SUMMARY
Systems based on the clustered, regularly interspaced, short palindromic repeat (CRISPR) and CRISPR‐associated proteins (Cas) have revolutionized genome editing in many organisms, including plants. Most CRISPR‐Cas strategies in plants rely on genetic transformation using Agrobacterium tumefaciens to supply the gene editing reagents, such as Cas nucleases or the synthetic guide RNA (sgRNA). While Cas nucleases are constant elements in editing approaches, sgRNAs are target‐specific and a screening process is usually required to identify those most effective. Plant virus‐derived vectors are an alternative for the fast and efficient delivery of sgRNAs into adult plants, due to the virus capacity for genome amplification and systemic movement, a strategy known as virus‐induced genome editing. We engineered Potato virus X (PVX) to build a vector that easily expresses multiple sgRNAs in adult solanaceous plants. Using the PVX‐based vector, Nicotiana benthamiana genes were efficiently targeted, producing nearly 80% indels in a transformed line that constitutively expresses Streptococcus pyogenes Cas9. Interestingly, results showed that the PVX vector allows expression of arrays of unspaced sgRNAs, achieving highly efficient multiplex editing in a few days in adult plant tissues. Moreover, virus‐free edited progeny can be obtained from plants regenerated from infected tissues or infected plant seeds, which exhibit a high rate of heritable biallelic mutations. In conclusion, this new PVX vector allows easy, fast and efficient expression of sgRNA arrays for multiplex CRISPR‐Cas genome editing and will be a useful tool for functional gene analysis and precision breeding across diverse plant species, particularly in Solanaceae crops.
Keywords: CRISPR‐Cas9, sgRNA expression, virus‐induced genome editing, multiplexing, Potato virus X, solanaceous plants, technical advance
Significance Statement
Plant virus‐derived vectors allow fast and efficient delivery of synthetic guide RNAs into adult plants for CRISPR‐Cas‐based genome editing. We engineered a Potato virus X vector for CRISPR‐Cas genome editing of solanaceous plants. This vector expresses unspaced arrays of synthetic guide RNAs and achieves multiplex editing in adult plant tissues in a few days. Virus‐free multiplex genome‐edited plants with biallelic mutations can be easily obtained from inoculated plants.
![]()
INTRODUCTION
Targeted gene editing of plant DNA is a valuable tool for basic and applied biology as it facilitates gene function studies and crop improvement (Chen et al., 2019; Pennisi, 2010). This can be performed by sequence‐specific nucleases that specifically bind to the user‐selected genomic region and induce DNA double‐strand breaks (DSBs) (Voytas, 2013), but recent emergence of tools based on bacterial clustered, regularly interspaced, short palindromic repeat (CRISPR) and CRISPR‐associated (Cas) protein systems have revolutionized targeted genome editing (Cong et al., 2013). Most common arrangements comprise the Cas9 endonuclease from Streptococcus pyogenes and a synthetic guide RNA (sgRNA), which combines the functions of CRISPR RNA (crRNA) and trans‐activating crRNA (tracrRNA). The sgRNA directs the Cas9 endonuclease to a target sequence complementary to 20 nucleotides preceding the 5′‐NGG‐3′ protospacer‐adjacent motif (PAM) required for Cas9 activity. The DSBs created by Cas9 activate the native host DNA repair mechanisms of non‐homologous end‐joining or homology‐directed repair. In plants, non‐homologous end‐joining occurs more frequently and results in small insertions or deletions (indels) that restore the integrity of the host DNA. These indels cause localized DNA disruption and have been used for sequence‐specific knockout of downstream gene products, such as proteins and long non‐coding RNAs (Ran et al., 2013; Schiml et al., 2014). Thus, the specificity and versatility provided by the CRISPR‐Cas tools allow for unprecedented, simple genome engineering of plant model species and economically important crops (Li et al., 2017, 2018; Nekrasov et al., 2013; Zhang et al., 2016).
A key advantage of CRISPR genome editing is the possibility to program the cellular pool of Cas9 proteins with several sgRNAs operating in parallel, a feature known as multiplexing. The multiplexing capacity of an editing tool determines the speed at which simultaneous modifications can be introduced in the genome and therefore the ability to perform comprehensive genome engineering. An efficient way to deliver several sgRNAs acting simultaneously in the cell consists of engineering long RNAs comprising several sgRNA units arrayed in tandem, which are later processed into single functional sgRNAs. Contrary to other CRISPR nucleases as Cas12a (Zetsche et al., 2017), Cas9 itself has not been described to process sgRNA arrays on its own. Therefore, functional tandem sgRNAs require processable spacers to be included in the array design. Spacers can be engineered involving either self‐cleavable ribozymes (Gao and Zhao, 2014; Xu et al., 2017), or other RNA motifs cleaved by trans‐acting RNases. For the second option, two main strategies have been described. One of them makes use of tRNAs as spacers, which are then removed by endogenous tRNA‐processing RNases, RNase P and RNase Z (Xie et al., 2015). In the second approach, spacers are small recognition sequences processed by a trans‐acting Csy4 RNase (Nissim et al, 2014; Tsai et al., 2014), which needs to be exogenously supplied, usually from a transgene.
CRISPR‐Cas approaches in plants have mainly focused on the delivery of the nucleases and sgRNAs by transformation technologies or transient delivery to protoplasts (Cong et al., 2013; Nekrasov et al., 2013). However, recent studies indicate that viral vectors may be most useful to express CRISPR‐Cas reagents, as they are in other biological systems (Platt et al., 2014; Senís et al., 2014; Lau and Suh, 2017; Xu et al., 2019), following the so‐called virus‐induced genome editing (VIGE). Several plant RNA virus‐based replicons have been tested as vectors for the delivery of sgRNAs to create gene knockouts and insertions, including Tobacco rattle virus (TRV) (Ali et al., 2015; Ellison et al., 2020), Tobacco mosaic virus (Cody et al., 2017), Pea early browning virus (Ali et al., 2018), Beet necrotic yellow vein virus (Jiang et al., 2019) and Barley stripe mosaic virus (Hu et al., 2019). Compared with delivery methods via Agrobacterium tumefaciens, plant virus‐mediated sgRNA delivery systems possess some advantages: (i) sgRNAs can accumulate to high levels owing to viral replication, which usually contributes to a higher genome editing efficiency; (ii) if the viral vector moves systemically, phenotypic alterations may appear in infected plants in a relatively short period after virus inoculation; and (iii) in the case of RNA viruses, they can produce a high rate of edited cells without the risk of integration of heterologous material into the plant genome, which avoids raising additional regulatory and ethical issues. As drawbacks, each viral vector has its own particularities based on virus molecular biology and is restricted to a specific host range. Consequently, there is a necessity to enlarge and improve the available toolbox for VIGE.
Potato virus X (PVX) is a member of the genus Potexvirus (family Alphaflexiviridae) that infects 62 plant species of 27 families (Edwardson and Christie, 1997), including important crops in the family Solanaceae, such as potato, tomato, pepper or tobacco, and is transmitted mechanically from plant to plant (Adams et al., 2004). PVX has a plus (+) single‐strand RNA genome 6.4‐kb in length, with a 5′‐methylguanosine cap, a polyadenylated 3′ tail, and five open reading frames (Loebenstein and Gaba, 2012). In this study, we describe a novel PVX‐based sgRNA delivery vector for CRISPR‐Cas genome editing in plants, which produces targeted indels with high efficiency (nearly 80%) in N. benthamiana plants previously transformed to express a Cas9 nuclease constitutively. We also demonstrate that PVX is a suitable vector for the simultaneous delivery of multiple sgRNAs per viral genome. Unexpectedly, we found that, in the absence of spacers and processing signals, sgRNAs engineered in tandem in the PVX genome are able to induce efficient gene editing. Finally, we demonstrate that virus‐free progeny efficiently carrying biallelic indels at the target genes are obtained from plants regenerated from infected tissue or from seeds of infected plants, strategies that could facilitate high‐throughput screenings.
RESULTS
Engineering a PVX‐based sgRNA delivery system for gene editing in plants
CRISPR‐Cas9 is a two‐component system that requires both Cas9 nuclease and a small sgRNA to perform gene editing. We aimed to develop a sgRNA delivery system based on PVX, an RNA virus that infects many plant species, particularly important crops in the family Solanaceae. To set up this system, we used an N. benthamiana transformed line that constitutively expresses a version of Streptococcus pyogenes Cas9 (SpCas9) under the control of Cauliflower mosaic virus 35S promoter and A. tumefaciens Nopaline synthase terminator (Bernabé‐Orts et al., 2019). More specifically, this line expresses a human codon‐optimized version of SpCas9 fused to a nuclear localization signal at the carboxyl terminus, which have been shown to improve Cas9 activity in plants (Yin et al., 2015). Our PVX‐based sgRNA delivery system (PVX::sgRNA) was designed using as a template a PVX mutant where the 29 initial codons of the coat protein (CP) are deleted and heterologous RNA expression is controlled by the subgenomic CP promoter. In turn, PVX CP is expressed from a heterologous promoter derived from that of CP of Bamboo mosaic virus (genus Potexvirus). These combined features provide remarkable increased stability in PVX recombinant clones (Dickmeis et al., 2014). The PVX::sgRNA system was constructed by cloning a cDNA corresponding to the 96‐nt sgRNA downstream the PVX CP promoter. The sgRNA consisted of a 20‐nt protospacer sequence specific to the target gene and a 76‐nt scaffold highly conserved in the CRISPR‐Cas9 system, also known as a direct repeat.
Xie et al. (2015) showed that the host endogenous tRNA‐processing system cleaves tandemly arrayed tRNA‐sgRNA constructs into sgRNAs, thus improving the efficiency of gene editing. Aiming to explore whether this strategy also applies to splice out the sgRNA from the viral subgenomic RNA that is transcribed from the PVX CP promoter, four different constructs were designed: (i) sgRNA flanked with tRNAs at both 5′ and 3′ ends (tR‐sgRNA‐tR); (ii) sgRNA flanked with tRNA only at 5′ end (tR‐sgRNA); (iii) sgRNA flanked with tRNA only at 3′ end (sgRNA‐tR); and (iv) sgRNA without any flanking tRNAs (sgRNA) (Figure 1).
Figure 1.
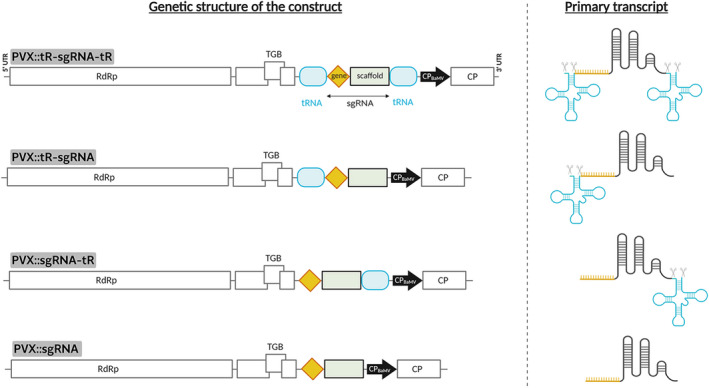
PVX vector to express sgRNAs for CRISPR‐Cas9‐based gene editing in Nicotiana benthamiana. Left panel, schematic representation of recombinant clones PVX::tR‐sgRNA‐tR, PVX::tR‐sgRNA, PVX::sgRNA‐tR and PVX::sgRNA, which express different versions of a sgRNA flanked or not with tRNAs to promote RNA processing by endogenous RNases. Right panel, structure of the subgenomic primary transcripts of each PVX recombinant clone. RdRp, RNA‐dependent RNA polymerase; TGB, triple gene block; and CP, coat protein, are represented by open boxes. Heterologous Bamboo mosaic virus CP promoter (CPBaMV) is represented by a black arrow. 5′ and 3′ untranslated regions (UTRs) are represented by lines. tRNA sequences are represented by round, light blue boxes. sgRNA consists of a gene‐specific 20‐nt protospacer (orange diamond) and a conserved 76‐nt scaffold (gray box). CPBaMV, tRNAs, protospacer and scaffold, not at scale.
PVX::sgRNA system can efficiently target host genes without tRNA‐mediated processing of the sgRNA
As a proof‐of‐concept of the PVX::sgRNA system, the N. benthamiana UDP‐xylosyltransferase 2B (NbXT2B) locus (SolGenomics Niben101Scf04551g02001.1) was selected as the target gene to be edited. Previous work assessed the efficiency of Cas9‐mediated gene editing of several N. benthamiana loci, including this gene, with different sgRNAs (Bernabé‐Orts et al., 2019). Loss of function of NbXT2B alters the glycosylation pattern in endogenous and recombinant proteins (Cavalier and Keegstra, 2006). Thus, NbXT2B‐specific sgRNA was cloned into the four different tRNA‐sgRNA constructs mentioned above to generate the PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR and PVX::sgXT2B derivatives (Figure 2a). Agrobacterium tumefaciens carrying the corresponding plasmids were then inoculated into leaves of Cas9 N. benthamiana. As a control for gene editing, additional plants were inoculated with A. tumefaciens carrying PVX::crtB. This is a visual tracker of PVX infection and movement previously developed, which results in a bright yellow pigmentation of infected tissue (Majer et al., 2017).
Figure 2.
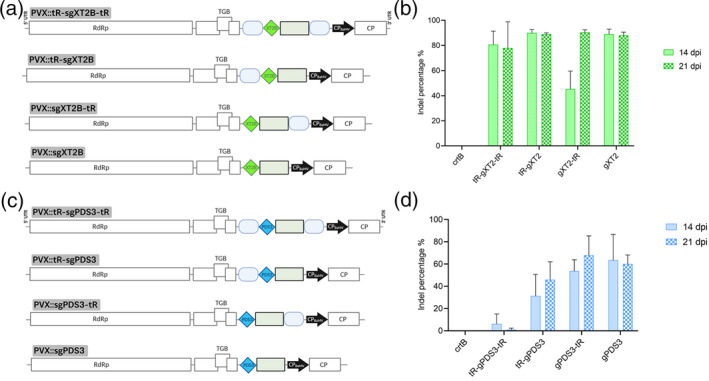
Single gene editing, with or without flanking tRNAs, using the PVX‐based system in Nicotiana benthamiana.
(a) Schematic representation of recombinant clones PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR and PVX::sgXT2B. NbXT2B protospacer is represented by a green diamond. Other details as in the legend to Figure 1.
(b) Inference of CRISPR edits (ICE) analysis of the first systemically infected upper leaf of N. benthamiana plants inoculated with PVX::sgXT2B derivatives at 14 days post‐inoculation (dpi) (n = 3) and 21 dpi (n = 3). PVX::crtB was used as a negative editing control.
(c) Schematic representation of recombinant clones PVX::tR‐sgPDS3‐tR, PVX::tR‐sgPDS3, PVX::sgPDS3‐tR and PVX::sgPDS3. NbPDS3 protospacer is represented by a blue diamond. Other details as in the legend to Figure 1.
(d) ICE analysis of the first systemically infected upper leaf of N. benthamiana plants inoculated with PVX::sgPDS3 derivatives at 14 dpi (n = 3) and 21 dpi (n = 3). (b,d) Columns and error bars represent average indels (%) and standard deviation, respectively.
At 7 days post‐inoculation (dpi), symptoms of PVX infection characterized by the appearance of vein banding, ring spots and leaf atrophy were observed in the upper non‐inoculated leaves of all PVX::sgRNA infiltrated N. benthamiana plants. These symptoms persisted and became more noticeable over time. However, symptoms of PVX::sgXT2B were slightly more intense than those of PVX::tR‐sgXT2B and PVX::sgXT2B‐tR were, and at the same time, symptoms of these two last recombinant clones were slightly more intense than those of PVX::tR‐sgXT2B‐tR were. Samples from the first systemically infected upper leaf were collected at 14 and 21 dpi. On the one hand, RNA was extracted and reverse transcription (RT)–polymerase chain reaction (PCR) analysis confirmed the presence of PVX in all inoculated plants. The insertion of the tRNA‐sgRNA region in the PVX progeny was also confirmed by RT‐PCR. These results indicated that, although addition of one or two tRNA sequences into the genome may be affecting virus fitness, they do not abolish PVX infectivity and systemic spread, and that the heterologous tRNA‐sgRNA region is conserved in PVX progeny over time. On the other hand, DNA was extracted from the leaf samples collected at 14 and 21 dpi and a 750‐bp fragment of the NbXT2B gene covering the Cas9 target site was amplified by PCR. Sanger sequencing of the PCR products and inference of CRISPR edits (ICE) analysis revealed an efficient genome editing in systemic leaves of all PVX::sgRNA‐inoculated plants (Figure 2b). No statistically significant differences were observed in NbXT2B gene editing among the four different PVX::sgRNA derivatives, with an average indel percentage ranging from 37% to 85%. These results suggest that all PVX::sgRNA derivatives are suitable for targeted editing of N. benthamiana genes in the presence of Cas9, regardless of the addition of a tRNA sequence at 5′ and/or 3′ ends of the sgRNA. To confirm this finding, the experiment with the four PVX::sgRNA derivatives was repeated following the same procedure, but this time selecting N. benthamiana Phytoene desaturase 3 (NbPDS3; Niben101Scf01283g02002.1) as the target gene (Figure 2c). PDS is a key enzyme of the carotenoid biosynthetic pathway in plants (Burch‐Smith et al., 2006). At 14 and 21 dpi, the first systemically infected upper leaf was sampled, following extraction of genomic DNA and PCR amplification of a 650‐bp fragment covering the Cas9 target site of NbPDS3. For this gene, ICE analysis displayed no editing for PVX::tR‐sgPDS3‐tR, a relatively low efficiency (25%) for PVX::tR‐sgPDS3 and high efficiency (68% and 73%) for PVX::sgPDS3‐tR and PVX::sgPDS3, respectively (Figure 2d). These results highlighted that editing efficiency depends on each particular sgRNA. Editing efficiency induced by the different versions of sgPDS3 was always lower than those induced by their sgXT2B counterparts, with negligible editing in the case of PVX::tR‐sgPDS3‐tR (Figure 2, compare panels b and d). Owing to its high editing efficiency and design simplicity, PVX::sgRNA constructs lacking tRNA sequences were used for subsequent experiments.
Efficient and easy editing of multiple genes from a single PVX construct
After the successful gene editing of endogenous genes in N. benthamiana, we wondered whether our PVX‐based vector could be feasible for the delivery of multiple sgRNA molecules from a single construct (i.e., multiplexing). The capacity to edit several genes at once using transient sgRNA delivery strategies remains of great interest for generating plants with multiple gene knockouts. In addition to the N. benthamiana genes tested through the single sgRNA strategy (NbXT2B and NbPDS3), we decided to include an additional target gene for the multiplexing. Based on results on editing efficiency obtained by Bernabé‐Orts et al. (2019), we selected N. benthamiana Flowering locus T (NbFT; Niben101Scf01519g10008.1). FT promotes the development of inflorescences from vegetative meristem, and loss of function of this gene results in late flowering (Wigge et al., 2005). Considering the finding that our PVX::sgRNA system can efficiently produce indels on target genes without any sgRNA processing, we hypothesized that multiple sgRNAs could be tandemly delivered within a single PVX construct. We further proposed to position the three sgRNA sequences next to each other, without any spacer sequence and under the control of the same CP promoter. Thus, using the PVX vector as a template, sgPDS3 and sgXT2B were positioned on the 5′ and 3′ ends of the sgRNA construct, respectively, and sgFT was included between them, creating the PVX::sgPDS3:sgFT:sgXT2B construct (Figure 3a). In addition, a PVX::sgFT construct harboring the single sgRNA for NbFT was designed to compare the editing efficiency between single and multiplex sgRNA strategies (Figure 3a). A. tumefaciens carrying either PVX::sgPDS3:sgFT:sgXT2B or PVX::sgFT were inoculated into Cas9 N. benthamiana leaves, and inoculation with PVX::crtB was used as a control for gene editing. Again, at 7 dpi, symptoms of PVX infection were observed in the upper non‐inoculated leaves of all PVX::sgRNA infiltrated N. benthamiana plants. Samples from the first systemically infected upper leaf were collected at 7, 14, 21 and 28 dpi.
Figure 3.
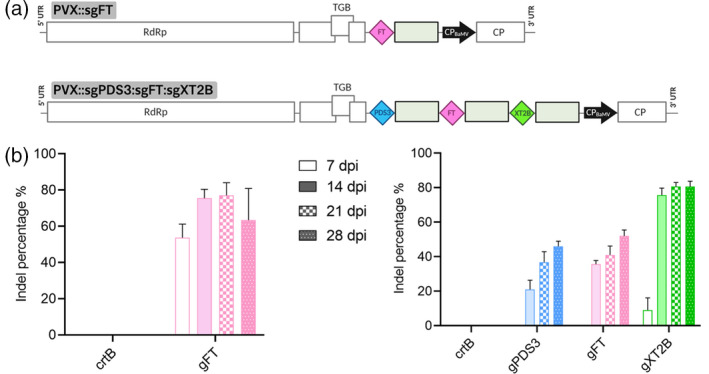
Single versus multiplex gene editing using the PVX‐based system in Nicotiana benthamiana.
(a) Schematic representation of recombinant clones PVX::sgFT (single sgRNA strategy) and PVX::sgPDS3:sgFT:sgXT2B (multiplex sgRNA strategy). Protospacers for NbPDS3, NbFT and NbXT2B are represented by blue, pink and green diamonds, respectively. Other details as in the legend to Figure 1.
(b) ICE analysis of the first systemically infected upper leaf of N. benthamiana plants (n = 4) inoculated with PVX::sgFT (left) and PVX::sgPDS3:sgFT:sgXT2B (right). PVX::crtB was used as a negative control. Columns and error bars represent average indels (%) and standard deviation, respectively. dpi, days post‐inoculation.
Following DNA extraction and PCR amplification of the target sites, NbPDS3, NbFT and NbXT2B amplicons were subjected to ICE analysis (Figure 3b). For PVX::sgPDS3:sgFT:sgXT2B infected leaves, at 7 dpi indels were detected only in NbXT2B gene but at a low percentage (9%). Remarkably, at 14 dpi gene editing was boosted as indels were detected in all three genes. A slight increase in gene editing was observed over time for NbPDS3 and NbFT (Figure 3b, right panel). Time‐course comparison with the single sgRNA construct, in the case of sgFT, showed that multiplexing lowers editing efficiency (Figure 3b, compare left and right graphs). These results indicate that various sgRNAs can be delivered simultaneously using the PVX‐based vector, although indel production is lower compared with single sgRNA delivery.
Heritable gene editing in progeny of plants regenerated from tissue infected with PVX::sgRNA vector
The above‐explained results demonstrate that the PVX‐based vector is suitable for the delivery of either single or multiple sgRNAs leading to efficient editing of target genes. It is well known that PVX is transmitted mainly by mechanical contact between infected and healthy plants, although transmission by zoospores of the fungus Synchytrium endobioticum has also been reported (Loebenstein and Gaba, 2012). Moreover, it is well established that this virus is not transmitted through seed or pollen. As NbXT2B appeared to be the most efficiently edited gene, seeds from PVX::sgXT2B‐infected plants were recovered. The progeny was then screened for the presence of PVX and heritability of editing in NbXT2B. All plants were virus‐free, but none of them carried indels at the target site of NbXT2B. These results indicate that PVX is unable to infect germline cells and therefore the progeny of transiently modified plants cannot inherit sequence modifications in the target genes.
We proposed that leaf tissue from plants infected with PVX::sgRNA could be regenerated into whole plants and screened for the presence of gene editing. We decided to test this possibility for plants modified at either single or multiple target genes. Thus, leaf discs from plants inoculated with PVX::sgXT2B or PVX::sgPDS3:sgFT:sgXT2B were collected at 14 and 21 dpi and regenerated following tissue culture. We next studied whether the regenerated plants, which still showed symptoms of virus infection, carried modifications in the target genes. ICE analysis of PCR products indicated a strong presence of indels in plants regenerated from leaf tissue that had been edited at either single or multiple genes, regardless of the sampling time (Figure 4). Interestingly, editing efficiency in regenerated plants was higher compared with that of parental tissues: from 77% to 95.8% for NbXT2B in single sgRNA delivery strategy (Figure 4a); and from 46% to 74% for NbPDS3, from 52% to 78.5% for NbFT, and from 52% to 94.5% for NbXT2B in multiple sgRNA delivery strategy, respectively (Figure 4b).
Figure 4.
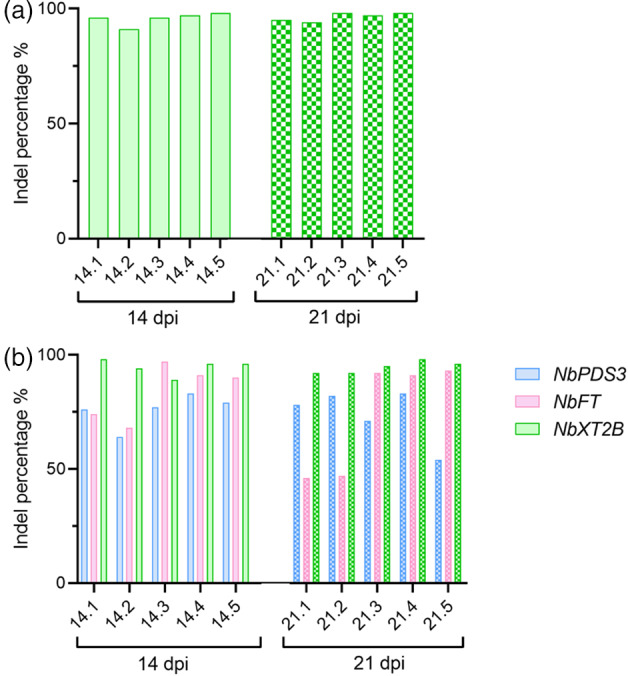
Gene editing in Nicotiana benthamiana plants regenerated from infected tissue. ICE analysis of plants regenerated from leaf discs infected with (a) PVX::sgXT2B or (b) PVX::sgPDS3:sgFT:sgXT2B. Individual plants regenerated from leaf discs collected at 14 dpi (n = 5) or 21 dpi (n = 5) are represented as 14.1–14.5 or 21.1–21.5, respectively. Columns represent average indels (%). dpi, days post‐inoculation
Subsequently, we wondered whether the gene editing observed in regenerated plants resulted in germline modifications that were transmitted to seedlings. Seeds were collected from plants 14.3 and 14.1, which had been regenerated from leaf discs infected with PVX::sgXT2B and PVX::sgPDS3:sgFT:sgXT2B, respectively (Figure 4). The absence of symptoms of PVX infection indicated that all progeny was virus‐free. ICE analysis of the target genes confirmed the inheritance of genome modifications (Figure 5a). Progeny derived from the single‐edited plant 14.3 contained mutations in both NbXT2B alleles (i.e., biallelic) (Figure 5b, left panel). In turn, genotyping of progeny derived from plant 14.1, which carried modifications in all target genes, indicated that both NbXT2B alleles were edited in all cases, while 20% and 30% of seedlings contained biallelic mutations for NbPDS3 and NbFT, respectively (Figure 5b, right panel). Only 40% of the progeny showed no modifications in neither of the two NbFT alleles (Figure 5b, right panel). Remarkably, with this small screening of only 10 plants from the progeny, we were able to find individuals with 100% biallelic mutations of all three target genes (Figure 5a, right panel, plant 14.1‐v).
Figure 5.
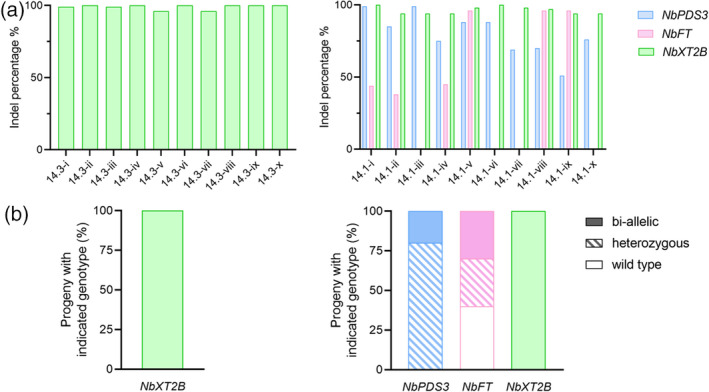
Heritable gene editing using the PVX‐based system in Nicotiana benthamiana.
(a) ICE analysis of the progeny of plants 14.3 (left) and 14.1 (right), regenerated from leaf discs infected with PVX::sgXT2B and PVX::sgPDS3:sgFT:sgXT2B, respectively. Individual plants of the progeny (n = 10) are numbered i–x. Columns represent average indels (%).
(b) Genotype analysis of the same progeny (n = 10) used to determine heritable gene editing in (a). Heterozygous progeny contain one wild‐type allele and one edited allele; biallelic progeny contain both alleles edited. Percentage of each genotype is represented in stacked columns and was determined by the fraction of progeny containing that genotype divided by total progeny assessed.
Heritable gene editing in seedlings from plants infected with the PVX::sgRNA vector
Using a TRV‐based VIGE vector, it has been recently reported that fusion of sequences that promote cell‐to‐cell mobility to sgRNAs allows direct editing of germline cells. This way, gene modifications are directly transmitted to seedling progeny of infected plants, avoiding the need for tissue culture (Ellison et al., 2020). Aiming to explore whether this strategy can be expanded to our PVX vector, we fused the 5′ 102‐nt Arabidopsis thaliana FT (truncated FT) coding region (Li et al., 2009) to the 3′ end of the NbXT2B sgRNA, creating the PVX::sgXT2B‐tFT construct (Figure 6a). Seeds were collected from Cas9 N. benthamiana plants inoculated with this vector. Genotype analyses indicated that 22% of seedlings contained biallelic mutations in NbXT2B (Figure 6b and Figure S3). A similar vector was constructed targeting Nb PDS3, also with the inclusion of the truncated FT extension. In this case, a different sgRNA was used (sgPDS3ii), which targeted simultaneously the two PDS3 homeologs present in the N. benthamiana genome (Niben101Scf01283g02002.1 and Niben101Scf14708g00023.1), thus aiming at complete suppression of PDS activity in tissues with biallelic mutations. Remarkably, the PVX::sgPDS3ii‐tFT vector induced a photobleaching phenotype that was first noticeable at 3 weeks after inoculation and increased intensity during the following weeks (Figure 6c).
Figure 6.
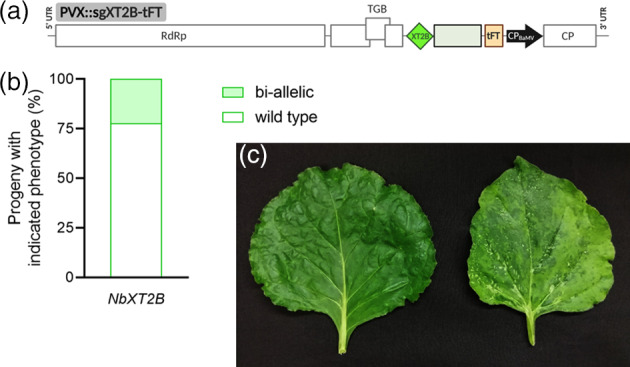
Heritable gene editing with a PVX vector expressing mobile sgRNAs.
(a) Schematic representation of recombinant clone PVX::sgXT2B‐tFT in which the sgRNA was modified by the 3′ addition of a truncated FT (tFT) motif (light orange box). Other details as in the legend to Figure 1.
(b) Genotype analysis of the seedling progeny (n = 36) from plants inoculated with PVX::sgXT2B‐tFT. Seeds were collected from the entire plants and pooled. Percentage of each genotype is represented in stacked columns and was determined as in Figure 5b.
(c) Phenotype of systemic leaves of Cas9 Nicotiana benthamiana plants infected with PVX::sgXT2B‐tFT (left) or PVX::sgPDS3ii‐tFT (right) 5 weeks after infiltration.
DISCUSSION
Targeted editing of plant genomes by CRISPR‐Cas9 technology is expected to play a crucial role in both basic genetic studies and crop improvement for 21st century agriculture. In this context, the use of plant viral vectors for the transient delivery of CRISPR‐Cas9 components has emerged as a promising strategy over conventional transformation technologies. The first VIGE reports focused on generation of gene knockout plant lines using sgRNA delivery systems based on geminiviruses (Baltes et al., 2014; Yin et al., 2015). Subsequent studies led to the development of several plant RNA virus‐based vectors with the same purpose (Ali et al., 2015, 2018; Cody et al., 2017; Ellison et al., 2020; Jiang et al., 2019; Ma et al., 2020). Here, we describe a PVX‐based sgRNA delivery vector that allows easy multiplexing for efficient targeted editing of the model species N. benthamiana as a novel approach to expand the current VIGE toolbox. PVX is a well‐known plant virus that is frequently used in biotechnological applications. It exhibits a wide host range, including important crops in the family Solanaceae (Adams et al., 2004).
First, we explored the potential of PVX for the transient delivery of sgRNA molecules into host plants. Several studies have documented that nucleotide overhangs on either 5′ or 3′ ends of the sgRNA negate Cas9 activity in vitro, which leads to a reduction of DSBs (Jinek et al., 2012; Mali et al., 2013; Dahlman et al., 2015; Zalatan et al., 2015). Considering that the host endogenous tRNA‐processing system can splice out tandemly arrayed tRNA‐sgRNA constructs into mature sgRNAs (Xie et al., 2015), we designed different PVX::sgRNA derivatives where the sgRNA was flanked or not with tRNAs (Figure 1). We hypothesized that, although host plant tRNA‐processing machinery cleaves some genomes, enough virus will survive to maintain infection. Of note, pre‐tRNA processing occurs in the nucleus and PVX replication in the cytoplasm. Certainly, the presence of one or two tRNA sequences in the viral genome had a minor effect on PVX infectivity and the ability to move systemically. Surprisingly, we also found that long sgRNA overhangs at both ends did not affect Cas9 catalytic activity in planta, as there were no substantial differences on editing efficiency of target genes among the PVX::sgRNA derivatives with no tRNA and those with tRNAs at either 5′ or 3′ side in most cases (Figure 2b and 2d). The capacity to produce biologically active sgRNAs without tRNA‐mediated processing follows findings from previous VIGE reports in which sgRNAs contained long 3′ extensions (Ali et al., 2018; Cody et al., 2017). Even more surprising is the observed ability of unspaced tandem sgRNA arrays to drive efficient editing, a feature earlier observed by Cody et al. (2017) in TMV‐based VIGE experiments using a smaller tandem comprising only two sgRNAs and, very recently, showed in a TRV‐based system (Ellison et al., 2020). To our knowledge, no other biological system has been described where Cas9 shows this level of permissiveness to the presence of 5′ extensions, neither are we aware of other systems where spacer‐less sgRNA arrays are fully functional for gene editing. These findings suggest that one or more of the following processes may be occurring in vivo: (i) Cas9 tolerates large, non‐complementary overhangs within the sgRNA protospacer (5′ end) in planta; (ii) conversely to what it is currently accepted, Cas9 is able to cleave sgRNA overhangs in planta precisely; (iii) endogenous RNases cleave sgRNA overhangs in planta; or (iv) because of imperfect replication or partial RNA degradation, the virus produces a relatively large population of subgenomic fragments and some of which, by chance, contain the correct 5′ ends. As the functionality of unspaced sgRNAs in Cas9 multiplexing has been only reported associated to viral vectors, we favor the last explanation, perhaps in combination with other mechanisms. For instance, direct repeats may protect sgRNA scaffold from degradation by unspecific plant RNases, favoring unspecific cleavage to take place in and around the protospacer and therefore increasing the relative concentration of correctly cleaved (functional) sgRNA species. Regardless of the precise mechanism underlying native sgRNA processing, our results reinforce the fact that sgRNAs can be launched from viral vectors without the need for processing spacer elements, which rather simplifies construct design.
One of the main advantages of VIGE is that high sgRNA levels are accumulated into host plants due to virus replication, which leads to a fast and efficient editing process not achievable with conventional delivery methods (Cody and Scholthof, 2019; Schmitz et al., 2020). In fact, we observed a high editing efficiency when single sgRNAs were delivered through PVX, reaching nearly 80% of indels (Figures 2b, 2d and Figure 3b, left panel).
The capacity to transmit genome modifications to the next generation is considered a desired feature of any VIGE system. Starting from infected tissue where single or multiple sgRNA had been delivered, we regenerated whole plants and screened them for the presence of modifications. The regenerated plants showed an increase on indel percentages compared with those observed on infected tissue, and most importantly, all of them carried biallelic mutations for NbXT2B gene (Figure 4). However, regenerated plants developed symptoms of viral infection, indicating that PVX was still present. It is well documented that PVX is unable to infect germline cells and therefore is not transmitted through true seed or pollen (Loebenstein and Gaba, 2012). Next, we collected seeds from regenerated plants edited in single or multiple target genes. We confirmed that virus‐free plants with heritable gene editing can be obtained from these seeds (Figure 5a). Heritable biallelic mutations were observed in the single sgRNA strategy in 100% of the progeny (Figure 5b, left panel). For the multiplexing strategy, the percentage of biallelic mutations ranged between 100% for NbXT2B and 20% and 30% for NbPDS3 and NbFT, respectively (Figure 5b, right panel). Remarkably, 100% biallelic mutations in all three genes were observed in individuals from the progeny (i.e., plant 14.1‐v, Figure 5a, right panel).
Progeny from regenerated plants is also the stage where Cas9 is usually segregated in traditional transgenesis approaches. Hence, PVX‐based VIGE offers a similar speed as transgenesis to obtain transgene‐free edited plants, but at a higher efficiency and now multiplexing capacity. Importantly, a recently published VIGE strategy, based on fusion of RNA sequences that promote cell‐to‐cell mobility, has shown to accelerate the recovery of mutant progeny (Ellison et al., 2020). We confirmed that this strategy can also be expanded to our PVX vector, as the addition of a truncated FT sequence to a sgRNA resulted in 22% biallelic mutations in seedlings (Figure 6). PVX‐sgRNAs can be a very useful tool when combined with transgenesis to enhance multigene editing efficiency. Often, multiplex editing with a transgenic construct miss some of the intended targets due to the limited capacity to predict the efficiency of sgRNAs. Provided that Cas9 is already present in parental lines, if missing targets are genotyped early in these lines, they could then be infected with multiplex PVX‐sgRNAs designed specifically to fill the editing gaps. This approach can be faster and more efficient than re‐transformation, and most importantly, it circumvents the need for a second selection marker.
In conclusion, the virus‐mediated genome editing system described here postulates PVX as a superior vector for the delivery of one or more sgRNAs without the need of processing elements, and therefore, leading to a highly efficient gene editing without the risk of integration of viral sequences into the plant genome. These modifications on the target genes are inherited to the next generations when plants are regenerated from infected tissue, while the viral vector is lost in the progeny from regenerated plants, or from seeds of infected plants. Furthermore, the wide range of host species that PVX can infect, including important crops in the family Solanaceae, highlights its potential for CRISPR‐Cas9‐based modification of economically relevant species. In addition, PVX is a preferred vector in plant biotechnology (Röder et al., 2019). Our findings expand the current knowledge about the VIGE toolbox and will contribute to future applications in plant functional genomics and agricultural biotechnology.
EXPERIMENTAL PROCEDURES
Design of sgRNAs and vector construction
Three N. benthamiana genes were chosen as targets for CRISPR‐Cas9‐mediated gene editing: NbFT, NbPDS3 and NbXT2B. The design of sgRNAs was performed using the CRISPR‐P online tool as described in Bernabé‐Orts et al. (2019). Target sequences for each gene are described in Table S1. Plasmid pPVX contains a full‐length PVX cDNA (GenBank accession number MT799816) flanked by the Cauliflower mosaic virus 35S promoter and A. tumefaciens Nopaline synthase terminator. In all PVX‐based recombinant clones, heterologous genes were expressed from the PVX CP promoter, and PVX CP was expressed from a heterologous promoter derived from Bamboo mosaic virus. The 29 initial codons of PVX CP were deleted (Dickmeis et al., 2014). Plasmids to express recombinant viruses PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR, PVX::sgXT2B, PVX::tR‐sgPDS3‐tR, PVX::tR‐sgPDS3, PVX::sgPDS3‐tR, PVX::sgPDS3, PVX::sgFT, PVX::sgPDS3:sgFT:sgXT2B, PVX::sgXT2B‐tFT and PVX::sgPDS3ii‐tFT were built by standard molecular biology techniques, including PCR amplification of target sgRNAs with high‐fidelity Phusion DNA polymerase (Thermo Scientific, Waltham, MA, USA) and Gibson DNA assembly with the NEBuilder HiFi DNA assembly master mix (New England Biolabs, Ipswich, MA, USA). Primers used for vector construction are listed in Tables S2 and S3. Recombinant virus PVX::crtB that expresses Pantoea ananatis phytoene synthase (crtB), which induces a distinctive yellow pigmentation in infected tissue (Majer et al., 2017), was used as a control for the experiments. The sequences of all the recombinant PVX‐derived clones were confirmed by standard DNA sequencing techniques and are included in Figure S1.
Plant growth conditions and inoculation
Transgenic Cas9‐expressing N. benthamiana plants (Figure S2) were grown in growth chambers at 25°C under a 16‐h day/8‐h night cycle. Fully expanded upper leaves from 4‐ to 6‐week‐old plants were used for inoculation of PVX::sgRNA constructs. Electrocompetent A. tumefaciens C58C1 containing the helper plasmid pCLEAN‐S48 (Thole et al., 2007) were transformed with plasmids containing the different PVX recombinant clones. Transformed cells were spread on 50 µg/L kanamycin, 50 µg/L rifampicin and 12.5 µg/L tetracycline Luria Bertani agar plates. Single colonies were grown about 24 h at 28°C in 10 ml Luria Bertani containing 50 µg/mL kanamycin. At optical density of 600 nm (OD600) 1‐2, cells were pelleted by centrifuging at 7200 g for 5 min and resuspended to an OD600 of 0.5 in infiltration buffer (10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES)‐NaOH, 10 mm MgCl2 and 150 µm acetosyringone, pH 5.6) (Bedoya et al., 2010). Resuspended bacteria were incubated at 28°C for 2 h. Two leaves per plant were agroinfiltrated on the abaxial side using a 1‐ml syringe. Control plants were inoculated with PVX::crtB following the same procedure. Immediately following agroinfiltration, plants were watered and transferred to a growth chamber under a 12‐h day/12‐h night and 25°C cycle. Samples from the first symptomatic systemic leaf were collected with a 1.2‐cm cork borer (approximately 100 mg of tissue), immediately frozen in liquid nitrogen and stored at −80ºC until use.
RT‐PCR analysis of PVX progeny
Leaf samples were ground with a VWR Star‐Beater for 1 min at 30 sec−1, homogenized in extraction buffer (4 m guanidinium thiocyanate, 0.1 m sodium acetate, 10 mm EDTA and 0.1 m 2‐mercaptoethanol, pH 5.5), and centrifuged at 15 000 g for 5 min. Next, the collected supernatants were mixed with 0.65 volumes of 96% ethanol and centrifuged at 15 000 g for 1 min. The remaining steps for RNA purification were performed using silica gel columns (Zymo Research, Irvine, CA, USA) that were finally eluted with 10 µl of 10 mm Tris‐HCl pH 8.5. Aliquots (1 µl) of RNA were subjected to RT with RevertAid reverse transcriptase (Thermo Scientific) using primer D2409 in a 10‐µl reaction, and 1‐µl aliquots of the products were subjected to PCR amplification with Thermus thermophilus DNA polymerase (Biotools, Madrid, Spain) using gene‐specific primers in 20‐µl reactions. Conditions were as previously described (Bedoya et al., 2010). Primers D2410 and D3436 were designed to amplify a 627‐nt region of PVX genome corresponding to CP open reading frames. Primers D1789 and D2069 were designed to amplify the heterologous sgRNA region. Primers used for RT and RT‐PCR are listed in Table S4. RT‐PCR products were analyzed by electrophoresis in 1% agarose gels in TAE buffer (40 mm Tris, 20 mm sodium acetate, and 1 mm EDTA, pH 7.2) and staining with ethidium bromide.
Detection of Cas9‐mediated gene editing
To purify DNA from leaf samples, the same procedure explained above for RNA was followed, except for not adding ethanol to the extract before loading the silica gel column. To confirm CRISPR‐Cas9‐mediated editing of the target genes, products covering NbFT, NbPDS3 and NbXT2B target sites were obtained by PCR amplification of 1 µl genomic DNA using high‐fidelity Phusion DNA polymerase and gene‐specific primers. The resulting PCR products were purified using silica gel columns after separation by electrophoresis in 1% agarose gels and subjected to Sanger sequencing. The presence of sequence modifications was analyzed using the ICE algorithm (https://www.synthego.com/products/bioinformatics/crispr‐analysis). Primers used for amplification of target genes and sequencing are listed in Table S5.
In vitro regeneration of edited Nicotiana benthamiana plants
Leaves from 4‐ to 5‐week‐old N. benthamiana plants were infiltrated with A. tumefaciens C58C1 cultures containing plasmids with the PVX recombinant clones PVX::sgXT2B and PVX::sgPDS3:sgFT:sgXT2B. As a control for gene editing, additional plants were infiltrated with PVX::crtB. At 14 and 21 dpi, the first upper leaf showing symptoms of viral infection was removed from each plant and surface‐sterilized by submersion in sterilization solution (10% bleach, 0.02% Nonidet P‐40) for 10 min. Leaves were then washed three times in sterile water. Sterilized leaves were cut into discs using a 1.2‐cm cork borer and plated on to regeneration media (1× Murashige and Skoog with vitamins Gamborg B5, 0.5 g/L MES, 20 g/L sucrose, 8 g/L phytoagar, 1 mg/L 6‐benzylaminopurine, 0.1 mg/L 1‐naphthaleneacetic acid and 50 µg/ml kanamycin, pH 5.7). Leaf discs were transferred to fresh plates every 2 weeks until shoots emerged. Shoots were cut and transferred to root induction media (1× Murashige and Skoog with vitamins, 0.5 g/L MES, 10 g/L sucrose, 6 g/L phytoagar, 0.1 mg/L 1‐naphthaleneacetic acid and 50 µg/ml kanamycin, pH 5.7). Regenerated plantlets with roots were transferred into soil and covered with plastic cups to keep moisture in. After 2 weeks, plastic cups were removed for plant normal growth.
AUTHOR CONTRIBUTIONS
All authors participated in work conception and design. MU, VA, MV and SS performed the experiments. All authors participated in result analyses. MU, DO and JAD wrote the manuscript with input from the rest of the authors.
CONFLICT OF INTERESTS
The authors declare that they have no competing interests
Supporting information
Figure S1. Full sequence of wild‐type Potato virus X (PVX; GenBank accession number MT799816) and its derived recombinant viruses PVX::crtB, PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR, PVX::XT2B, PVX::tR‐sgPDS3‐tR, PVX::tR‐sgPDS3, PVX::sgPDS3‐tR, PVX::sgPDS3, PVX::sgFT, PVX::sgPDS3:sgFT:sgXT2B, PVX::sgXT2B‐tFT and PVX::sgPDS3ii‐tFT.
Figure S2. Sequence of the transgenes inserted into Nicotiana benthamiana to generate a line that constitutively expresses a human codon‐optimized Streptococcus pyogenes Cas9.
Figure S3. Sequence electropherogram of a biallelic edited plant derived from parental plants inoculated with PVX::sgXT2B‐tFT in which a 1‐bp insertion was detected.
Table S1. N. benthamiana genes targeted by the PVX‐based CRISPR/Cas9 system. PAMs are highlighted on gray background.
Table S2. Primers used for the construction of recombinant viruses.
Table S3. Primer combinations used for the construction of recombinant viruses.
Table S4. Primers used for PVX diagnosis by RT‐PCR.
Table S5. Primers used for Cas9‐sgRNA gene editing analysis.
ACKNOWLEDGEMENTS
This work was supported by grants BIO2017‐83184‐R and PID2019‐108203RB‐I00 from Ministerio de Ciencia e Innovación (Spain) through the Agencia Estatal de Investigación (co‐financed European Regional Development Fund), and H2020‐760331 Newcotiana from the European Commission. M.U. and S.S. are the recipients of fellowships FPU17/05503 and BES‐2017‐0890098, respectively, from Ministerio de Ciencia e Innovación (Spain).
DATA AVAILABILITY STATEMENT
All data relating to this manuscript can be found within the manuscript and its supplementary files.
REFERENCES
- Adams, M.J. , Antoniw, J.F. , Bar‐Joseph, M. , Brunt, A.A. , Candresse, T. , Foster, G.D. et al. (2004) The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Archives of Virology, 149, 1045–1060. [DOI] [PubMed] [Google Scholar]
- Ali, Z. , Abul‐faraj, A. , Li, L. , Ghosh, N. , Piatek, M. , Mahjoub, A. et al. (2015) Efficient virus‐mediated genome editing in plants using the CRISPR/Cas9 system. Molecular Plant, 8, 1288–1291. [DOI] [PubMed] [Google Scholar]
- Ali, Z. , Eid, A. , Ali, S. & Mahfouz, M.M. (2018) Pea early‐browning virus‐mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Research, 244, 333–337. [DOI] [PubMed] [Google Scholar]
- Baltes, N.J. , Gil‐Humanes, J. , Cermak, T. , Atkins, P.A. & Voytas, D.F. (2014) DNA replicons for plant genome engineering. The Plant Cell, 26, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoya, L. , Martínez, F. , Rubio, L. & Daròs, J.A. (2010) Simultaneous equimolar expression of multiple proteins in plants from a disarmed potyvirus vector. Journal of Biotechnology, 150, 268–275. [DOI] [PubMed] [Google Scholar]
- Bernabé‐Orts, J.M. , Casas‐Rodrigo, I. , Minguet, E.G. , Landolfi, V. , Garcia‐Carpintero, V. , Gianoglio, S. et al. (2019) Assessment of Cas12a‐mediated gene editing efficiency in plants. Plant Biotechnology Journal, 17, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch‐Smith, T.M. , Schiff, M. , Liu, Y. & Dinesh‐Kumar, S.P. (2006) Efficient virus‐induced gene silencing in Arabidopsis. Plant Physiology, 142, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier, D.M. & Keegstra, K. (2006) Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. Journal of Biological Chemistry, 281, 34197–34207. [DOI] [PubMed] [Google Scholar]
- Chen, K. , Wang, Y. , Zhang, R. , Zhang, H. & Gao, C. (2019) CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annual Review of Plant Biology, 70, 667–697. [DOI] [PubMed] [Google Scholar]
- Cody, W.B. & Scholthof, H.B. (2019) Plant Virus Vectors 3.0: Transitioning into Synthetic Genomics. Annual review of Phytopathology, 57, 211–230. [DOI] [PubMed] [Google Scholar]
- Cody, W.B. , Scholthof, H.B. & Mirkov, T.E. (2017) Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant Physiology, 175, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L. , Ran, F.A. , Cox, D. , Lin, S. , Barretto, R. , Habib, N. et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science, 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman, J.E. , Abudayyeh, O.O. , Joung, J. , Gootenberg, J.S. , Zhang, F. & Konermann, S. (2015) Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nature Biotechnology, 33, 1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmeis, C. , Fischer, R. & Commandeur, U. (2014) Potato virus X‐based expression vectors are stabilized for long‐term production of proteins and larger inserts. Biotechnology Journal, 9, 1369–1379. [DOI] [PubMed] [Google Scholar]
- Edwardson, J.R. & Christie, R.G. (1997) Viruses infecting peppers and other solanaceous crops. Volume 1, Agricultural Experiment Station, University of Florida. [Google Scholar]
- Ellison, E.E. , Nagalakshmi, U. , Gamo, M.E. , Huang, P.J. , Dinesh‐Kumar, S. & Voytas, D.F. (2020) Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nature plants, 6, 620–624. [DOI] [PubMed] [Google Scholar]
- Gao, Y. & Zhao, Y. (2014) Self‐processing of ribozyme‐flanked RNAs into guide RNAs in vitro and in vivo for CRISPR‐mediated genome editing. Journal of Integrative Plant Biology, 56, 343–349. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Li, S. , Li, Z. , Li, H. , Song, W. , Zhao, H. et al. (2019) A barley stripe mosaic virus‐based guide RNA delivery system for targeted mutagenesis in wheat and maize. Molecular Plant Pathology, 20, 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. , Zhang, C. , Liu, J.Y. , Guo, Z.H. , Zhang, Z.Y. , Han, C.G. et al. (2019) Development of Beet necrotic yellow vein virus‐based vectors for multiple‐gene expression and guide RNA delivery in plant genome editing. Plant Biotechnology Journal, 17, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. & Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science., 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.H. & Suh, Y. (2017) In vivo genome editing in animals using AAV‐CRISPR system: Applications to translational research of human disease. F1000Research, 6, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhang, K. , Zeng, X. , Jackson, S. , Zhou, Y. & Hong, Y. (2009) A cis element within Flowering locus T mRNA determines its mobility and facilitates trafficking of heterologous viral RNA. Journal of Virology, 83, 3540–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhang, H. , Si, X. , Tian, Y. , Chen, K. , Liu, J. et al. (2017) Generation of thermosensitive male‐sterile maize by targeted knockout of the ZmTMS5 gene. Journal of Genetics and Genomics, 44, 465–468. [DOI] [PubMed] [Google Scholar]
- Li, T. , Yang, X. , Yu, Y. , Si, X. , Zhai, X. , Zhang, H. et al. (2018) Domestication of wild tomato is accelerated by genome editing. Nature Biotechnology, 36(12), 1160–1163. [DOI] [PubMed] [Google Scholar]
- Loebenstein, G. & Gaba, V. (2012) Viruses of Potato. Adv. Virus Res., 84, 209–246. [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, X. , Liu, H. & Li, Z. (2020) Highly efficient DNA‐free plant genome editing using virally delivered CRISPR–Cas9. Nature Plants, 6, 773–779. [DOI] [PubMed] [Google Scholar]
- Majer, E. , Llorente, B. , Rodríguez‐Concepción, M. & Daròs, J.A. (2017) Rewiring carotenoid biosynthesis in plants using a viral vector. Scientific Reports, 7, 41645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P. , Yang, L. , Esvelt, K.M. , Aach, J. , Guell, M. , DiCarlo, J.E. , et al. (2013) RNA‐guided human genome engineering via Cas9. Science., 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov, V. , Staskawicz, B. , Weigel, D. , Jones, J.D.G. & Kamoun, S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nature Biotechnology, 31, 691–693. [DOI] [PubMed] [Google Scholar]
- Nissim, L. , Perli, S.D. , Fridkin, A. , Perez‐Pinera, P. & Lu, T.K. (2014) Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Molecular Cell, 54, 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. (2010) Sowing the seeds for the ideal crop. Science, 327, 802–803. [DOI] [PubMed] [Google Scholar]
- Platt, R.J. , Chen, S. , Zhou, Y. , Yim, M.J. , Swiech, L. , Kempton, H.R. et al. (2014) CRISPR‐Cas9 knockin mice for genome editing and cancer modeling. Cell, 159, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A. , Hsu, P.D. , Wright, J. , Agarwala, V. , Scott, D.A. & Zhang, F. (2013) Genome engineering using the CRISPR‐Cas9 system. Nature Protocols, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder, J. , Dickmeis, C. & Commandeur, U. (2019) Small, smaller, nano: New applications for potato virus X in nanotechnology. Frontiers in Plant Science, 10, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml, S. , Fauser, F. & Puchta, H. (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. The Plant Journal, 80, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Schmitz, D.J. , Ali, Z. , Wang, C. , Aljedaani, F. , Hooykaas, P.J.J. , Mahfouz, M. et al. (2020) CRISPR/Cas9 mutagenesis by translocation of Cas9 protein into plant cells via the Agrobacterium type IV secretion system. Frontiers in Genome Editing, 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senís, E. , Fatouros, C. , Große, S. , Wiedtke, E. , Niopek, D. , Mueller, A.‐K. et al. (2014) CRISPR/Cas9‐mediated genome engineering: an adeno‐associated viral (AAV) vector toolbox. Biotechnology Journal, 9, 1402–1412. [DOI] [PubMed] [Google Scholar]
- Thole, V. , Worland, B. , Snape, J.W. & Vain, P. (2007) The pCLEAN dual binary vector system for Agrobacterium‐mediated plant transformation. Plant Physiol., 145, 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, S.Q. , Wyvekens, N. , Khayter, C. , Foden, J.A. , Thapar, V. , Reyon, D. et al. (2014) Dimeric CRISPR RNA‐guided FokI nucleases for highly specific genome editing. Nature Biotechnology, 32, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D.F. (2013) Plant genome engineering with sequence‐specific nucleases. Annual Review of Plant Biology, 64, 327–350. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A. , Kim, M.C. , Jaeger, K.E. , Busch, W. , Schmid, M. , Lohmann, J.U. et al. (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science, 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. & Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proceedings of the National Academy of Sciences of the United States of America, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C.L. , Ruan, M.Z.C. , Mahajan, V.B. & Tsang, S.H. (2019) Viral Delivery Systems for CRISPR. Viruses, 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Zhao, L. , Gao, Y. , Xu, J. & Han, R. (2017) Empower multiplex cell and tissue‐specific CRISPR‐mediated gene manipulation with self‐cleaving ribozymes and tRNA. Nucleic Acids Research, 45, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, K. , Han, T. , Liu, G. , Chen, T. , Wang, Y. , Yu, A.Y.L. et al. (2015) A geminivirus‐based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Scientific Reports, 5, 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan, J.G. , Lee, M.E. , Almeida, R. , Gilbert, L.A. , Whitehead, E.H. , La Russa, M. et al. (2015) Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell, 160, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B. , Heidenreich, M. , Mohanraju, P. , Fedorova, I. , Kneppers, J. , Degennaro, E.M. et al. (2017) Multiplex gene editing by CRISPR‐Cpf1 using a single crRNA array. Nature Biotechnology, 35, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. et al. (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nature Communications, 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Full sequence of wild‐type Potato virus X (PVX; GenBank accession number MT799816) and its derived recombinant viruses PVX::crtB, PVX::tR‐sgXT2B‐tR, PVX::tR‐sgXT2B, PVX::sgXT2B‐tR, PVX::XT2B, PVX::tR‐sgPDS3‐tR, PVX::tR‐sgPDS3, PVX::sgPDS3‐tR, PVX::sgPDS3, PVX::sgFT, PVX::sgPDS3:sgFT:sgXT2B, PVX::sgXT2B‐tFT and PVX::sgPDS3ii‐tFT.
Figure S2. Sequence of the transgenes inserted into Nicotiana benthamiana to generate a line that constitutively expresses a human codon‐optimized Streptococcus pyogenes Cas9.
Figure S3. Sequence electropherogram of a biallelic edited plant derived from parental plants inoculated with PVX::sgXT2B‐tFT in which a 1‐bp insertion was detected.
Table S1. N. benthamiana genes targeted by the PVX‐based CRISPR/Cas9 system. PAMs are highlighted on gray background.
Table S2. Primers used for the construction of recombinant viruses.
Table S3. Primer combinations used for the construction of recombinant viruses.
Table S4. Primers used for PVX diagnosis by RT‐PCR.
Table S5. Primers used for Cas9‐sgRNA gene editing analysis.
Data Availability Statement
All data relating to this manuscript can be found within the manuscript and its supplementary files.


