry of BAL cells
After collection and red blood cell lysis, BAL cells were blocked with anti‐mouse CD16/32 Antibody (Clone 93, BioLegend) for 10 minutes on ice and then, stained with FITC‐anti‐NK1.1 (Clone PL136, Thermo Fisher) and APC‐anti‐CD3 (Clone 17A2, Thermo Fisher) for 30 minutes on ice. After two washes with flow cytometry buffer, cells were analyzed on a CytoFLEX LX (Beckman Coulter, Indianapolis IN). Detailed information on the antibodies is listed in Supplementary Table 3.
2.12. Immunofluorescence
The BAL cell slides were prepared with cytospin (Hettich ROTOFIX 32A centrifuge, Germany) and cells were fixed with freshly made 4% PFA solution for 20 minutes at room temperature. Following blocking with 2.5% normal goat serum at room temperature for 1 hour, the slides were incubated with primary antibody at 4°C overnight. Negative control slides were incubated with the recombinant rabbit IgG. Conjugated secondary antibodies were used to apply the fluorescence dye. Slides were mounted after counterstaining with DAPI. A confocal microscope (Leica, Germany) was used to observe and document the staining. Detailed information on the antibodies is listed in Supplementary Table 3.
2.13. mRNA‐sequencing of BAL cells
After BAL cell collection and red blood cell lysis, magnetic antibody‐mediated positive selection was then performed to deplete the samples for neutrophils using the MojoSort Mouse Ly‐6g Selection Kit (BioLegend) and RNA was isolated (RNeasy Mini Kit, Qiagen). The RNA‐seq was performed at the UTHealth CPRIT Cancer Genomics Core. The libraries were prepared with KAPA mRNA Hyper Prep Kit (KK8581, Roche Holding AG, Switzerland). The pooled libraries went for the paired‐end 75‐cycle sequencing on an Illumina NextSeq 550 System (Illumina, Inc., USA) using High Output Kit v2.5 (#20024907, Illumina, Inc., USA). Bases with quality scores < 20 and adapter sequences were removed from raw data with Cutadapt (v1.15), 67 followed by alignment of clean RNA‐seq reads to GRCm38 with STAR (v2.5.3a). 68 Gene abundance was counted by HTseq‐count uniquely mapped reads number with default parameter using GencodeM15. Genes with > 5 reads in at least one sample were included for differential expression analysis by DESeq2 software. 69 The P‐values were adjusted with Benjamini and Hochberg’s approach 70 to control for false discovery rate (FDR). Only the genes with fold change > 2 and FDR < 0.05 were considered as differentially expressed genes. Standard Gene Ontology (GO) and KEGG pathway enrichment analyses were performed using the online tool WebGestalt (v0.4.3). 71 Upregulated differentially expressed genes were matched to cell‐type ontology using the CellKb database (v2.0) software. The sequencing data is available in the NCBI’s Gene Expression Omnibus (GEO) repository (GSE167333).
2.14. Antibody‐mediated neutralization of CCL2
For CCL2 neutralization, mice were treated with intraperitoneal injections of 10 μg/g of body weight of anti‐mouse CCL2 (clone 2H5, Cat# BE0185) or control IgG (Cat# BE0091). Both antibodies were purchased from BioXCell (Lebanon, NH).
2.15. Profiling for cytokine and chemokine expression in mouse BAL fluid
To screen for expression of cytokine and chemokines in the BAL fluid of mice, we performed the Proteome Profiler Mouse Cytokine Panel A (R&D Systems, Minneapolis, MN) using 100 μL of mouse BAL fluid. Dot blot membranes were imaged and densitometry was performed using ImageJ software (National Institutes of Health).
2.16. Chromatin immunoprecipitation‐quantitative PCR
After treatment of 1 mg/ml LPS to hMDMs for 8 hours, cells were cross‐linked by 1% methanol‐free formaldehyde solution (Thermo Fisher Scientific) for 8 minutes (room temperature). Formaldehyde was then quenched by 140 mM glycine. Nuclei were isolated with lysis buffer I (50 mM HEPES‐KOH pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP‐40, 0.25% Triton X‐100, 1 mM PMSF, 1 μg/mL leupeptin, and 1X PIC) by incubating for 10 minutes with rotation (4˚C). The isolated nuclei were washed with lysis buffer II (10 mM Tris‐HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 mM PMSF, and 1X PIC) by incubating for 10 minutes with rotation (4˚C). The nuclei were then sheared in lysis buffer III (10 mM Tris‐HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% sarkosyl, 1 mM PMSF, and 1X PIC) by 30 sec‐on/off cycles for 8 min with the S220 focused‐ultrasonicator (Covaris, Woburn, MA). Samples were precleared with protein A/G UltraLink Resin (Thermo Scientific) at 4°C with rotation for 1 hour. After preclearing, 10% of lysate was saved as an input sample, and antibodies, including isotype control IgG (Abcam) and HIF1A (Novus),) were added to each 45% of lysate. After incubation with rotation at 4°C overnight, A/G UltraLink resin was added to the lysate for conjugation with antibody and beads. The samples were allowed to rotate at 4°C for 4 hours. The beads conjugated with antibody were washed twice with high salt buffer (20 mM Tris‐HCl pH 7.5, 500 mM NaCl, 2 mM EDTA, 1% Triton X‐100, 1 mM PMSF, and 1X PIC) and twice with LiCl buffer (20 mM Tris‐HCl pH 7.5, 250 mM LiCl, 2 mM EDTA, 0.5% NP‐40, 1 mM PMSF, and 1X PIC). The beads‐antibody conjugates were washed once with TE buffer (10 mM Tris‐HCl pH 7.5, 1 mM EDTA, and 1X PIC). After removal of washing buffer, the conjugates were re‐suspended in 100 μL of elution buffer (10 mM Tris‐HCl pH 7.5, 5 mM EDTA, and 0.5% sodium dodecyl sulfate), and incubated at 65°C for 15 hours for reverse cross‐linking. The enriched chromatin and 10% input were purified using Qiagen PCR purification kit (Qiagen). The efficiency of ChIP was verified by qPCR. Primers flanking the hypoxia response element −303 bp upstream of the NTN1 transcriptional start site were used for qPCR (Forward: TCCTCCTCCTCTTCCTCACG, Reverse: CTCTAACCCAGCCTGATGGC). ChIP‐qPCR was performed using SYBR green supermix (Qiagen). ChIP fold‐enrichment was calculated by the ratio of HIF1A‐enriched signals normalized to input and isotype control IgG‐enriched signal normalized to input. The PCR cycling conditions were as follows: 95°C for 15 minutes and 35 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds.
2.17. Statistical analysis
Data analysis was performed using GraphPad Prism (version 7, San Diego, CA). Data were summarized as mean and standard deviation (SD). We performed Grubb’s test to check for outliers. No outlier was detected. Normality of data was evaluated using Shapiro‐Wilk test. F test and Brown‐Forsythe test were conducted to evaluate equal variances. For normally distributed data, we used the unpaired t test to compare means when equal‐variance assumption held and Welch’s t test when variances were significantly different. Mann‐Whitney test was used to compare medians when data of two groups had very different locations and dispersions. One‐way ANOVA was performed for K‐sample setting and two‐way ANOVA was performed for experimental data of two factors. P‐values for multiple comparisons were corrected using the Dunnett’s method when comparing to a control group, the Bonferroni method in analysis of other experimental data and the Benjamini‐Hochberg FDR method for gene screening. For time‐to‐mortality data, Kaplan‐Meier method was used to estimate survival probabilities and the log‐rank test was used to compare survival outcomes of two samples. We reported two‐sided P‐values and P‐values less than .05 were considered as statistically significant. Data are reported as mean ± standard deviation. All statistical analyses were reviewed by a faculty‐level biostatistician prior to the submission of the manuscript.
3. RESULTS
3.1. Netrin‐1 is upregulated in endotoxin stimulated myeloid cells
Previous studies have demonstrated that NGPs play significant roles in regulating the inflammatory processes of macrophages. 50 , 56 , 58 , 59 , 72 , 73 Along these lines of evidence, we hypothesized that NGPs may be upregulated by macrophages in response to inflammatory stimuli as a potential endogenous anti‐inflammatory mechanism. To investigate the induction of NGP expression in macrophages, we treated human monocyte‐derived macrophages (MΦs) with LPS for 8 hours and then, performed an RT‐qPCR array for NGP gene expression (Figure 1A). NTN1 (the gene encoding for netrin‐1) was the highest expressed NGP in the array reaching statistical significance, and the result was further confirmed using an alternate TaqMan‐based qPCR assay (Figures 1B,C). Besides the upregulation of NTN1, cell lysates also demonstrated increased levels of netrin‐1 protein following LPS exposure (Figures 1D,E). In addition, we investigated the expression of netrin‐1 in LPS‐treated human PMNs and found increased levels of netrin‐1 transcript and protein levels (Figure 1F‐I). Together these results demonstrate that netrin‐1 expression is significantly induced in myeloid cells during treatment with LPS.
FIGURE 1.
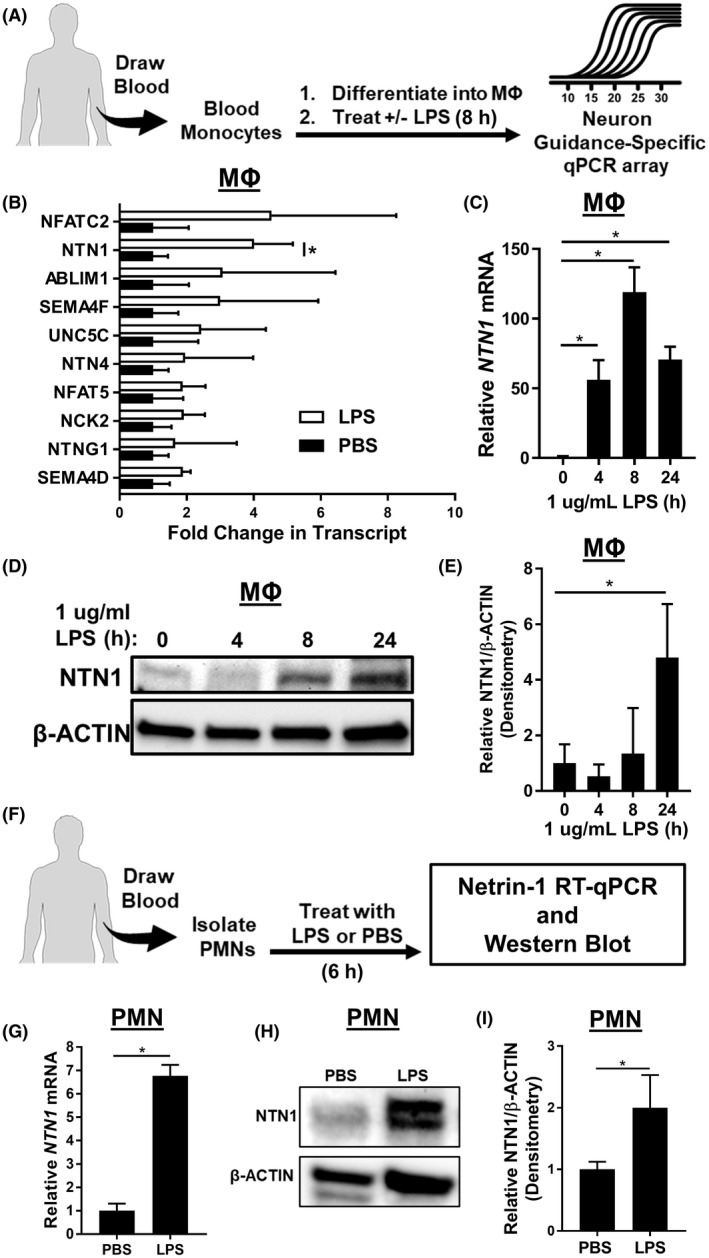
Netrin‐1 is significantly upregulated in endotoxin‐treated myeloid cells. A, Monocytes were isolated from blood of healthy donors and differentiated into macrophages (MΦs) then treated with LPS (1 µg/mL for 8 hours). RNA was isolated and used to perform a neuron guidance‐specific RT‐qPCR array. B, The top upregulated genes from the array demonstrated the netrin‐1 gene (NTN1) as the top‐statistically significant induced gene (n = 4, Bonferroni adjusted unpaired or Welch’s t test). C, PCR confirmation of NTN1 upregulation in LPS stimulated human blood‐derived MΦs (n = 3 per group, one‐way ANOVA with Dunnett’s post hoc tests). D and E, Representative image and quantification for Western blot for netrin‐1 protein expression in LPS‐treated MΦs (n = 3 per group, one‐way ANOVA with Dunnett’s post hoc tests). F, Peripheral polymorphonuclear neutrophils (PMNs) were isolated from healthy donors and treated with 1 µg/mL of LPS for 8 hours. G, NTN1 RT‐qPCR using RNA isolation from LPS‐treated PMNs (n = 3 per group, unpaired t test). H and I, Representative image and quantification for Western blot for netrin‐1 protein expression in LPS‐treated PMNs (n = 3, unpaired t test). All data are represented as mean ± SD; *P‐value < .05
3.2. Myeloid cells express high levels of netrin‐1 during LPS‐induced lung injury
In order to investigate whether netrin‐1 is expressed in myeloid cells during sepsis‐associated lung inflammation, we utilized a murine model of LPS‐induced lung injury, which results in significant recruitment and activation of leukocytes. 74 , 75 , 76 , 77 , 78 After intratracheal (i.t.) instillation of a weight‐based dose of LPS (Figure 2A), mice had the highest measured peak weight loss and BAL cell counts 3 days after LPS challenge (Figure 2B,C). Because peak leukocyte infiltration was observed 3 days post LPS instillation, we performed western blot analysis for netrin‐1 protein expression in lung tissue lysates collected at this time point. Netrin‐1 protein was significantly upregulated in lung tissue lysates (Figure 2D,E). Next, we further characterized the localization of netrin‐1 in the lungs of mice with LPS‐induced lung injury by immunohistochemistry staining for netrin‐1. Netrin‐1 staining was observed primarily in alveolar airspace infiltrating leukocytes (Figure 2F). Furthermore, we quantified the netrin‐1 expression on the surface of naïve alveolar macrophages (Ly6G‐ F4/80+, collected from i.t. PBS‐treated controls), BAL macrophages (Ly6G− F4/80+), and PMNs (Ly6G+ F4/80−) isolated from mice 3 days after i.t. instillation of LPS by flow cytometry. Netrin‐1 levels were significantly elevated in BAL macrophages after i.t. LPS instillation when compared to naïve alveolar macrophages (Figure 2G,H). Interestingly, netrin‐1 expression was significantly reduced in infiltrating PMNs when compared with both naïve alveolar macrophages and macrophages collected from i.t. LPS‐treated mice (Figure 2G,H). Finally, immunofluorescence staining of BAL cells demonstrated macrophage dominant netrin‐1 expression 3 days after i.t. LPS treatment (Figure 2I). Taken together, these data indicate that netrin‐1 is highly expressed in BAL macrophages during i.t. LPS‐induced lung injury.
FIGURE 2.
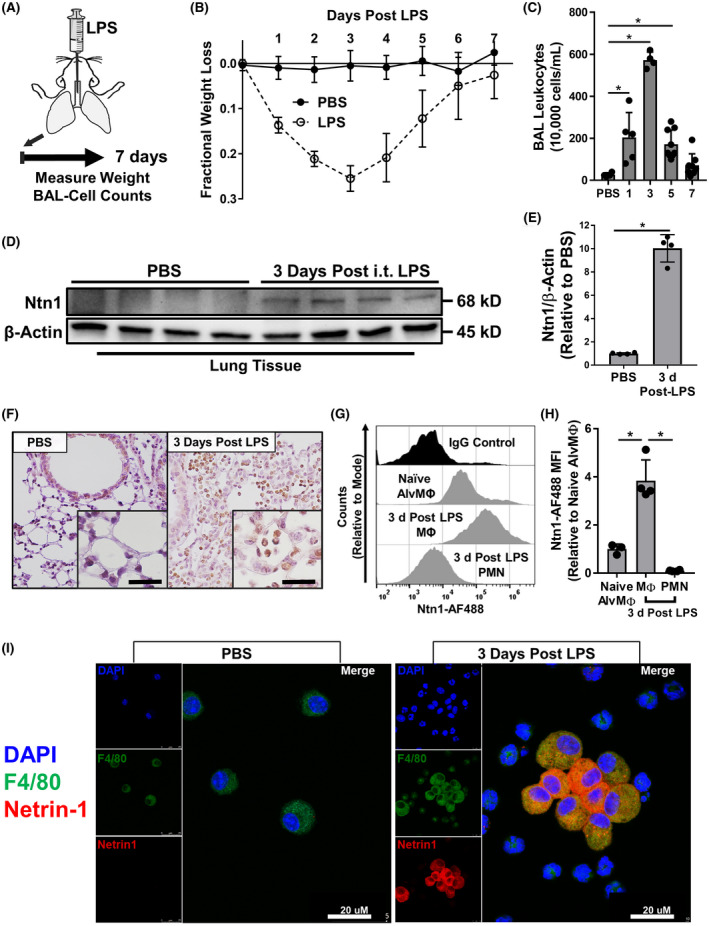
Myeloid cells recruited during endotoxin‐induced lung injury have high levels of netrin‐1. A, Schematic for LPS‐induced lung injury in mice. Mice were administered intratracheal instillation of LPS (3.75 μg/g of body weight) or PBS for control and then, monitored for weight loss or collected 1, 3, 5, or 7 days later for quantification of BAL leukocytes. B, Fractional weight loss measured in mice during LPS‐induced lung injury (n = 10 in the PBS group, n = 29 in the LPS group). C, BAL leukocytes counts in mice collected 1, 3, 5, or 7 days after LPS‐induced lung injury (n = 4‐9 per group, one‐way ANOVA with Bonferroni post hoc tests). D and E, Image and densitometry quantification for Western blot for netrin‐1 protein expression in lung tissue isolated from mice 3 days after LPS or PBS instillation (n = 4 mice per group, Mann‐Whitney test). F, Representative immunohistochemistry staining for netrin‐1 in formalin‐fixed paraffin‐embedded lung from mice 3 days after LPS or PBS instillation (n = 3 replicates per group, images are magnified 40× with 120× inserts). G, Representative flow cytometry histogram counts for netrin‐1 expression in PBS control BAL alveolar macrophages (Naïve AlvMΦ, Ly6G‐ F4/80+) or in BAL macrophages (MΦ, Ly6G‐ F4/80+) and neutrophils (PMN, Ly6G+ F4/80‐) from mice 3 days after LPS instillation (n = 3‐4 mice per group, counts relative to mode). H, Netrin‐1 flow cytometry mean fluorescent intensity quantification (n = 3‐4 per group, one‐way ANOVA with Bonferroni post hoc tests). I, Representative immunofluorescence staining of BAL leukocytes isolated 3 days after LPS or PBS instillation demonstrating co‐staining for F4/80 with netrin‐1 (n = 3 mice per group). All data are represented as mean ± SD; *P‐value < .05
3.3. Netrin‐1 expression in myeloid cells is dependent on HIF‐1α
Previous work has demonstrated that HIF‐1α directly targets the promoter of the netrin‐1 to induce its transcription. 47 Consistently, we identified an essential DNA‐binding element for HIF‐1α, named hypoxia response element (HRE; 5′‐GCGTG‐3′), 79 located 303 nucleotide base pairs upstream from the transcriptional start site of netrin‐1 (Figure 3A). To investigate the role of HIF‐1α in the expression of netrin‐1 in LPS‐stimulated human monocyte‐derived MΦs, we performed chromatin immunoprecipitation (ChIP) quantitative PCR to assess HIF‐1α binding to the HRE located in the netrin‐1 promoter. After stimulation with LPS for 8 hours, we observed a significant increase in HIF‐1α association to the netrin‐1 promoter compared with PBS‐treated controls (Figure 3B). Next, we sought to determine if the expression of netrin‐1 in BAL leukocytes is dependent on HIF‐1α during LPS‐induced lung injury in vivo. To address this question, we utilized Hif1aloxp/loxp LysM Cre mice that contain a conditional deletion for Hif1a in myeloid cells. 80 On day 3 after LPS instillation, BAL leukocytes isolated from Hif1aloxp/loxp LysM Cre mice had a significant reduction in netrin‐1 transcript levels when compared to LysM Cre control mice (Figure 3C). Similarly, immunohistochemistry staining revealed abrogated netrin‐1 protein in BAL cells collected from Hif1aloxp/loxp LysM Cre mice on day 3 after LPS‐induced lung injury (Figure 3D). In order to assess for potential nonmyeloid contributions of netrin‐1 during lung inflammation, we measured the protein level of netrin‐1 in total lung tissue of Hif1aloxp/loxp LysM Cre mice 3 days after LPS instillation. As we previously observed in C57BL/6 mice, netrin‐1 expression is increased in lung tissue of LysM Cre control mice 3 days after LPS instillation, which was completely abolished in Hif1aloxp/loxp LysM Cre mice (Figure 3E, F). These results indicate that netrin‐1 expression in infiltrating myeloid cells during LPS‐induced lung injury is dependent on HIF‐1α and support the importance of myeloid cells in netrin‐1 induction during lung inflammation.
FIGURE 3.
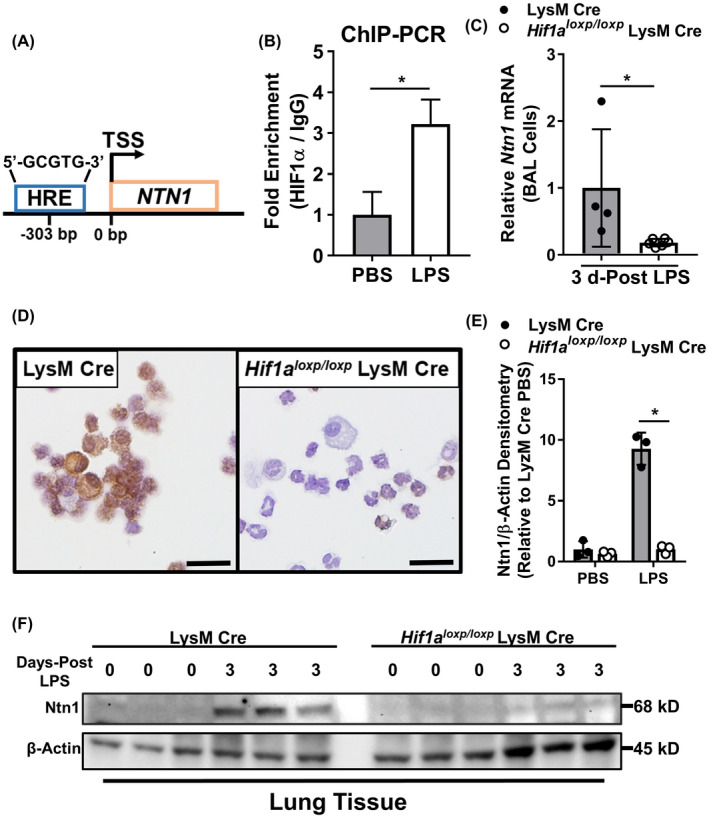
LPS‐induced netrin‐1 expression in myeloid cells is regulated by HIF‐1α. A, Schematic illustrating the location of a hypoxia response element (HRE) 303 base pairs upstream of the transcriptional start site (TSS) of NTN1. B, Human monocyte‐derived MΦs were treated with LPS (1 μg/mL) for 8 hours and then, fixed to induce DNA‐protein cross‐linking. Chromatin Immunoprecipitation (ChIP)‐PCR targeting a sequence proximal to the HRE was performed to demonstrate fold enrichment for HIF‐1α association with the promoter of NTN1 compared with IgG isotype control. Data are normalized relative to PBS‐treated MΦs (n = 3, unpaired t test). C, Relative expression of Ntn1 in BAL cells collected from mice with conditional genetic deletion of Hif1α in myeloid cells (Hif1α loxp/loxp LysM Cre) 3 days after onset of LPS‐induced lung injury compared with LysM Cre mice for control (n = 4‐6 mice per group, Mann‐Whitney test). D, Representative immunohistochemistry staining for netrin‐1 protein in BAL cells isolated from Hif1α loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced lung injury compared with LysM Cre mice (n = 3). E and F, Densitometry and western blot image and quantification for netrin‐1 protein expression in lung tissue of LysM Cre and Hif1α loxp/loxp LysM Cre 3 days after the onset of LPS‐induced lung injury (n = 3 mice per group, two‐way ANOVA with Bonferroni post hoc tests). All data are represented as mean ± SD; *P‐value < .05
3.4. Netrin‐1 deletion in myeloid cells results in exacerbated LPS‐induced lung injury
In order to investigate the functional role for myeloid cell‐derived netrin‐1 during LPS‐induced lung injury, we crossbred Ntn1loxp/loxp mice with LysM Cre mice to generate offspring that are deficient for netrin‐1 in myeloid cells (Ntn1loxp/loxp LysM Cre). We confirmed the knockout efficiency of Ntn1loxp/loxp LysM Cre mice by measuring significantly reduced transcript levels of Ntn1 in bone marrow cells and in infiltrating BAL cells 3 days after LPS instillation when compared with LysM Cre control mice (Supplementary Figure 2A, B). Following LPS instillation, Ntn1loxp/loxp LysM Cre mice had statistically increased mortality and delayed recovery of weight loss in surviving mice compared to LysM Cre mice (Figure 4A, B). Increased albumin concentration was observed in bronchoalveolar lavage fluid (BALF) from Ntn1loxp/loxp LysM Cre mice at 3 days post LPS instillation, suggesting elevated pulmonary edema (Figure 4C). Additionally, we observed elevated BAL neutrophil counts and BALF IL‐1β and IL‐6 protein levels in Ntn1loxp/loxp LysM Cre mice 3 days after LPS instillation compared with LysM Cre controls, suggesting increased levels of pulmonary inflammation in these animals (Figure 4D‐F). Finally, blinded pathological scoring of lung tissue harvested from mice 3 days after LPS instillation suggested increased lung pathology in Ntn1loxp/loxp LysM Cre mice (Figure 4G, H). Altogether, netrin‐1 deletion in myeloid cells results in a worsened outcome, increased inflammatory markers and lung pathology during LPS‐induced lung injury.
FIGURE 4.
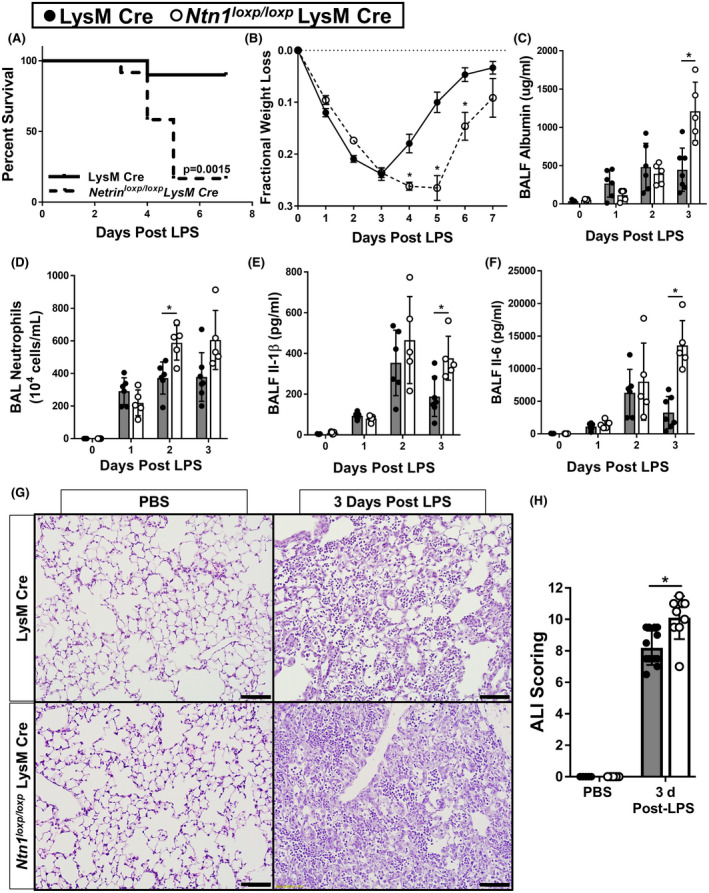
Netrin‐1 deletion in myeloid cells exacerbates LPS‐induced lung injury. A, Mortality of mice with conditional deletion of Ntn1 in myeloid cells (Ntn1loxp/loxp LysM Cre) compared with LysM Cre control mice in LPS‐induced lung injury (n = 10 mice in LysM Cre group, n = 12 mice in Ntn1loxp/loxp LysM Cre group, Kaplan‐Meier curves, log‐rank test). B) Fractional weight loss in Ntn1loxp/loxp LysM Cre mice compared with LysM Cre controls (n = 10‐12 mice per group, Bonferroni‐adjusted Welch’s or unpaired t test). C, Pulmonary edema was assessed by enzyme‐linked immunosorbent assay (ELISA) quantification of bronchoalveolar lavage fluid (BALF) albumin in Ntn1loxp/loxp LysM Cre and LysM Cre mice with LPS‐induced lung injury at indicated time points (n = 5‐7 mice per group, Bonferroni‐adjusted unpaired t test). D, Quantification of BAL neutrophils in Ntn1loxp/loxp LysM Cre and LysM Cre mice with LPS‐induced lung injury at indicated time points (n = 5‐7 mice per group, Bonferroni‐adjusted unpaired t tests). E, F, ELISA quantification of inflammatory cytokines, IL‐1β and IL‐6, in BALF collected from Ntn1loxp/loxp LysM Cre and LysM Cre mice with LPS‐induced lung injury at indicated time points (n = 5‐7 mice per group, Bonferroni‐adjusted unpaired t test). G and H, Representative sections and lung injury scoring of hematoxylin and eosin stained lung tissue collected from Ntn1loxp/loxp LysM Cre and LysM Cre mice 3 days after onset of LPS‐induced lung injury (4× magnification, n = 5‐13 mice per group, unpaired t test). All data are represented as mean ± SD. *P‐value < .05
3.5. Increased NK cell infiltration in Ntn1loxp/loxp LysM Cre mice during LPS‐induced lung injury
Based on the profound increase in lung inflammation in Ntn1loxp/loxp LysM Cre mice following i.t. LPS instillation, we next set out to gain mechanistic insight on the regulatory role of myeloid‐derived netrin‐1 by transcriptomic approach. Because we initially demonstrated an insignificant contribution of neutrophils in netrin‐1 expression, we performed mRNA sequencing using RNA isolated from BAL cells collected on day 3 after LPS instillation that was depleted of neutrophils (Figure 5A). Differential gene regulation analysis revealed 105 downregulated (fold change < 0.5) and 145 upregulated (fold change >2) statistically significant (False Discovery Rate < 0.05) genes (Figure 5B). Gene Ontology and KEGG pathway enrichment analysis of downregulated genes did not identify any enriched pathways, but upregulated genes revealed an increase in multiple pathways, with the highest enrichment being in NK cell‐mediated cytotoxicity (Figure 5C). To determine if the enrichment in the NK cell‐mediated cytotoxicity KEGG pathway was attributed to an increased number of NK cells in the total BAL cells, we first analyze the upregulated genes using CellKb, which uses a rank‐biased overlap method to match the upregulated genes to cell‐type marker sets that are published in the literature (CellKb, https://cellkb.combinatics.com/). We found that 87 of the 145 upregulated genes were matched to NK cell gene sets, with an expression‐weighted match score of 66.31, both of which were the strongest signals among other cell types that were matched (Figure 5D). Interestingly, the macrophage cell type represented the weakest match result from the CellKb algorithm (Figure 5D), suggesting differential gene regulation was not a result of transcriptomic changes between Ntn1loxp/loxp LysM Cre and LysM Cre control macrophages. In order to investigate whether there is an increase in NK cell numbers in the alveolar airspace during LPS‐induced lung injury in Ntn1loxp/loxp LysM Cre mice, we performed flow cytometry using BAL cells collected 3 days after LPS instillation and gated for NK cells (CD3− NK1.1+). Consistent with our transcriptomic results, we observed an increased percentage and the total number of NK cells in the BAL of Ntn1loxp/loxp LysM Cre mice when compared with LysM Cre controls (Figure 5E‐G). Altogether, these results demonstrate that myeloid cell‐derived netrin‐1 plays a critical role in NK cell recruitment during LPS‐induced lung injury.
FIGURE 5.
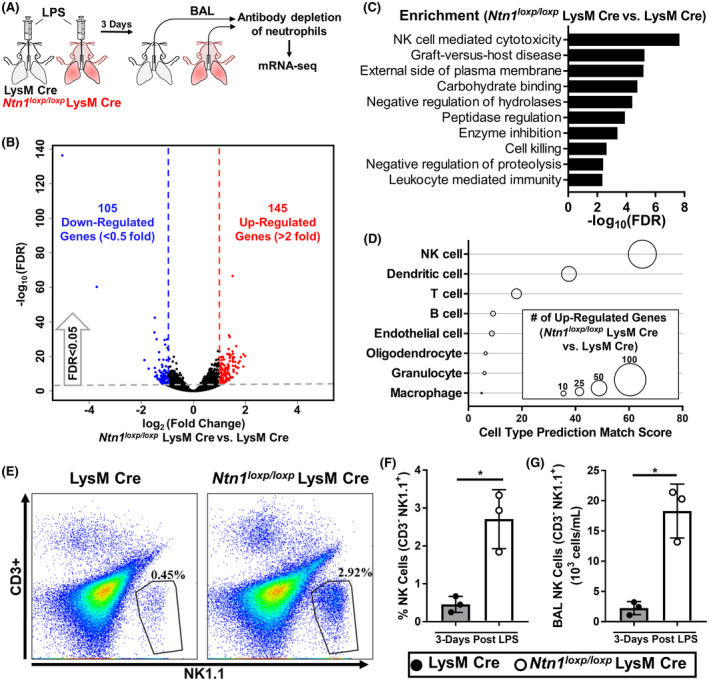
Ntn1loxp/loxp LysM Cre mice display increased natural killer (NK) cell infiltration in the airway during LPS‐induced lung injury. A, Experimental scheme: bronchia alveolar lavage (BAL) cells were collected from LysM Cre and Ntn1loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced lung injury and then, depleted for neutrophils before RNA isolation. RNA was then submitted for analysis via mRNA‐seq. B, Volcano plot of upregulated and downregulated genes in BAL cells collected from Ntn1loxp/loxp LysM Cre mice when compared with LysM Cre controls (n = 3 mice the Ntn1loxp/loxp LysM Cre group, n = 4 in LysM Cre group, the threshold for FDR was < 0.05, thresholds for fold change were < 0.5 or > 2). C, Pathway enrichment analysis for upregulated genes in BAL cells collected from Ntn1loxp/loxp LysM Cre mice when compared with LysM Cre controls. D, CellKb analysis was used to match upregulated genes with cell‐type marker sets that are published in the literature. Diameters of circles indicate the number of genes matching to each cell‐type marker set and the prediction match score is weighted by the fold enrichment of the corresponding matching genes. E, F, and G, Representative gating and quantification (percentage and total) for NK cells (CD3‐ NK1.1+) in the BAL of LysM Cre and Ntn1loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced lung injury (n = 3 mice per group, unpaired t test). All data are represented as mean ± SD; *P‐value < .05
3.6. CCL2 elevation in Ntn1loxp/loxp LysM Cre mice is associated with increased NK cell levels and inflammation
After having shown an increased NK cell infiltration in Ntn1loxp/loxp LysM Cre mice during LPS‐induced inflammation, we subsequently pursued studies to address a potential mechanistic cause. Previous studies of lung inflammation have demonstrated a role for chemokine‐mediated recruitment of NK cells. 81 , 82 , 83 We, therefore, hypothesized that chemoattractant signal is responsible for the increased accumulation of NK cells in Ntn1loxp/loxp LysM Cre mice during LPS‐induced lung injury. To identify potential chemokine or cytokine mediators, we performed a membrane‐based antibody array to measure relative levels of cytokines and chemokines in the BALF of Ntn1loxp/loxp LysM Cre mice and LysM Cre mice 3 days after intratracheal LPS instillation. We found protein expression of CCL2, a chemoattractant for NK cells, 81 , 84 , 85 to be the most elevated cytokine or chemokine in BALF of Ntn1loxp/loxp LysM Cre mice (Figure 6A, B). Using ELISA, we confirmed the elevation of CCL2 in the BALF of Ntn1loxp/loxp LysM Cre mice compared with LysM Cre mice 3 days after intratracheal LPS instillation (Figure 6C). Consequently, we pursued the notion that CCL2 inhibition might reverse the elevated NK cell recruitment and lung inflammation in Ntn1loxp/loxp LysM Cre mice. Thus, we treated Ntn1loxp/loxp LysM Cre mice with intraperitoneal neutralizing antibodies against CCL2 on days 1 and 2 after intratracheal instillation of LPS, and then, assessed NK cell recruitment and lung inflammation on day 3 (Figure 6D). Compared with IgG‐treated controls, Ntn1loxp/loxp LysM Cre mice treated with CCL2 neutralizing antibodies had a statistically significant reduction in total BAL NK cells via flow cytometry (Figure 6E). Ntn1loxp/loxp LysM Cre mice treated with CCL2 neutralizing antibodies also had a significant reduction in infiltrating BAL neutrophils and in alveolar barrier permeability, as indicated by reduced BALF albumin concentration (Figure 6F, G). Upon histological evaluation of hematoxylin and eosin‐stained lung tissue, there was a statistically significant reduction in lung pathology as determined by blinded pathological scoring (Figure 6H, I). Taken together, these studies support the role of myeloid‐derived netrin‐1 in limiting CCL2‐mediated NK cell recruitment during lung inflammation.
FIGURE 6.
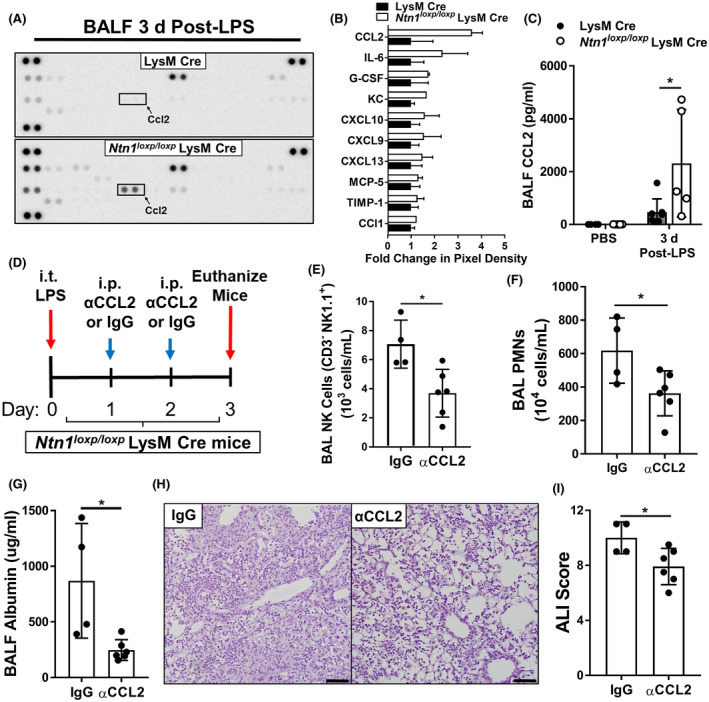
C‐C motif chemokine ligand 2 (CCL2) neutralization in Ntn1loxp/loxp LysM Cre mice reduces natural killer (NK) cell infiltration in the airway and improves LPS‐induced lung injury. Using Bronchoalveolar lavage fluid (BALF) collected from LysM Cre and Ntn1loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced lung injury, we performed a membrane‐based sandwich immunoassay to profile for changes in inflammatory cytokines and chemokines. A, Representative dot‐blot of the membrane‐based immunoassay. B, Quantified pixel density of the top 10 upregulated cytokines/chemokines in Ntn1loxp/loxp LysM Cre mice relative to LysM Cre controls (n = 2 per group). C, CCL2 upregulation was confirmed in BALF of Ntn1loxp/loxp LysM Cre mice relative to LysM Cre mice 3 days after onset of LPS‐induced lung injury using enzyme‐linked immunosorbent assay (ELISA) (n = 4‐7 mice per group, Bonferroni‐adjusted unpaired t test). D, Experimental scheme: Ntn1loxp/loxp LysM Cre were given LPS‐induced lung injury and then, subsequently given intraperitoneal (i.p.) injections of neutralizing CCL2 antibodies (αCCL2) or IgG isotype control (10 μg/g of body weight) on days 1 and 2 after the onset of lung inflammation. Mice were subsequently euthanized for analysis on day 3 after the onset of lung inflammation. E, Quantification of total NK cells (CD3‐ NK1.1+) in the BAL collected from αCCL2‐treated Ntn1loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced inflammation was measured by flow cytometry (n = 4‐6 mice per group, unpaired t test). F, Quantification of total polymorphonuclear neutrophils (PMNs) in the BAL collected from αCCL2‐treated Ntn1loxp/loxp LysM Cre mice 3 days after onset of LPS‐induced lung injury (n = 4‐6 mice per group, unpaired t test). G, Pulmonary edema was assessed by enzyme‐linked immunosorbent assay (ELISA) quantification of bronchoalveolar lavage fluid (BALF) albumin in αCCL2‐treated Ntn1loxp/loxp LysM Cre mice (n = 4‐6 mice per group, Mann‐Whitney test). H and I, Representative hematoxylin and eosin staining and lung pathology scoring of lung sections from αCCL2‐treated Ntn1loxp/loxp LysM Cre mice (n = 4‐6 mice per group, unpaired t test). All data are represented as mean ± SD; *P‐value < .05
4. DISCUSSION
An increasingly recognized class of immune‐modulators is the NGPs. 43 The present studies aim at investigating the role of myeloid cell‐derived NGPs during sepsis‐associated lung inflammation. Through a qPCR‐based screen, we identified netrin‐1 to be highly upregulated in LPS stimulated hMDMs, as well as in peripheral circulating neutrophils. During LPS‐induced lung injury in mice, pulmonary netrin‐1 was significantly elevated 3 days after the onset of inflammation, which coincided with the peak infiltration of immune cells into the alveolar airspace. Using immunohistochemistry and flow cytometry, we localized netrin‐1 expression to infiltrating immune cells, particularly in macrophages. Subsequent transcriptional studies identified HIF‐1α as a key transcriptional factor for netrin‐1 regulation, and transgenic mice lacking HIF‐1α in the myeloid compartment failed to induce netrin‐1 during LPS‐induced lung injury. Furthermore, the deletion of netrin‐1 in the myeloid cell compartment (Ntn1loxp/loxp LysM Cre) resulted in worsened outcomes during LPS‐induced lung injury in mice, indicating a protective role for myeloid cell‐derived netrin‐1. Surprisingly, increased NK cell and CCL2 accumulation are observed in Ntn1loxp/loxp LysM Cre mice. Lastly, when Ntn1loxp/loxp LysM Cre mice were treated with CCL2 neutralizing antibodies, we observed a reduction in NK cell recruitment and improvement in lung inflammation. Together, these data reveal myeloid‐derived netrin‐1 as a critical negative feedback mechanism during lung inflammation.
Our findings that myeloid‐derived netrin‐1 protects mice against lung inflammation are in alignment with other studies. Netrin‐1 has been reported to dampen inflammation in several models of inflammatory conditions including peritonitis, 56 acute lung injury 46 , 86 kidney ischemia‐reperfusion injury, 58 , 87 arthritis, 88 and colitis. 89 Mechanistically, netrin‐1 serves to limit inflammation by regulation of leukocyte migration and accumulation 88 , 90 , 91 , 92 and is also shown to facilitate the active resolution of inflammation by promoting pro‐resolving mediator expression during peritonitis 61 and liver ischemia‐reperfusion injury. 93 Ex vivo netrin‐1 treatment of macrophages and endogenous overexpression of netrin‐1 in vivo was found to promote the transition to an anti‐inflammatory M2‐like phenotype macrophage, which acts to limit inflammation and encourage wound healing. 58 , 87 , 92 Previous studies have also demonstrated that netrin‐1 is expressed by myeloid cells and plays a functional role in disease conditions. For example, macrophage‐derived netrin‐1 has been shown to promote retention of adipose tissue macrophages leading to insulin resistance, 94 promote the development of atherosclerosis, 48 and to support the progression of aortic aneurysms. 95 Thus, myeloid cell‐derived netrin‐1 plays diverse regulatory roles under different inflammatory conditions, such as in the case of adenosine signaling. 6 , 96
Several previous studies have demonstrated hypoxia and HIF‐1α‐dependent expression of netrin‐1 during inflammatory conditions. HIF‐1α‐dependence netrin‐1 induction in mucosal epithelial cells has been illustrated in models of hypoxia‐induced mucosal inflammation. 47 In addition, macrophages found in hypoxic regions of atherosclerotic show increased netrin‐1 level in both human and mouse studies, and HIF activation induces netrin‐1 expression in macrophages in a HIF‐1α‐dependent manner. 97 Besides HIF‐1α‐dependent induction, netrin‐1 expression is also controlled by NF‐κB, and NF‐ κB can act as both a transcriptional activator 98 and repressor. 46 Furthermore, vagal nerve innervation has also been shown to regulate pulmonary expression of netrin‐1 as unilateral vagotomy decreases netrin‐1 expression in the lung. 61 However, the neuronal stimulation of netrin‐1 expression in myeloid cells is unknown.
Netrin‐1 imparts cellular functions through interacting with its receptors. The best described netrin‐1 receptors during inflammatory regulation are uncoordinated receptor 5 (UNC5b), the neogenin receptor, and the adenosine 2B (A2B) receptor. 46 , 47 , 49 , 56 Netrin‐1 interacting with UNC5b on leukocytes has been implicated in regulating migratory functions as well as dampening inflammatory cytokine expression. 48 , 94 , 95 , 99 The A2B receptor is involved in regulating inflammation in response to elevated extracellular adenosine levels that are commonly present during settings of tissue injury and has also been demonstrated to be a critical mediator for netrin‐1 regulation of inflammation. 77 , 91 , 100 , 101 , 102 Previous studies have shown that the A2B receptor mediates netrin‐1‐induced expression of resolution mediators to promote regeneration during liver inflammation, 93 dampening of inflammation during peritonitis, 56 attenuate experimental colitis, 89 and limit pulmonary inflammation. 46 In contrast with UNC5b and the A2B receptor, the netrin‐1 receptor, neogenin, has been shown to promote acute inflammation and inhibit monocyte polarization toward anti‐inflammatory and pro‐resolution phenotypes. 103 , 104
The present studies have established an exciting new link between myeloid‐derived netrin‐1 and NK cell migration during endotoxin‐induced lung injury. Indeed, others have reported NK cells as drivers of immune‐pathology in several models of organ injuries. 105 , 106 , 107 , 108 For example, tissue‐resident NK cells promote ischemic kidney injury by mediating local responses. 106 During LPS‐induced lung injury, it was shown that NK cell depletion in mice resulted in reduced chemokine‐mediated neutrophil recruitment and improved outcomes. 107 Additionally, NK cells were reported to be primary drivers of immune‐pathology during a model of interstitial pneumonia. 108 In models of viral infection, NK cells were implicated as the critical players in causing excessive inflammation leading to bystander lung tissue injury which was reversed by antibody‐mediated depletion. 109 , 110 Interestingly, pre‐infecting mice with Klebsiella pneumoniae provided protection in subsequent lethal challenges of influenza virus, which was attributed to K. pneumoniae‐conditioned reduction of NK cell recruitment and inflammation. 111 Consistent with our findings of CCL2‐driven NK cell accumulation in Ntn1loxp/loxp LysM Cre mice, previous work has implicated CCL2 in the recruitment of NK cells during a model of invasive pulmonary aspergillosis infection. 81
Our studies have several limitations. Ntn1loxp/loxp LysM Cre mice have a deletion of netrin‐1 in macrophage and neutrophil populations, as well as in other granulocytes. 112 , 113 Thus, the profound phenotype in these mice could be mediated by several cell types together. To further dissect the specific role of netrin‐1 in macrophages and neutrophils during endotoxin‐induced lung injury, Ntn1loxp/loxp mice could be crossbred with hCD68‐rtTA/ Teto‐Cre mice, and MRP8‐Cre mice, respectively. Furthermore, the contribution of myeloid netrin‐1 in NK cell recruitment was only indicated in in vivo studies. Further in vitro experiments will facilitate the determination of direct or indirection interaction between these two cell populations. Finally, although we have identified the upregulation of netrin‐1 in human neutrophils and macrophages during inflammatory stimulation, the functional role of netrin‐1 in NK cell recruitment during human endotoxin‐induced lung injury was not explored in our study. Additional studies employing lung tissues and cells isolated from respiratory washout from patients suffering from lung gram‐negative bacteria infection or sepsis would be crucial to illustrate this particular interaction.
Altogether, these findings highlight a novel role of myeloid cell‐derived netrin‐1 during pulmonary inflammation in limiting excessive inflammation (Figure 7). The impact of myeloid‐derived netrin‐1 is achieved by orchestrating CCL2‐mediated NK cell infiltration during lung inflammation. Our studies indicate a novel mechanism for the immune‐modulatory effect of netrin‐1 in myeloid cells, particularly during lung injury. Our studies have also implied a functional role of myeloid‐derived netrin‐1 in NK cell recruitment, which contributes to the complexity of netrin‐1 biology for the field. Moreover, our studies suggest that myeloid‐derived netrin‐1 might play an important role in other diseases where NK cells play major functional roles, for example, cancer, 114 autoimmune diseases, 115 and viral infection. 116 Thus, investigating how myeloid‐derived netrin‐1 contributes to the pathophysiology of these conditions could be of great interest to the field. Finally, future work should focus on dissecting the target cells of myeloid‐derived netrin‐1 as the sources of CCL2, as well as the receptor that mediates its immune‐modulatory effects.
FIGURE 7.
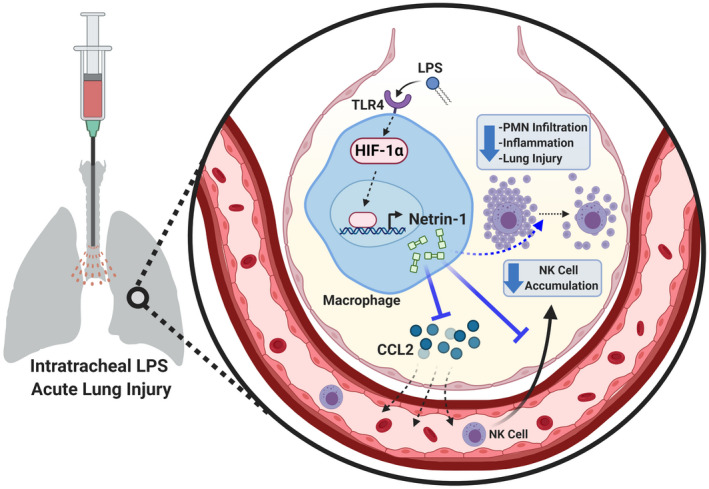
Proposed mechanism for myeloid cell‐derived Netrin‐1 during LPS‐induced acute lung injury. During acute lung injury initiated by intratracheal injection of LPS, macrophages are activated through toll‐like receptor 4. Netrin‐1 is expressed by activated macrophages via stabilization of the transcription factor, hypoxia inducible factor‐1 alpha (HIF‐1α), which binds to and activates the promoter of Netrin‐1. Myeloid‐derived Netrin‐1 plays a critical role in regulating chemokine (C‐C motif) ligand 2 (CCL2) and subsequently limits the recruitment of pro‐inflammatory natural killer (NK) cells. The ultimate effect of regulating NK cell accumulation by myeloid‐derived Netrin‐1 is to dampen inflammation, polymorphonuclear neutrophil (PMN) accumulation, and lung injury. Created with BioRender.com
CONFLICT OF INTEREST
The authors declare no competing or financial interests.
AUTHOR CONTRIBUTIONS
N. K. Berg and J. Li designed and performed the experiments and wrote and revised the manuscript. B. Kim, T. Mills, and X. Li performed the experiments and analyzed the data. G. Pei and Z. Zhao processed and analyzed the mRNA‐sequencing data. X. Zhang analyzed and performed the statistical analyses for the data. W. Ruan designed the experiments and analyzed the data. H. K. Eltzschig and X. Yuan supervised the experimental design and data analysis, and finalized the manuscript.
Supporting information
Fig S1‐S2
table S1‐S3
ACKNOWLEDGMENTS
This work was supported by the Training Interdisciplinary Pharmacology Scientists (TIPS) Program (NIH Grant No. T32GM120011 from the National Institutes of Health, Bethesda, Maryland; to N. K. Berg); unrestricted grant from the American Thoracic Society (New York, New York; to X. Yuan); grant No. 19CDA34660279, an American Heart Association (Dallas, Texas; to X. Yuan) Career Development Award; grant No. CA‐622265; to X. Yuan, an American Lung Association (Chicago, Illinois; to X. Yuan) Catalyst Award; grant No. 1UL1TR003167–01 to X. Yuan, a Center for Clinical and Translational Sciences, McGovern Medical School (Houston, Texas; to X. Yuan) Pilot Award; a Parker B. Francis Fellowship (Kansas City, Missouri; to X. Yuan); and National Institutes of Health (Bethesda, Maryland) grant Nos. R01HL154720, R01DK122796, R01DK109574 and R01HL133900; and Department of Defense, grant number: W81XWH2110032 (to H. K. Eltzschig). The sequencing data were generated by the UTHealth Cancer Genomics Core supported by the Cancer Prevention and Research Institute of Texas (CPRIT RP180734). The authors acknowledge Yanyu Wang, Ph.D. for critically reviewing the data presented in this manuscript.
Berg NK, Li J, Kim B, et al. Hypoxia‐inducible factor‐dependent induction of myeloid‐derived netrin‐1 attenuates natural killer cell infiltration during endotoxin‐induced lung injury. The FASEB Journal. 2021;35:e21334. 10.1096/fj.202002407R
Nathaniel K. Berg and Jiwen Li are the authors contributed equally to this work.
Contributor Information
Jiwen Li, Email: lijiwen@zju.edu.cn.
Holger K. Eltzschig, Email: Holger.Eltzschig@uth.tmc.edu.
Xiaoyi Yuan, Email: Xiaoyi.Yuan@uth.tmc.edu.
REFERENCES
- 1. Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377:1904‐1905. [DOI] [PubMed] [Google Scholar]
- 2. Lee LK, Medzikovic L, Eghbali M, Eltzschig HK, Yuan X. The Role of MicroRNAs in acute respiratory distress syndrome and sepsis, from targets to therapies: a narrative review. Anesthesia & Analgesia. 2020;131(5):1471–1484. 10.1213/ane.0000000000005146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428‐435. [DOI] [PubMed] [Google Scholar]
- 4. Shen H, Kreisel D, Goldstein DR. Processes of sterile inflammation. J Immunol. 2013;191:2857‐2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Millien VO, Lu W, Mak G, et al. Airway fibrinogenolysis and the initiation of allergic inflammation. Ann Am Thorac Soc. 2014;11(Suppl 5):S277‐S283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Le TT, Berg NK, Harting MT, Li X, Eltzschig HK, Yuan X. Purinergic signaling in pulmonary inflammation. Front Immunol. 2019;10:1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hong MJ, Gu BH, Madison MC, et al.. Protective role of gammadelta T cells in cigarette smoke and influenza infection. Mucosal Immunol. 2018;11:894‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA. 2016;315:801‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matthay MA, Zemans RL, Zimmerman GA, et al. Acute respiratory distress syndrome. Nat Rev Dis Primers. 2019;5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rabb H, Griffin MD, McKay DB, et al. Acute dialysis quality initiative consensus XWG: inflammation in AKI: current understanding, key questions, and knowledge gaps. J Am Soci Nephrol: JASN. 2016;27:371‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arslan F, De Kleijn DP, Pasterkamp G. Innate immune signaling in cardiac ischemia. Nat Rev Cardiol. 2011;8:292. [DOI] [PubMed] [Google Scholar]
- 12. Harari OA, Liao JK. NF‐κB and innate immunity in ischemic stroke. Ann N Y Acad Sci. 2010;1207:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhai Y, Busuttil RW, Kupiec‐Weglinski JW. Liver ischemia and reperfusion injury: new insights into mechanisms of innate—adaptive immune‐mediated tissue inflammation. Am J Transplant. 2011;11:1563‐1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eltzschig HK, Weissmuller T, Mager A, Eckle T. Nucleotide metabolism and cell‐cell interactions. Methods Mol Biol. 2006;341:73‐87. [DOI] [PubMed] [Google Scholar]
- 15. Bhavani S, Yuan X, You R, Shan M, Corry D, Kheradmand F. Loss of peripheral tolerance in emphysema. phenotypes, exacerbations, and disease progression. Ann Am Thorac Soc. 2015;12(Suppl 2):S164‐S168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan X, Chang CY, You R, et al. Cigarette smoke‐induced reduction of C1q promotes emphysema. JCI Insight. 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu W, You R, Yuan X, et al. The microRNA miR‐22 inhibits the histone deacetylase HDAC4 to promote T(H)17 cell‐dependent emphysema. Nat Immunol. 2015;16:1185‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poth JM, Brodsky K, Ehrentraut H, Grenz A, Eltzschig HK. Transcriptional control of adenosine signaling by hypoxia‐inducible transcription factors during ischemic or inflammatory disease. J Mol Med (Berl). 2013;91:183‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shan M, Yuan X, Song LZ, et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med. 2012;4:117ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yuan X, Berg N, Lee JW,. MicroRNA miR‐223 as regulator of innate immunity. J Leukoc Biol. 2018;104:515‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deaglio S, Dwyer KM, Gao W, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257‐1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eltzschig HK, Sitkovsky MV, Robson SC. Purinergic signaling during inflammation. N Engl J Med. 2012;367:2322‐2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hasko G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Afonina IS, Zhong Z, Karin M, Beyaert R. Limiting inflammation—the negative regulation of NF‐κB and the NLRP3 inflammasome. Nat Immunol. 2017;18:861‐869. [DOI] [PubMed] [Google Scholar]
- 25. Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti‐inflammatory and pro‐resolution lipid mediators. Nat Rev Immunol. 2008;8:349‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Serhan CN. Pro‐resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colgan SP. Resolvins resolve to heal mucosal wounds. Proc Natl Acad Sci USA. 2020;117:10621‐10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kominsky DJ, Keely S, MacManus CF, et al. An endogenously anti‐inflammatory role for methylation in mucosal inflammation identified through metabolite profiling. J Immunol. 2011;186:6505‐6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacManus CF, Campbell EL, Keely S, Burgess A, Kominsky DJ, Colgan SP. Anti‐inflammatory actions of adrenomedullin through fine tuning of HIF stabilization. FASEB J. 2011;25:1856‐1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Körner A, Schlegel M, Theurer J, et al. Resolution of inflammation and sepsis survival are improved by dietary Ω‐3 fatty acids. Cell Death Differ. 2018;25:421‐431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watanabe S, Alexander M, Misharin AV, Budinger GRS. The role of macrophages in the resolution of inflammation. J Clin Invest. 2019;129:2619‐2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Serhan CN, Levy BD. Resolvins in inflammation: emergence of the pro‐resolving superfamily of mediators. J Clin Invest. 2018;128:2657‐2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krishnamoorthy N, Abdulnour RE, Walker KH, Engstrom BD, Levy BD. Specialized proresolving mediators in innate and adaptive immune responses in airway diseases. Physiol Rev. 2018;98:1335‐1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sham HP, Walker KH, Abdulnour RE, et al. 15‐epi‐Lipoxin A4, resolvin D2, and resolvin D3 induce NF‐kappaB regulators in bacterial pneumonia. J Immunol. 2018;200:2757‐2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duvall MG, Bruggemann TR, Levy BD. Bronchoprotective mechanisms for specialized pro‐resolving mediators in the resolution of lung inflammation. Mol Aspects Med. 2017;58:44‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abdulnour RE, Howrylak JA, Tavares AH, et al. Phospholipase D isoforms differentially regulate leukocyte responses to acute lung injury. J Leukoc Biol. 2018;103:919‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sugimoto MA, Sousa LP, Pinho V, Perretti M, Teixeira MM. Resolution of inflammation: what controls its onset? Front Immunol. 2016;7:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nemeth ZH, Lutz CS, Csoka B, et al. Adenosine augments IL‐10 production by macrophages through an A2B receptor‐mediated posttranscriptional mechanism. J Immunol. 2005;175:8260‐8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389‐1399. [DOI] [PubMed] [Google Scholar]
- 40. Serafini T, Kennedy TE, Gaiko MJ, Mirzayan C, Jessell TM, Tessier‐Lavigne M. The netrins define a family of axon outgrowth‐promoting proteins homologous to C. elegans UNC‐6. Cell. 1994;78:409‐424. [DOI] [PubMed] [Google Scholar]
- 41. Varadarajan SG, Kong JH, Phan KD, et al. Netrin1 produced by neural progenitors, not floor plate cells, is required for axon guidance in the spinal cord. Neuron. 2017;94(790‐799):e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brunet I, Gordon E, Han J, et al. Netrin‐1 controls sympathetic arterial innervation. J Clin Invest. 2014;124:3230‐3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mirakaj V, Rosenberger P. Immunomodulatory functions of neuronal guidance proteins. Trends Immunol. 2017;38:444‐456. [DOI] [PubMed] [Google Scholar]
- 44. Shimizu I, Yoshida Y, Moriya J, et al. Semaphorin3E‐induced inflammation contributes to insulin resistance in dietary obesity. Cell Metabol. 2013;18:491‐504. [DOI] [PubMed] [Google Scholar]
- 45. Movassagh H, Saati A, Nandagopal S, et al. Chemorepellent semaphorin 3E negatively regulates neutrophil migration in vitro and in vivo. J Immunol. 2017;198:1023‐1033. [DOI] [PubMed] [Google Scholar]
- 46. Mirakaj V, Thix CA, Laucher S, et al. Netrin‐1 dampens pulmonary inflammation during acute lung injury. Am J Respir Crit Care Med. 2010;181:815‐824. [DOI] [PubMed] [Google Scholar]
- 47. Rosenberger P, Schwab JM, Mirakaj V, et al. Hypoxia‐inducible factor‐dependent induction of netrin‐1 dampens inflammation caused by hypoxia. Nat Immunol. 2009;10:195‐202. [DOI] [PubMed] [Google Scholar]
- 48. van Gils JM, Derby MC, Fernandes LR, et al. The neuroimmune guidance cue netrin‐1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat Immunol. 2012;13:136‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ly NP, Komatsuzaki K, Fraser IP, et al. Netrin‐1 inhibits leukocyte migration in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:14729‐14734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu JY, Feng L, Park H‐T, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948‐952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aherne CM, Collins CB, Eltzschig HK. Netrin‐1 guides inflammatory cell migration to control mucosal immune responses during intestinal inflammation. Tissue Barriers. 2013;1:e24957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Granja T, Köhler D, Mirakaj V, Nelson E, König K, Rosenberger P. Crucial role of Plexin C1 for pulmonary inflammation and survival during lung injury. Mucosal Immunol. 2014;7:879‐891. [DOI] [PubMed] [Google Scholar]
- 53. König K, Granja T, Eckle VS, et al. Inhibition of plexin C1 protects against hepatic ischemia‐reperfusion injury. Crit Care Med. 2016;44:e625‐e632. [DOI] [PubMed] [Google Scholar]
- 54. Köhler D, Granja T, Volz J, et al. Red blood cell‐derived semaphorin 7A promotes thrombo‐inflammation in myocardial ischemia‐reperfusion injury through platelet GPIb. Nat Commun. 2020;11:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ly NP, Komatsuzaki K, Fraser IP, et al. Netrin‐1 inhibits leukocyte migration in vitro and in vivo. Proc Natl Acad Sci USA. 2005;102:14729‐14734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mirakaj V, Gatidou D, Pötzsch C, König K, Rosenberger P. Netrin‐1 signaling dampens inflammatory peritonitis. J Immunol. 2011;186:549‐555. [DOI] [PubMed] [Google Scholar]
- 57. van Gils JM, Derby MC, Fernandes LR, et al. The neuroimmune guidance cue netrin‐1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat Immunol. 2012;13:136‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ranganathan PV, Jayakumar C, Mohamed R, Dong Z, Ramesh G. Netrin‐1 regulates the inflammatory response of neutrophils and macrophages, and suppresses ischemic acute kidney injury by inhibiting COX‐2‐mediated PGE2 production. Kidney Int. 2013;83:1087‐1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ranganathan PV, Jayakumar C, Ramesh G. Netrin‐1‐treated macrophages protect the kidney against ischemia‐reperfusion injury and suppress inflammation by inducing M2 polarization. Am J Physiol‐Renal Physiol. 2013;304:F948‐F957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schlegel M, Köhler D, Körner A, et al. The neuroimmune guidance cue netrin‐1 controls resolution programs and promotes liver regeneration. Hepatology. 2016;63:1689‐1705. [DOI] [PubMed] [Google Scholar]
- 61. Mirakaj V, Dalli J, Granja T, Rosenberger P, Serhan CN. Vagus nerve controls resolution and pro‐resolving mediators of inflammation. J Exp Med. 2014;211:1037‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bin JM, Han D, Lai Wing Sun K, et al. Complete loss of netrin‐1 results in embryonic lethality and severe axon guidance defects without increased neural cell death. Cell Rep. 2015;12:1099‐1106. [DOI] [PubMed] [Google Scholar]
- 63. Ryan HE, Poloni M, McNulty W, et al. Hypoxia‐inducible factor‐1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010‐4015. [PubMed] [Google Scholar]
- 64. Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265‐277. [DOI] [PubMed] [Google Scholar]
- 65. Cramer T, Yamanishi Y, Clausen BE, et al. HIF‐1α is essential for myeloid cell‐mediated inflammation. Cell. 2003;112:645‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang Y‐L, Tang G‐J, Wu Y‐L, Yien H‐W, Lee T‐S, Kou YR. Exacerbation of wood smoke‐induced acute lung injury by mechanical ventilation using moderately high tidal volume in mice. Respir Physiol Neurobiol. 2008;160:99‐108. [DOI] [PubMed] [Google Scholar]
- 67. Martin M. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.J. 2011;17:10‐12. [Google Scholar]
- 68. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. Ser B (Methodological). 1995;57:289‐300. [Google Scholar]
- 71. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199‐W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mirakaj V, Jennewein C, König K, Granja T, Rosenberger P. The guidance receptor neogenin promotes pulmonary inflammation during lung injury. FASEB J. 2012;26:1549‐1558. [DOI] [PubMed] [Google Scholar]
- 73. Mirakaj V, Brown S, Laucher S, et al. Repulsive guidance molecule‐A (RGM‐A) inhibits leukocyte migration and mitigates inflammation. Proc Natl Acad Sci. 2011;108:6555‐6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kabir K, Gelinas J‐P, Chen M, et al. Characterization of a murine model of endotoxin‐induced acute lung injury. Shock. 2002;17:300‐303. [DOI] [PubMed] [Google Scholar]
- 75. Ngamsri K‐C, Wagner R, Vollmer I, Stark S, Reutershan J. Adenosine receptor A1 regulates polymorphonuclear cell trafficking and microvascular permeability in lipopolysaccharide‐induced lung injury. J Immunol. 2010;185:4374‐4384. [DOI] [PubMed] [Google Scholar]
- 76. Schingnitz U, Hartmann K, Macmanus CF, et al. Signaling through the A2B adenosine receptor dampens endotoxin‐induced acute lung injury. J Immunol (Baltimore, Md: 1950). 2010;184:5271‐5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ehrentraut H, Westrich JA, Eltzschig HK, Clambey ET. Adora2b adenosine receptor engagement enhances regulatory T cell abundance during endotoxin‐induced pulmonary inflammation. PLoS ONE. 2012;7:e32416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schingnitz U, Hartmann K, Macmanus CF, et al. Signaling through the A2B adenosine receptor dampens endotoxin‐induced acute lung injury. J Immunol. 2010;184:5271‐5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dengler VL, Galbraith MD, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cramer T, Yamanishi Y, Clausen BE, et al. HIF‐1alpha is essential for myeloid cell‐mediated inflammation. Cell. 2003;112:645‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Morrison BE, Park SJ, Mooney JM, Mehrad B. Chemokine‐mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J Clin Invest. 2003;112:1862‐1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jiang D, Liang J, Hodge J, et al. Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest. 2004;114:291‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Carlin LE, Hemann EA, Zacharias ZR, Heusel JW, Legge KL. Natural killer cell recruitment to the lung during influenza A virus infection is dependent on CXCR3, CCR5, and virus exposure dose. Front Immunol. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maghazachi AA. Role of chemokines in the biology of natural killer cells. Curr Top Microbiol Immunol. 2010;341:37‐58. [DOI] [PubMed] [Google Scholar]
- 85. Robertson MJ. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71:173‐83. [PubMed] [Google Scholar]
- 86. Mutz C, Mirakaj V, Vagts DA, et al. The neuronal guidance protein netrin‐1 reduces alveolar inflammation in a porcine model of acute lung injury. Crit Care (London, England). 2010;14:R189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ranganathan PV, Jayakumar C, Ramesh G. Netrin‐1‐treated macrophages protect the kidney against ischemia‐reperfusion injury and suppress inflammation by inducing M2 polarization. Am J Physiol Renal Physiol. 2013;304:F948‐F957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mediero A, Wilder T, Ramkhelawon B, Moore KJ, Cronstein BN. Netrin‐1 and its receptor Unc5b are novel targets for the treatment of inflammatory arthritis. FASEB J. 2016;30:3835‐3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aherne CM, Collins CB, Masterson JC, et al. Neuronal guidance molecule netrin‐1 attenuates inflammatory cell trafficking during acute experimental colitis. Gut. 2012;61:695‐705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Taylor L, Brodermann MH, McCaffary D, Iqbal AJ, Greaves DR. Netrin‐1 Reduces monocyte and macrophage chemotaxis towards the complement component C5a. PLoS ONE. 2016;11:e0160685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Schingnitz U, Hartmann K, MacManus CF, et al. Signaling through the A2B adenosine receptor dampens endotoxin‐induced acute lung injury. J Immunol. 2010;184:5271‐5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang Y, Chen P, Di G, Qi X, Zhou Q, Gao H. Netrin‐1 promotes diabetic corneal wound healing through molecular mechanisms mediated via the adenosine 2B receptor. Sci Rep. 2018;8:5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schlegel M, Köhler D, Körner A, Granja T, Straub A, Giera M, et al. The neuroimmune guidance cue netrin‐1 controls resolution programs and promotes liver regeneration. Hepatology. 2016;63:1689‐1705. [DOI] [PubMed] [Google Scholar]
- 94. Ramkhelawon B, Hennessy EJ, Ménager M, et al. Netrin‐1 promotes adipose tissue macrophage retention and insulin resistance in obesity. Nat Med. 2014;20:377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hadi T, Boytard L, Silvestro M, et al. Macrophage‐derived netrin‐1 promotes abdominal aortic aneurysm formation by activating MMP3 in vascular smooth muscle cells. Nat Comm. 2018;9:5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karmouty‐Quintana H, Xia Y, Blackburn MR. Adenosine signaling during acute and chronic disease states. J Mol Med (Berl). 2013;91:173‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ramkhelawon B, Yang Y, van Gils JM, et al. Hypoxia Induces Netrin‐1 and Unc5b in Atherosclerotic Plaques; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Paradisi A, Maisse C, Bernet A, et al. NF‐kappaB regulates netrin‐1 expression and affects the conditional tumor suppressive activity of the netrin‐1 receptors. Gastroenterology. 2008;135:1248‐1257. [DOI] [PubMed] [Google Scholar]
- 99. Tadagavadi RK, Wang W, Ramesh G. Netrin‐1 regulates Th1/Th2/Th17 cytokine production and inflammation through UNC5B receptor and protects kidney against ischemia‐reperfusion injury. J Immunol. 2010;185:3750‐3758. [DOI] [PubMed] [Google Scholar]
- 100. Koscsó B, Trepakov A, Csóka B, et al. Stimulation of A2B adenosine receptors protects against trauma‐hemorrhagic shock‐induced lung injury. Purinergic Signal. 2013;9:427‐432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Koscsó B, Csóka B, Kókai E, et al. Adenosine augments IL‐10‐induced STAT3 signaling in M2c macrophages. J Leukoc Biol. 2013;94:1309‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Haskó G, Csóka B, Németh ZH, Vizi ES, Pacher P. A(2B) adenosine receptors in immunity and inflammation. Trends Immunol. 2009;30:263‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. König K, Gatidou D, Granja T, Meier J, Rosenberger P, Mirakaj V. The axonal guidance receptor neogenin promotes acute inflammation. PLOS ONE. 2012;7:e32145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schlegel M, Körner A, Kaussen T, et al. Inhibition of neogenin fosters resolution of inflammation and tissue regeneration. J Clin Invest. 2019;129:2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Beldi G, Wu Y, Banz Y, et al. Natural killer T cell dysfunction in CD39‐null mice protects against concanavalin A‐induced hepatitis. Hepatology. 2008;48:841‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Victorino F, Sojka DK, Brodsky KS, et al. Tissue‐resident NK cells mediate ischemic kidney injury and are not depleted by anti‐asialo‐GM1 antibody. J Immunol. 2015;195:4973‐4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hoegl S, Ehrentraut H, Brodsky KS, et al. NK cells regulate CXCR2+ neutrophil recruitment during acute lung injury. J Leuk Biol. 2017;101:471‐480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Okamoto M, Kato S, Oizumi K, et al. Interleukin 18 (IL‐18) in synergy with IL‐2 induces lethal lung injury in mice: a potential role for cytokines, chemokines, and natural killer cells in the pathogenesis of interstitial pneumonia. Blood. 2002;99:1289‐1298. [DOI] [PubMed] [Google Scholar]
- 109. Abdul‐Careem MF, Mian MF, et al. Critical role of natural killer cells in lung immunopathology during influenza infection in mice. J Infect Dis. 2012;206:167‐177. [DOI] [PubMed] [Google Scholar]
- 110. Li F, Zhu H, Sun R, Wei H, Tian Z. Natural killer cells are involved in acute lung immune injury caused by respiratory syncytial virus infection. J Virol. 2012;86:2251‐2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang J, Li F, Sun R, Gao X, Wei H, Tian Z. Klebsiella pneumoniae alleviates influenza‐induced acute lung injury via limiting NK cell expansion. J Immunol. 2014;193:1133‐1141. [DOI] [PubMed] [Google Scholar]
- 112. Abram CL, Roberge GL, Hu Y, Lowell CA. Comparative analysis of the efficiency and specificity of myeloid‐Cre deleting strains using ROSA‐EYFP reporter mice. J Immunol Methods. 2014;408:89‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shi J, Hua L, Harmer D, Li P, Ren G. Cre driver mice targeting macrophages. Methods Mol Biol. 2018;1784:263‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19:200‐218. [DOI] [PubMed] [Google Scholar]
- 115. Gianchecchi E, Delfino DV, Fierabracci A. NK cells in autoimmune diseases: linking innate and adaptive immune responses. Autoimmun Rev. 2018;17:142‐154. [DOI] [PubMed] [Google Scholar]
- 116. Hammer Q, Ruckert T, Romagnani C. Natural killer cell specificity for viral infections. Nat Immunol. 2018;19:800‐808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
table S1‐S3


