Abstract
The International human leukocyte antigen (HLA) and Immunogenetics Workshops (IHIWs) have fostered international collaborations of researchers and experts in the fields of HLA, histocompatibility and immunology. These IHIW collaborations have comprised many projects focused on achieving a variety of specific goals. The international and collaborative nature of these projects necessitates the collection and analysis of complex data generated in multiple laboratories, often using multiple methods of acquisition. Collection and storage of these data in a consistent way adds value to IHIW projects, which can be extended to future work. DNA‐based genotyping data, especially HLA genotyping data, can be transmitted in the form of a Histoimmunogenetics Markup Language (HML) document. HML facilitates clear communication of a genotype and supporting metadata, such as, sequencing platform, laboratory assays, consensus sequence, and interpretation. Sequence information can be reported relative to known reference sequences, which add meaning and context to genotypes. Selecting the correct reference sequence for a given allele sequence is nuanced, and guidelines have emerged through collaborative community efforts such as Data Standards Hackathons. Here, we describe the guidelines established for the selection of reference sequences to be used in transmission of HLA (and MICA/MICB) genotyping data for the 18th IHIW.
Keywords: IHIW, HLA, HML, reference sequence, workshop
1. INTERNATIONAL HLA & IMMUNOGENETICS WORKSHOP
The International human leukocyte antigen (HLA) and Immunogenetics Workshops (IHIWs) are a recurring international collaboration of researchers and domain experts who come together to collaborate and find consensus on important goals in immunogenetics. The IHIWs have a long history of defining standards and establishing community consensus, 1 particularly in identifying and naming reagents and immunological molecules. For example, the now well‐established and recognized HLA Nomenclature was originally established in the context of the third such HLA workshop. 2
The 18th IHIW is scheduled for May, 2022 in Amsterdam, the Netherlands. This workshop consists of three main components, comprising a variety of projects with varied goals and milestones. For many of these projects, collection of DNA sequences of HLA or related histocompatibility sequences at a variety of resolutions is essential. In the most recent 17th IHIW, significant and valuable efforts were made in collecting and cataloging HLA genotyping data, 3 and provided valuable context for planning and defining data collection for the 18th IHIW.
To promote consistent analysis and allow for reinterpretation of immunogenetic data, it is necessary to define standard techniques for collecting and sharing genotyping data. Histoimmunogenetics Markup Language (HML) 4 is the official format for submitting sequence‐based HLA genotyping data to the 18th IHIW. HML is an XML‐based[https://www.w3.org/2001/XMLSchema] format that allows transmission of a genotyping result, as well as details of the sequencing platform used, biological assays performed, generated consensus sequences, and inferred genotype in a single document.
1.1. Selection of standard reference sequences
HLA genotypes are commonly identified using their official allele names, 5 which facilitates concise and effective communication and interpretation of the genotype. In some cases, especially when describing novel polymorphism, it is valuable to communicate additional detail and resolution by providing a consensus sequence associated with an HLA genotype. This genotype can be even further enriched by specifying specific variants 6 in its consensus sequence in comparison to a known, well‐defined reference sequence. These reference sequences are often, especially in the case of HLA, the sequence of another allele from the same genetic locus, but artificially constructed or inferred sequences can also be used. Matching of the consensus sequence to known references, especially in the exonic regions, provides greater context to the consensus sequence, facilitates clear re‐interpretation of the sequence, and ensures that both the submitter and consumer of the genotype understand the data in the context of known HLA reference sequences.
The guidelines for selecting standard reference sequences were developed as a collaborative community effort. Workshop participants and experts discussed standards over the course of several Data Standards Hackathons (DaSH) meetings, organized by the Center for International Blood and Marrow Transplant Research (CIBMTR) [https://github.com/nmdp-bioinformatics/dash10]. HLA genotyping assay vendors, immunogenetics researchers, and data‐scientists discussed the challenges in adopting HML and in selecting appropriate references, and consensus for choosing high‐quality reference sequences was established and recorded. A set of specific reference sequences for each HLA locus, and for specific allele‐families within a locus, were previously selected for the 17th IHIW. 3
The official repository for HLA sequences is the IPD‐IMGT/HLA Database. 7 The World Health Organization Nomenclature Committee for Factors of the HLA System (Nomenclature Committee) 5 curates and categorizes high‐quality, well‐defined HLA gene sequences that are commonly used as genotyping references. Various groups have identified a subset of these sequences as Common [Intermediate] and Well‐Documented (CWD/CIWD) alleles, 8 , 9 , 10 , 11 , 12 , 13 establishing them as more prevalent within certain global populations. The IPD‐IMGT/HLA Database is updated every 3 months; each database release contains newly identified allele sequences, as well as extensions of and corrections to previously known sequences. The IPD‐IMGT/HLA Database is the standard reference repository for IHIW efforts.
For 18th IHIW efforts, the 17th IHIW reference sequence selection guidelines were used to automate the selection of a set of sequences to be used as standard references for each IPD‐IMGT/HLA Database release. This process was facilitated by Python (v3.6.0) scripts, which parse the hla.xml files containing the curated HLA sequences provided by the IPD‐IMGT/HLA Database and assemble the available allele sequence with their corresponding gene feature annotations. With the exception of HLA‐DPB1 alleles (as described below), allele sequences were clustered by allele group as indicated by the first field of HLA nomenclature, and were analyzed to determine if the sequence can be considered as a reference allele candidate. Alleles were minimally required to have continuous sequence for every exon, the intervening introns, and the 5′ and 3′ UTR regions. The lowest‐numbered HLA allelic designation with a full‐length sequence in each allele group was selected as the standard reference for that group (e.g. HLA‐A*01:01:01:01 was selected as a standard reference for the HLA‐A*01 allele group).
References were selected to represent each of the 19 HLA genes, as well as the MICA and MICB genes when these sequences are available (Figure 1). For each gene, one allele was designated as a locus‐level reference, either because it had the longest 5′ and 3′ UTR sequences, or because it is used by the IPD‐IMGT/HLA Database as an alignment reference. In most cases, reference sequences were identified for all allele groups with an available full‐length sequence, based on the first field of the HLA allele name. Single references were provided for each of the HLA‐DMA, ‐DMB, ‐DOA, ‐DOB, ‐DRA, ‐E, ‐F, and ‐G, genes, the alleles of which are not currently named using multiple distinct allele groups. Similarly, a single reference was provided for the MICA and MICB genes. In the case of HLA‐DPB1, as the first field nomenclature does not indicate serological or protein similarity, four reference sequences were provided, to represent each of the four major allele categories as defined by Cano and Fernandez‐Vina. 14
FIGURE 1.
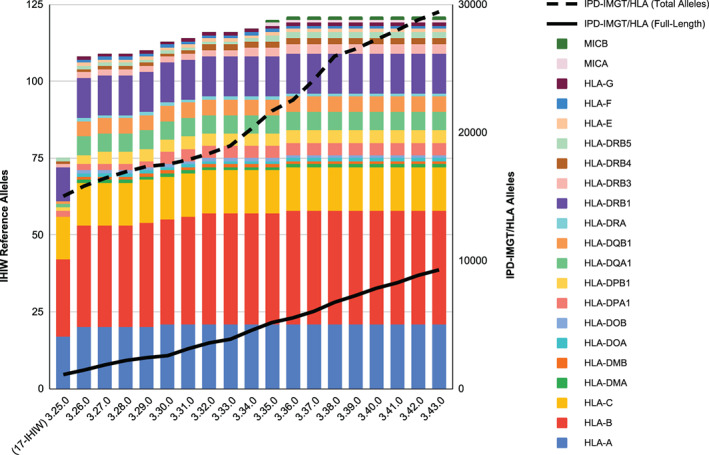
Number of designated IHIW references per IPD‐IMGT/HLA Database release version. Columns represent individual release versions of the IPD‐IMGT/HLA Database. 3.25.0 was the designated release version for the 17th IHIW, and 3.39.0 is the first release version for the 18th IHIW. The left axis indicates the number of designated IHIW reference sequences, and colors indicate references for individual genes. The right axis indicates the number of available alleles for successive IPD‐IMGT/HLA Database releases, where the number of full‐length sequences in hla.xml is plotted as a solid black line, and the total number of alleles including incomplete sequences is plotted as a dashed black line. Although the IPD‐IMGT/HLA Database allele count is continuously increasing, the number of representative IHIW reference sequences remains relatively constant
In a few specific cases, allele sequences were manually selected, to extend and improve the automated allele selection. For instance, HLA‐B*15:10:01:01 is included in addition to HLA‐B*15:01:01:01 group reference, due to the historical use of HLA‐B*15:10 as a B70 serotype reference. 15 HLA‐DRB1*14:54:01:01 was similarly included, due to its frequent occurrence in the population and use as a DR14 reference. 16 , 17 Alleles that were selected due to availability of extended UTR sequence include HLA‐B*27:05:02:01, HLA‐B*56:01:01:02, HLA‐C*03:03:01:01, HLA‐DRB1*08:02:01:01, HLA‐DRB1*15:01:01:02.
Reference sequence lists were compiled for IPD‐IMGT/HLA Database releases 3.26.0 through 3.43.0. Release 3.39.0 is the earliest version under which genotype data can be submitted for the 18th IHIW. Successive IPD‐IMGT/HLA Database releases provide additional full‐length sequences, which is reflected in the catalog of corresponding standard IHIW reference sequences. The sequence additions, extensions of previous references, and sequence replacements between IPD‐IMGT/HLA Database releases are shown in Table 1, and illustrated in Figure 1. In some cases, allele sequences were removed, due to either the identification of errors in the source sequence, or due to replacement with a more suitable reference sequence. In these cases, removed sequences were replaced by a suitable group‐level reference sequence when possible. Additional details on the changes between revisions, and the C[I]WD designations of individual allele sequences, can be found in Supporting Information S1.
TABLE 1.
Extension of reference sequence list per release of IPD‐IMGT/HLA
| IPD‐IMGT/HLA release Version | # Ref Seqs | Additions for this version |
|---|---|---|
| 3.25.0 | 75 | 17th IHIW Reference Collection |
| 3.26.0 | 108 |
Added references for Loci: HLA‐DMA, HLA‐DMB, HLA‐DOA, HLA‐DOB, HLA‐DRA, HLA‐E, HLA‐F, HLA‐G Added references for DPB1 categories HLA‐DPB1*03:01:01, ‐DPB1*04:01:01:01 Added references for Allele Groups: HLA‐A*26, ‐A*43, ‐A*69, HLA‐B*39, ‐B*41, ‐B*50, ‐B*52, ‐B*55, ‐B*56, ‐B*58, ‐B*59, ‐B*78, HLA‐DQA1*02, ‐DQA1*03, ‐DQA1*04, ‐DQA1*05, ‐DQA1*06, HLA‐DQB1*03, ‐DQB1*04, ‐DQB1*05, ‐DQB1*06, HLA‐DRB1*10, ‐DRB1*16, HLA‐DRB3*02 Removed: HLA‐B*40:305 (replaced by HLA‐B*41:01:01) Replaced: HLA‐C*17:01:01:01 (erroneous) replaced by HLA‐C*17:01:01:02 HLA‐C*03:02:02:01 replaced with HLA‐C*03:03:01:01 |
| 3.27.0 | 109 |
Added references for DPB1 category: HLA‐DPB1*01:01:01 Extended: HLA‐A*69:01:01:01, HLA‐B*07:02:01:01, ‐B*08:01:01:01, ‐B*13:01:01, ‐B*14:01:01, ‐B*15:01:01:01, ‐B*18:01:01:01. ‐B*37:01:01, ‐B*38:01:01, ‐B*40:01:02:01, ‐B*42:01:01, ‐B*44:02:01:01, ‐B*46:01:01, ‐B*48:01:01, ‐B*49:01:01, ‐B*50:01:01:01, ‐B*51:01:01:01, ‐B*52:01:01:01, ‐B*57:01:01, ‐B*58:01:01:01, ‐B*67:01:01 Replaced: HLA‐A*74:02:01:02 replaced with HLA‐A*74:01:01 |
| 3.28.0 | 109 |
Extended: HLA‐C*01:02:01:01, ‐C*02:02:02:01, ‐C*03:03:01:01, ‐C*04:01:01:01, ‐C*05:01:01:01, ‐C*06:02:01:01, ‐C*07:01:01:01, ‐C*08:01:01, ‐C*12:02:02:01, ‐C*14:02:01:01, ‐C*16:01:01:01, ‐C*17:01:01:02 Replaced: HLA‐B*27:02:01:01 replaced with HLA‐B*27:05:02:01 |
| 3.29.0 | 110 |
Added reference for Allele Group: HLA‐B*82:01 Replaced: HLA‐DPA1*01:03:01:02 replaced with HLA‐DPA1*01:03:01:01 HLA‐DPA1*02:01:02 replaced with HLA‐DPA1*02:01:01:01 HLA‐DRB1*08:03:02 replaced with HLA‐DRB1*08:02:01:01 HLA‐DRB1*09:21 replaced with HLA‐DRB1*09:01:02:01 HLA‐DRB1*14:05:01 replaced with HLA‐DRB1*14:54:01:01 Extended: HLA‐C*03:03:01:01, HLA‐B*13:01:01:01, ‐B*40:01:02:01 |
| 3.30.0 | 113 |
Added reference for Allele Group: HLA‐A*36:01 HLA‐DPA1*03:01:02, ‐DPA1*04:02 Extended: HLA‐B*45:01:01, ‐B*47:01:01:03, ‐B*55:01:01, ‐B*67:01:01, ‐B*82:01, HLA‐C*02:02:02:01, ‐C*03:03:01:01, ‐C*04:01:01:01, ‐C*08:01:01:01, ‐C*14:02:01:01, ‐C*15:02:01:01, ‐C*17:01:01:02, HLA‐DPA1*02:01:01:01, HLA‐DQA1*01:01:01:01 |
| 3.31.0 | 114 |
Added reference for Allele Group: HLA‐B*81:01 Extended: HLA‐DQA1*05:01:01:01 |
| 3.32.0 | 116 |
Added reference for Allele Group: HLA‐B*83:01, HLA‐DRB4*02:01 N |
| 3.33.0 | 116 | (No major changes) |
| 3.34.0 | 117 |
Added reference for Allele Group: HLA‐DRB3*03:01:01 Replaced: HLA‐B*56:01:01:02 replaced with HLA‐B*56:01:01:01 HLA‐DPA1*04:02 replaced with HLA‐DPA1*04:01 Extended: HLA‐A*69:01:01:01, HLA‐C*18:01, HLA‐DPA1*02:01:01:01, HLA‐DQA1*04:01:01:01 |
| 3.35.0 | 120 |
Added references for Loci: MICA*001, MICB*004:01:01 Added reference for Allele Group: HLA‐DRB5*02:02:01 Replaced: HLA‐DPA1*03:01:02 replaced with HLA‐DPA1*03:01:01 Extended: HLA‐B*14:01:01:01, ‐B*41:01:01:01, HLA‐DRB3*01:01:02:01 |
| 3.36.0 | 121 |
Added reference: HLA‐B*15:10:01:01 (B70 Reference) Extended: HLA‐A*36:01,‐A*74:01:01:01 HLA‐B*52:01:01:01, ‐B*53:01:01:01, ‐B*54:01:01, ‐B*73:01, ‐B*81:01:01, |
| 3.37.0 | 121 | (No major changes) |
| 3.38.0 | 121 |
Extended: HLA‐DRB1*04:01:01:01 |
| 3.39.0 | 121 |
Extended: HLA‐A*43:01 |
| 3.40.0 | 121 |
Replaced: HLA‐B*78:01:01:01 (erroneous) replaced with HLA‐B*78:01:01:02 |
| 3.41.0 | 121 |
Extended: HLA‐C*12:02:02:01 |
| 3.42.0 | 121 | (No major changes) |
| 3.43.0 | 121 | (No major changes) |
Note: For each release of the IPD‐IMGT/HLA Database, the number of designated full‐length reference sequences is shown. Major reference additions, removals, replacements, and extensions between versions are detailed.
The selected IHIW reference sequences are provided on the Anthony Nolan HLA Informatics Group (ANHIG) Github repository [https://github.com/ANHIG/IMGTHLA/tree/Latest/ihiw/hml]. The data for each release are provided as a text file containing allele names, accession numbers and the associated reference type for each designated sequence. The Python code that selects the HLA alleles is available in the IHIW Github repository [https://github.com/IHIW/bioinformatics/tree/master/reference_alleles/generate_references], alongside scripts that collect C[I]WD designations for Supporting Information S1C . Linux command line scripts are provided that illustrate how the reference generators and validations are executed. As new IPD‐IMGT/HLA Database versions are released, new reference lists will be added to the ANHIG Github repository, up to and after the 18th IHIW.
In addition, guidelines for creating high‐quality, interpretable HML documents are also provided on the IHIW Github repository [https://github.com/IHIW/bioinformatics/blob/master/reference_alleles/18IHIWS_Vendor_Genotyping_Requirements.md]. These guidelines indicate techniques and minimal standards that can indicate the best way to use these reference sequences in generating HML genotyping documents. Vendors of HLA genotyping software and kits are encouraged to refer to these guidelines when creating HML documents containing HLA genotypes.
1.2. Validation
The panel of selected reference sequences is intended to provide a practical set of sequences that is usable for laboratories and vendors in creating clear and interpretable genotyping documents. While interpretation and adoption is a primary goal, the ease of generation of a genotyping report, as well as clarity and accuracy is improved if sequence polymorphism can be represented with minimal variants. With the aim of examining the coverage of the selected reference sequence panel, a validation strategy was devised to compare known full‐length sequences against their corresponding ''best'' matching reference. Full‐length sequences and their corresponding reference sequences were examined to determine if they share a common allele group, with the conjecture that high amounts of group‐level deviations might indicate that the selected reference panel does not sufficiently represent known polymorphism. BLAST 18 was used to align all known full‐length sequences from successive releases of the IPD‐IMGT/HLA Database against the chosen reference panel. An alignment score was calculated for each full‐length sequence against the selected references, and the sequence with the highest alignment score was selected as the ''best'' match for that sequence. Alleles were categorized based on whether they matched the selected reference at allele‐group level.
The validation results, including the chosen best‐matching reference for each full‐length sequence are provided for IPD‐IMGT/HLA Database release 3.43.0 in Supporting Information S2 . In summary, of the 9271 full‐length sequences tested, 100% matched best with a reference sequence for the same gene, and 8219 (88.7%) matched a reference sequence in the same allele‐group. Of the 1052 group‐level mismatches, 501 were in the HLA‐DPB1, MICA, and MICB genes, for which allele group‐level references were not provided. When these sequences are excluded, 93.4% of full‐length sequences identified a group‐level matching reference sequence as the closest match, indicating that in most cases the selected reference sequences provide sufficient variety to provide a similar reference sequence to potential submitted sequences.
2. DISCUSSION
The designated reference sequences presented here are aimed to provide a useful and unambiguous tool for reporting HLA genotyping. The selection of specific alleles is intended to maximize usability; it should allow the representation of a majority of known polymorphism, but provide some restriction to improve clarity and ease of interpretation. The designated IHIW list is a subset of the sequences in the hla.xml file provided by the IPD‐IMGT/HLA Database, and in many cases the IHIW references are also used as standard alignment references in IPD‐IMGT/HLA. In this way it is intended to complement and extend the use of the standard repository of HLA sequences. As the IPD‐IMGT/HLA Database releases more full length sequences (Figure 1), our understanding of HLA polymorphism improves, and more refined approaches to selecting reference sequences can be implemented. Increased availability of full‐length sequences and understanding of the variation of gene structure of other polymorphic genes, especially KIR, will allow the extension of these efforts to other valuable areas.
The best reference sequences to use to align an HLA genotyping result can be chosen by any of a number of selection strategies. In theory, any reference sequence, even an empty sequence or a poly‐A sequence, could be used, and the variants defined against it can still communicate the same sequence. However, it is more useful to define an HLA sequence in comparison with a known and stable standard reference, especially those sequences designated as C[I]WD, to communicate information about the newly genotyped sequence. Comparison against known sequence allows more intuitive interpretation, especially in the context of HLA variation.
Identification of the best reference allele based on alignment scores, as was done in the validation of these selected references, is a practical initial step, but may not fully indicate the sequence behavior or encoded protein. The selected references found in the validation should not be treated as objectively correct, but may provide insight into which reference would be useful in that instance. Historically, HLA allele sequences have often been aligned against a reference that matches in the polymorphic region that encodes the antigen presentation domain, exon 2 (class II), or exons 2 and 3 (class I). Targeting this region is primarily due to its importance in antigen presentation, but it is also due to historical difficulties in obtaining full‐length HLA allele sequences. In the case of HLA‐DRB1, full‐length allele sequences can be 10–16 kb in length. Due to the size of full‐length sequences, and the presence of STR regions and recombination sites within introns, 19 , 20 , 21 , 22 amplifying and sequencing the gene from 5′UTR ‐ 3′UTR with a single amplicon is a particular challenge, often resulting in laboratories ignoring the HLA‐DRB1 intron sequences in favor of the (likely) more important exons.
Selecting a reference sequence based on homology to the antigen presentation regions provides important information on the resulting molecule, but in some cases another reference sequence can be found with fewer sequence differences and a better alignment score, regardless of homology in exons 2 and 3. One such case was observed in the validation ( Supporting Information S2 ), where HLA‐A*24:02:01:01 was the closest‐matched reference sequence to HLA‐A*02:19. As has been previously observed, 23 alignments of only exons 2 and 3 show that, as expected, HLA‐A*02:19 is more similar to HLA‐A*02:01:01:01 than HLA‐A*24:02:01:01. However, alignments of the full genomic sequences showed extensive intronic variation between HLA‐A*02:19 and HLA‐A*02:01:01:01. This may suggest a historical recombination which resulted in hybrid allele sequence. 24 In any case, although choosing HLA‐A*24:02:01:01 as a reference sequence results in fewer and simpler sequence variants, it does not communicate this allele's encoded protein, and it's subsequent inclusion in the HLA‐A*02 allele group. As our understanding of intronic polymorphism is extended in the IPD‐IMGT/HLA Database, it is likely that additional reference sequences may be needed to represent major patterns of variation in intron and UTR sequences.
For submission of sequences to the 18th IHIW, in particular HML genotyping documents, vendors are encouraged to use a valid, well‐documented, full‐length reference sequence selected from the lists described here. Vendors may use their own tools to select which reference from the list is best suited for alignment of their submitted sequence.
CONFLICT OF INTEREST
Matthias Niemann is an employee of PIRCHE AG which runs the PIRCHE web‐portal. The authors have declared no conflicting interests.
AUTHOR CONTRIBUTIONS
Evaluation of available sequences and defining the strategy for reference selection was developed by Benedict M. Matern, Steven J. Mack, Kazutoyo Osoegawa, Martin Maiers, James Robinson and Eric Spierings. Scripts for parsing hla.xml and selecting references was created by Benedict M. Matern and Matthias Niemann. Integration of reference scripts and hosting of reference lists in the IPD‐IMGT/HLA Database repositories was facilitated by James Robinson. IHIW 18 organization and establishment of the IHIW database is done by Eric Spierings, Sebastiaan Heidt, Matthias Niemann and Benedict M. Matern. Manuscript was drafted by Benedict M. Matern, and all authors contributed to evaluation and revision of the text.
Supporting information
Table S1
Table S2
Appendix S1: Supporting Information
ACKNOWLEDGMENTS
Thanks to all the attendees and participants of the Data Standards Hackathons, especially Dr. Bob Milius and Dr. Loren Gragert, who provided valuable context in discussions on HML and reference sequences. Thanks also to the 18th IHIW coordinators and database team for assistance in scripting and validating the selection of reference sequences. This work was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) grant R01AI128775 (SM), N00014‐20‐1‐2832 from the US Office of Naval Research (ONR) (MM), by a grant of the International HLA & Immunogenetics Workshop Foundation and by an internal grant from the UMC Utrecht. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIAID, NIH, ONR, United States government, UMC Utrecht or government of the Netherlands.
Matern BM, Mack SJ, Osoegawa K, et al. Standard reference sequences for submission of HLA genotyping for the 18th International HLA and Immunogenetics Workshop. HLA. 2021;97:512–519. 10.1111/tan.14259
Funding information HLA & Immunogenetics Workshop Foundation; National Institutes of Health; Universitair Medisch Centrum Utrecht; US Office of Naval Research
DATA AVAILABILITY STATEMENT
All allele sequences are available in the IPD‐IMGT/HLA Database. Details of the designated 18th IHIW reference sequences are provided as a supplement to the manuscript.
REFERENCES
- 1. Thorsby E. A short history of HLA. Tissue Antigens. 2009;74(2):101‐116. 10.1111/j.1399-0039.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 2. WHO Nomenclature Committee for Factors of the HL‐A System . Nomenclature for factors of the HL‐A system. Bull World Health Organ. 1968;39(3):483‐486. [PMC free article] [PubMed] [Google Scholar]
- 3. Chang C‐J, Osoegawa K, Milius RP, et al. Collection and storage of HLA NGS genotyping data for the 17th international HLA and immunogenetics workshop. Hum Immunol. 2018;79(2):77‐86. 10.1016/j.humimm.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Milius RP, Heuer M, Valiga D, et al. Histoimmunogenetics markup language 1.0: reporting next generation sequencing‐based HLA and KIR genotyping. Hum Immunol. 2015;76(12):963‐974. 10.1016/j.humimm.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marsh SGE, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75(4):291‐455. 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564‐569. 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 7. Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD‐IMGT/HLA Database. Nucleic Acids Res. 2019;48(D1):D948‐D955. 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cano P, Klitz W, Mack SJ, et al. Common and well‐documented HLA alleles: report of the ad‐hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68(5):392‐417. 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 9. Mack SJ, Cano P, Hollenbach JA, et al. Common and well‐documented HLA alleles: 2012 update to the CWD catalogue. Tissue Antigens. 2013;81(4):194‐203. 10.1111/tan.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hurley CK, Kempenich J, Wadsworth K, et al. Common, intermediate and well‐documented HLA alleles in world populations: CIWD version 3.0.0. HLA. 2020;95(6):516‐531. 10.1111/tan.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanchez‐Mazas A, Nunes JM, Middleton D, et al. Common and well‐documented HLA alleles over all of Europe and within European sub‐regions: a catalogue from the European Federation for Immunogenetics. HLA. 2017;89(2):104‐113. 10.1111/tan.12956. [DOI] [PubMed] [Google Scholar]
- 12. Eberhard H‐P, Schmidt AH, Mytilineos J, Fleischhauer K, Müller CR. Common and well‐documented HLA alleles of German stem cell donors by haplotype frequency estimation. HLA. 2018;92(4):206‐214. 10.1111/tan.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Y, Li J, Mao W, et al. HLA common and well‐documented alleles in China. HLA. 2018;92(4):199‐205. 10.1111/tan.13358. [DOI] [PubMed] [Google Scholar]
- 14. Cano P, Fernandez‐Vina M. Two sequence dimorphisms of DPB1 define the immunodominant serologic epitopes of HLA‐DP. Hum Immunol. 2009;70(10):836‐843. 10.1016/j.humimm.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez SG, Bei M, Inamdar A, Stewart D, Johnson AH, Hurley CK. Molecular and serological characterization of HLA‐B71 in association with different class I haplotypes or in different ethnic groups. Tissue Antigens. 1996;47(1):58‐62. 10.1111/j.1399-0039.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]
- 16. Creary LE, Gangavarapu S, Mallempati KC, et al. Next‐generation sequencing reveals new information about HLA allele and haplotype diversity in a large European American population. Hum Immunol. 2019;80(10):807‐822. 10.1016/j.humimm.2019.07.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Osoegawa K, Mallempati KC, Gangavarapu S, et al. HLA alleles and haplotypes observed in 263 US families. Hum Immunol. 2019;80(9):644‐660. 10.1016/j.humimm.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403‐410. 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19. Klasberg S, Surendranath V, Lange V, Schöfl G. Bioinformatics strategies, challenges, and opportunities for next generation sequencing‐based HLA genotyping. Transfus Med Hemother. 2019;46(5):312‐325. 10.1159/000502487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albrecht V, Zweiniger C, Surendranath V, et al. Dual redundant sequencing strategy: full‐length gene characterisation of 1056 novel and confirmatory HLA alleles. HLA. 2017;90(2):79‐87. 10.1111/tan.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kotsch K, Blasczyk R. The noncoding regions of HLA‐DRB uncover interlineage recombinations as a mechanism of HLA diversification. J Immunol. 2000;165(10):5664‐5670. 10.4049/jimmunol.165.10.5664. [DOI] [PubMed] [Google Scholar]
- 22. Hosomichi K, Jinam TA, Mitsunaga S, Nakaoka H, Inoue I. Phase‐defined complete sequencing of the HLA genes by next‐generation sequencing. BMC Genomics. 2013;14(355):1–16. 10.1186/1471-2164-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lazaro A, Hou L, Tu B, et al. Full gene HLA class I sequences of 79 novel and 519 mostly uncommon alleles from a large United States registry population. HLA. 2018;92(5):304‐309. 10.1111/tan.13377. [DOI] [PubMed] [Google Scholar]
- 24. Robinson J, Guethlein LA, Cereb N, et al. Distinguishing functional polymorphism from random variation in the sequences of >10,000 HLA‐A, ‐B and ‐C alleles. PLoS Genet. 2017;13(6):e1006862. 10.1371/journal.pgen.1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Appendix S1: Supporting Information
Data Availability Statement
All allele sequences are available in the IPD‐IMGT/HLA Database. Details of the designated 18th IHIW reference sequences are provided as a supplement to the manuscript.


