Abstract
Background
Equal dosage of X-linked genes between males and females is maintained by the X-inactivation of the second X chromosome in females through epigenetic mechanisms. Boys with aneuploidy of the X chromosome exhibit a host of symptoms such as low fertility, musculoskeletal anomalies, and cognitive and behavioral deficits that are presumed to be caused by the abnormal dosage of these genes. The objective of this pilot study is to assess the relationship between CpG methylation, an epigenetic modification, at several genes on the X chromosome and behavioral dysfunction in boys with supernumerary X chromosomes.
Results
Two parental questionnaires, the Behavior Rating Inventory of Executive Function (BRIEF) and Child Behavior Checklist (CBCL), were analyzed, and they showed expected differences in both internal and external behaviors between neurotypical (46,XY) boys and boys with 49,XXXXY. There were several CpGs in AR and MAOA of boys with 49,XXXXY whose methylation levels were skewed from levels predicted from having one active (Xa) and three inactive (Xi) X chromosomes. Further, methylation levels of multiple CpGs in MAOA showed nominally significant association with externalizing behavior on the CBCL, and the methylation level of one CpG in AR showed nominally significant association with the BRIEF Regulation Index.
Conclusions
Boys with 49,XXXXY displayed higher levels of CpG methylation at regulatory intronic regions in X-linked genes encoding the androgen receptor (AR) and monoamine oxidase A (MAOA), compared to that in boys with 47,XXY and neurotypical boys. Our pilot study results suggest a link between CpG methylation levels and behavior in boys with 49,XXXXY.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-021-01123-4.
Keywords: Sex chromosome aneuploidies; DNA methylation; MAOA; 47,XXY; 49,XXXXY; Executive functioning; Externalizing disorders; Internalizing disorders
Background
47,XXY (Klinefelter syndrome or KS) is an X and Y chromosomal variation that results from the addition of an extra X chromosome, affecting 1 out of every 660 live male births [1–3]. 49,XXXXY is a more severe variant of 47,XXY that occurs in 1 out of every 85,000 to 100,000 live male births [4–6]. Both sex chromosome aneuploidies (SCA) commonly exhibit testicular dysgenesis and hypergonadotropic hypogonadism attributable to the one or more additional X chromosomes, and the decreased levels of testosterone in these males also may affect neurocognitive development [7–10]. Behaviorally, patients with 47,XXY may have elevated levels of anxiety, peer social interactional differences, and internalizing problems when compared to neurotypical standards, according to the Child Behavior Checklist (CBCL) [11]. Boys with 49,XXXXY have exhibited elevated scores on similar domains of the CBCL, including anxiety, internalizing problems, thought problems, and externalizing problems to an increased extent than as described for 47,XXY [11].
Previous studies suggest the benefit of hormonal replacement therapy (HRT) on neurocognitive, neurobehavioral, and neuromotor capabilities for boys with 47,XXY and 49,XXXXY. Pediatric endocrinologists currently prescribe HRT, specifically testosterone, to males with 47,XXY and 49,XXXXY based on physical examination. Neurodevelopmental investigations on these males showed that infants with 47,XXY who receive early hormonal treatment (EHT) showed improvement in Full-Scale IQ, Verbal IQ, and speech and language development when compared to untreated boys [12]. In addition, parents of treated boys reported significantly fewer behavioral concerns along with improved social behavioral and initiation skills [13]. Males with 47,XXY who were treated with HRT experienced fewer social, thought, and affective problems when compared to the untreated group [14]. In a recent study, infants with 49,XXXXY who received EHT scored higher on the Bayley Scales of Infant Development compared to their untreated counterparts [7]. While such studies suggest global improvement in these populations with testosterone treatment, the etiology of these behaviors has not been investigated from a genetic perspective [13, 15].
It has been hypothesized that the behavioral variation between these patients may be attributed to the presence and skewed inactivation of their additional X chromosomes. In the neurotypical 46,XX female, one of the two X chromosomes is inactivated in order to silence the transcriptional activity of the second sex chromosome. This X inactivation (Xi) allows the female to maintain an appropriately equal expression of sex-linked genes compared to her neurotypical 46,XY male counterpart. Inactivation is a complex process that depends on genes on autosomes that are activated by the presence of two X chromosomes and involves, in part, the transcription of the XIST (X-inactive specific transcript) RNA from the inactive X chromosome [16]. The inactive X chromosome is also associated with specific DNA methylation patterns that play a key role in the silencing of its genes [17, 18]. DNA methylation regulates gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factors to DNA [19]. DNA methylation is therefore a powerful mechanism for the regulation of gene transcription.
When there are more than two X chromosomes, such as in 49,XXXXY syndrome, the typical pattern of DNA methylation might be skewed on the inactive X chromosome. Such aberrant X chromosome methylation patterns are likely to impact gene expression, possibly resulting in specific clinical phenotypes [20]. For instance, fragile X syndrome is caused by CGG expansion and hypermethylation at the FMR1 (fragile X mental retardation 1) gene promoter, resulting in transcriptional repression [21]. The FMRP protein encoded by FMR1 helps regulate synaptic plasticity, which is the hallmark of learning and memory. As such, fragile X syndrome is characterized by severe intellectual disability, autism spectrum disorder (ASD), language delay, sensory hyperarousal and anxiety [22]. While fragile X syndrome exemplifies the effects of hypermethylation of genes on the X chromosome, hypomethylation of genes on additive X chromosomes could also have potentially powerful phenotypic effects by disrupting the typical dosage of transcriptional activity.
This pilot study is the first to explore DNA methylation on genes on the X chromosome associated with behavior in patients with 47,XXY and 49,XXXXY and the possible interaction between HRT and DNA methylation levels. The behavior of patients with 47,XXY and 49,XXXXY is assessed using the BRIEF (Behavior Rating Inventory of Executive Function) and CBCL (Child Behavioral Checklist). These behavioral assessment results are then compared to epigenetic analyses of degree of DNA methylation at several X-linked genes. Our approach has the potential to elucidate the complex interaction between gene and behavior in individuals with multiple X chromosomes.
Results
BRIEF analysis
Detailed information on the participants, such as cohort size, diagnosis, and exclusion criteria, is provided in Materials and methods (Sect. 5.1). For the first analysis, individuals with 47,XXY were compared to individuals with 49,XXXXY using the BRIEF, which is a questionnaire administered to the participants’ parents and includes three scales to measure behavioral regulation, five scales for metacognition, and a composite score that combines all scales. The mean age of the twenty-six participants with 47,XXY (mean = 159.35 months; SD = 42.22) for whom BRIEF was available was significantly higher than the group of eleven participants with 49,XXXXY (mean = 102.27 months, SD = 26.10, P = 0.001). To control for this age disparity between groups, a subgroup of the youngest participants with 47,XXY (mean age = 119.18 months, SD = 25.01) was created. The mean age of this group of eleven younger participants with 47,XXY was not significantly different than the group of eleven participants with 49,XXXXY (P = 0.14). In the group of eleven younger participants with 47,XXY, none of the mean averages for the BRIEF Composite scales (i.e., Behavioral Regulation Index, Metacognition Index, and Global Executive Composite) were significantly different from a test value of T = 50 (T = 52.5 to 53.4). In contrast, mean averages for all of the BRIEF Composite scales were significantly higher than the test value of T = 50 in the 49,XXXXY group (T = 62.0 to 66.6, P < 0.01 for all comparisons using the Student’s T-test), suggesting a significantly higher level of parent-reported executive dysfunction in these individuals. Moreover, when the 47,XXY and 49,XXXXY groups were compared, significant differences were identified on all BRIEF categories (Table 1).
Table 1.
Standardized measures (mean, SD) of behavior rating inventory of executive functions (BRIEF)
| Test | Mean 47,XXY (SD) | Mean 49,XXXXY (SD) | Significance | Cohen's d |
|---|---|---|---|---|
| BRIEF Behavioral Regulation Index (T) | 52.5 (15.2) | 66.6 (13.9) | 0.034 | 0.97 |
| BRIEF Metacognition Index (T) | 52.5 (12.6) | 62.0 (8.0) | 0.049 | 0.90 |
| BRIEF Global Executive Composite (T) | 53.4 (14.0) | 64.7 (9.4) | 0.037 | 0.95 |
T = T-score; SD = standard deviation; mean scores for the 49,XXXXY group were all significantly different from the test value T = 50 (P < 0.01)
CBCL analysis
For the second analysis, 47,XXY and 49,XXXXY groups were compared using the CBCL, which is a questionnaire administered to the participants’ parents to measure the expression of internalizing problems such as anxiety and depression, externalizing problems, and other social problems. Once again, the mean age of the twenty-nine participants with 47,XXY (mean = 146.68 months; SD = 42.53) with available CBCL data was significantly higher (P = 0.003) than that of the group of thirteen participants with 49,XXXXY (mean = 104.94 months, SD = 29.06). To control for this age disparity between groups, a subgroup of the thirteen youngest participants with 47,XXY (mean age = 108.08 months; SD = 26.74) was created so that the mean age difference between the two groups was not significant (P = 0.78). In the thirteen individuals with 47,XXY, mean values for the CBCL Internalizing and Externalizing Problems scales (T = 55.7 and 53.9, respectively) were not elevated when compared to a reference value of T = 50, but the mean value for the Total Problems scale was significantly elevated (T = 58.9, P = 0.02). In contrast, mean values for the 49,XXXXY group were significantly higher than the test value of T = 50 for all three CBCL composite scales (T = 64.7 to 68.8, P < 0.001). Moreover, mean values for the two groups were significantly different for each of the composite scales on the CBCL, indicating significantly higher parent reports of internalizing, externalizing, and total behavioral problems in the 49,XXXXY group (Table 2).
Table 2.
Standardized measures (mean, SD) of child behavior checklist (CBCL)
| Test | Mean 47,XXY (SD) | Mean 49,XXXXY (SD) | Significance | Cohen's d |
|---|---|---|---|---|
| CBCL Internalizing Problems (T) | 55.7 (13.1) | 66.2 (10.4) | 0.034 | 0.89 |
| CBCL Externalizing Problems (T) | 53.92 (9.6) | 64.7 (8.6) | 0.006 | 1.18 |
| CBCL Total Problems (T) | 58.9 (11.8) | 68.8 (8.2) | 0.020 | 0.97 |
T = T-score; SD = standard deviation; mean scores for the 49,XXXXY group were all significantly different from the test value T = 50 (P < 0.001)
DNA methylation analysis
To identify potential mechanisms that underlie the behavioral deficits in the individuals with 49,XXXXY, bisulfite pyrosequencing assays were conducted to compare saliva DNA between the 49,XXXXY and neurotypical 46,XY groups for methylation differences in a small selection of genes on the X chromosome. Since we had no a priori knowledge of specific CpG targets that have been implicated for individuals with 49,XXXXY, we focused on several X-linked candidate genes that were involved in neurodevelopment and behavior. Further, we chose CpGs adjacent to glucocorticoid response elements (GREs) given the role of glucocorticoids in neurodevelopment and behavior. Only one significant but subtle difference was observed at one of the two neighboring CpGs in the methyl-CpG binding protein 2 (MeCP2) gene (P = 3.5 × 10−4, Fig. 1a). At a GRE in the first intron of the androgen receptor (AR), significant decreases in DNA methylation were observed in three of the four CpG examined, although the standard deviations were high in both groups (P < 0.02, Fig. 1b). Interestingly, significant increase in DNA methylation was observed at all three CpGs at an intronic GRE in the monoamine oxidase A gene (MAOA) (P < 0.003, Fig. 1c). We also targeted a region adjacent to a CpG island in the first intron of MAOA, since its CpG methylation has been associated with MAOA enzymatic activity in the brain [30] and thus may play a role in the transcriptional regulation of MAOA. Six–eightfold increases in methylation were observed at each of the seven CpGs examined between the 49,XXXXY and 46,XY groups (P < 0.001, Fig. 1d).
Fig. 1.
Methylation levels at candidate X-linked loci between 46,XY and 49,XXXXY. Genomic DNA extracted from saliva were used for bisulfite pyrosequencing of putative regulatory regions at three genes involved in neurodevelopment and neurotransmission: methyl CpG-binding protein 2 (MeCP2) (a), androgen receptor (AR) (b), and the third (c) and first (D) introns of monoamine oxidase A (MAOA). Error bars are presented as mean ± STD. *P < 0.05; **P < 0.01; and ***P < 0.001
X chromosome methylation versus copy number analysis
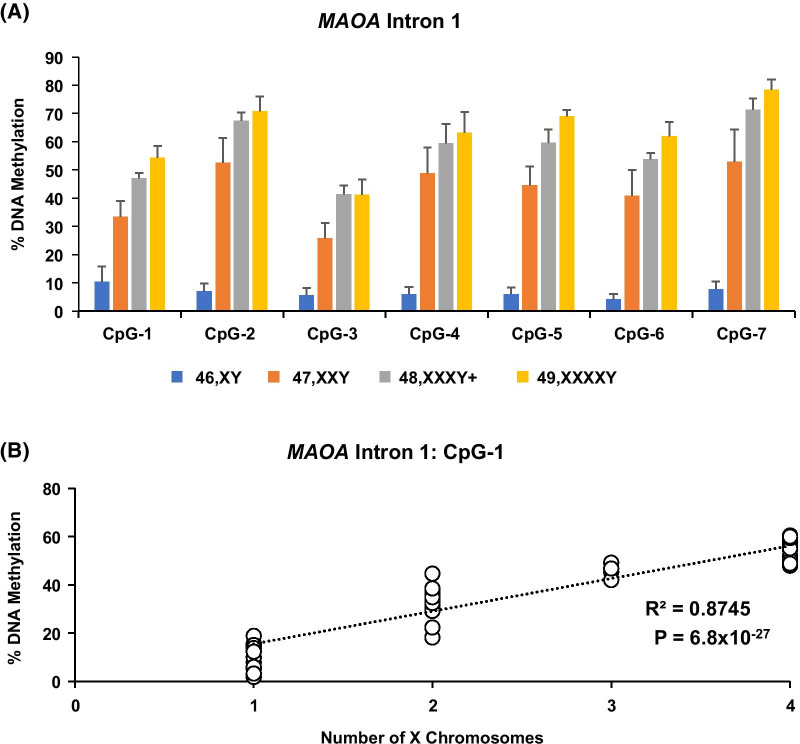
To assess whether the significant MAOA methylation differences in Fig. 1d between neurotypical 46,XY and 49,XXXXY individuals were dependent on the number of X chromosomes, we compared CpG methylation at the MAOA first intron in males with different numbers of X chromosomes. We also chose another region within AR whose CpG methylation levels have been linked to gene regulation [31, 32]. Similar to individuals with 49,XXXXY, methylation levels for individuals with 47,XXY and 48,XXXY (for MAOA only) at both regions were higher at each CpG compared to the neurotypical 46,XY males in a copy number-dependent manner. For instance, CpG-1 in MAOA showed a copy number-dependent increase in DNA methylation from 10.4% in 46,XY to 54.3% in 49,XXXXY samples (Fig. 2a). The relationship between X chromosome number and DNA methylation was significant (R2 = 0.87, P = 6.8 × 10−27, Fig. 2b). Similar results were obtained for other MAOA CpGs tested. Similar methylation patterns (Fig. 3a) and statistical significance (R2 = 0.85, P = 9.4 × 10−9, Fig. 3b) were observed for CpG-1 of the AR locus. For the AR analysis, the samples also included DNA collected from the mothers of probands. These parental samples were included as 46,XX to compare against the 47,XXY samples, and similar DNA methylation levels were observed in both groups (Fig. 3a). We also sought to compare observed methylation values at MAOA and AR with hypothetical methylation values for the active and the inactive X chromosomes derived from 46XY, 46,XX, and 47,XXY samples. For the majority of the CpGs in both MAOA and AR, DNA methylation levels in 47,XXY and 49,XXXXY groups for both genes and 48,XXXY for MAOA were consistent with there being only one active X chromosome. For example, MAOA CpG-2 in the 46,XY was 6.9%, and this number was used to assign a value of 6.9% to the active X chromosome (Xa) in the 47,XXY group. An assumption was made that the second X chromosome in the 47,XXY sample was inactive (Xi) with an estimated 98.1% methylation [(98.1% + 6.9%)/2 = 52.5%] based on the experimentally obtained CpG-2 methylation (52.5%) in the 47,XXY samples. These derived values for Xa and Xi for CpG-2 were then used to assign hypothetical values to each X chromosome in the 48,XXXY and 49,XXXXY groups to assess whether the observed methylation levels corresponded to specific numbers of Xa and Xi in each group. For CpG-2, the estimated methylation values for 47,XaXiXiY and 49, XaXiXiXiY were 67.7% and 75.3%, respectively. Notably, these estimated values were similar to the observed pyrosequencing values at CpG-2 for 47,XXXY and 49,XXXXY groups at 67.5% and 70.8%, respectively. However, some CpGs showed methylation levels that were consistent with a scenario where all of the X chromosomes have been inactivated, i.e., 47,XiXiXiY or 49,XiXiXiXiY. For instance, the observed methylation levels at CpG-1 of MAOA in 49,XXXXY was 54.3%, which showed no statistically significant deviation from the 56.4% methylation predicted from all four X chromosomes being inactive (Table 3). P values for the differences between the hypothetical combinations of Xa/Xi and observed methylation levels are provided in Additional file 3: Table 2.
Fig. 2.
X chromosome number-dependent increase in MAOA methylation levels in 46,XY; 47,XXY; 48,XXXY+; and 49,XXXXY individuals. Bisulfite pyrosequencing of saliva DNA shows a dose-dependent increase in DNA methylation in intron 1 of the MAOA gene (a). Linear regression analysis for CpG-1 showed a strong correlation between observed DNA methylation and X chromosome number (b). The 48,XXXY + group consists of two individuals with 48,XXXY and one with 49,XXXYY karyotype. Groups with different numbers of X chromosomes (1–4) are: 46,XY; 47,XXY; 48,XXXY + ; and 49,XXXXY, respectively. Error bars are presented as mean ± STD
Fig. 3.
X chromosome number-dependent increase in AR methylation levels in 47,XXY and 49,XXXXY probands and their 46,XY or 46,XX parents and siblings. Bisulfite pyrosequencing of saliva DNA shows a dose-dependent increase in DNA methylation in exon 1 CAG tandem repeat of the AR gene (a). Linear regression analysis showed a strong correlation between observed DNA methylation levels and X chromosome number. Graph shows the regression results for CpG-1 (b). Note that the 46,XX and 47,XXY show the same methylation levels, suggesting that the second X chromosome in the XXY probands are inactivated. Groups with different numbers of X chromosomes (1–4) are: 46,XY; 46,XX; 47,XXY; and 49,XXXXY, respectively. Error bars are presented as mean ± STD
Table 3.
Predicted and observed methylation levels for XY, XXY, XXXY, and XXXXY at the MAOA and AR loci
| XY | XXY | XXXY* | XXXXY | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xa/Obs | Xa | Xi | Obs | XaXaXa | XaXaXi | XaXiXi | XiXiXi | Obs | XaXaXaXa | XaXaXaXi | XaXaXiXi | XaXiXiXi | XiXiXiXi | Obs | |
| MAOA | |||||||||||||||
| CpG-1 | 10.4 | 10.4 | 56.4 | 33.4 | 10.4 | 25.8 | 41.1 | 56.4 | 47.2 | 10.4 | 21.9 | 33.4 | 44.9 | 56.4 | 54.3 |
| CpG-2 | 6.9 | 6.9 | 98.1 | 52.5 | 6.9 | 37.3 | 67.7 | 98.1 | 68.4 | 6.9 | 29.7 | 52.5 | 75.3 | 98.1 | 70.8 |
| CpG-3 | 5.6 | 5.6 | 45.9 | 25.7 | 5.6 | 19.0 | 32.5 | 45.9 | 39.6 | 5.6 | 15.7 | 25.7 | 35.8 | 45.9 | 41.2 |
| CpG-4 | 5.9 | 5.9 | 91.8 | 48.9 | 5.9 | 34.6 | 63.2 | 91.8 | 59.9 | 5.9 | 27.4 | 48.9 | 70.3 | 91.8 | 63.1 |
| CpG-5 | 6.0 | 6.0 | 83.0 | 44.5 | 6.0 | 31.7 | 57.3 | 83.0 | 61 | 6.0 | 25.2 | 44.5 | 63.7 | 83.0 | 69.0 |
| CpG-6 | 4.2 | 4.2 | 77.5 | 40.9 | 4.2 | 28.7 | 53.1 | 77.5 | 53.0 | 4.2 | 22.5 | 40.9 | 59.2 | 77.5 | 62.0 |
| CpG-7 | 7.6 | 7.6 | 98.1 | 52.9 | 7.6 | 37.8 | 67.9 | 98.1 | 72.2 | 7.6 | 30.2 | 52.9 | 75.5 | 98.1 | 78.3 |
| AR | |||||||||||||||
| CpG-1 | 3.0 | 3.0 | 34.2 | 18.6 | NA | NA | NA | NA | NA | 3.0 | 10.8 | 18.6 | 26.4 | 34.2 | 31.0 |
| CpG-2 | 4.6 | 4.6 | 37.1 | 20.8 | NA | NA | NA | NA | NA | 4.6 | 12.7 | 20.8 | 28.9 | 37.1 | 28.4 |
| CpG-3 | 7.1 | 7.1 | 82.7 | 44.9 | NA | NA | NA | NA | NA | 7.1 | 26.0 | 44.9 | 63.8 | 82.7 | 54.9 |
| CpG-4 | 10.7 | 10.7 | 38.6 | 24.7 | NA | NA | NA | NA | NA | 10.7 | 17.7 | 24.7 | 31.6 | 38.6 | 36.5 |
*Methylation levels for XXXY were not available (NA) for AR, as there were no XXY probands. Xa denotes active X chromosome, and Xi denotes inactive X chromosome. Numbers in bold denote the predicted methylation values of Xa and Xi combinations that gave the closest value to that observed (Obs) experimentally by pyrosequencing. For XY, Xa was based on observed methylation at the X chromosome. For XXY, Xi was calculated based on Xa values from XY. For XXXY and XXXXY, all of the Xa and Xi were predicted based on values from XY and XXY and then compared to actually observed values from XXXY and XXXXY by pyrosequencing. All observed methylation vales have been adjusted by effects of testosterone
CpG methylation at autosomes
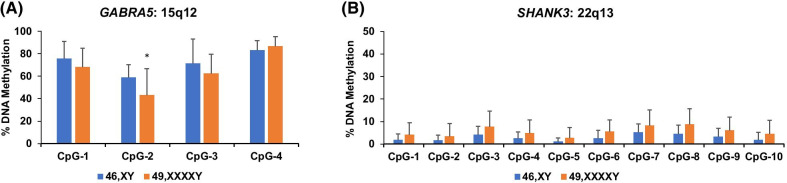
DNA methylation was also investigated at the GABA receptor 5 (GABRA5)/GABA receptor 3 (GABRB3) and SHANK3 loci that are located on chromosomes 15q12 and 22q13, respectively. These loci were chosen based on their association with neurodevelopmental disorders such as autism [33–36] and served as negative controls to the significant across-group methylation differences observed on the X chromosome. There were no significant differences in CpG methylation between the 46,XY and 49,XXXXY groups at these two loci, although one CpG in GABRA5 was slightly lower in methylation for 49,XXXXY (CpG-2, P = 0.04, Fig. 4a, b).
Fig. 4.
Methylation levels at candidate autosomal loci between 46,XY and 49,XXXXY. Genomic DNA extracted from saliva was used for bisulfite pyrosequencing of putative regulatory regions at two genes involved in neurodevelopment and neurotransmission: GABA type A receptor alpha5 subunit (GABRA5) on 15q12 (a) and SH3 multiple ankyrin repeat domains 3 (SHANK3) on 22q13 (b). Error bars are presented as mean ± STD. *P < 0.05
Association between methylation and behavioral assessment
To assess whether behavioral differences between individuals with 49,XXXXY and neurotypical 46,XY were associated with methylation, BRIEF and CBCL scores were compared against MAOA and AR CpG methylation levels. The greatest association was observed with the CBCL externalizing behavior, where a 10% increase in methylation at MAOA CpG-2, CpG-3, CpG-4, and CpG-7 was significantly associated with at least a 2.5-point increase on the externalizing behavior scale (Table 4). The associations with the CBCL internalizing subscale were not significant, and only marginal significance was achieved for CpG-1 (P = 0.08), CpG-2 (P = 0.06), and CpG-7 (P = 0.08) on the CBCL total scale. There were no statistically significant associations between MAOA methylation and any of the BRIEF subscales. For the CpGs in AR, only CpG-1 was significantly associated (P = 0.03) with the BRIEF Behavioral Regulation Index, with a 10% increase in methylation correlating to a 15.9-point (95% CI: 6.3, 25.4 points) increase in this scale. Similar to the MAOA CpG sites, there was only marginal significance with the CBCL total scale for CpG-1 (estimated 9.9-point increase, P = 0.07) and CpG-2 (estimated 13.3-point increase, P = 0.09).
Table 4.
Association between DNA methylation and behavior reports in individuals with 49,XXXXY
| CBCL total problems (95% CI) | P value | CBCL externalizing problems (95% CI) | P value | CBCL internalizing problems (95% CI) | P value | |
|---|---|---|---|---|---|---|
| MAOA CpG-1 | 0.32 (− 0.03, 0.67) | 0.08 | 0.30 (− 0.03, 0.62) | 0.08 | 0.32 (− 0.09, 0.73) | 0.13 |
| MAOA CpG-2 | 0.27 (0.0002, 0.54) | 0.06 | 0.31 (0.07, 0.55) | 0.02 | 0.16 (− 0.16, 0.49) | 0.33 |
| MAOA CpG-3 | 0.33 (− 0.07, 0.73) | 0.12 | 0.40 (0.04, 0.76) | 0.04 | 0.23 (− 0.24, 0.70) | 0.35 |
| MAOA CpG-4 | 0.25 (− 0.06, 0.56) | 0.13 | 0.32 (0.04, 0.60) | 0.03 | 0.12 (− 0.25, 0.50) | 0.52 |
| MAOA CpG-5 | 0.20 (− 0.07, 0.47) | 0.17 | 0.24 (− 0.004, 0.49) | 0.06 | 0.15 (− 0.17, 0.47) | 0.38 |
| MAOA CpG-6 | 0.22 (− 0.07, 0.50) | 0.15 | 0.26 (− 0.001, 0.52) | 0.06 | 0.15 (− 0.18, 0.49) | 0.38 |
| MAOA CpG-7 | 0.22 (− 0.02, 0.45) | 0.08 | 0.27 (0.06, 0.47) | 0.02 | 0.13 (− 0.15, 0.40) | 0.38 |
| AR CpG-1 | 0.99 (− 0.14, 1.84) | 0.07 | 1.05 (− 0.002, 2.11) | 0.11 | 0.56 (− 0.28, 1.41) | 0.25 |
| AR CpG-2 | 1.33 (− 0.10, 2.57) | 0.09 | 1.38 (− 0.16, 2.92) | 0.14 | 0.71 (− 0.51, 1.93) | 0.31 |
| AR CpG-3 | 0.36 (− 0.44, 1.16) | 0.42 | 0.32 (− 0.63, 1.26) | 0.54 | 0.19 (− 0.48, 0.86) | 0.61 |
| AR CpG-4 | 1.16 (− 0.46, 2.78) | 0.22 | 1.13 (− 0.84, 3.09) | 0.31 | 0.68 (− 0.74, 2.10) | 0.39 |
| BRIEF Behavioral Regulation Index (95% CI) | P value | BRIEF Metacognition Index (95% CI) | P value | BRIEF Global Executive Composite (95% CI) | P value | |
|---|---|---|---|---|---|---|
| MAOA CpG-1 | 0.23 (− 0.23, 0.68) | 0.33 | 0.05 (− 0.32, 0.42) | 0.80 | 0.10 (− 0.30, 0.50) | 0.63 |
| MAOA CpG-2 | 0.23 (− 0.14, 0.60) | 0.24 | 0.03 (− 0.28, 0.33) | 0.86 | 0.09 (− 0.24, 0.41) | 0.60 |
| MAOA CpG-3 | 0.25 (− 0.30, 0.79) | 0.39 | 0.06 (− 0.39, 0.50) | 0.81 | 0.11 (− 0.36, 0.59) | 0.65 |
| MAOA CpG-4 | 0.31 (− 0.11, 0.74) | 0.16 | 0.03 (− 0.33, 0.39) | 0.87 | 0.12 (− 0.26, 0.50) | 0.54 |
| MAOA CpG-5 | 0.10 (− 0.26, 0.47) | 0.58 | − 0.02 (− 0.32, 0.27) | 0.88 | 0.01 (− 0.31, 0.32) | 0.97 |
| MAOA CpG-6 | 0.21 (− 0.17, 0.59) | 0.29 | 0.03 (− 0.28, 0.35) | 0.85 | 0.08 (− 0.26, 0.42) | 0.65 |
| MAOA CpG-7 | 0.18 (− 0.14, 0.51) | 0.28 | 0.02 (− 0.25, 0.28) | 0.90 | 0.07 (− 0.22, 0.35) | 0.66 |
| AR CpG-1 | 1.59 (0.63, 2.54) | 0.03 | 0.97 (0.14, 1.80) | 0.08 | 1.11 (− 0.06, 2.28) | 0.13 |
| AR CpG-2 | 1.99 (0.41, 3.58) | 0.07 | 1.22 (− 0.05, 2.48) | 0.13 | 1.40 (− 0.32, 3.12) | 0.19 |
| AR CpG-3 | 0.69 (− 0.40, 1.79) | 0.28 | 0.45 (− 0.33, 1.22) | 0.32 | 0.42 (− 0.63, 1.47) | 0.48 |
| AR CpG-4 | 2.17 (0.26, 4.07) | 0.09 | 1.55 (0.30, 2.80) | 0.07 | 1.55 (− 0.44, 3.53) | 0.20 |
The P value threshold for statistical significance after Bonferroni correction for the number of CpG sites tested was 0.005
Impact of testosterone on behavior and methylation
Males with 49,XXXXY typically undergo testosterone treatment (T) to promote optimal sexual and neurocognitive development [37]. First, scores on the CBCL and BRIEF were compared between the 49,XXXXY no-T and 49,XXXXY T groups. The 49,XXXXY T group showed a lower average score on the CBCL Total and the Externalizing Problems subscale, but not with any of the three BRIEF subscales (Table 5). For instance, receiving testosterone was associated with, on average, an 8.4-point reduction in the CBCL Total Problems score (P = 0.04). History of testosterone treatment was then compared with MAOA methylation. When examining the effect of different testosterone doses on changes in methylation, there were two nominally significant.
Table 5.
Effect of testosterone replacement therapy on behavior in individuals with 49,XXXXY
| Received T | Did not receive T | P value | |
|---|---|---|---|
| CBCL total problems | 58.66 (40.0, 82.0) | 67.10 (49.0, 78.0) | 0.04 |
| CBCL externalizing problems | 54.3 (34.0, 75.0) | 62.5 (48.0, 79.0) | 0.04 |
| CBCL internalizing problems | 57.6 (34.0, 92.0) | 62.9 (45.0, 75.0) | 0.24 |
| BRIEF BRI | 57.2 (39.0, 88.0) | 60.9 (50.0, 87.0) | 0.54 |
| BRIEF MI | 56.7 (35.0, 75.0) | 64.9 (50.0, 79.0) | 0.11 |
| BRIEF global | 58.3 (36.0, 78.0) | 64.7 (51.0, 85.0) | 0.24 |
MAOA CpG sites that increased in methylation following EHT (< 60 months of age). Specifically, CpGs 2 and 4 both showed an increase of 6.5% (P = 0.01) and 6.4% (P = 0.047) in methylation, respectively. In contrast, individuals undergoing hormonal treatment later in life (> 60 months of age) did not show any significant MAOA CpG methylation differences compared to those that did not receive treatment (Table 6). To compare the effect of testosterone administered at different age on DNA methylation, we also examined methylation changes using those who received EHT (< 60 months) as reference. Methylation levels in individuals with 49,XXXXY who received HBT showed significantly higher methylation levels at MAOA CpG-7 (5.76%, P = 0.02). Although there was a general trend of increasing MAOA DNA methylation in individuals receiving testosterone treatment after 6 years of age, none of the increases were significant with the exception of CpG-3 (10.05%, P = 0.04, Table 7). We noticed a greater increase in AR methylation compared to MAOA methylation in individuals receiving testosterone later in life, although there were no nominally significant differences in DNA methylation at the AR locus with the exception of CpG-4 in those receiving TRT after 11 years of age (28.37%, P = 0.05). Despite several promising CpGs that increase in DNA methylation with treatment, none of the P values for the HBT and TRT groups compared to the EHT group were statistically significant following correction for the multiple CpGs tested. When modeling the effect of testosterone and DNA methylation on CBCL or BRIEF outcomes, there was no statistically significant interactive effect between the two variables for any CpG sites.
Table 6.
Effect of testosterone replacement therapy on DNA methylation in individuals with 49,XXXXY
| MAOA | Did not receive T | Received < 60 (N = 11) | Booster 60–107 (N = 4) | Megabooster 108–131 (N = 3) | 132 + (N = 3) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Methylation % change (95% CI) | P value | Methylation % change (95% CI) | P value | Methylation % change (95% CI) | P value | Methylation % change (95% CI) | P value | ||
| CpG-1 | Ref | 2.365 (− 1.15, 5.88) | 0.20 | − 1.95 (− 6.50, 2.60) | 0.41 | 2.60 (− 3.49, 8.69) | 0.41 | − 0.53 (− 6.73, 5.66) | 0.87 |
| CpG-2 | Ref | 6.52 (1.59, 9.24) | 0.01 | 1.07 (− 4.62, 6.77) | 0.72 | − 0.27 (− 7.92, 7.37) | 0.95 | 6.93 (− 0.06, 13.91) | 0.07 |
| CpG-3 | Ref | 4.02 (− 0.36, 8.40) | 0.09 | − 4.44 (− 10.07, 1.19) | 0.14 | 0.99 (− 6.99, 8.97) | 0.81 | 1.67 (− 6.28, 9.63) | 0.069 |
| CpG-4 | Ref | 6.41 (0.50, 12.32) | 0.047 | 1.83 (− 6.41, 10.07) | 0.67 | − 3.34 (− 14.32, 7.63) | 0.56 | 6.45 (− 4.24, 17.15) | 0.25 |
| CpG-5 | Ref | 0.38 (− 1.57, 2.32) | 0.71 | 0.13 (− 2.33, 2.59) | 0.92 | 1.75 (− 1.45, 4.94) | 0.30 | − 0.42 (− 3.70, 2.86) | 0.81 |
| CpG-6 | Ref | 2.11 (− 2.24, 6.46) | 0.35 | − 2.00 (− 7.55, 3.53) | 0.49 | − 0.79 (− 8.29, 6.71) | 0.84 | 5.10 (− 2.06, 12.25) | 0.18 |
| CpG-7 | Ref | − 0.95 (− 4.18, 2.28) | 0.57 | 2.74 (− 1.17, 6.66) | 0.19 | 1.14 (− 4.33, 6.61) | 0.69 | 1.93 (− 3.50, 7.35) | 0.50 |
Ref = reference point for T treatment. The numbers in the top row denote age in months of T treatment. The P value threshold for statistical significance after Bonferroni correction for the number of CpG sites tested was 0.005
Table 7.
Timing of testosterone replacement therapy on DNA methylation in individuals with 49,XXXXY
| MAOA | Received < 60 (N = 11) | Booster 60–107 (N = 4) | Megabooster 108–131 (N = 3) | 132 + (N = 3) | |||
|---|---|---|---|---|---|---|---|
| % Methylation change (95% CI) | P value | % Methylation change (95% CI) | P value | % Methylation change (95% CI) | P value | ||
| CpG-1 | Ref | 0.37 (− 6.58, 7.32) | 0.92 | 4.04 (− 2.45, 10.53) | 0.25 | − 6.13 (− 2.45, 14.70) | 0.19 |
| CpG-2 | Ref | − 1.17 (− 7.11, 4.77) | 0.71 | 2.49 (− 3.29, 8.27) | 0.42 | 5.82 (− 1.39, 13.04) | 0.15 |
| CpG-3 | Ref | − 1.83 (− 9.38, 5.72) | 0.64 | 3.14 (− 4.24, 10.52) | 0.42 | 10.05 (1.85, 18.24) | 0.04 |
| CpG-4 | Ref | − 0.9 (− 6.89, 6.71) | 0.98 | − 0.33 (− 7.13, 6.46) | 0.93 | 4.38 (− 4.38, 13.13) | 0.35 |
| CpG-5 | Ref | 1.56 (− 2.16, 5.27) | 0.43 | 2.09 (− 1.52, 5.71) | 0.28 | 4.06 (− 0.47, 8.58) | 0.11 |
| CpG-6 | Ref | − 3.88 (− 13.61, 5.84) | 0.45 | 0.23 (− 9.79, 10.25) | 0.97 | 5.61 (− 7.45, 18.66) | 0.42 |
| CpG-7 | Ref | 5.76 (1.61, 9.91) | 0.02 | 0.75 (− 4.71, 6.20) | 0.79 | 0.73 (− 6.65, 8.10) | 0.85 |
| AR | |||||||
| CpG-1 | Ref | 6.46 (− 5.49, 18.42) | 0.31 | − 4.61 (− 16.80, 7.78) | 0.49 | − 2.18 (− 19.12, 14.77) | 0.81 |
| CpG-2 | Ref | 3.97 (− 14.25, 22.19) | 0.69 | 9.43 (− 8.01, 26.86) | 0.31 | 17.53 (− 4.76, 39.81) | 0.15 |
| CpG-3 | Ref | − 0.34 (− 15.22, 14.53) | 0.97 | 8.39 (− 5.55, 22.32) | 0.27 | 16.43 (− 0.84, 33.71) | 0.09 |
| CpG-4 | Ref | 4.26 (− 18.00, 26.52) | 0.72 | 12.41 (− 8.65, 33.47) | 0.28 | 28.37 (3.78, 52.96) | 0.05 |
Ref = reference point for T treatment. The numbers in the top row denote age in months of T treatment
1Confidence intervals were estimated using the likelihood ratio, while P values were calculated using the Wald test. As a result, due to our small sample size and the estimated effect being very close to the 0.05 threshold, at times the confidence interval does not reject the null hypothesis. The P value threshold for statistical significance after Bonferroni correction for the number of CpG sites tested was 0.005
Discussion
The objective of this pilot study was to assess the relationship among the number of supernumerary X chromosomes, CpG methylation at different loci on the X chromosome, and parent report of behavior in males. In two parental questionnaires, the BRIEF and CBCL, significant differences were observed in both internalizing and externalizing behaviors between the 49,XXXXY group and both the 47,XXY and neurotypical 46,XY groups, which is consistent with a previous study by our group [15]. However, there were no significant differences between the 47,XXY and the 46,XY groups on the BRIEF and CBCL, which is in contrast to another study [11, 38]. The extra three X chromosomes in 49,XXXXY were known to cause greater effects on gene expression and behavior than the one extra X chromosome in 47,XXY, as supported by our study.
For males with supernumerary X chromosomes, increased gene expression could be a result of incomplete inactivation of the X chromosomes. Elevated expression of genes on the X chromosome due to duplication at different X-linked loci is associated with neurodevelopmental disorders. For example, duplications of Xq28 that include the MeCP2 gene have been described in male patients that present with severe developmental delay and neurological effects [39]. Another duplication, at the Xq12-q13.3 position, was associated with developmental delay, autistic features, and increased dosage of genes in the duplicated region [40]. We studied X chromosome inactivation indirectly by measuring DNA methylation at different locations. Considering that the inactive X chromosome has much higher levels of methylation than the active X chromosome, each additional inactive X chromosome is expected to increase the observed methylation at each gene.
Due to its involvement in the oxidative deamination of monoamines such as dopamine, norepinephrine, and serotonin, MAOA is a noteworthy candidate to investigate the role of DNA methylation in behavior. It has been shown that genetic variants linked to low MAOA activity have been associated with increases in aggression and antisocial behavior in neurotypical 46, XY males exposed to childhood trauma [41, 42]. There is also genetic evidence for MAOA in attention deficit hyperactivity disorder (ADHD) [43], a disorder observed in individuals with 47,XXY [44]. In fact, studies have shown that MAOA may be one of the genes targeted by methylphenidate or Ritalin [45]. Other studies found MAOA hypomethylation in female patients with panic disorder when compared to neurotypical counterparts [46, 47]. In the current study, CpG methylation was much higher in the MAOA intronic region in the 49,XXXXY group compared to the 46,XY controls. This finding was expected due to the extra number of X chromosomes that are presumed to have been inactivated, in part, by DNA methylation. However, we found evidence of multiple loci where methylation levels were different from expected. For instance, several intronic CpGs in MAOA did not display the expected greater methylation levels in the 49,XXXXY group compared to the 46,XY controls, whereas others reflected methylation levels that were consistent with all four of the X chromosomes being methylated and inactive. A study investigating the same intronic CpGs as ours reported a significant inverse correlation between methylation levels and MAOA enzymatic activity in the brain [30]. If these CpGs can mediate transcription, then increase in methylation of these CpGs would decrease MAOA gene activity, consistent with the above studies that examined low MAOA activity, aggression, and antisocial behavior. Studies of additional regulatory regions as well as gene expression are needed to delineate the functional consequences of DNA methylation in MAOA.
Methylation levels at AR were also investigated in our study due to its reported influence on male behavior. The androgen receptor is necessary for mediating androgen signaling to shape the male sexual phenotype. Polymorphisms in the androgen receptor gene have also been associated with aggression [48, 49]. Similar to the MAOA locus, methylation levels at one region in AR displayed the expected increased levels of methylation in boys with extra X chromosomes but another region did not. The less-than-expected methylation levels evident at some loci of the AR gene may be associated with the upregulation of AR expression as a compensatory response to hypogonadism.
The MeCP2 gene encodes a protein that regulates gene expression and maintains normal CNS functioning by binding to methylated CpGs [50]. Mutations in MeCP2 causes Rett syndrome, which is characterized by impairments in language, loss of motor coordination, and severe autistic features [51]. A recent study reported that the inactivation of the MeCP2 gene was also associated with an increase in anxiety-driven behavior in males [52]. The MeCP2 locus also did not display drastically elevated methylation levels in the 49,XXXXY group. However, only two CpGs in an intronic GRE were tested to demonstrate that not all X-linked regions show X chromosome number-dependent increase in DNA methylation. A thorough investigation of additional CpGs is needed.
The effect of testosterone treatment on behavior and DNA methylation was also assessed. Receiving testosterone was associated with lower average CBCL total scores and CBCL externalizing subscales, suggesting that treatment may have a positive effect on mental health. The beneficial effects associated with testosterone and behavior have been reported for more than a decade. Notably, testosterone treatment regulates both MAOA and AR expression [53–55] with one study reporting an interaction between testosterone treatment and an allelic variant in MAOA [56]. Generally, DNA methylation seems to be responsive to hormone treatment. In individuals with gender dysphoria who received cross-sex hormone therapy (CHT), those who underwent 12 months of estrogenic treatment to transition from male to female underwent an increase in methylation of the AR gene. Individuals who transitioned from female to male and underwent 12 months of testosterone treatment showed an increase in estrogen receptor alpha (ESR1) methylation [57]. Our findings suggest that hormone treatment can impact epigenetic mechanisms, although it is unclear whether this is a causal relationship. Our findings in this pilot study that testosterone treatment was associated with an increase in DNA methylation in several CpGs in MAOA and AR are preliminary and warrant further interrogation in a larger study of boys with 47,XXY as well as 48,XXXY and 49,XXXXY.
Several notable studies have investigated DNA methylation levels in individuals with Klinefelter syndrome [20, 58–60]. These studies were genome-wide in design and have identified hundreds of syndrome-relevant genes or regions that were differentially methylated between 47,XXY and their 46,XY counterpart. However, there are no studies that have examined DNA methylation in 49,XXXY, except for a study that investigated the FMR1 CGG repeat methylation in the context of a screening tool for newborns [61]. To date, our study is one of the first to focus on DNA methylation and have attempted to link methylation with behavior and androgen treatment in individuals with 49,XXXXY.
The current study has several limitations. First, although DNA methylation can provide a mechanistic understanding of how likely a gene is to be expressed, it is unclear whether any of the specific CpG methylation differences observed among the groups may account for differential gene expression. However, we state that the methylation levels, especially at the MAOA locus, have been linked with MAOA activity, suggesting that they may play a functional role in the transcriptional activity of MAOA. Second, we cannot conclude from statistical association that MAOA or AR methylation is responsible for the behavioral deficits observed in the 49,XXXXY boys, although it is intriguing to consider. On the X chromosome alone, there are a myriad of genes that can affect behavior, and there are numerous CpGs that may potentially regulate the function of any single gene. Given the relatively small number of CpGs tested in this study, it is probable that other loci may play a role on behavior as well, which will be investigated in future studies. An animal model such as one that employs the disruption of MAOA activity may contribute to augmenting our understanding of these very important interactions. Third, another limitation stems from the subjective BRIEF and CBCL scores provided by the participants’ parents. More objective assessments, such as those administered for neurocognitive evaluations, are needed to better understand correlative relationships between behavioral scores and biological variables such as DNA methylation. We estimated the values of Xa and Xi based on experimental data from 46,XY and 47,XXY with the assumption that the Xa and Xi had the identical methylation levels across the different groups. It may be the case that both active and inactive X chromosomes harbor variable methylation levels in the different groups tested. Such a finding would undermine our assumption. However, our results show that methylation levels of many of the CpGs obey our assumption. Given the large number of neurodevelopmental genes that reside on the X chromosome and their potential to affect the transcription of genes across the genome, it is likely that autosomal CpGs undergo epigenetic changes with extra number of X chromosomes. A comprehensive genome-wide approach is needed and will be utilized in future investigations. We acknowledge the limitations associated with using saliva for molecular analysis, as it does not provide a sterile medium for reliably extracting full-length mRNA [62]. This meant that we could not easily assay for gene expression of MAOA, AR, or MeCP2, which is an endeavor much more easily accomplished with blood samples. Finally, we were unable to determine whether there was a statistical interaction between testosterone and MAOA methylation on the behavioral outcomes, as we were underpowered due to low sample size. However, these limitations provide fertile ground for future investigations into the intriguing relationship between genes, hormones, and behavior in the supernumerary X disorders.
Conclusions
Our study provides preliminary evidence indicating that boys with multiple supernumerary X chromosomes and their resulting behavioral issues likely involve some genes on the X chromosome that are aberrantly methylated due to incomplete X chromosome inactivation. These findings warrant a more comprehensive and detailed investigation to understand the relationship between specific genes, biological underpinnings of the testosterone deficiency, the inactivation of X chromosomes, and the behavioral presentations.
Materials and methods
Participants
In this study, there were 29 boys with 47,XXY and 27 boys with 49,XXXXY who were referred by their physicians or ancillary healthcare providers for comprehensive neurodevelopmental evaluations. Inclusion for this study required confirmation of 47,XXY or 49,XXXXY diagnosis via karyotype. Three individuals with three X chromosomes (two with XXXY and one with XXXYY) were also included only for the epigenetic analysis at MAOA. Males who were found to be born premature (i.e., ≥ 37 weeks), have mosaicism, copy-number variants, and/or other co-existing genetic disorders were excluded from the study. Participants were thoroughly evaluated by their pediatric endocrinologist including necessary laboratory tests and physical examination. The endocrinologist then administered HRT to the participant on an individual basis. Fourteen neurotypical boys (46, XY) were also included in this study to serve as controls. Not all participants submitted saliva samples for DNA methylation analysis.
Behavioral report measures
The school-aged version of the Child Behavior Checklist (CBCL) [23] is a behavioral assessment for children ranging between 6 and 18 years of age. This form contains 118 questions in which a parent rates the child’s behavior over the past two months using a 3-point scale (1 = not true, 2 = somewhat true, 3 = very true). The CBCL measures a child’s expression of internalizing problems (anxious, withdrawn, depressed), externalizing problems (aggression, rule-breaking behavior) and other problems (attention, social problems, thought problems) in terms of a raw score. This raw score is then converted to a standardized percentile in which scores at the 50th percentile are considered average. Any score below the 93rd percentile is considered normal, scores between the 93rd and 97th percentiles are borderline, and any score above the 97th percentile is clinical. Borderline and clinical scores imply significant behavioral deviation compared to a normative population.
The school-aged edition of the Behavior Rating Inventory of Executive Function-2 (BRIEF-2) [24] is an 86 question-long form designed to assess the behavior of children between ages 5 and 18. This form is intended to be completed by both parents and teachers. The BRIEF-2 uses three scales to measure behavioral regulation (inhibit, shift, and emotional control), and five scales to measure metacognition (initiate, working memory, plan/organize, organization of materials, and monitor). The global executive composite combines all these scales to arrive at a behavioral summary score. T-scores between 60 and 64 are considered slightly elevated, T-scores from 65 to 69 are considered at risk of clinical elevation, and T-scores above 70 are considered clinically elevated in terms of executive function.
DNA extraction from saliva
Each of the participants provided ~ 1 mL of saliva sample in an Oragene Discover salivette tube (DNAGenotek, Ottawa, Ontario, Canada). DNA was extracted using the prepIT-L2P saliva DNA isolation kit according to the manufacturer’s instructions (DNAGenotek). Briefly, saliva samples were incubated at 50 °C for 1 h, after which the PT-L2P solution was added at 1/25 of the saliva volume, and samples were vortexed and incubated on ice for 10 min. After the centrifugation of impurities at 15,000 × g, the supernatant was mixed with 1.2 times the volume of 100% EtOH, and saliva DNA was precipitated at room temperature. Following centrifugation at 15,000 × g, the supernatant was discarded, and the DNA pellet was washed once with 70% EtOH. The dried DNA pellet was resuspended in 100 μL of TE buffer, and DNA quantity was obtained by a Qubit 2 Fluorometer (ThermoFisher Scientific, Waltham, MA). For each sample, 500 ng of DNA was used for subsequent bisulfite conversion, PCR amplification, and methylation analysis by pyrosequencing.
Bisulfite pyrosequencing assays for DNA methylation
DNA (500 ng) was first bisulfite-converted using the EZ DNA Methylation-Gold Kit, according to the manufacturer's protocol (Zymo Research, Irvine, CA). Primers were designed against regions as described in Additional file 2: Table 1. Whenever possible, we included regions experimentally verified as binding sites for the glucocorticoid receptor, or glucocorticoid response elements (GREs) [25], since anxiety is a common feature in the SCA disorder [11, 15]. For each region, 2 sets of primers were designed. Thermocycling was performed using the Veriti thermal cycler (Life Technologies, Carlsbad, CA), and 25 ng of bisulfite-treated DNA was used with the first outer set of primers. An additional nested PCR was performed with 2 µL of the first PCR reaction and one biotinylated primer (other primer being unmodified). Amplification for both PCR steps consisted of 40 cycles (94ºC for 1 min, 53ºC for 30 s, 72 °C for 1 min). PCR products were confirmed on agarose gels. Pyro Gold reagents were used to prepare samples for pyrosequencing according to manufacturer's instructions (Qiagen, Germantown, MD). For each sample, biotinylated PCR product was mixed with streptavidin-coated sepharose beads (GE Healthcare, Chicago, IL), binding buffer, and Milli-Q water, and shaken at room temperature. A vacuum preptool was used to isolate the sepharose bead-bound single-stranded PCR products. PCR products were then released into a PSQ HS 96-plate containing pyrosequencing primers in annealing buffer. Pyrosequencing reactions were performed on the PyroMark 96MD System according to the manufacturer’s protocol (Qiagen). CpG methylation quantification was performed with the Pyro Q-CpGt 1.0.9 software (Qiagen). An internal quality-control step was used to disqualify any assays that contained unconverted DNA. Percentage of methylation at each CpG as determined by pyrosequencing was compared among different groups.
Statistical analysis
X chromosome copy number analysis
We estimated the expected level of DNA methylation at MAOA and AR CpG sites by using the observed methylation in XY and multiplying it by the potential number of inactivated X chromosomes: mj = n * m1, where m is the estimated average level of methylation at the CpG site, j is the total number of X chromosomes in a given sample, n is the number of inactivated X chromosomes (max(n) = j), and m1 is the average level of methylation observed among samples from neurotypical 46,XY individuals. We then determined the predicted methylation levels from the number of inactive X chromosomes that were closest to those experimentally obtained by bisulfite pyrosequencing. The observed vs. expected values were tested using the Kolmogorov–Smirnov test, with lack of statistical significance indicating that the observed was similar to the expected values. Finally, observed values were also adjusted by testosterone treatment. Same conclusions were reached by both sets of values, and therefore, only the adjusted methylation values are reported.
Methylation correlation with behavior
We tested whether methylation was correlated with behavior across the CBCL: (1) total problems and (2) internalizing and (3) externalizing subscales using ordinary least-squares regression. This was also done across the BRIEF subscales: (1) the behavioral regulation index, (2) the metacognition index, and (3) the global executive functioning composite. These models were conducted without adjustment and with adjustment for aneuploidy status, with status in the model defined as having 2 X chromosomes (47,XXY) to having 4 (49,XXXXY) to assess whether this differential aneuploidy accounted for the relationship between differential methylation and behavior.
Testosterone treatment’s correlation with methylation
We assessed whether testosterone treatment had an impact on methylation, focusing on MAOA due to its prior association with behavior [26–28]. Seventeen individuals with 47,XXY received testosterone treatment (47,XXY T group), whereas three individuals with 47,XXY did not (47,XXY no-T group). Fifteen individuals with 49,XXXXY received testosterone treatment (49,XXXXY T group), and twelve individuals with 49,XXXXY did not (49,XXXXY no-T group). Individuals with no definitive treatment status were excluded from analysis. Testosterone treatment (T) includes early hormonal treatment (EHT), hormonal booster treatment (HBT), and testosterone replacement therapy (TRT). EHT is defined as three intramuscular injections of 25 mg testosterone enanthate, typically administered to boys between 4 and 60 months of age. HBT encompasses three intramuscular injections of 50 mg testosterone enanthate administered to boys between 5 and 8 years of age. TRT, beginning at puberty and continuing for life, involves either daily application of testosterone using Androgel or weekly or monthly injections.
Testosterone treatment’s correlation with behavior
We tested for an association between testosterone treatment and scores on the CBCL and BRIEF subscales using ordinary least squares regression. In a separate model, we also tested whether there was an interactive effect between receiving any T and differential methylation at the MAOA CpG sites.
Multiple testing correction
We used a Bonferroni correction for methylation analyses using the number of CpG sites to control for the Type I error rate. With an intended alpha threshold of 0.05, across 11 CpG sites, we used a corrected alpha threshold of 0.005. Given prior studies showing overlap in the psychodevelopmental constructs the BRIEF and CBCL measure [29], we considered statistical analyses with these outcomes as one experiment and did not apply a correction.
Supplementary Information
Additional file 1: Supplementary Table 1. Primers used for bisulfite pyrosequencing.
Additional file 2: Supplementary Table 2. P-values of one-sample Wilcoxon Signed rank test (non-parametric test of means) between the predicted and observed methylation levels for 46,XY, 47,XXY, 48,XXXY, and 49,XXXXY at the MAOA locus.
Additional file 3. Behavioral and epigenetic database used for analysis.
Acknowledgements
We are grateful to the study participants with 47,XXY and 49,XXXXY and their families for their time and support.
Abbreviations
- ADHD
Attention deficit hyperactivity disorder
- AR
Androgen receptor
- ASD
Autism spectrum disorder
- BRIEF
Behavior Rating Inventory of Executive Function
- CBCL
Child Behavior Checklist
- CHT
Cross-sex hormone therapy
- EHT
Early hormonal treatment
- ESR1
Estrogen receptor alpha
- FMR1
Fragile X mental retardation 1
- FMRP
Fragile X mental retardation protein
- GABRA5
GABA receptor subunit alpha 5
- GABRB3
GABA receptor subunit beta 3
- GRE
Glucocorticoid response element
- HBT
Hormonal booster treatment
- HRT
Hormonal replacement therapy
- KS
Klinefelter syndrome
- MAOA
Monoamine oxidase A
- MeCP2
Methyl-CpG binding protein 2
- SCA
Sex chromosome aneuploidy
- SHANK3
SH3 multiple ankyrin repeat domain 3
- TRT
Testosterone replacement treatment
Authors' contributions
A.Z., J.B., and C.S.S. conceptualized the work. R.S.L. conducted epigenetic analysis and was a major contributor in writing the manuscript. S.Q.S. collected behavioral and biological specimen and was a significant contributor in writing the manuscript. H.M.G.D. analyzed patient behavioral data. J.L.C. also conducted epigenetic analysis. P.L. also collected behavioral and biological specimen. A.Z., J.B., A.G., and C.S.S. were responsible for critical revision of the article and final approval of the version to be published. All authors read and approved the final manuscript.
Funding
This work was supported by the Focus Foundation (S.Q.S., P.L., and C.S.) and the James Wah Mood Disorders Scholar Fund via the Charles T. Bauer Foundation (R.S.L.).
Availability of data and materials
The datasets used and/or analyzed during the current study are available as Additional file 3: Behavioral and epigenetic databased used for analysis.
Declarations
Ethics approval and consent to participate
Informed consent was obtained from the caregivers of each participant. This study protocol was approved by the Western Institutional Review Board (#20081226).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bandmann HJ, Breit R, Perwein E. Klinefelter's syndrome. Berlin: Springer; 1984. [Google Scholar]
- 2.Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus. Denmark Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 3.Savic I. Advances in research on the neurological and neuropsychiatric phenotype of Klinefelter syndrome. Curr Opin Neurol. 2012;25:138–143. doi: 10.1097/WCO.0b013e32835181a0. [DOI] [PubMed] [Google Scholar]
- 4.Kleczkowska A, Fryns JP, Van den Berghe H. X-chromosome polysomy in the male. The Leuven experience 1966–1987. Hum Genet. 1988;80:16–22. doi: 10.1007/BF00451449. [DOI] [PubMed] [Google Scholar]
- 5.Linden MG, Bender BG, Robinson A. Sex chromosome tetrasomy and pentasomy. Pediatrics. 1995;96:672–682. [PubMed] [Google Scholar]
- 6.Samango-Sprouse C, Kirkizlar E, Hall MP, Lawson P, Demko Z, Zneimer SM, Curnow KJ, Gross S, Gropman A. Incidence of X and Y chromosomal aneuploidy in a large child bearing population. PLoS ONE. 2016;11:e0161045. doi: 10.1371/journal.pone.0161045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gropman AL, Porter GF, Lasutschinkow PC, Sadeghin T, Tipton ES, Powell S, Samango-Sprouse CA. Neurocognitive development and capabilities in boys with 49,XXXXY syndrome. Am J Med Genet A 2020. [DOI] [PubMed]
- 8.Gropman AL, Rogol A, Fennoy I, Sadeghin T, Sinn S, Jameson R, Mitchell F, Clabaugh J, Lutz-Armstrong M, Samango-Sprouse CA. Clinical variability and novel neurodevelopmental findings in 49, XXXXY syndrome. Am J Med Genet A. 2010;152A:1523–1530. doi: 10.1002/ajmg.a.33307. [DOI] [PubMed] [Google Scholar]
- 9.Milani D, Bonarrigo F, Avignone S, Triulzi F, Esposito S. 48, XXXY/49, XXXXY mosaic: new neuroradiological features in an ultra-rare syndrome. Ital J Pediatr. 2015;41:50. doi: 10.1186/s13052-015-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visootsak J, Rosner B, Dykens E, Tartaglia N, Graham JM., Jr Behavioral phenotype of sex chromosome aneuploidies: 48, XXYY, 48, XXXY, and 49, XXXXY. Am J Med Genet A. 2007;143A:1198–1203. doi: 10.1002/ajmg.a.31746. [DOI] [PubMed] [Google Scholar]
- 11.Samango-Sprouse C, Lasutschinkow P, Powell S, Sadeghin T, Gropman A. The incidence of anxiety symptoms in boys with 47, XXY (Klinefelter syndrome) and the possible impact of timing of diagnosis and hormonal replacement therapy. Am J Med Genet A. 2019;179:423–428. doi: 10.1002/ajmg.a.61038. [DOI] [PubMed] [Google Scholar]
- 12.Samango-Sprouse CA, Sadeghin T, Mitchell FL, Dixon T, Stapleton E, Kingery M, Gropman AL. Positive effects of short course androgen therapy on the neurodevelopmental outcome in boys with 47, XXY syndrome at 36 and 72 months of age. Am J Med Genet A. 2013;161A:501–508. doi: 10.1002/ajmg.a.35769. [DOI] [PubMed] [Google Scholar]
- 13.Samango-Sprouse C, Stapleton EJ, Lawson P, Mitchell F, Sadeghin T, Powell S, Gropman AL. Positive effects of early androgen therapy on the behavioral phenotype of boys with 47, XXY. Am J Med Genet C Semin Med Genet. 2015;169:150–157. doi: 10.1002/ajmg.c.31437. [DOI] [PubMed] [Google Scholar]
- 14.Samango-Sprouse C, Stapleton E, Chea S, Lawson P, Sadeghin T, Cappello C, de Sonneville L, van Rijn S. International investigation of neurocognitive and behavioral phenotype in 47, XXY (Klinefelter syndrome): predicting individual differences. Am J Med Genet A. 2018;176:877–885. doi: 10.1002/ajmg.a.38621. [DOI] [PubMed] [Google Scholar]
- 15.Lasutschinkow PC, Gropman AL, Porter GF, Sadeghin T, Samango-Sprouse CA. Behavioral phenotype of 49, XXXXY syndrome: presence of anxiety-related symptoms and intact social awareness. Am J Med Genet A. 2020;182:974–986. doi: 10.1002/ajmg.a.61507. [DOI] [PubMed] [Google Scholar]
- 16.Willard HF, Salz HK. Remodelling chromatin with RNA. Nature. 1997;386:228–229. doi: 10.1038/386228a0. [DOI] [PubMed] [Google Scholar]
- 17.Grant SG, Chapman VM. Mechanisms of X-chromosome regulation. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- 18.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–396. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 19.Fasolino M, Zhou Z. The Crucial Role of DNA Methylation and MeCP2 in Neuronal Function. Genes (Basel) 2017; 8. [DOI] [PMC free article] [PubMed]
- 20.Zhang X, Hong D, Ma S, Ward T, Ho M, Pattni R, Duren Z, Stankov A, Bade Shrestha S, Hallmayer J, et al. Integrated functional genomic analyses of Klinefelter and Turner syndromes reveal global network effects of altered X chromosome dosage. Proc Natl Acad Sci U S A. 2020;117:4864–4873. doi: 10.1073/pnas.1910003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018; 172:979–92 e6. [DOI] [PMC free article] [PubMed]
- 22.Saldarriaga W, Tassone F, Gonzalez-Teshima LY, Forero-Forero JV, Ayala-Zapata S, Hagerman R. Fragile X syndrome. Colomb Med (Cali) 2014;45:190–198. doi: 10.25100/cm.v45i4.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achenbach T. Manual for Child Behavior Checklist/ 4–18 and 1991 Profile.: Burlington: University of Vermont Department of Psychiatry, 1991.
- 24.Gioia GA, Isquith PK, Guy SC, Kenworthy L. TEST REVIEW behavior rating inventory of executive function. Child Neuropsychol. 2000;6:235–238. doi: 10.1076/chin.6.3.235.3152. [DOI] [PubMed] [Google Scholar]
- 25.Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2004;101:15603–15608. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckholtz JW, Meyer-Lindenberg A. MAOA and the neurogenetic architecture of human aggression. Trends Neurosci. 2008;31:120–129. doi: 10.1016/j.tins.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 27.Marquez C, Poirier GL, Cordero MI, Larsen MH, Groner A, Marquis J, Magistretti PJ, Trono D, Sandi C. Peripuberty stress leads to abnormal aggression, altered amygdala and orbitofrontal reactivity and increased prefrontal MAOA gene expression. Transl Psychiatry. 2013;3:e216. doi: 10.1038/tp.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjoberg RL, Ducci F, Barr CS, Newman TK, Dell'osso L, Virkkunen M, Goldman D. A non-additive interaction of a functional MAO-A VNTR and testosterone predicts antisocial behavior. Neuropsychopharmacology. 2008;33:425–430. doi: 10.1038/sj.npp.1301417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Familiar I, Ruisenor-Escudero H, Giordani B, Bangirana P, Nakasujja N, Opoka R, Boivin M. Use of the behavior rating inventory of executive function and child behavior checklist in Ugandan children with HIV or a history of severe Malaria. J Dev Behav Pediatr. 2015;36:277–284. doi: 10.1097/DBP.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shumay E, Logan J, Volkow ND, Fowler JS. Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics. 2012;7:1151–1160. doi: 10.4161/epi.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 32.Vottero A, Capelletti M, Giuliodori S, Viani I, Ziveri M, Neri TM, Bernasconi S, Ghizzoni L. Decreased androgen receptor gene methylation in premature pubarche: a novel pathogenetic mechanism? J Clin Endocrinol Metab. 2006;91:968–972. doi: 10.1210/jc.2005-2354. [DOI] [PubMed] [Google Scholar]
- 33.Braat S, Kooy RF. The GABAA receptor as a therapeutic target for neurodevelopmental disorders. Neuron. 2015;86:1119–1130. doi: 10.1016/j.neuron.2015.03.042. [DOI] [PubMed] [Google Scholar]
- 34.Lutz AK, Pfaender S, Incearap B, Ioannidis V, Ottonelli I, Fohr KJ, Cammerer J, Zoller M, Higelin J, Giona F, et al. Autism-associated SHANK3 mutations impair maturation of neuromuscular junctions and striated muscles. Sci Transl Med 2020; 12. [DOI] [PubMed]
- 35.Pagani M, Bertero A, Liska A, Galbusera A, Sabbioni M, Barsotti N, Colenbier N, Marinazzo D, Scattoni ML, Pasqualetti M, et al. Deletion of autism risk gene Shank3 disrupts prefrontal connectivity. J Neurosci. 2019;39:5299–5310. doi: 10.1523/JNEUROSCI.2529-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzilli R, Delfino M, Elia J, Benedetti F, Alesi L, Chessa L, Mazzilli F. Testosterone replacement in 49, XXXXY syndrome: andrological, metabolic and neurological aspects. Endocrinol Diabetes Metab Case Rep. 2016;2016:150114. doi: 10.1530/EDM-15-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross JL, Roeltgen DP, Kushner H, Zinn AR, Reiss A, Bardsley MZ, McCauley E, Tartaglia N. Behavioral and social phenotypes in boys with 47, XYY syndrome or 47 XXY Klinefelter Syndrome. Pediatrics. 2012;129:769–778. doi: 10.1542/peds.2011-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandewalle J, Van Esch H, Govaerts K, Verbeeck J, Zweier C, Madrigal I, Mila M, Pijkels E, Fernandez I, Kohlhase J, et al. Dosage-dependent severity of the phenotype in patients with mental retardation due to a recurrent copy-number gain at Xq28 mediated by an unusual recombination. Am J Hum Genet. 2009;85:809–822. doi: 10.1016/j.ajhg.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaya N, Colak D, Albakheet A, Al-Owain M, Abu-Dheim N, Al-Younes B, Al-Zahrani J, Mukaddes NM, Dervent A, Al-Dosari N, et al. A novel X-linked disorder with developmental delay and autistic features. Ann Neurol. 2012;71:498–508. doi: 10.1002/ana.22673. [DOI] [PubMed] [Google Scholar]
- 41.Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Taylor A, Poulton R. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 42.Godar SC, Fite PJ, McFarlin KM, Bortolato M. The role of monoamine oxidase A in aggression: current translational developments and future challenges. Prog Neuropsychopharmacol Biol Psychiatry. 2016;69:90–100. doi: 10.1016/j.pnpbp.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das M, Bhowmik AD, Sinha S, Chattopadhyay A, Chaudhuri K, Singh M, Mukhopadhyay K. MAOA promoter polymorphism and attention deficit hyperactivity disorder (ADHD) in Indian children. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:637–642. doi: 10.1002/ajmg.b.30385. [DOI] [PubMed] [Google Scholar]
- 44.Cederlof M, Ohlsson Gotby A, Larsson H, Serlachius E, Boman M, Langstrom N, Landen M, Lichtenstein P. Klinefelter syndrome and risk of psychosis, autism and ADHD. J Psychiatr Res. 2014;48:128–130. doi: 10.1016/j.jpsychires.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Guimaraes AP, Zeni C, Polanczyk G, Genro JP, Roman T, Rohde LA, Hutz MH. MAOA is associated with methylphenidate improvement of oppositional symptoms in boys with attention deficit hyperactivity disorder. Int J Neuropsychopharmacol. 2009;12:709–714. doi: 10.1017/S1461145709000212. [DOI] [PubMed] [Google Scholar]
- 46.Domschke K, Tidow N, Kuithan H, Schwarte K, Klauke B, Ambree O, Reif A, Schmidt H, Arolt V, Kersting A, et al. Monoamine oxidase A gene DNA hypomethylation—a risk factor for panic disorder? Int J Neuropsychopharmacol. 2012;15:1217–1228. doi: 10.1017/S146114571200020X. [DOI] [PubMed] [Google Scholar]
- 47.Ziegler C, Richter J, Mahr M, Gajewska A, Schiele MA, Gehrmann A, Schmidt B, Lesch KP, Lang T, Helbig-Lang S, et al. MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry. 2016;6:e773. doi: 10.1038/tp.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butovskaya ML, Lazebny OE, Vasilyev VA, Dronova DA, Karelin DV, Mabulla AZ, Shibalev DV, Shackelford TK, Fink B, Ryskov AP. Androgen receptor gene polymorphism, aggression, and reproduction in Tanzanian foragers and pastoralists. PLoS ONE. 2015;10:e0136208. doi: 10.1371/journal.pone.0136208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maney DL. Polymorphisms in sex steroid receptors: from gene sequence to behavior. Front Neuroendocrinol. 2017;47:47–65. doi: 10.1016/j.yfrne.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Na ES, Monteggia LM. The role of MeCP2 in CNS development and function. Horm Behav. 2011;59:364–368. doi: 10.1016/j.yhbeh.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 52.Philippe TJ, Vahid-Ansari F, Donaldson ZR, Le Francois B, Zahrai A, Turcotte-Cardin V, Daigle M, James J, Hen R, Merali Z, et al. Loss of MeCP2 in adult 5-HT neurons induces 5-HT1A autoreceptors, with opposite sex-dependent anxiety and depression phenotypes. Sci Rep. 2018;8:5788. doi: 10.1038/s41598-018-24167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krongrad A, Wilson CM, Wilson JD, Allman DR, McPhaul MJ. Androgen increases androgen receptor protein while decreasing receptor mRNA in LNCaP cells. Mol Cell Endocrinol. 1991;76:79–88. doi: 10.1016/0303-7207(91)90262-Q. [DOI] [PubMed] [Google Scholar]
- 54.Ou XM, Chen K, Shih JC. Glucocorticoid and androgen activation of monoamine oxidase A is regulated differently by R1 and Sp1. J Biol Chem. 2006;281:21512–21525. doi: 10.1074/jbc.M600250200. [DOI] [PubMed] [Google Scholar]
- 55.Wolf DA, Herzinger T, Hermeking H, Blaschke D, Horz W. Transcriptional and posttranscriptional regulation of human androgen receptor expression by androgen. Mol Endocrinol. 1993;7:924–936. doi: 10.1210/mend.7.7.8413317. [DOI] [PubMed] [Google Scholar]
- 56.Wagels L, Votinov M, Hupen P, Jung S, Montag C, Habel U. Single-dose of testosterone and the MAOA VNTR polymorphism influence emotional and behavioral responses in men during a non-social frustration task. Front Behav Neurosci. 2020;14:93. doi: 10.3389/fnbeh.2020.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aranda G, Fernandez-Rebollo E, Pradas-Juni M, Hanzu FA, Kalko SG, Halperin I, Mora M. Effects of sex steroids on the pattern of methylation and expression of the promoter region of estrogen and androgen receptors in people with gender dysphoria under cross-sex hormone treatment. J Steroid Biochem Mol Biol. 2017;172:20–28. doi: 10.1016/j.jsbmb.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Sharma A, Jamil MA, Nuesgen N, Schreiner F, Priebe L, Hoffmann P, Herns S, Nothen MM, Frohlich H, Oldenburg J, et al. DNA methylation signature in peripheral blood reveals distinct characteristics of human X chromosome numerical aberrations. Clin Epigenetics. 2015;7:76. doi: 10.1186/s13148-015-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skakkebaek A, Nielsen MM, Trolle C, Vang S, Hornshoj H, Hedegaard J, Wallentin M, Bojesen A, Hertz JM, Fedder J, et al. DNA hypermethylation and differential gene expression associated with Klinefelter syndrome. Sci Rep. 2018;8:13740. doi: 10.1038/s41598-018-31780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Viana J, Pidsley R, Troakes C, Spiers H, Wong CC, Al-Sarraj S, Craig I, Schalkwyk L, Mill J. Epigenomic and transcriptomic signatures of a Klinefelter syndrome (47, XXY) karyotype in the brain. Epigenetics. 2014;9:587–599. doi: 10.4161/epi.27806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inaba Y, Herlihy AS, Schwartz CE, Skinner C, Bui QM, Cobb J, Shi EZ, Francis D, Arvaj A, Amor DJ, et al. Fragile X-related element 2 methylation analysis may provide a suitable option for inclusion of fragile X syndrome and/or sex chromosome aneuploidy into newborn screening: a technical validation study. Genet Med. 2013;15:290–298. doi: 10.1038/gim.2012.134. [DOI] [PubMed] [Google Scholar]
- 62.Park NJ, Li Y, Yu T, Brinkman BM, Wong DT. Characterization of RNA in saliva. Clin Chem. 2006;52:988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Primers used for bisulfite pyrosequencing.
Additional file 2: Supplementary Table 2. P-values of one-sample Wilcoxon Signed rank test (non-parametric test of means) between the predicted and observed methylation levels for 46,XY, 47,XXY, 48,XXXY, and 49,XXXXY at the MAOA locus.
Additional file 3. Behavioral and epigenetic database used for analysis.
Data Availability Statement
The datasets used and/or analyzed during the current study are available as Additional file 3: Behavioral and epigenetic databased used for analysis.