Summary
The woody stems of coniferous gymnosperms produce specialised compression wood to adjust the stem growth orientation in response to gravitropic stimulation. During this process, tracheids develop a compression‐wood‐specific S2L cell wall layer with lignins highly enriched with p‐hydroxyphenyl (H)‐type units derived from H‐type monolignol, whereas lignins produced in the cell walls of normal wood tracheids are exclusively composed of guaiacyl (G)‐type units from G‐type monolignol with a trace amount of H‐type units. We show that laccases, a class of lignin polymerisation enzymes, play a crucial role in the spatially organised polymerisation of H‐type and G‐type monolignols during compression wood formation in Japanese cypress (Chamaecyparis obtusa).
We performed a series of chemical‐probe‐aided imaging analysis on C. obtusa compression wood cell walls, together with gene expression, protein localisation and enzymatic assays of C. obtusa laccases.
Our data indicated that CoLac1 and CoLac3 with differential oxidation activities towards H‐type and G‐type monolignols were precisely localised to distinct cell wall layers in which H‐type and G‐type lignin units were preferentially produced during the development of compression wood tracheids.
We propose that, not only the spatial localisation of laccases, but also their biochemical characteristics dictate the spatial patterning of lignin polymerisation in gymnosperm compression wood.
Keywords: compression wood, fluorescence‐tagged monolignols, gravitropism, laccase, lignin, peroxidase, reaction wood
Introduction
In response to gravitropic stimulation, the woody stems and branches of trees can change their growth orientation even after elongation growth has ended. This process is achieved chiefly by the generation of asymmetric mechanical stress by the development of specialised secondary xylem, the so‐called ‘reaction wood’ (Timell, 1986; Yamamoto et al., 2002; Alméras & Clair, 2016; Groover, 2016). In coniferous gymnosperms, reaction wood is more specifically termed ‘compression wood’ because it commonly develops on the underside (compression side) of leaning stems or branches and generates compressive growth stresses (Timell, 1986). The resultant longitudinal stresses lead to restoration of vertical growth or reinforcement to maintain nonvertical growth against gravity (Brown, 1971; Timell, 1986; Hejnowicz, 1997; Yamamoto, 1998; Yamamoto et al., 2002). To this end, tracheids, which comprise the bulk of cells in gymnosperm wood tissues, show marked changes with regard to their cell wall anatomy; typically, tracheids in compression wood are thick walled with altered layer configurations, almost circular in cross‐section and with large intercellular spaces, compared with tracheids in normal wood (Timell, 1986; Donaldson, 2001; Kim et al., 2010; Donaldson & Knox, 2012). At the molecular level, the chemical composition and supramolecular structures (for example, orientation of cellulose microfibrils) of the cell wall biopolymers are greatly modified in compression wood tracheids. In particular, the content of lignin, a complex aromatic polymer of secondary cell walls, is notably elevated and the types of structural units constituting the polymer are uniquely modified (Timell, 1986; Donaldson, 2001; Brennan et al., 2012; Zhang et al., 2017).
Lignin is a ubiquitous, major component of the secondary cell wall in xylem of vascular plants, including that of tracheids in gymnosperms. The polymer is embedded and tightly associated with wall polysaccharides, that is cellulose and hemicelluloses, to confer essential mechanical and physicochemical properties to the cells (Boerjan et al., 2003). Recent studies on lignin biosynthesis and bioengineering are motivated not only by their fundamental importance to plant development and physiology, but also by their agro‐industrial importance in the context of plant‐derived biomass utilisation (Umezawa, 2018; Mahon & Mansfield, 2019). The primary building blocks of lignin polymers are monolignols, that is p‐coumaryl (PA), coniferyl (CA) and sinapyl (SA) alcohols (Fig. 1a), which give rise to p‐hydroxyphenyl (H)‐type, guaiacyl (G)‐type and syringyl (S)‐type lignin polymer units, respectively (Ralph et al., 2019). After their synthesis by the cinnamate/monolignol pathway in the cytoplasm (Dixon & Barros, 2019), monolignols are transported to the apoplast cell wall domains (Perkins et al., 2019), where they are polymerised into lignin polymer units by wall‐localised oxidation systems, such as laccase/O2 and peroxidase/H2O2, by combinatorial radical coupling (Tobimatsu & Schueltz, 2019). As the key determinant of the chemical structure and physicochemical properties of the final lignin polymers, monolignol composition is highly variable among plant species, and tissue and cell types, and even among independent cell wall layers within a single cell (Boerjan et al., 2003; Ralph et al., 2019). Currently, however, the mechanisms underlying such spatiotemporal control of lignin polymerisation in planta, that is how plants coordinate when, where and which types of monolignols are assembled for lignification, remain largely elusive (Barros et al., 2015; Meents et al., 2018; Tobimatsu & Schueltz, 2019).
Fig. 1.
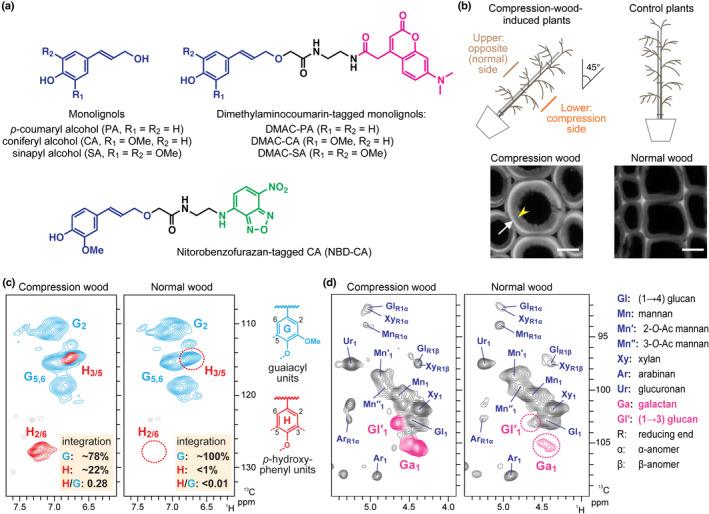
Compression wood formation in inclined Chamaecyparis obtusa seedlings. (a) Monolignols and fluorescence‐tagged monolignols used in this study. (b) Illustrations showing induction of compression wood formation in 3‐yr‐old C. obtusa seedlings. Presented are confocal laser microscopic images (showing lignin autofluorescence) of tracheids developed in compression and normal wood. White arrow and yellow arrowhead indicate the highly lignified S2 (S2L, located in outer S2, oS2) and less lignified inner S2 (iS2) layers, respectively. Bars, 10 μm. (c, d) Partial short‐range 1H–13C correlation (HSQC) NMR spectra of cell wall samples from compression and normal wood. Aromatic (c) and sugar anomeric (d) subregions are shown. Signals from characteristic p‐hydroxyphenyl (H)‐type lignin units (c), and (1→4)‐β‐d‐galactan and (1→3)‐β‐d‐glucan sugar units (d) produced in compression wood cell walls are highlighted.
Compression wood formation in gymnosperms represents a pre‐eminent example of an elaborately controlled lignification process in planta. In normal wood of gymnosperms, tracheid cell walls contain lignins almost exclusively composed of G‐type units derived from polymerisation of G‐type monolignol CA with typically only trace amounts (< 3%) of H‐type units derived from H‐type monolignol PA (Fig. 1a) (Ralph et al., 2006). The cell walls of compression wood tracheids, by contrast, show higher lignin contents than tracheids of normal wood, and their lignins contain considerably higher amounts of H‐type units (Timell, 1986; Donaldson, 2001; Brennan et al., 2012; Zhang et al., 2017), which may comprise as much as 30% of the total lignin units present in bulk compression wood samples, depending on the compression wood severity (Timell, 1986; Ralph et al., 2006). Accumulated data from chemical imaging studies strongly indicate that H‐type lignin units are specifically enriched in the compression‐wood‐specific S2L layer (located in the outer S2, oS2 layers), where the increased lignin depositions are most prominent, whereas the typical G‐type lignins are present across the secondary cell walls, in typical tracheids produced in compression wood (Fukushima & Terashima, 1991; Donaldson, 2001; Tokareva et al., 2007; Donaldson & Radotic, 2013; Zhang et al., 2017). Recent genetic studies have reported that some genes encoding enzymes and transcriptional factors associated with secondary cell wall formation, monolignol biosynthesis and monolignol polymerisation are upregulated in compression‐wood‐forming tissues; this is in general consistent with the increased extent of lignification in compression wood compared with normal wood (Allona et al., 1998; Whetten et al., 2001; Bedon et al., 2007; Koutaniemi et al., 2007; Yamashita et al., 2008, 2009; Ramos et al., 2012; Villalobos et al., 2012; Li et al., 2013; Sato et al., 2013, 2014; Hiraide et al., 2014, 2016; Cruz et al., 2019). However, the molecular mechanisms that control heterologous distribution of H‐type and G‐type lignin units in compression wood cell walls remain unknown.
An increasing body of evidence emphasises the role of monolignol‐oxidising enzymes as the crucial determinant of the spatial patterning of lignin deposition in planta. In the inflorescence stems of Arabidopsis (Arabidopsis thaliana), specific laccase and peroxidase enzymes are precisely localised to apoplastic cell wall domains before lignin deposition (Schuetz et al., 2014; Chou et al., 2018; Hoffmann et al., 2020). For example, it was shown that Arabidopsis LACCASE4 (AtLac4) protein secreted to the secondary cell wall domains of fibre cells is tightly anchored to the polysaccharide‐rich cell wall matrix and therefore cannot be remobilised once secreted (Chou et al., 2018). By contrast, monolignols are likely to be highly mobile after export to the apoplast for lignification (Tobimatsu et al., 2013, 2014; Lion et al., 2017). Collectively, together with similar models proposed for lignification in other specific tissues (Lee et al., 2013, 2018, 2019), precise localisation of lignin polymerisation machinery including monolignol‐oxidising enzymes in specific cell wall domains may play a crucial role in guiding the spatial distribution of the final lignin polymers at the tissue and subcellular levels (Schuetz et al., 2014; Chou et al., 2018; Hoffmann et al., 2020).
In this study, we report evidence showing that laccases also play an important role in spatial organisation of the lignin polymers in gymnosperm compression wood. Based on a series of imaging analyses of tilted stems of Japanese cypress (Chamaecyparis obtusa) seedlings, we demonstrated that specific laccases showing preferential monolignol polymerisation activities toward PA and CA are localised in distinct cell wall layers, which are coincidentally considered to be enriched with H‐type and G‐type lignin polymer units, respectively, in the developing compression wood tracheids. We then identified two C. obtusa laccase isoforms, that is C. obtusa LACCASE1 (CoLac1) (Hiraide et al., 2014, 2016) and LACCASE3 (CoLac3), which are most likely to be responsible for the depositions of H‐type and G‐type lignin units, respectively, in compression wood cell walls, based on gene expression patterns, protein localisation and recombinant enzyme assays. Intriguingly, the recombinant CoLac1 and CoLac3 laccase proteins displayed differential monolignol‐oxidation activities towards PA and CA, in line with their implicated roles in the formation of H‐type and G‐type lignin polymer unis. Collectively, we propose that not only the spatial localisation of laccases, but also their biochemical characteristics, dictate the spatial patterning of lignin polymerisation in gymnosperm compression wood.
Materials and Methods
Plant materials and sample preparations
In this study, 3‐yr‐old Chamaecyparis obtusa seedlings used for stem imaging were cultivated at the Kitashirakawa Experimental Station of the Field Science Education and Research Center of Kyoto University, Japan (35°N, 136°E). To induce compression wood formation, seedlings were artificially inclined (c. 45° from the vertical) in late May. After 1 month, stem blocks containing differentiating compression wood xylem were harvested and stored at −30°C until used for further analysis. Stem blocks from noninclined seedlings grown side‐by‐side with the inclined seedlings were used for preparation of normal wood control samples in imaging studies. The field‐grown C. obtusa tree (c. 12 cm trunk diameter), used for the preparation of crude protein fractions for enzymatic assay, was cultivated at the Kamigamo Experimental Station of the Field Science Education and Research Center of Kyoto University, Japan (35°N, 135°E). To induce compression wood formation, a rope was secured to bend the stem at c. 20° from the vertical in late May. After 1 month, differentiating xylem tissues were collected using a scraper from the compression side of the stem, immediately frozen in liquid nitrogen, and stored at −30°C until protein extraction. Xylem tissues collected from the normal‐wood‐forming opposite side of the same bent tree’s stem were used for preparation of opposite wood control samples in the enzymatic assay. Suspension‐cultured cells of tobacco BY‐2 (Nicotiana tabacum L. ‘Bright Yellow‐2’) were cultured and maintained in modified Linsmaier & Skoog medium as described previously (Nagata et al., 1992).
Cell wall imaging using fluorescence‐tagged monolignols
Serial transverse sections (60 µm thick) were cut from stem block samples of debarked compression and normal wood using a sliding microtome. The sections were washed in 100 ml of 20 mM Na‐acetate buffer (pH 5.0), and then treated with 10 µM dimethylaminocoumarin (DMAC)‐tagged or nitrobenzofurazan (NBD)‐tagged monolignol probes (Fig. 1a) (Tobimatsu et al., 2013) in the presence of catalase (1900 U, from bovine liver; Nacalai Tesque), glucose (5 mM)/glucose oxidase (100 U, from Aspergillus niger; Nacalai Tesque), or exogeneous H2O2 (0.5 mM), in 20 mM Na‐acetate buffer (pH 5.0) for 1 h at room temperature. For the dual‐labelling experiments, sections were sequentially treated with DMAC‐PA (10 µM, for 1 h) and then with NBD‐CA (10 µM for 0.5 h), or in reverse order, in the presence of catalase in 20 mM Na‐acetate buffer (pH 5.0). The labelled sections were thoroughly washed with ethanol to remove nonincorporated probes. The incorporated probes were visualised using a Leica TCS SPE confocal laser microscope (CLSM) (Leica Microsystems) with excitation/emission at 405/440–480 nm for DMAC fluorescence and/or at 488/500–550 nm for NBD fluorescence. Heat‐treated (85°C for 1 h) control sections were subjected to the same labelling treatments and CLSM imaged with the same laser strength and emission gain. Lignin autofluorescence images of nonlabelled sections (Fig. 1b) were collected with excitation/emission at 405/415–600 nm. For the quantitative image analysis, the mean grey value was determined using Imagej software (National Institutes of Health; http://rsb.info.nih.gov/ij/) for differentiating xylem areas (tangential × radial direction = 2 × 10 cells for compression wood and 2 × 6 cells for normal wood) or for the distinct lignifying areas.
Other methods
Description of the methods used for histochemical, chemical and nuclear magnetic resonance (NMR) analyses of the cell walls, protein sequence and gene expression analyses, immunolocalisation, expression of recombinant laccases, preparation of crude protein extracts, western blotting, monolignol‐oxidation assay and statistical analysis, as well as sequence accession numbers can be found in Supporting Information Methods S1. Primer sequences are listed in Table S1.
Results
Cell wall structural analysis of compression wood in inclined C. obtusa seedlings
In this study, 3‐yr‐old C. obtusa seedlings were used for all the imaging experiments conducted. To induce compression wood formation, the seedlings were artificially inclined at c. 45° from the vertical, as illustrated in Fig. 1(b). Transverse stem sections prepared from the lower side (compression side) of the inclined stems were examined using CLSM to visualise lignin autofluorescence. As anticipated, transverse stem sections from the compression‐wood‐induced plants displayed round‐shaped tracheids with thick secondary walls composed of highly lignified S2L (located in oS2) and less lignified inner S2 (iS2) layers (Fig. 1b), all of which are typical anatomical features of gymnosperm compression wood (Timell, 1986; Donaldson, 2001). To determine the chemical composition of compression and normal wood of the C. obtusa seedlings, cell wall residue (CWR) samples were subjected to two‐dimensional (2D) heteronuclear single‐quantum correlation (HSQC) NMR analysis (Kim & Ralph, 2010) (Fig. 1c,d) and a series of chemical analyses (Table S2). As expected, the compression wood cell walls showed higher lignin content compared with the normal wood cell walls (Table S2), and contained considerable amounts of H‐type lignin polymer units, which was evident by the appearance of characteristic C2–H2/C6–H6 correlations from H‐type aromatic nuclei (H2/6) at δC/δH ≈ 127.5/7.20 (Brennan et al., 2012; Takeda et al., 2018) in the 2D HSQC NMR spectra (Fig. 1c) as well as the appearance of H‐type monomeric compounds released upon thioacidolysis (Table S2). The 2D NMR, based on volume integration of the aromatic contours, estimated that H‐type lignin units accounted for 22% of the total G‐type and H‐type units in the compression wood cell walls, whereas thioacidolysis determined that H‐type monomers accounted for 15% of the total G‐type and H‐type monomers released from the cell walls. By contrast, both NMR and thioacidolysis determined that incorporation of H units in the control normal wood cell walls was almost negligible (Fig. 1c; Table S2). In addition, NMR analysis determined that the compression wood cell walls were enriched with (1→4)‐β‐d‐galactans and (1→3)‐β‐d‐glucans (Fig. 1d), consistent with previous observations on gymnosperm compression wood (Kim et al., 2010; Brennan et al., 2012; Donaldson & Knox, 2012). Neutral sugar analysis corroborated the increment of galactans in compression wood cell walls (Table S2). Taken together, the present data affirmed that typical compression wood tracheids showing the characteristic anatomical and biochemical features were developed in the C. obtusa seedling samples, which were used for further imaging analyses.
Laccase and peroxidase activities in lignifying C. obtusa compression wood
At the onset of the current imaging study, to visualise laccase/O2 and peroxidase/H2O2 activities in differentiating xylem of the C. obtusa compression and normal wood, we used the histochemical reagent 3,3′‐diaminobenzidine (DAB) to detect phenol oxidation activity by the coloration produced upon DAB oxidation (Ranocha et al., 1999; Hiraide et al., 2016). To selectively visualise laccase/O2 and peroxidase/H2O2 activities, DAB was applied to the freshly cut transverse sections in the presence of catalase to suppress peroxidase/H2O2 by removing endogenous H2O2 for selective visualisation of laccase/O2 activity (LACs), glucose/glucose oxidase to suppress laccase/O2 by decomposing O2 for selective visualisation of peroxidase/H2O2 activity (PRXs), or exogenous H2O2 for visualisation of both laccase/O2 and peroxidase/H2O2 activities (LACs + PRXs) (Hiraide et al., 2016). The application of DAB under the described conditions resulted in the appearance of positive, dark‐brown coloration in the differentiating xylem on the inner side of the cambial zone, where lignification typically occurs during secondary xylem development. However, no positive coloration was observed in the background control (heat control) sections (Fig. 2).
Fig. 2.
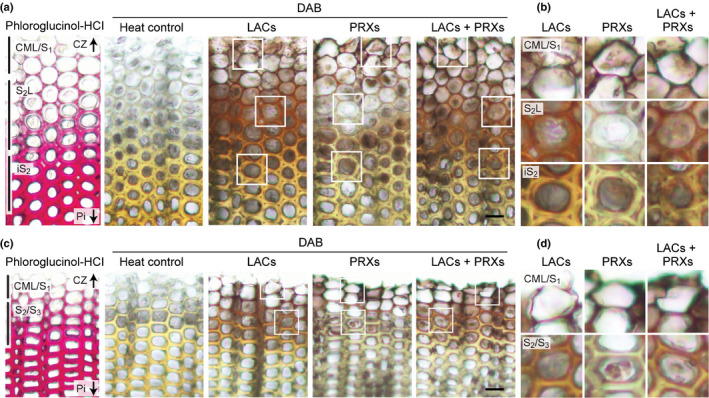
Visualisation of laccase and peroxidase activities in differentiating compression and normal wood tracheids of Chamaecyparis obtusa seedlings. Serial transverse sections from compression (a, b) and normal (c, d) wood tissues were visualised by administering DAB in the presence of catalase for detection of laccase/O2 (LACs), glucose and glucose oxidase for peroxidase/H2O2 (PRXs), and exogeneous H2O2 for both laccase/O2 and peroxidase/H2O2 (LACs + PRXs) activities. Background control sections (heat control) were obtained by heating sections at 85°C for 1 h before administering DAB. Regions that showed positive DAB staining in different cell wall compartments and arrows indicating directions to cambium zone (CZ) and pith (Pi) are indicated on the sections stained with phloroglucinol‐HCl. Images in (b) and (d) are magnified views of the boxed regions in (a) and (c), respectively. Bars, 20 μm.
In both compression and normal wood sections imaged under the LACs + PRXs treatment, positive DAB coloration was first detectable on the compound middle lamella (CML; including the cell corner regions) in tracheids at an early stage of differentiation adjacent to the cambium zone (CZ). With increasing maturity of the tracheids, that is with increasing distance from CZ, positive DAB staining spread to the secondary cell walls (Fig. 2b,d). Distribution of laccase/O2 activity visualised under the LACs treatment was similar to that of the total phenol oxidation activity visualised under the LACs + PRXs treatment: in both compression and normal wood sections, laccase/O2 activity was detected across the lignifying CML and the secondary wall regions. By contrast, peroxidase/H2O2 activity visualised under the PRXs treatment was confined to the lignifying CML, and substantially weaker signals were detected in the lignifying secondary cell wall regions, in both compression and normal wood sections (Fig. 2). Collectively, these observations suggested that laccases contributed to lignification across the CML and the secondary walls of differentiating tracheids, whereas peroxidases predominantly contributed to lignification in the CML, in compression and normal wood of C. obtusa seedlings.
Visualisation of lignification in C. obtusa compression wood with fluorescence‐tagged monolignols
Next, we assessed monolignol‐structure‐dependent lignification patterns in C. obtusa compression wood using fluorescence‐tagged monolignols (Fig. 1a), which have been shown to be useful in monitoring active lignification sites through metabolic incorporation in various plant systems (Tobimatsu et al., 2011, 2013; Schuetz et al., 2014; Takenaka et al., 2018; Lee et al., 2019).
Laccase‐dependent incorporation of DMAC‐tagged H‐type, G‐type and S‐type monolignol probes
Given that DAB staining showed that laccase(s), rather than peroxidase(s), were the predominant source of phenol oxidation activity in compression wood (Fig. 2), we first investigated laccase‐dependent incorporation of DMAC‐tagged H‐type (DMAC‐PA) and G‐type (DMAC‐CA) monolignol probes, which mimic canonical H‐type (PA) and G‐type (CA) monolignols (Fig. 1a), respectively, by administering them to the stem sections in the presence of catalase (Fig. 3). Strikingly, the compression wood sections labelled with DMAC‐PA exhibited intense fluorescence signals specifically in the lignifying S2L layer, but much weaker or negligible signals were detected in the lignifying CML/S1 and iS2 regions (Fig. 3a). By contrast, the compression wood sections labelled with DMAC‐CA exhibited stronger signals in the lignifying CML/S1 and iS2 regions, with apparently weaker signals detected in the lignifying S2L layer (Fig. 3a). These results suggested that, compared with laccase(s) in lignifying CML and iS2, laccase(s) in lignifying S2L may possess a considerably higher PA‐polymerisation activity for the preferential deposition of H‐type lignin polymer units. The two monolignol probes also showed distinct incorporation patterns in the normal wood sections. Under the same microscopic conditions, the sections treated with DMAC‐PA exhibited weak DMAC fluorescence signals, whereas, by contrast, the sections treated with DMAC‐CA displayed intense signals in all lignification sites, that is in lignifying CML/S1 and S2/S3 (Fig. 3b). Therefore, laccase(s) in normal wood may preferentially accept CA over PA for the exclusive deposition of G‐type lignin polymer units. Quantitative analysis of DMAC fluorescence affirmed our assessment for these contrasting incorporation patterns of DMAC‐PA and DMAC‐CA in compression and normal wood sections (Fig. 3c,d).
Fig. 3.
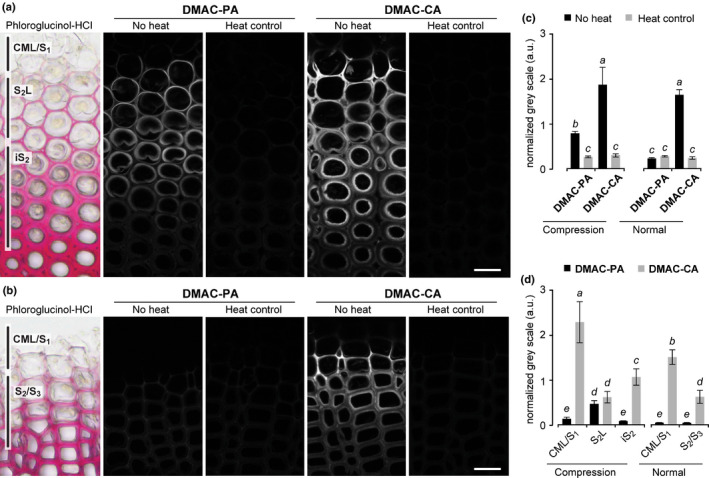
Laccase‐dependent fluorescence‐tagged monolignol incorporation into differentiating compression and normal wood tracheids of Chamaecyparis obtusa seedlings. (a, b) Compression (a) and normal (b) wood sections were labelled with DMAC‐tagged monolignols (DMAC‐PA and DMAC‐CA) in the presence of catalase, and then DMAC fluorescence was visualised with a confocal microscope. Sections were stained with phloroglucinol‐HCl, and background controls visualised after heat treatment are shown. Regions where the incorporation of fluorescence‐tagged monolignols was detected in different cell wall compartments are indicated on the sections stained with phloroglucinol‐HCl. Bars, 20 μm. (c, d) Normalised DMAC florescence determined for the whole imaged regions (c) and separately for where the incorporation of fluorescence‐tagged monolignols was detected in different cell wall compartments (d) as indicated in the sections stained by phloroglucinol‐HCl in (a) and (b). Values are based on the mean fluorescence intensity in grey levels ± SD (n = 15). Different letters indicate significant differences (analysis of variance (ANOVA) with post‐hoc Tukey–Kramer test, P < 0.05).
We also tested incorporation of an S‐type monolignol probe, DMAC‐SA, that mimics the natural S‐type monolignol, SA (Fig. 1a), which is a common lignin monomer in angiosperms but not in typical gymnosperms including C. obtusa used in this study (Wagner et al., 2015). Consequently, the DMAC‐SA probe showed a similar incorporation pattern to that of the G‐type DMAC‐CA probe in compression and normal wood of the C. obtusa seedlings (Fig. S1). Therefore, C. obtusa laccases that preferentially incorporate CA in compression and normal wood may have a capability to polymerise SA, albeit that C. obtusa, consistent with other gymnosperms, does not naturally incorporate S‐type lignin units from SA, as shown by the present NMR and chemical analysis data (Fig. 1c; Table S2).
Co‐visualisation of laccase‐dependent incorporation of H‐ and G‐type monolignol probes
To further evaluate the contrasting incorporation patterns of H‐type and G‐type monolignol probes by laccase in compression wood, we performed a dual‐labelling experiment using DMAC‐PA and NBD‐CA for simultaneous visualisation of their incorporation by detection of the distinguishable DMAC and NBD fluorescence. Before this experiment, we performed a single‐labelling experiment using NBD‐CA under the same conditions we used for the single‐labelling experiment with DMAC‐CA as described earlier (Fig. 3a). As expected, NBD‐CA showed essentially the same incorporation patterns to those of DMAC‐CA, giving rise to strong fluorescence in CML/S1 and iS2 with weaker fluorescence in S2L (Fig. S2). This result affirmed that the difference in fluorophore, that is DMAC versus NBD, does not considerably affect the CA‐dependent probe‐incorporation patterns (Tobimatsu et al., 2011; Tobimatsu et al., 2013). Dual labelling was then performed by incubating the wood sections sequentially with DMAC‐PA and then with NBD‐CA in the presence of catalase. Consistent with the single‐labelling experiments (Fig. 3a), the incorporation of DMAC‐PA and NBD‐CA was majorly localised to the lignifying S2L, and CML/S1 plus iS2 regions, respectively, in compression wood (Fig. 4). Intriguingly, however, administrating the two monolignol probes in reverse order, that is first with NBD‐CA and then with DMAC‐PA, considerably reduced incorporation of DMAC‐PA into the lignifying S2L layer in compression wood (Fig. S3). It is plausible that NBD‐CA that had been administrated and partially incorporated into S2L before administrating DMAC‐PA may block incorporation of DMAC‐PA by physically covering the surface of wall‐localised laccases, and/or biochemically hinder oxidation of DMAC‐PA. This result supports the view that, in addition to substrate specificity of wall‐localised laccases, substrate availability is also an important factor affecting the incorporation of different lignin units in cell walls.
Fig. 4.
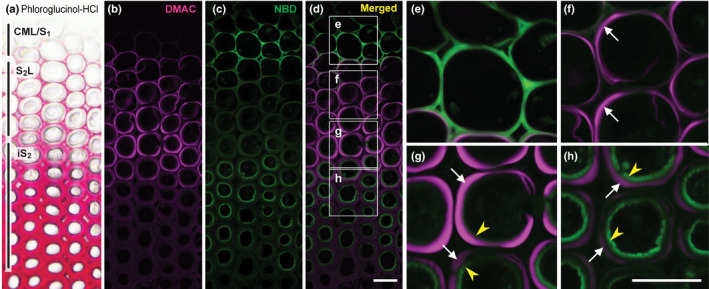
Simultaneous visualisation of laccase‐dependent DMAC‐PA and NBD‐CA incorporation into differentiating compression wood tracheids of Chamaecyparis obtusa seedlings. (a) A control section stained with phloroglucinol‐HCl. (b–h) Sections were labelled sequentially with DMAC‐tagged p‐coumaryl alcohol (DMAC‐PA) and NBD‐tagged coniferyl alcohol (NBD‐CA) probes in the presence of catalase, and then visualised as DMAC (b) and NBD (c) fluorescence and merged (d–h). Images in (e–h) are magnified views of the boxed regions in (d). White arrows and yellow arrowheads indicate the S2L and iS2 layers, respectively. Regions where the incorporation of fluorescence‐tagged monolignols was detected in different cell wall compartments are indicated in (a). Bars, 20 μm.
Peroxidase‐dependent incorporation of H‐, G‐ and S‐type monolignol probes
We also visualised peroxidase‐dependent lignification patterns using DMAC‐tagged monolignol probes. As described for DAB staining (Fig. 2), the monolignol probes were administered to the stem sections in the presence of glucose/glucose oxidase to suppress laccase/O2 activity (PRXs), and then visualised under the same microscopic conditions used for visualisation of laccase activity (LACs) (Figs 5, S4). Under the PRXs treatment, incorporation of DMAC‐CA (Fig. 5), as well as that of DMAC‐SA (Fig. S4), was generally lower compared with those observed under the LACs treatment in all lignifying regions including the CML and secondary cell wall regions. Conversely, notable incorporation of DMAC‐PA under the PRXs treatment was observed especially in the lignifying CML/S1 regions in both compression wood and normal wood sections. In particular, the DMAC‐PA incorporation into CML/S1 in compression wood was clearly more conspicuous under the PRXs treatment than that under the LACs treatment (Fig. 5a,c). These results suggested that peroxidase(s) may also contribute to polymerisation of PA, particularly in CML/S1, during compression wood formation in C. obtusa.
Fig. 5.
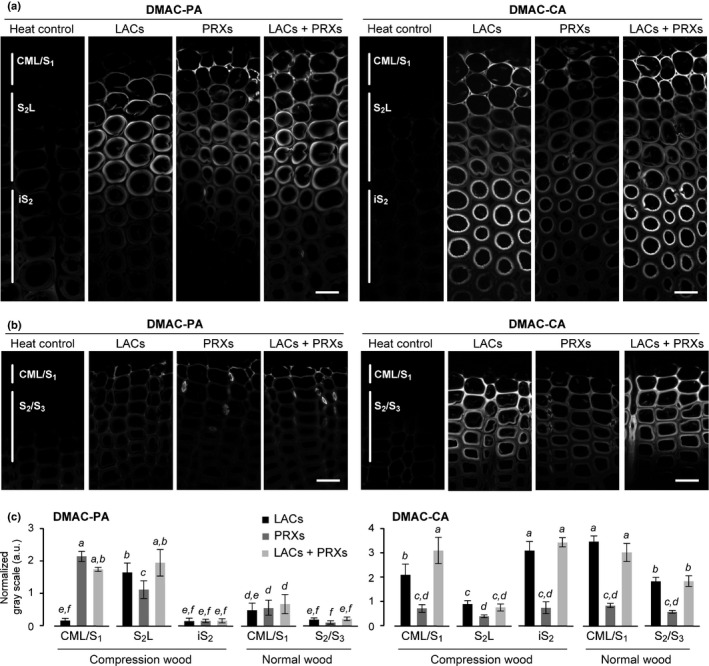
Comparison of laccase‐mediated and peroxidase‐mediated incorporation of fluorescence‐tagged monolignols into differentiating compression and normal wood tracheids of Chamaecyparis obtusa seedlings. (a, b) Serial transverse sections from compression (a) and normal (b) wood tissues were labelled with DMAC‐tagged monolignols (DMAC‐PA and DMAC‐CA) in the presence of catalase for detection of laccase/O2 (LACs), glucose and glucose oxidase for peroxidase/H2O2 (PRXs), and with H2O2 for both laccase/O2 and peroxidase/H2O2 (LACs + PRXs) oxidation activities. Regions where the incorporation of fluorescence‐tagged monolignols was detected are indicated on the heat control sections. Bars, 20 μm. (c) Normalised DMAC florescence determined for where the incorporation of fluorescence‐tagged monolignols was detected in different cell wall compartments. Values are based on the mean fluorescence intensity in grey levels ± SD (n ≧ 5). Different letters indicate significant differences (ANOVA with post‐hoc Tukey–Kramer test, P < 0.05).
Expression and localisation of laccases involved in lignification of C. obtusa compression wood
To further investigate the potential roles of laccases during compression wood formation, we characterised C. obtusa laccases with regard to their gene expression, protein localisation and enzymatic activity. Previously, two C. obtusa laccases, that is CoLac1 (AB762662) and CoLac2 (AB762663), were cloned from cDNA libraries prepared from differentiating C. obtusa compression and normal wood tissues (Hiraide et al., 2014). By searching for laccase sequences against an RNA‐seq contig sequence dataset (DRA001036) (Sato et al., 2014), we identified an additional candidate laccase sequence, from this point forward referred to as CoLac3 (LC494389), that was potentially involved in lignification during compression wood formation; the full‐length coding sequence of CoLac3 was obtained using a rapid amplification of cDNA ends (RACE)‐PCR approach (see Methods S1). A phylogenetic tree was constructed for the three C. obtusa laccase proteins together with 17 Arabidopsis laccases and ZmLac3, a laccase from maize (Zea mays) for which X‐ray crystal structures were recently solved (Xie et al., 2020) (Fig. 6a). The three candidate C. obtusa laccases, that is CoLac1, CoLac2 and CoLac3, clustered together with the lignin‐specific laccases of Arabidopsis, that is AtLac4, AtLac11, and AtLac17 (Berthet et al., 2011; Zhao et al., 2013). Multiple sequence analysis indicated that, whereas the three C. obtusa laccases contained the conserved copper‐binding domain motifs essential for laccase function (Cesarino et al., 2013; Wang et al., 2015a, 2015; Wang et al., 2015b), there were some variations in the substrate‐binding loops and the residues that may interact with monolignol substrates (Xie et al., 2020) (Fig. S5).
Fig. 6.
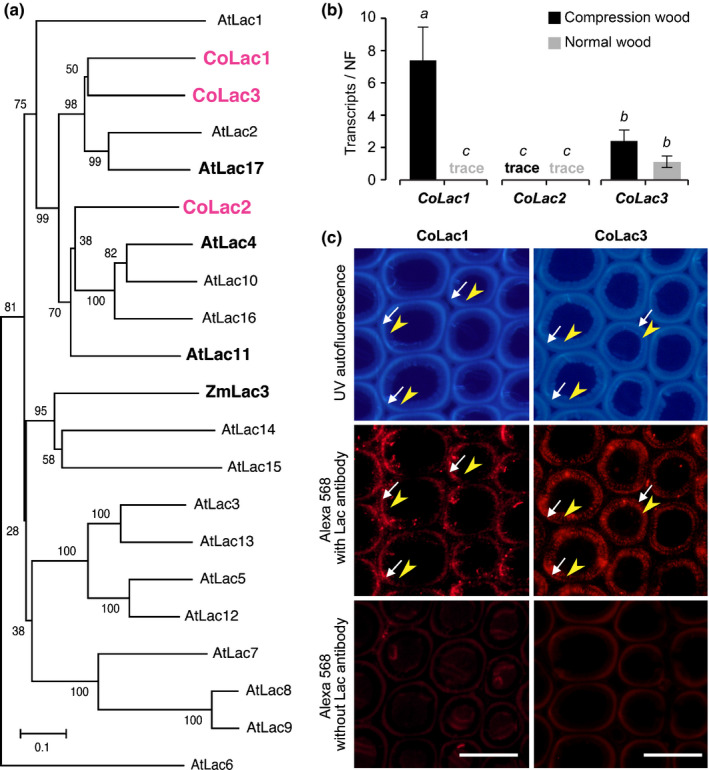
Laccases responsible for lignification in Chamaecyparis obtusa compression wood. (a) Phylogenetic analysis of laccase proteins. An unrooted phylogenetic tree was built using neighbour‐joining method. Bootstrapping with 1000 replicates was performed. Bar, 0.1 substitutions per site. (b) Transcript abundance of CoLac1, CoLac2 and CoLac3 in differentiating compression and normal wood tracheids of C. obtusa seedlings as determined using quantitative real‐time PCR. The normalisation factor (NF) for calibration of transcript abundance was determined as described in Methods S1. Values are the mean ± SD for three independently analysed plants. Different letters indicate significant differences (ANOVA with post‐hoc Tukey–Kramer test, P < 0.05). (c) Immunolocalisation of CoLac1 and CoLac3 in compression wood cell walls of C. obtusa seedlings. Sections were visualised using autofluorescence from deposited lignin and Alexa 568 secondary antibody fluorescence from CoLac1 and CoLac3 epitopes. Control sections visualised without anti‐CoLac1 or anti‐CoLac2 treatment are also shown. White arrows and yellow arrowheads indicate the S2L and iS2 layers, respectively. Bars, 20 μm.
The gene expression of CoLac1, CoLac2 and CoLac3 in differentiating compression and normal wood xylem was studied using a quantitative real‐time PCR (qRT‐PCR) approach. As previously determined (Hiraide et al., 2014), CoLac1 was highly expressed in compression wood but poorly expressed in normal wood (Fig. 6b), which supported the hypothesis that CoLac1 may be specifically involved in compression wood lignification. The newly characterised CoLac3 was expressed in both compression and normal wood at comparable levels (Fig. 6b), suggesting that, together with CoLac1, CoLac3 may play a role in compression wood lignification. By contrast, the transcript level of CoLac2 was substantially low in both compression and normal wood (Hiraide et al., 2014) (Fig. 6b), which suggested that CoLac2 is unlikely to play an important role in lignification, at least in the wood samples tested in this study.
To further clarify the involvement of CoLac1 and CoLac3 in compression wood lignification, we examined their immunolocalisation in transverse sections of developing compression wood from C. obtusa seedling stems. Both anti‐CoLac1 and anti‐CoLac3 antibodies (Fig. S6) showed distinct immunofluorescence signals in the secondary cell wall regions of differentiating tracheids in compression wood (Fig. 6c). Close comparison of epitope immunofluorescence and lignin autofluorescence of the sections, revealed that the CoLac1 epitope was localised specifically in the S2L layer, whereas the CoLac3 epitope clearly localised in the both S2L and iS2 layers (Fig. 6c). We also performed CoLac3 immunolocalisation experiments in normal wood sections, in which, consistent with the gene expression data (Fig. 6b), CoLac3 epitope signals were detected across the lignifying secondary cell walls (Fig. S7). By contrast, a previous study detected negligible immunolocalisation signals from the CoLac1 epitope in normal wood tracheids (Hiraide et al., 2016).
Enzyme assays of recombinant C. obtusa laccases
The differential capabilities of lignifying cell walls in compression wood to incorporate H‐type and G‐type monolignols might be correlated with the different substrate specificities of CoLac1 and CoLac3 specifically localised in these distinct cell wall domains. To test this possibility, we performed an enzymatic assay for recombinant laccase proteins. Recombinant laccases harbouring C‐terminal hexa‐histidine (His6)‐tags, that is rCoLac1, rCoLac2 and rCoLac3, corresponding to the native CoLac1, CoLac2 and CoLac3 enzymes, respectively, were expressed in tobacco BY‐2 cells under the control of the Cauliflower mosaic virus 35S promoter (Sato & Whetten, 2006). Given that several attempts to purify His6‐tag recombinant proteins retaining proper enzyme activities failed, we used crude cell‐wall‐bound protein fractions extracted from two independent transformed cell lines for each construct (a and b) and a control wild‐type cell line (WT) for comparative enzyme assays. The presence of the desired recombinant laccases in the protein extracts was confirmed using western blot analysis with an anti‐His6 antibody (Fig. 7a). Notably, under denaturing conditions in SDS‐PAGE, all the three recombinant laccases were detected at molecular sizes (80–100 kDa) larger than those predicted based on their amino acid sequences (61–64 kDa), which could be attributed to post‐translational glycosylations common in typical plant laccases (Turlapati et al., 2011).
Fig. 7.
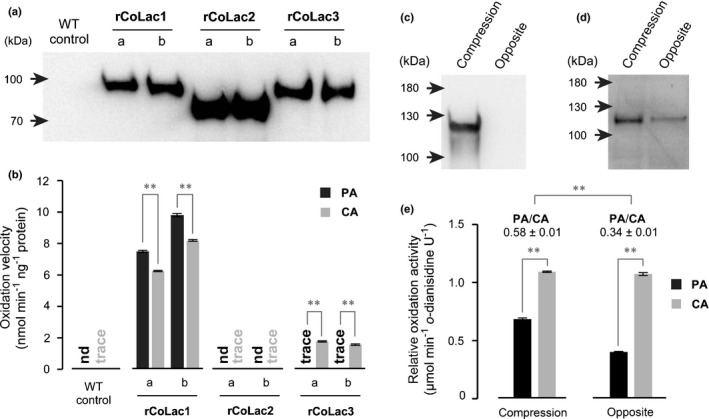
Enzyme assays for recombinant and native Chamaecyparis obtusa laccases. (a, b) Recombinant C. obtusa laccase proteins, rCoLac1, rCoLac2 and rCoLac3, were expressed in tobacco BY‐2 cells. Cell‐wall‐bound protein fractions prepared from two independent transformed lines for each construct (a, b) and control wild‐type cell lines (WT) were used for the laccase activity assay using PA and CA as substrates. Western blots for protein detection using an anti‐His6 antibody (a) and monolignol‐oxidation velocities of rCoLac1, rCoLac2 and rCoLac3 protein preparations (b) are shown. Data in (b) are expressed as the mean ± SD for three independent reaction runs. Asterisks indicate significant difference between PA and CA oxidation velocities (Student’s t‐test; **, P < 0.01). nd, not detected. (c–e) Monolignol‐oxidation activities of cell‐wall‐bound protein fractions prepared from differentiating compression wood tissues of a bent mature C. obtusa tree. Protein fractions from normal‐wood‐forming opposite wood tissues of the same tree were used as a control. Western blots using anti‐CoLac1 (c) and anti‐CoLac3 (d) antibodies, and relative monolignol‐oxidation activities of the cell‐wall‐bound protein fractions (e) are shown. Data in (e) are expressed as oxidation velocities relative to o‐dianisidine used as a reference substrate, and values are the mean ± SD for three independent reaction runs. Asterisks indicate significant difference between PA and CA oxidation velocities and PA/CA oxidation ratios between the compression wood and opposite wood control proteins (Student’s t‐test; **, P < 0.01).
A monolignol‐oxidation assay was performed by incubating the recombinant laccase protein extracts with PA or CA in the presence of catalase (Fig. 7b) (Sato & Whetten, 2006). Under the current assay conditions, the wild‐type control protein extracts showed negligibly low monolignol‐oxidation activities. The rCoLac2 extracts also did not show any oxidation activity either for PA or CA. Conversely, the rCoLac1 extracts oxidised PA and CA efficiently at comparable reaction velocities (with c. 1.2 of the PA/CA oxidation velocity ratio), which suggested that CoLac1 may show comparable substrate specificities towards PA and CA. In contast, the rCoLac3 extracts appeared to oxidise CA efficiently, but barely oxidise PA under the same assay conditions (with < 0.05 of the PA/CA oxidation velocity ratio) (Fig. 7b). Therefore, unlike CoLac1, CoLac3 is likely to show considerably lower substrate specificity toward PA than toward CA. Overall, the notably higher PA‐oxidation activity of CoLac1 compared with that of CoLac3 was consistent with our earlier finding that this enzyme was specifically expressed in compression wood (Fig. 6b), and precisely localised in the S2L layer of differentiating tracheids (Fig. 6c), where the H‐type fluorescence‐tagged monolignol probe was preferentially incorporated (Figs 3, 4, 5).
Enzyme assays of crude enzyme preparations from C. obtusa compression wood
We also performed a monolignol‐oxidation assay for crude enzyme preparations extracted from developing C. obtusa compression wood tissues. In this particular experiment, we used a mature bent C. obtusa tree (c. 12 cm trunk diameter) to prepare crude protein fractions from differentiating compression wood tissues as well as from normal‐wood‐forming opposite wood tissues as a control (Fig. S8a). Consistent with the gene expression data (Fig. 6b), immunoblotting with anti‐CoLac1 and anti‐CoLac3 antibodies demonstrated the presence and absence of CoLac1 in the cell‐wall‐bound protein fractions from compression wood and opposite wood control tissues, respectively, and also the presence of CoLac3 in the cell‐wall‐bound protein fractions from both compression wood and opposite wood (Figs 7c,d, S8b,c). Native CoLac1 and CoLac3 were detected in SDS‐PAGE at molecular sizes (c. 120 kDa) even larger than those determined for rCoLac1 and rCoLac3 expressed in tobacco BY‐2 cells (c. 100 kDa) (Fig. 7a), suggesting different extents of post‐translational glycosylations in tobacco and C. obtusa cells. Consequently, the monolignol‐oxidation assay determined that, although both the compression wood and the opposite wood control samples showed PA and CA oxidation activities, the PA/CA activity ratio of the compression wood sample (c. 0.58) was higher than that of the opposite wood sample (c. 0.34) (Fig. 7e), which might be attributed to the presence and absence of CoLac1 with significant PA‐oxidation activity in compression wood and normal‐wood‐forming opposite wood, respectively.
Discussion
The mechanisms underlying the spatiotemporal control of lignin polymerisation in planta remain elusive. Our current view envisions that specific laccase and/or peroxidase enzymes are secreted to facilitate the spatial control of lignin polymerisation at subcellular levels (Schuetz et al., 2014; Chou et al., 2018; Tobimatsu & Schuetz, 2019; Hoffmann et al., 2020). However, to what degree the substrate specificities of the lignin polymerisation enzymes influence the incorporation of different monolignol types into distinct lignin polymer types remains largely unknown. Together with previous findings (Hiraide et al., 2014, 2016), our current data demonstrated that precise localisation of a laccase, CoLac1, with a prominent PA‐polymerisation ability facilitates the S2L‐specific deposition of H‐type lignin units during compression wood formation in the coniferous gymnosperm C. obtusa. Similarly, it was recently shown that, in the seed coat of Cleome hassleriana, the seed‐coat‐specific deposition of atypical catechol (C)‐type lignin polymers (catechyl lignin) is mediated by a laccase, ChLac8, which shows a preferential oxidation ability towards C‐type lignin monomer (caffeyl alcohol) (Wang et al., 2020). Collectively, these findings provide evidence that not only the spatial localisation of laccases, but also their biochemical characteristics, impact the spatial patterning of lignin polymerisation in planta.
While the respective roles of the two monolignol‐oxidising enzymes, that is laccases and peroxidases, in lignification have long been debated, an increasing body of evidence has suggested that both contribute to lignification and their functions are likely to be spatially and/or developmentally separated (Tobimatsu & Schuetz, 2019). In accordance with this suggestion, the present data support the hypothesis that xylem lignification in the conifer C. obtusa is cooperatively facilitated by laccase and peroxidase enzymes in a spatiotemporally specific manner. Nevertheless, the present imaging analysis using the DAB histochemical reagent and fluorescence‐tagged monolignols suggested that a majority of the monolignol‐oxidising activities detected in the tracheid secondary cell wall regions can be attributed to laccases rather than peroxidases in both compression and normal wood. Both imaging approaches detected laccase‐dependent monolignol‐oxidation activities at all tracheid lignification stages in CML as well as in the major secondary cell wall layers (Figs 2, 5). Peroxidase‐dependent monolignol‐oxidation activities were similarly intensely detected in CML (and also possibly in S1), but were generally weaker compared with the laccase‐dependent activities in the major secondary cell wall regions of tracheids in compression and normal wood (Figs 2, 5). Therefore, although laccases may contribute in all lignification stages of differentiating tracheids across the CML and secondary cell wall regions, contributions of peroxidases may be confined to the early stage of tracheid lignification in the CML regions.
A striking difference was observed between the distinct secondary cell wall layers in the C. obtusa compression wood with respect to their capabilities to incorporate PA and CA upon lignification. Laccase‐dependent PA‐polymerisation activity was distinctly higher in the lignifying S2L layer compared with that detected in other lignifying regions, where, by contrast, CA‐polymerisation activity was stronger (Figs 3, 4). This result supports our notion that different laccase isoforms that show preferential monolignol‐oxidising activities toward PA and CA are present in S2L and in other lignifying regions, to facilitate the deposition of H‐type and G‐type lignin units, respectively. The present imaging data obtained using fluorescence‐tagged monolignols are accordant with a previous observation by Fukushima & Terashima (1991) who reported preferential incorporation of p‐glucocoumaryl alcohol (a glycoside of PA), a potential H‐type lignin precursor (Fukushima et al., 1997; Tsuyama et al., 2019), into CML and S2L of compression wood, but only into CML in normal wood of Pinus thunbergii, as determined using microautoradiography.
We have identified two C. obtusa laccase isoforms, that is CoLac1 and CoLac3, which may, at least partially, provide the monolignol‐oxidation activities for the preferential deposition of H‐type and G‐type lignin units, respectively, during compression wood formation. Congruous with previous findings (Hiraide et al., 2014, 2016), CoLac1 is a compression‐wood‐specific laccase that is specifically expressed during compression wood formation (Fig. 6b), precisely positions in the S2L cell wall layer (Fig. 6c), and displays a prominent capability to oxidise PA for generation of H‐type lignin units (Fig. 7). Conversely, CoLac3, a newly identified C. obtusa laccase isoform, is likely to play a role as a more general lignin laccase responsible for generation of G‐type lignin units from CA, because it is more widely expressed and localised in both compression and normal wood cell walls (Fig. 6b,c) and its recombinant protein displayed considerably poorer oxidation activity toward PA than towards CA, at least under the tested conditions (Fig. 7). It should be noted, however, that not only PA but also CA can be incorporated into the compression wood S2L, although the laccase‐dependent CA‐polymerisation activity in S2L seems to be considerably less compared with those in CML/S1 and iS2 (Figs 3d, 5c). The incorporation of CA in S2L can be operated by either or both CoLac1 and CoLac3 as they are both localised in lignifying S2L (Fig. 6c) and display CA oxidation activity (Fig. 7b).
Several previous studies of compression wood, including those in loblolly pine (Pinus taeda L.) (Allona et al., 1998), Sitka spruce (Picea sitchensis) (MacDougall 2000), Norway spruce (Picea abies) (Koutaniemi et al., 2007) and maritime pine (Pinus pinaster) (Villalobos et al., 2012), have reported upregulation of specific laccase genes during compression wood formation. It is tempting to test whether these compression‐wood‐associated laccases also exhibit similar S2L‐specific wall localisation and/or preferential PA‐oxidisation activities as has been found for CoLac1 in C. obtuse. The reason for the distinct PA/CA oxidation capabilities of CoLac1 and CoLac3 is currently unclear. As PA typically shows a higher redox potential than that of CA due to the electron‐donating effect of the methoxyl group present in CA but absent in PA aromatic systems (Kobayashi et al., 2005; Tobimatsu et al., 2008), the activated copper reaction centre of CoLac1 may somehow exhibit a higher redox potential compared with that of the activated CoLac3 reaction centre to withdraw an electron from the electron‐poor PA aromatic system. In addition, catalytic activity of laccases in general may be affected by the size and shape of the substrate‐binding site near the copper reaction centre (Mayer & Staples, 2002; Frasconi et al., 2010). Indeed, contributions of the methoxy group to monolignol–laccase interactions are suggested in the recently solved crystal structure of ZmLac3 (Xie et al., 2020). Further genetic and biochemical characterisation of CoLac1 and CoLac3, as well as CoLac2, which appeared to be almost inactive in oxidising monolignols under the present assay conditions, is needed to elucidate their precise functions.
It is plausible that the S2L‐specific deposition of H‐type lignin polymers in compression wood is facilitated not only by the localisation of CoLac1 but also by an increased supply of PA. The biosynthesis of PA can be promoted over the biosynthesis of CA upon downregulation of some specific monolignol biosynthetic enzymes, including p‐COUMAROYL ESTER 3‐HYDROXYLASE (C3′H) (Ralph et al., 2006; Bonawitz et al., 2014; Takeda et al., 2018), p‐HYDROXYCINNAMOYL‐CoA: SHIKIMATE p‐HYDROXYCINNAMOYL TRANSFERASE (HCT) (Wagner et al., 2007) and CAFFEOYL‐CoA O‐METHYLTRANSFERASE (CCoAOMT) (Wagner et al., 2011), which are involved in the aromatic hydroxylation and O‐methylation steps along the cinnamate/monolignol pathway. An earlier gene expression study, however, determined that these enzymes (at least for the tested isoforms) were highly expressed together with other major lignin biosynthetic enzymes with no clear relationship with intensification of the S2L layer in the differentiating compression wood tissues of C. obtusa (Hiraide et al. 2014), which might be due at least partially to CA biosynthesis, which is continuously operative to generate G‐type lignin units, along with H‐type units from PA, across the secondary wall layers, including S2L during compression wood formation. There is therefore a need for further high‐resolution analysis of spatiotemporal gene expression profiles of monolignol biosynthetic genes to clarify how the biosynthesis and supply to the cell walls of PA and CA are precisely controlled during compression wood formation. Future studies may also apply advancing genetic and transgenic approaches in gymnosperms (Chang et al., 2018) to further investigate these aspects.
Author contributions
HH, YT, AY and KT conceived research and designed experiments. HH, YT, AY, PYL, MK, YM and KF performed experiments and analysed data. HH, YT, AY and KT wrote the manuscript with help from all other authors.
Supporting information
Fig. S1 Additional wood sections labelled with DMAC‐SA.
Fig. S2 Additional wood sections labelled with NBD‐CA.
Fig. S3 Additional wood sections labelled sequentially with NBD‐CA and DMAC‐PA.
Fig. S4 Additional wood sections labelled with DMAC‐SA.
Fig. S5 Multiple alignment of laccase proteins.
Fig. S6 Immunoblotting with anti‐CoLac1 and anti‐CoLac3 antibodies.
Fig. S7 Immunolocalisation of CoLac3 in normal wood.
Fig. S8 Immunodetection of laccases in crude enzyme extracts.
Methods S1 Additional experimental procedures.
Table S1 Primers used in this study.
Table S2 Chemical analysis data of C. obtusa cell wall samples.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Hiroyuki Yamamoto and Masato Yoshida for providing C. obtusa wood cDNA samples and their helpful support and suggestions. We also thank Hironori Kaji and Ayaka Maeno for their support in NMR experiments, and Tatsuya Awano for his support in wood imaging experiments. This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant‐in‐Aid for JSPS Fellows (#JP17J08050 to HH) and JSPS KAKENHI grants (#JP16H06198 and #JP20H03044 to YT and #JP15H02454 to KT). Part of this study was conducted using the facilities in the DASH/FBAS at RISH and the NMR spectrometer in the JURC at ICR, Kyoto University.
References
- Allona I, Quinn M, Shoop E, Swope K, St Cyr S, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R et al. 1998. Analysis of xylem formation in pine by cDNA sequencing. Proceedings of the National Academy of Sciences, USA 95: 9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alméras T, Clair B. 2016. Critical review on the mechanisms of maturation stress generation in trees. Journal of the Royal Society Interface 13: 20160550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E. 2015. The cell biology of lignification in higher plants. Annals of Botany 115: 1053–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedon F, Grima‐Pettenati J, Mackay J. 2007. Conifer R2R3‐MYB transcription factors: sequence analyses and gene expression in wood‐forming tissues of white spruce (Picea glauca). BMC Plant Biology 7: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S, Demont‐Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S et al. 2011. Disruption of LACCASE4 and 17 results in tissue‐specific alterations to lignification of Arabidopsis thaliana stems. The Plant Cell 23: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M et al. 2014. Disruption of Mediator rescues the stunted growth of a lignin‐deficient Arabidopsis mutant. Nature 509: 376–380. [DOI] [PubMed] [Google Scholar]
- Brennan M, McLean JP, Altaner CM, Ralph J, Harris PJ. 2012. Cellulose microfibril angles and cell‐wall polymers in different wood types of Pinus radiata . Cellulose 19: 1385–1404. [Google Scholar]
- Brown CL. 1971. Trees, structure and function. Berlin, Germany: Springer‐Verlag, 98–99. [Google Scholar]
- Cesarino I, Araújo P, Sampaio Mayer JL, Vicentini R, Berthet S, Demedts B, Vanholme B, Boerjan W, Mazzafera P. 2013. Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S: G ratio of Arabidopsis lac17 mutant. Journal of Experimental Botany 64: 1769–1781. [DOI] [PubMed] [Google Scholar]
- Chang S, Mahon EL, MacKay HA, Rottmann WH, Strauss SH, Powell WA, Coffey, V , Lu H, Mansfield, SD et al. 2018. Genetic engineering of trees: progress and new horizons. In Vitro Cellular & Developmental Biology ‐ Plant 54: 341–376. [Google Scholar]
- Chou YE, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL. 2018. Distribution, mobility, and anchoring of lignin‐related oxidative enzymes in Arabidopsis secondary cell walls. Journal of Experimental Botany 69: 1849–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz N, Méndez T, Ramos P, Urbina D, Vega A, Gutiérrez RA, Moya‐León MA, Herrera R. 2019. Induction of PrMADS10 on the lower side of bent pine tree stems: potential role in modifying plant cell wall properties and wood anatomy. Scientific Reports 9: 18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Barros J. 2019. Lignin biosynthesis: old roads revisited and new roads explored. Open Biology 9: 190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LA. 2001. Lignification and lignin topochemistry ‐ an ultrastructural view. Phytochemistry 57: 859–873. [DOI] [PubMed] [Google Scholar]
- Donaldson LA, Knox JP. 2012. Localisation of cell wall polysaccharides in normal and compression wood of radiata pine: relationships with lignifications and microfibril orientation. Plant Physiology 158: 642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LA, Radotic K. 2013. Fluorescence lifetime imaging of lignin autofluorescence in normal and compression wood. Journal of Microscopy 251: 178–187. [DOI] [PubMed] [Google Scholar]
- Frasconi M, Favero G, Boer H, Koivula A, Mazzei F. 2010. Kinetic and biochemical properties of high and low redox potential laccases from fungal and plant origin. Biochimica et Biophysica Acta (BBA) ‐ Proteins and Proteomics 1804: 899–908. [DOI] [PubMed] [Google Scholar]
- Fukushima K, Taguchi S, Matsui N, Yasuda S. 1997. Distribution and seasonal changes of monolignol glucosides in Pinus thunbergii . Mokuzai Gakkaishi 43: 254–259. [Google Scholar]
- Fukushima K, Terashima N. 1991. Heterogeneity in formation of lignin. Wood Science and Technology 25: 259–270. [Google Scholar]
- Groover A. 2016. Gravitropisms and reaction woods of forest trees – evolution, functions and mechanisms. New Phytologist 211: 790–802. [DOI] [PubMed] [Google Scholar]
- Hejnowicz Z. 1997. Graviresponses in herbs and trees: a major role for the redistribution of tissue and growth stresses. Planta 203: S136–S146. [DOI] [PubMed] [Google Scholar]
- Hiraide H, Yoshida M, Ihara K, Sato S, Yamamoto H. 2014. High lignin deposition on the outer region of the secondary wall middle layer in compression wood matches the expression of a Laccase gene in Chamaecyparis obtusa . Journal of Plant Biology Research 3: 87–100. [Google Scholar]
- Hiraide H, Yoshida M, Sato S, Yamamoto H. 2016. In situ detection of laccase activity and immunolocalisation of a compression‐wood‐specific laccase (CoLac1) in differentiating xylem of Chamaecyparis obtusa . Functional Plant Biology 43: 542–552. [DOI] [PubMed] [Google Scholar]
- Hoffmann N, Benske A, Betz H, Schuetz M, Samuels LA. 2020. Laccases and peroxidases co‐localize in lignified secondary cell walls throughout stem development. Plant Physiology 184: 806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ralph J. 2010. Solution‐state 2D NMR of ball‐milled plant cell wall gels in DMSO‐d 6/pyridine‐d 5 . Organic & Biomolecular Chemistry 8: 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Awano T, Yoshinaga A, Takabe K. 2010. Immunolocalization of β‐1‐4‐galactan and its relationship with lignin distribution in developing compression wood of Cryptomeria japonica . Planta 232: 109–119. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Taguchi H, Shigematsu M, Tanahashi M. 2005. Substituent effects of 3,5‐disubstituted p‐coumaryl alcohols on their oxidation using horseradish peroxidase–H2O2 as the oxidant. Journal of Wood Science 51: 607–614. [Google Scholar]
- Koutaniemi S, Warinowski T, Kärkönen A, Alatalo E, Fossdal CG, Saranpää P, Laakso T, Fagerstedt KV, Simola LK, Paulin L et al. 2007. Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real‐time RT‐PCR. Plant Molecular Biology 65: 311–328. [DOI] [PubMed] [Google Scholar]
- Lee M‐H, Jeon HS, Kim SH, Chung JH, Roppolo D, Lee H‐J, Cho HJ, Tobimatsu Y, Ralph J, Park OK. 2019. Lignin‐based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO Journal 38: e101948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. 2013. A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412. [DOI] [PubMed] [Google Scholar]
- Lee Y, Yoon TH, Lee J, Jeon SY, Lee JH, Lee MK, Chen H, Yun J, Oh SY, Wen X et al. 2018. A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173: 1468–1480, e1469. [DOI] [PubMed] [Google Scholar]
- Li X, Yang X, Wu HX. 2013. Transcriptome profiling of radiata pine branches reveals new insights into reaction wood formation with implications in plant gravitropism. BMC Genomics 14: 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lion C, Simon C, Huss B, Blervacq A‐S, Tirot L, Toybou D, Spriet C, Slomianny C, Guerardel Y, Hawkins S et al. 2017. BLISS: A bioorthogonal dual‐labeling strategy to unravel lignification dynamics in plants. Cell Chemical Biology 24: 326–338. [DOI] [PubMed] [Google Scholar]
- Mahon EL, Mansfield SD. 2019. Tailor‐made trees: engineering lignin for ease of processing and tomorrow’s bioeconomy. Current Opinion in Biotechnology 56: 147–155. [DOI] [PubMed] [Google Scholar]
- Mayer AM, Staples RC. 2002. Laccase: new functions for an old enzyme. Phytochemistry 60: 551–565. [DOI] [PubMed] [Google Scholar]
- McDougall GJ. 2000. A comparison of proteins from the developing xylem of compression and non‐compression wood of branches of Sitka spruce (Picea sitchensis) reveals a differentially expressed laccase. Journal of Experimental Botany 51: 1395–1401. [PubMed] [Google Scholar]
- Meents MJ, Watanabe Y, Samuels AL. 2018. The cell biology of secondary cell wall biosynthesis. Annals of Botany 121: 1107–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Nemoto Y, Hasezawa S. 1992. Tobacco BY‐2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. International Review of Cytology 132: 1–30. [Google Scholar]
- Perkins M, Smith RA, Samuels L. 2019. The transport of monomers during lignification in plants: anything goes but how? Current Opinion in Biotechnology 56: 69–74. [DOI] [PubMed] [Google Scholar]
- Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. 2006. Effects of coumarate 3‐hydroxylase down‐regulation on lignin structure. Journal of Biological Chemistry 281: 8843–8853. [DOI] [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W. 2019. Lignin structure and its engineering. Current Opinion in Biotechnology 56: 240–249. [DOI] [PubMed] [Google Scholar]
- Ramos P, Le Provost G, Gantz C, Plomion C, Herrera R. 2012. Transcriptional analysis of differentially expressed genes in response to stem inclination in young seedlings of pine. Plant Biology 14: 923–933. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McDougall G, Hawkins S, Sterjiades R, Borderies G, Stewart D, Cabanes‐Macheteau M, Boudet AM, Goffner D. 1999. Biochemical characterization, molecular cloning and expression of laccases ‐ a divergent gene family ‐ in poplar. European Journal of Biochemistry 259: 485–495. [DOI] [PubMed] [Google Scholar]
- Sato S, Hiraide H, Yoshida M, Yamamoto H. 2013. Changes in xylem tissue and laccase transcript abundance associated with posture recovery in Chamaecyparis obtusa saplings growing on an incline. Functional Plant Biology 40: 637–643. [DOI] [PubMed] [Google Scholar]
- Sato S, Yoshida M, Hiraide H, Ihara K, Yamamoto H. 2014. Transcriptome analysis of reaction wood in gymnosperms by next‐generation sequencing. American Journal of Plant Sciences 5: 2785–2798. [Google Scholar]
- Sato Y, Whetten RW. 2006. Characterization of two laccases of loblolly pine (Pinus taeda) expressed in tobacco BY‐2 cells. Journal of Plant Research 119: 581–588. [DOI] [PubMed] [Google Scholar]
- Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL, Samuels AL. 2014. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiology 166: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Tobimatsu Y, Karlen SD, Koshiba T, Suzuki S, Yamamura M, Murakami S, Mukai M, Hattori T, Osakabe K et al. 2018. Downregulation of p‐COUMAROYL ESTER 3‐HYDROXYLASE in rice leads to altered cell wall structures and improves biomass saccharification. The Plant Journal 95: 796–811. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Watanabe Y, Schuetz M, Unda F, Hill JL, Phookaew P, Yoneda A, Mansfield SD, Samuels L, Ohtani M et al. 2018. Patterned deposition of xylan and lignin is independent from that of the secondary wall cellulose of Arabidopsis xylem vessels. The Plant Cell 30: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timell TE. 1986. Compression wood in gymnosperms, vol. 1. Berlin, Germany: Springer. [Google Scholar]
- Tobimatsu Y, Davidson CL, Grabber JH, Ralph J. 2011. Fluorescence‐tagged monolignols: synthesis and application to studying in vitro lignification. Biomacromolecules 12: 1752–1761. [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Schuetz M. 2019. Lignin polymerization: how do plants manage the chemistry so well? Current Opinion in Biotechnology 56: 75–81. [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Takano T, Kamitakahara H, Nakatsubo F. 2008. Studies on the dehydrogenative polymerizations of monolignol β‐glycosides. Part 3: Horseradish peroxidase–catalyzed polymerizations of triandrin and isosyringin. Journal of Wood Chemistry and Technology 28: 69–83. [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W et al. 2013. Visualization of plant cell wall lignification using fluorescence‐tagged monolignols. The Plant Journal 76: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Wouwer DVd, Allen E, Kumpf R, Vanholme B, Boerjan W, Ralph J. 2014. A click chemistry strategy for visualization of plant cell wall lignification. Chemical Communications 50: 12262–12265. [DOI] [PubMed] [Google Scholar]
- Tokareva EN, Pranovich AV, Fardim P, Daniel G, Holmbom B. 2007. Analysis of wood tissues by time‐of‐flight secondary ion mass spectrometry. Holzforschung 61: 647–655. [Google Scholar]
- Tsuyama T, Matsushita Y, Fukushima K, Takabe K, Yazaki K, Kamei I. 2019. Proton gradient‐dependent transport of p‐glucocoumaryl alcohol in differentiating xylem of woody plants. Scientific Reports 9: 8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlapati PV, Kim K‐W, Davin LB, Lewis NG. 2011. The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233: 439–470. [DOI] [PubMed] [Google Scholar]
- Umezawa T. 2018. Lignin modification in planta for valorization. Phytochemistry Reviews 17: 1305–1327. [Google Scholar]
- Villalobos DP, Díaz‐Moreno SM, Said el SS, Cañas RA, Osuna D, Van Kerckhoven SH , Bautista R, Claros MG, Cánovas FM, Cantón FR. 2012. Reprogramming of gene expression during compression wood formation in pine: coordinated modulation of S‐adenosylmethionine, lignin and lignan related genes. BMC Plant Biology 12: 100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr K, Nanayakkara B, Te Kiri L. 2007. Exploring lignification in conifers by silencing hydroxycinnamoyl‐CoA:shikimate hydroxycinnamoyltransferase in Pinus radiata . Proceedings of the National Academy of Sciences, USA 104: 11856–11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Torr K, Donaldson L, Pears L, Ralph J. 2011. CCoAOMT suppression modifies lignin composition in Pinus radiata . The Plant Journal 67: 119–129. [DOI] [PubMed] [Google Scholar]
- Wagner A, Tobimatsu Y, Phillips L, Flint H, Geddes B, Lu F, Ralph, J . 2015. Syringyl lignin production in conifers: proof of concept in a pine tracheary element system. Proceedings of the National Academy of Sciences, USA 112: 6218–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Feng J, Jia W, Chang S, Li S, Li Y. 2015a. Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnology for Biofuels 8: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhuo C, Xiao X, Wang X, Docampo‐Palacios ML, Chen F, Dixon RA. 2020. Substrate specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for C‐lignin biosynthesis in the seed coat of Cleome hassleriana . The Plant Cell 32: 3825–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bouchabke‐Coussa O, Lebris P, Antelme S, Soulhat C, Gineau E, Dalmais M, Bendahmane A, Morin H, Mouille G et al. 2015. LACCASE5 is required for lignification of the Brachypodium distachyon culm. Plant Physiology 168: 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten R, Sun YH, Zhang Y, Sederoff R. 2001. Functional genomics and cell wall synthesis in loblolly pine. Plant Molecular Biology 47: 275–291. [PubMed] [Google Scholar]
- Xie T, Liu Z, Wang G. 2020. Structural basis for monolignol oxidation by a maize laccase. Nature Plants 6: 231–237. [DOI] [PubMed] [Google Scholar]
- Yamamoto H. 1998. Generation mechanism of growth stresses in wood cell walls: roles of lignin deposition and cellulose microfibril during cell wall maturation. Wood Science and Technology 32: 171–182. [Google Scholar]
- Yamamoto H, Yoshida M, Okuyama T. 2002. Growth stress controls negative gravitropism in woody plant stems. Planta 216: 280–292. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Yoshida M, Yamamoto H, Okuyama T. 2008. Screening genes that change expression during compression wood formation in Chamaecyparis obtusa . Tree Physiology 28: 1331–1340. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Yoshida M, Yamamoto H. 2009. Relationship between development of compression wood and gene expression. Plant Science 176: 729–735. [Google Scholar]
- Zhang M, Lapierre C, Nouxman NL, Nieuwoudt MK, Smith BG, Chavan RR, McArdle BH, Harris PJ. 2017. Location and characterization of lignin in tracheid cell walls of radiata pine (Pinus radiata D. Don) compression woods. Plant Physiology and Biochemistry 118: 187–198. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin YB, Fu CX, Yun JF, Shao H, Wang XQ, Wang ZY, Dixon RA. 2013. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. The Plant Cell 25: 3976–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Additional wood sections labelled with DMAC‐SA.
Fig. S2 Additional wood sections labelled with NBD‐CA.
Fig. S3 Additional wood sections labelled sequentially with NBD‐CA and DMAC‐PA.
Fig. S4 Additional wood sections labelled with DMAC‐SA.
Fig. S5 Multiple alignment of laccase proteins.
Fig. S6 Immunoblotting with anti‐CoLac1 and anti‐CoLac3 antibodies.
Fig. S7 Immunolocalisation of CoLac3 in normal wood.
Fig. S8 Immunodetection of laccases in crude enzyme extracts.
Methods S1 Additional experimental procedures.
Table S1 Primers used in this study.
Table S2 Chemical analysis data of C. obtusa cell wall samples.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


