Abstract
Muller glial cells (MGCs) are responsible for the homeostatic and metabolic support of the retina. Despite the importance of MGCs in retinal disorders, reliable and accessible human cell sources to be used to model MGC‐associated diseases are lacking. Although primary human MGCs (pMGCs) can be purified from post‐mortem retinal tissues, the donor scarcity limits their use. To overcome this problem, we developed a protocol to generate and bank human induced pluripotent stem cell‐derived MGCs (hiMGCs). Using a transcriptome analysis, we showed that the three genetically independent hiMGCs generated were homogeneous and showed phenotypic characteristics and transcriptomic profile of pMGCs. These cells expressed key MGC markers, including Vimentin, CLU, DKK3, SOX9, SOX2, S100A16, ITGB1, and CD44 and could be cultured up to passage 8. Under our culture conditions, hiMGCs and pMGCs expressed low transcript levels of RLPB1, AQP4, KCNJ1, KCJN10, and SLC1A3. Using a disease modeling approach, we showed that hiMGCs could be used to model the features of diabetic retinopathy (DR)‐associated dyslipidemia. Indeed, palmitate, a major free fatty acid with elevated plasma levels in diabetic patients, induced the expression of inflammatory cytokines found in the ocular fluid of DR patients such as CXCL8 (IL‐8) and ANGPTL4. Moreover, the analysis of palmitate‐treated hiMGC secretome showed an upregulation of proangiogenic factors strongly related to DR, including ANG2, Endoglin, IL‐1β, CXCL8, MMP‐9, PDGF‐AA, and VEGF. Thus, hiMGCs could be an alternative to pMGCs and an extremely valuable tool to help to understand and model glial cell involvement in retinal disorders, including DR.
Keywords: diabetes, disease modeling, dyslipidemia, iPSC, Muller glial cells, retinopathy, stem cells
Main Points
We developed a protocol to generate and bank human iPSC‐derived Müller Glial cells (hiMGCs).
hiMGCs showed phenotypic characteristics and transcriptomic profile of primary MGCs.
hiMGCs can be used to model the features of diabetic retinopathy.
1. INTRODUCTION
Diabetic retinopathy (DR) remains a major cause of visual loss in the working‐age population in industrialized countries, and current treatments are not fully satisfactory (Hernandez, Simo‐Servat, Bogdanov, & Simo, 2017). DR is classically considered a microangiopathy. The most significant sign in the early stages of the disease is the breakdown of the blood–retinal barrier (BRB) leading to plasma leakage in the retina and microvascular abnormalities. In addition to these clinically characterizable signs, neurodegenerative processes and the release of pro‐inflammatory cytokines have also been described in early DR (Rubsam, Parikh, & Fort, 2018). Accordingly, treatments targeting inflammation reduce diabetes‐induced neurodegeneration and vascular remodeling (Semeraro et al., 2019). Although it has often been considered that DR pathogenesis was the consequence of a cell exposure to high glucose concentrations, two large clinical studies (FIELD and ACCORD) assessing lipid‐lowering drugs have shown a reduction in DR progression, independently of the glycemic control (Group et al., 2010; Keech et al., 2007). Thus, although often underestimated compared to glucose‐focused studies, the role of dyslipidemia is now widely investigated in DR (Chang & Wu, 2013; Eid et al., 2019; Hammer & Busik, 2017; Zhou, Wang, Shi, & Yin, 2018).
Müller glial cells (MGCs) are responsible for the homeostatic and metabolic support of the retina. MGCs are intercalated between the vasculature and the neurons so that they contribute to the BRB and play a critical role in response to plasma leakage and vascular changes (Rungger‐Brandle, Dosso, & Leuenberger, 2000; Tout, Chan‐Ling, Holländer, & Stone, 1993). Cross‐talks between MGCs and the microglia further drive neuroinflammation that contributes to DR development (Abcouwer, 2017). Free fatty acids (FFAs) found in the plasma induce an inflammatory response in retinal cells (Capozzi, Hammer, McCollum, & Penn, 2016; Capozzi, McCollum, Cousins, & Penn, 2016; Chen, Jump, Grant, Esselman, & Busik, 2003; Mohamed et al., 2014). Primary MGCs (pMGCs) exposed to linoleic and oleic acids overexpress inflammatory mediators involved in DR (Capozzi, McCollum, et al., 2016). More recently, the analysis of pMGC transcriptome has highlighted the changes induced by palmitic acid (PA), the plasma levels of which being the highest in diabetic patients, in multiple pathways activated in DR such as inflammation and angiogenesis (Capozzi, Giblin, & Penn, 2018).
Despite the importance of MGCs in retinal pathologies, reliable and easily expandable human cell sources to be used to model retinal disorders are lacking. Although, pMGCs can be selected and amplified from post‐mortem retinal tissues (Hicks & Courtois, 1990), the generation of large numbers of cells is limited by the scarcity of human donors and their limited replication capacities. To overcome this limitation, Limb et al have immortalized retinal cells from post‐mortem eye donors and characterized the spontaneously immortalized MIO‐M1 Muller cell line (Limb, Salt, Munro, Moss, & Khaw, 2002). They have shown that MIO‐M1 expressed markers of immature neurons and maintained a capacity to differentiate into more mature neurons when engrafted in a host retina (Lawrence et al., 2007). Recently, human embryonic stem cells have been used to generate retinal glial cells positive for GFAP, a marker expressed by astrocytes and gliotic MGCs (Chung et al., 2019; Dahl, 1979; Sarthy, Fu, & Huang, 1991). Human induced‐pluripotent stem cell (hiPSC)‐derived retinal organoids (ROs) can be used to produce MGCs (Eastlake et al., 2019; Reichman, Terray, et al., 2014). The generation of hiPSC‐derived MGCs (hiMGCs) could help to develop cells that can be used in cell therapy or in drug development for multifactorial diseases such as glaucoma, age‐related macular degeneration or DR. During the retinal development, all retinal cell types are generated sequentially and MGCs are the last cells to be born (Turner & Cepko, 1987). Similarly, among ROs, retinal progenitors give rise to post‐mitotic retinal neurons (retinal ganglion cells, amacrine cells, horizontal cells, bipolar cells, cone, and rod photoreceptors) and MGCs (Reichman, Terray, et al., 2014; Slembrouck‐Brec, Nanteau, Sahel, Goureau, & Reichman, 2018). Recently, an improvement in retinal ganglion cell function has been shown after transplantation of fresh hiMGCs isolated from ROs (Eastlake et al., 2019), but no hiMGC‐based model has been described so far.
In this study, we described an innovative and scalable process to generate MGCs from hiPSCs in order to model DR in vitro. The transcriptome analysis of three hiMGCs derived from iPSCs from three different donors showed that they had phenotypic and transcriptomic characteristics of human pMGCs. We found an induction of inflammatory and angiogenic responses in hiMGCs and pMGCs cultured in the presence of PA rather than glucose. This new cell‐based model could be used to develop in vitro models of multifactorial diseases such as DR to identify innovative treatments.
2. RESULTS
2.1. Generation of hiPSC‐derived MGCs
The aim was to select and amplify iPSC‐derived MGCs from ROs based on the isolation of glial cells from post‐mitotic retinal tissues (Hicks & Courtois, 1990). ROs were obtained using our previously described method for adherent hiPSC culture (Figure 1a) from hiPSCs of three genetically independent donors (Table 1) (Reichman et al., 2017; Slembrouck‐Brec et al., 2019). After more than 100 days in culture, ROs contained cells positive for key MGC markers such as Glutamine synthetase (GS)/Sry‐box transcription factor 9 (SOX9) and Sry‐box transcription factor 2 (SOX2)/Vimentin (VIM) and the positivity persisted until D245 (Figure 1b) (Eastlake et al., 2019; Reichman, Terray, et al., 2014; Roesch et al., 2008). To select MGCs, ROs between D160 and D210 were dissociated using a papain‐based protocol and seeded in culture flasks noted as passage 0 (P0) (Figure 1c). Dissociated cells were cultured in MGC medium as previously described for adult MGC isolation from post‐mortem human retinas to positively select mitotically active hiMGCs from post‐mitotic neurons (Hicks & Courtois, 1990). After two runs of expansion (P0 and P1), 10 dissociated ROs generated up to 6 million hiMGCs at P1 within 14 days that could be cryopreserved in 15 cryovials of 375,000 cells before use. One thawed vial could be expanded 12 times to obtain 4.5 million hiMGCs at P3 within 2 weeks. To ensure that cryopreservation did not modify hiMGC phenotype, fresh or thawed hiMGCs at P2 were stained to confirm the presence of markers specific of MGCs. The immunostaining analysis revealed that both fresh and thawed hiMGC‐1 at P2 similarly expressed VIM, GS and SOX9 (Figure S1). To analyze hiMGC phenotype stability after serial passages, hiMGC‐1 were grown as a non‐confluent monolayer from P2 to P8 and analyzed by immunofluorescence using the same panel of MGC‐specific markers (Figure S2). Between P2 and P8, hiMGCs remained positive for VIM, GS, and SOX9. From P4, the shape of hiMGCs progressively elongated and their appearance was very similar to that of pMGCs (Figure 1d and Figure S2). Remarkably, GFAP, a marker only expressed by retinal astrocytes under physiological conditions and by MGCs in pathological situations was absent in hiMGCs from P4 to P8 (Figure 1d and Figure S2a–g) while pMGCs expressed it from P2 to P8 (Figure S2h–n). Thus, from P2 to P6, hiMGCs showed phenotypical features of MGCs found in vivo. The absence of detectable GFAP expression in hiMGCs suggested that these cells could be an interesting alternative to pMGCs for in vitro studies (Figure 1c).
FIGURE 1.
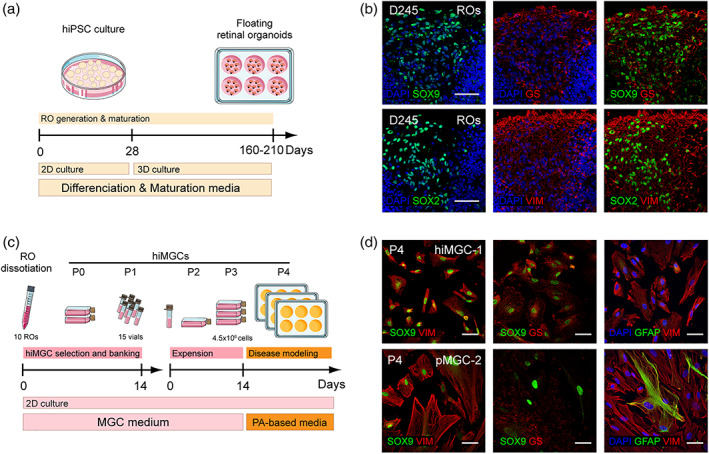
Generation and characterization of hiMGCs. (a) Schematic representation of RO generation and maturation. (b) Representative micrographs of the immunohistochemical detection of the following MGC makers: GS, SOX9 SOX2, and VIM in cryostated ROs at D245 (hiPSC clone AHF1pi2) showing the presence of MGCs in ROs. (c) Schematic representation of hiMGC selection, banking, and amplification. hiMGCs at P4 were used for palmitate‐based assays. (d) Representative micrographs of the immunohistochemical detection of the following MGC markers: SOX9 and VIM (left), SOX9 and GS (middle), and GFAP and VIM (right) P4 hiMGCs (top panels) and pMGCs (bottom panels) showing that the immunohistochemical labeling of MGC markers was similar between hiMGCs at P4 and pMGCs. Nuclei were stained with DAPI. GS, glutamine synthetase; hiMGCs, human iPSC‐derived MGCs; hiPSC, human induced‐pluripotent stem cell; MGC, Muller glial cell; pMGCs, primary MGCs; P, passage; PA, palmitate; RO, retinal organoid; VIM, vimentin. Scale bar = 50 μm [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
Tissue and cell origin of hiMGCs and pMGCs
| MGCs | |||
|---|---|---|---|
| Name | Origin | ||
| Tissues/cells | hiPSC clones | Comments/references | |
| pMGC‐1 | 65 yo male retina | – | 16 hr post mortem |
| pMGC‐2 | 82 yo male retina | – | 12 hr post mortem |
| pMGC‐3 | 83 yo female retina | – | 30 hr post mortem |
| hiMGC‐1 | Adult dermal fibroblasts | AHF1pi2 | Reichman, Terray, et al. (2014) and Reichman et al. (2017) |
| hiMGC‐2 | Neonatal foreskin | FS2pi3 | Reichman et al. (2017) |
| hiMGC‐3 | pMGC‐3 | 5f | Slembrouck‐Brec et al. (2019) |
Abbreviations: hiMGCs, human iPSC‐derived MGCs; hiPSC, human induced‐pluripotent stem cell; MGC, Muller Glial Cell; pMGCs, primary MGCs; yo, years old.
2.2. Transcriptome analysis of hiPSC‐derived MGCs
A transcriptome analysis was then performed to compare the three independent hiPSCs to their derivative hiMGCs and to the three human pMGCs (Table 1). The first two principal components (PC1 and PC2) of the PC analysis (PCA) revealed that iPSCs, hiMGCs and pMGCs were separated into three cell population clusters (Figure 2a). PC1 represented 83% of the observed variance between the samples and allowed differentiating hiPSCs from pMGCs and hiMGCs. PC2 represented 8% of the variance and allowed differentiating pMGCs from hiMGCs. The expression level of 5,841 transcripts was above a selected cutoff of 40 transcripts per kilobase million (TPM) in at least one of the three cell populations. To compare the expression of individual transcripts between the three cell populations, we plotted the log2 TPM expression level of each transcript in hiMGCs and hiPSC relative to its expression level in pMGCs (Figure 2b). The overall profile of log2 TPM expression in hiMGCs was similar to that found in pMGCs while the log2 TPM expression profile in hiPSCs strongly differed from that found in pMGCs (Figure 2b). The PCA and the RNAseq analysis thus showed that hiMGCs had lost the pluripotent stemness features of hiPSCs and were very similar to pMGCs.
FIGURE 2.
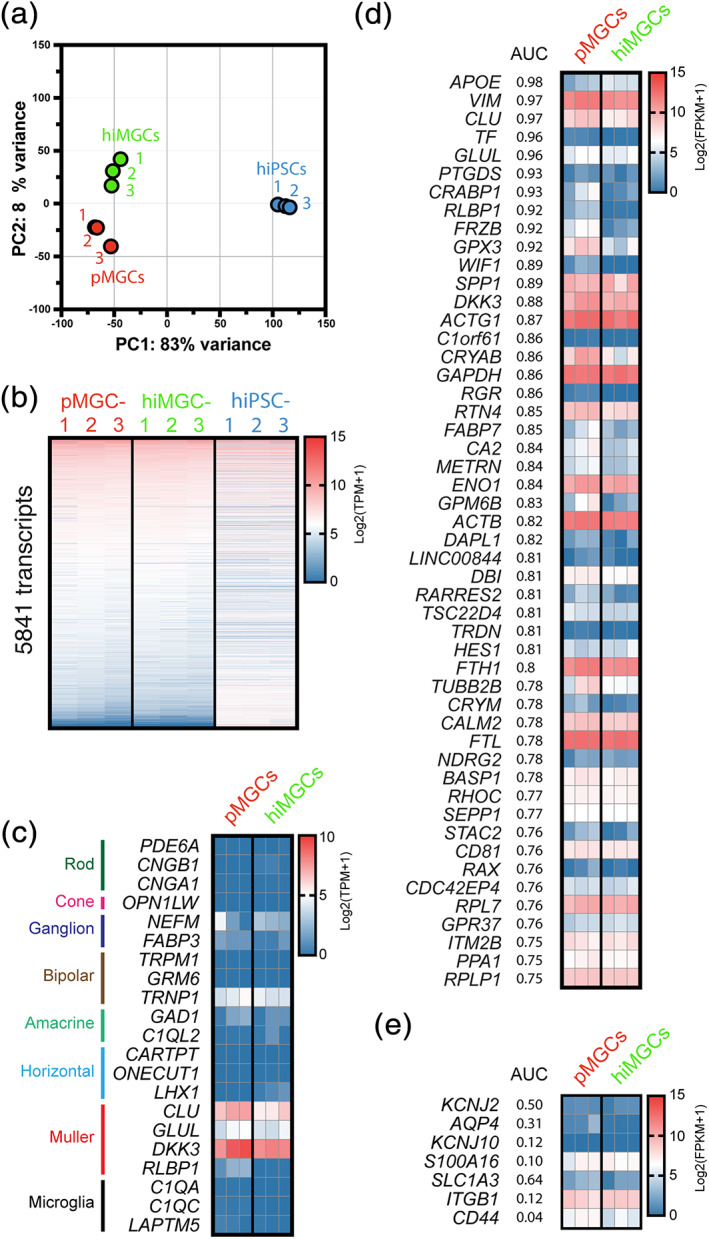
The gene expression profile of hiMGCs is very close to that of pMGCs. (a) PCA of normalized counts of three iPSCs (blue dots), three pMGCs (red dots), and three hiMGCs (green dots). The first two PC revealed that iPSCs, pMGCs, and hiMGCs clustered in three highly homogeneous cell populations. PC1 allowed differentiating hiPSCs from pMGCs and hiMGCs and PC2 allowed differentiating pMGCs from hiMGCs. (b) Heatmap representation of TPM values of the 5,841 genes with an expression level >40 TPM in pMGCs, hiMGCs and hiPSCs. Genes were ranked according to their expression level in pMGCs. In line with the PCA, the overall log2 (TPM + 1) expression profile of hiMGCs was similar to that of pMGCs while the log2 (TPM + 1) expression profile of hiPSCs highly differed. (c) Heatmap representation of the expression level of neuronal, glial and microglial markers in pMGCs and hiMGCs. MGC markers, including CLU, GLUL, DKK3, and RLBP1 were similarly expressed in hiMGCs and pMGCs. (d) Heatmap representation of the expression levels in pMGCs and hiMGCs of the 50 genes with the highest AUC enrichment score in hMacroglia. The AUC scores have been determined by Menon et al for the cluster of hMacroglia from a human retina and downloaded from a public repository (GSE137537, GSE137847). All genes were similarly expressed in hiMGCs and pMGCs. (e) Heatmap representation of the expression level of 7 classical MGC markers not found in the 50 genes with the highest AUC in hMacroglia. All genes were similarly expressed in hiMGCs and pMGCs. The expression levels in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with Hoang et al study). AUC, area under the curve; FPKM, fragment per kilobase million; hiPSCs, human induced pluripotent stem cells; hiMGCs, human iPSC‐derived MGCs; hMacroglia, human macroglial cell cluster determined by Menon and al and containing Muller cells and astrocytes; PC, principal component; PCA, principal component analysis; pMGCs, primary MGCs; TPM, transcripts per kilobase million. TPM and FPKM are represented in b, c d and e as their log2 (value +1) to have positive values [Color figure can be viewed at wileyonlinelibrary.com]
To further identify MGC transcript profile, we then investigated the expression of commonly used neuronal, MGC and microglial markers (Figure 3c) (Hu et al., 2019; Lukowski et al., 2019; Menon et al., 2019; Peng et al., 2019; A. P. Voigt et al., 2019; Yan et al., 2020) and compared it to the expression levels of recently published transcriptomes: a human single macroglial cell (hMacroglia, including MGCs and astrocytes) transcriptome performed by Menon et al on human retina and the bulk RNA‐seq transcriptome of cell‐sorted mouse MGCs (mMGCs) performed by Hoang et al (Figure S3) (Hoang et al., 2020; Menon et al., 2019).
FIGURE 3.
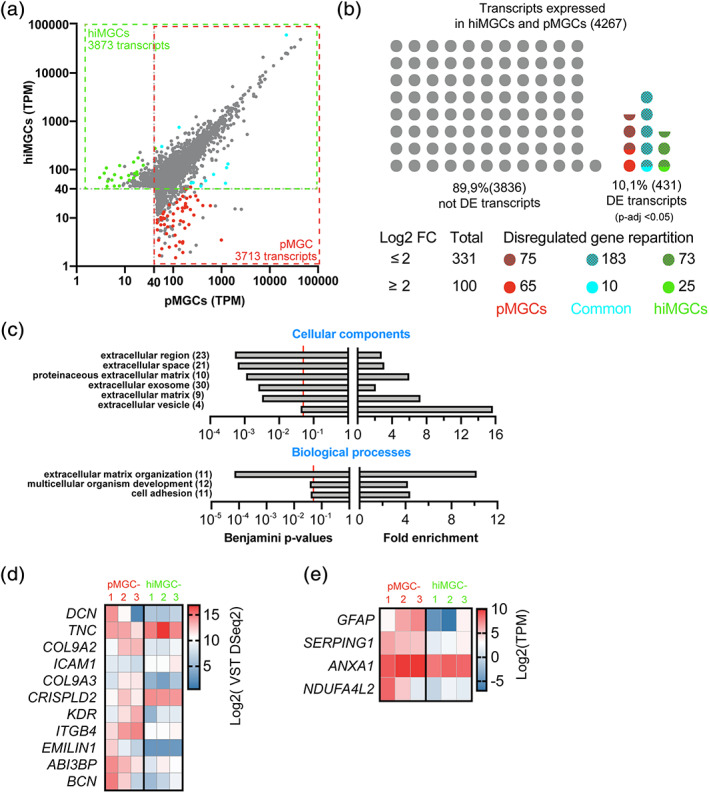
Genes differentially expressed between hiMGCs and pMGCs. (a) Scatter plot of all the transcripts detected in hiMGCs (Y axis) or pMGCs (X axis) with a mean TPM value >40. Grey dots represent genes that were not differentially expressed (DESeq2, p‐value >.05). Colored dots represent genes differentially expressed with a log2 FC ≥2. Green and red dots represent genes only expressed in hiMGCs and pMGCs, respectively. Blue dots represent genes differentially expressed in both groups of cells. (b) Schematic representation of the distribution of the 4,267 genes not differentially expressed (grey dots) or differentially expressed (colored dots) between hiMGCs and pMGCs. Red and hatched red dots represent the 75 and 65 genes differentially expressed (p < .05) found only in pMGCs (Log2 FC ≥2 or <2, respectively). Blue and hatched blue dots represent the 183 and 10 genes differentially expressed (p < .05) found in pMGCs and hiMGCs (log2 FC ≥2 or <2, respectively). Green and hatched green dots represent the 73 and 25 genes differentially expressed (p < .05) found only in hiMGCs (log2 FC ≥2 or <2, respectively). (c) GO enrichment analysis (upper panel, cellular components; lower panel, biological components) of the 100 genes differentially expressed between hiMGCs and pMGCs with a log2 FC ≥2. Left panels represent the Benjamini statistical p‐values for the enrichment test; right panel represents the fold enrichment. Extracellular components were highly represented in the cellular component analysis and the GO term of “extracellular matrix organization” was associated with the highest fold enrichment and the lowest Benjamini p‐values among the biological processes. (Numbers) indicate the total number of transcripts identified for each GO term (d) Heatmap representation of the log2 VST DESeq2 values for the 11 genes differentially expressed involved in the “extracellular matrix organization” in the three pMGCs and the three hiMGCs. (e) Heatmap representation of the log2 TPM expression of four genes differentially expressed found in MGCs from AIR retinas in the three pMGCs and the three hiMGCs. AIR, autoimmune retinitis; hiMGCs, human induced‐pluripotent stem cell‐derived MGCs; pMGCs, primary Müller glial cells; TPM, transcripts per kilobase million; VST, variance stabilizing transformation [Color figure can be viewed at wileyonlinelibrary.com]
MGC markers such as CLU, GLUL and DKK3 were expressed in hiMGCs and pMGCs. Surprisingly, RLBP1 was not detected in hiMGCs and pMGCs (Figure 1c), faintly expressed in hMacroglia while it has been previously detected in mMGCs by bulk RNAseq analysis (Figure S3). As expected, all other retinal neuronal and microglial markers were not detected in hiMGCs, and pMGCs (Figure 2c) at the exception of TRNP1. Indeed, TRNP1, a marker of bipolar cells, was expressed in hiMGCs and pMGCs while it was not expressed in hMacroglia or mMGCs (Figure S3). Thus, the transcriptome analysis showed a very similar transcriptome profile between hiMGCs and pMGCs.
To analyze the expression of a higher number of MGC makers, we selected 50 genes from the hMacroglia scRNAseq analysis showing the highest area under the curve (AUC) scores in the hMacroglia cluster and we determined the expression of these 50 transcripts in hiMGCs and pMGCs (Figure 2d) and in hMacroglia and mMGCs (Figure S4). This gene list included the 50 best transcripts allowing differentiating hMacroglia from other retinal cells and unsurprisingly, it included six classical MGC markers (APOPE, VIM, CLU, GLUL, RLBP1, DKK3; Grosche et al., 2016; Roesch et al., 2008; Shinoe et al., 2010). The heatmap analysis showed that hiMGCs and pMGCs had close transcript profiles (Figure 2d). However, hiMGCs, pMGCs, hMacroglia, and mMGCs expressed different transcript profiles, showing the limitation of data comparison from different studies (Figure S4). Conversely, we analyzed the expression of the 50 genes with the lowest AUC scores. The expression level of these genes was low, except for HSP90AA1, MFGE8, and GNB1 that were expressed in hiMGCs, pMGCs and in mMGCs (Figure S5).
To further identify genes preferentially expressed in MGCs, we analyzed the expression level of seven classical MGC markers not found in Figure 2d: KCNJ2, AQP4, KCNJ10, S100A16, ITGB1, and CD44. All these transcripts were similarly expressed in hiMGCs and pMGCs (Figure 2e). The expression level of S100A16, ITGB1, and CD44 was similar between hiMGCs, pMGCs and mMGCs. The expression level of AQP4, KCNJ10, and SLC1A3 was below our detection threshold of 40 TPM in hiMGCs and pMGCs while these transcripts were detected in mMGCs. Finally, the expression level of KCNJ2 was low in all studied MGCs (Figure S4).
2.3. Differential transcript expression between hiMGCs and pMGCs
To comprehensively analyze differences in transcriptome profiles between hiMGCs and pMGC, a differential expression analysis using DESeq2 was performed (Love, Huber, & Anders, 2014). In both hiMGCs and pMGCs, the expression level of 4,267 transcripts was above the selected cutoff of 40 TPM. In hiMGCs and in pMGCs, the mean expression level of respectively 3,873 and 3,713 transcripts was above the threshold (Figure 3a). The analysis showed that 89.9% of the expressed transcripts (3,836 out of the 4,267 transcripts) were not differentially expressed between hiMGCs and pMGCs (Figure 3b) and Supplementary Table S1). Only 10.1% of transcripts (431 out of the 4,267) were significantly differentially expressed with a p‐adj value ≤.05 (Figure 3b and Table S2). Then, an enrichment analysis of these 431 differentially expressed genes (DEGs) using pathway classifications from the Reactome database was performed (Fabregat et al., 2017) and showed that the differences in the expression of these transcripts between hiMGCs and pMGCs were not restricted to a particular class of pathways (Figure S6a). To limit our analysis to the most relevant changes, we selected only the 2.3% of DEGs with a log2 FC > 2 (100 out of the 4,267 transcripts). Among them, 25 and 65 transcripts were only expressed in hiMGCs and in pMGCs, respectively (Figure 3b, bottom panel). The Reactome pathway analysis of these 100 transcripts suggested that proteins involved in extracellular matrix organization could be particularly represented in transcripts with a log2 FC >2 (Figure S6 b). Then we identified enriched GO terms in these 100 transcripts. A DAVID v6.8 analysis (Huang da, Sherman, & Lempicki, 2009) did not show any statistically significant enrichment in the GO annotation class of molecular processes. In contrast extracellular components were highly represented in the cellular component analysis (Figure 3c). This over‐representation was further confirmed with the identification of the GO term of extracellular matrix organization associated with the highest fold enrichment and the lowest Benjamini p‐values among all the biological processes (Figure 3c). The expression of 11 DEGs related to the GO term of extracellular matrix organization (DCN, TNC, COL9A2, ICAM1, COL9A3, CRISPLD2, KDR, ITGB4, EMILIN1, ABI3BP, and BCN) was further analyzed. EMILIN1 was the gene the most differentially expressed (FC = 6.6; Figure 3d). To confirm this observation, we obtained independent cDNA from unrelated cultures of pMGCs and hiMGCs at P4 and quantified the expression of EMILIN1, collagens (COL9A3 and COL9A2), integrin (ITGB4) and cellular adhesion molecules (ICAM1) by RT‐qPCR. The selected genes were similarly expressed in independent culture batches (Figure S7).
The immunostaining analysis showed that from P2 to P8, pMGC‐2 were positive for GFAP while the transcript was not expressed by hiMGC‐1 (Figure 1 and Figure S2h–n). Accordingly GFAP expression was also elevated in pMGC‐2 and pMGC‐3 while our three hiMGCs did not express the protein (Figure 3e), suggesting that these cells could be gliotic. To further characterize hiMGCs, the expression level of transcripts recently identified by scRNAseq as differentially expressed between peripheral MGCs from healthy subjects (six transcripts) and peripheral MGCs from patients with auto‐immune retinitis (AIR) (nine transcripts) was analyzed (Andrew P. Voigt et al., 2020). In line with GFAP expression in pMGCs, the expression level of two AIR‐specific genes (ANXA1 and SERPING1) was significantly higher in pMGCs compared to hiMGCs (log2 FC of 1.22 and 2.97, respectively; Figure 3e). Surprisingly, the expression level of NDUFA4L2, a transcript specifically expressed in MGCs from healthy retinas, was higher in pMGCs compared to hiMGCs (log2 FC of 3.85; Figure 3e). The 12 remaining transcripts were similarly expressed in pMGCs and hiMGCs (Table S3).
Thus, the immunofluorescence and transcriptome analyses showed that hiMGCs derived from various somatic cell types formed a very homogeneous group of cells that were transcriptionally and phenotypically very close to pMGCs.
2.4. Disease Modeling
Given the involvement of MGCs in retinal disorders and in particular in early DR and diabetic macular edema pathogenesis, the response of hiMGCs to a DR‐related stress and their use to model DR features were assessed. Human pMGCs have recently been shown to upregulate the expression of genes involved in the inflammatory and angiogenic pathways (ATF3, CXCL8 (IL‐8), CXCL2, and ANGPTL4) after PA exposure (Capozzi et al., 2018). PA is the most studied and abundant saturated FFA in the blood. Its concentration in the blood of healthy subjects is around 350 μM and increases up to 600 μM in the plasma of diabetic patients (Liu et al., 2010; P. S. Patel et al., 2010). At P4, hiMGC‐1 were treated for 24 hr with normal glucose concentrations (NG, 5.6 mM) and exposed to increasing concentrations of PA. The relative expression level of these DR‐related genes was then assessed by RT‐qPCR and normalized to that of each gene in hiMGC‐1 cultured with NG in the absence of PA (Figure 4a). Low PA concentration (5 μM) did not induced any significant changes in the expression of ATF3, CXCL8, CXCL2, and ANGPTL4 while high PA concentrations (250 and 500 μM) upregulated the expression of these genes (Figure 4a). To assess the response of hiMGCs and pMGCs to different glycemic and lipid conditions, hiMGCs and pMGCs were cultured with NG or high glucose concentrations (HG, 25 mM) in the absence or in the presence of 500 μM of PA. The relative expression of the selected genes in the six cell lines is represented as a heatmap in Figure 4b according to the culture conditions. The expression of ATF3, CXCL8, CXCL2, and ANGPTL4 was strongly upregulated in all hiMGCs and pMGCs in response to PA with NG and HG (Figure 4b and Figure S8). HG alone did not change the regulation of these genes in hiMGCs and pMGCs (Figure 4b and Figure S8). Our results showed a similar physiological response between hiMGCs and pMGCs.
FIGURE 4.
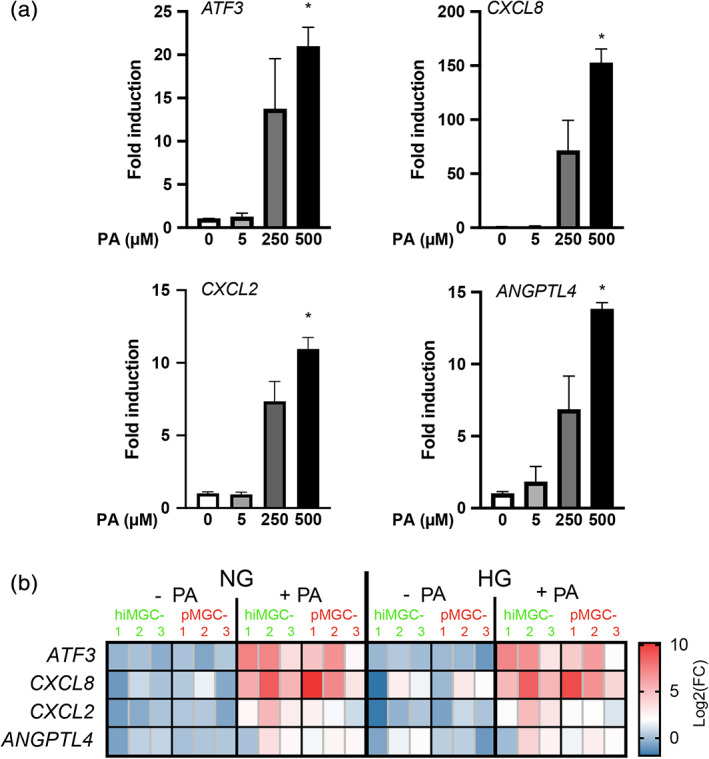
Response of hiMGCs and pMGCs to a DR‐related stress. Gene expression level analyzed by RT‐qPCR after 18 hr of cell incubation with normal or high glucose concentrations ± palmitic acid. (a) Graph bar representation of the mean (± SEM) relative expression level of ATF3, CXCL8 (IL‐8), CXCL2, and ANGPTL4 in hiMGC‐1 cultured with NG in the presence of increasing PA concentration (0, 5, 250, or 500 μM). Low concentration of PA (5 μM) did not induce any significant changes in the expression level of ATF3, CXCL8, CXCL2, and ANGPTL4 while high concentrations (250 and 500 μM) induced an increase in the expression level of these genes. The expression level represented is the mean of n = 3 independent points. Two tailed Kruskal‐Wallis non‐parametric test (4 groups, 12 values), with p = .0064; p = .0014; p = .0020; p = .0064 followed by Dunn's multiple comparisons tests, * p = .0382; p = .0197; p = .0382; p = .0064, respectively (b) Heatmap representation of the Log2 relative expression level of ATF3, CXCL8 (IL‐8), CXCL2, and ANGPTL4 transcripts in the three pMGCs and the three hiMGCs incubated with NG, HG, NG + PA and HG + PA. The expression of ATF3, CXCL8, CXCL2, and ANGPTL4 was strongly induced in all hiMGCs and pMGCs in response to PA with NG and HG. HG alone did not induce the regulation of these genes in hiMGCs and pMGCs. DR, diabetic retinopathy; HG, high glucose concentration (25 mM); hiMGCs: human iPSC‐derived Müller glial cells; NG, normal glucose concentration (5 mM); pMGCS, primary Müller glial cells; PA, palmitate [Color figure can be viewed at wileyonlinelibrary.com]
To further support the potential use of these new hiMGCs to model DR, their capacity to release cytokines involved in vessel remodeling was investigated. The PA‐induced regulation of hiMGC was independent of glucose stimulation. Therefore, we only compared cells cultured with NG versus NG + PA thereafter. In the subsequent experiments, hiMGC‐1 was chosen as a prototypic cell line. The release of 55 proteins with vascular remodeling properties was quantified in the supernatant of PA‐treated hiMGCs using an angiogenic blot assay (Figure 5a). After 24 hr of culture with NG + PA, the expression of 41 angiogenic proteins was detectable in hiMGC supernatant, suggesting potential vascular remodeling capacities (Figure 5b). PA regulated 46% (19) of the released angiogenic proteins tested and among them, 84% (16) were proangiogenic and 16% (3) were anti‐angiogenic. The 16 pro‐angiogenic factors, including growth factors (EG‐VEGF, FGF1, and VEGF), cytokines (CXCL8, CXCL16, IL‐1β, and MIP1α), and proteins involved in mural‐cell maintenance (ANG1, ANG2) and matrix remodeling (MMP‐9, and PDGF‐AA) were all up‐regulated (Figure 5c) and only the anti‐angiogenic proteins ADAMTS1 and CXCL4 were up‐regulated (Figure 5d). DPP‐IV (adenosine deaminase complexing protein 2) was the only protein down‐regulated by PA (Figure 5d). DPP‐IV is an exopeptidase with known activities on various substrates, including incretins but also cytokines such as CCL2 and IL‐1ß. DPP‐IV inhibitors are primarily used as glucose‐lowering drugs due to their effect on GLP‐1, however they also promote angiogenesis in a model of ischemia (Lei et al., 2017). Our results indicated that all hiMGCs responded to a DR‐related stress by upregulating the expression of pro‐inflammatory genes and by releasing factors with vascular remodeling properties.
FIGURE 5.
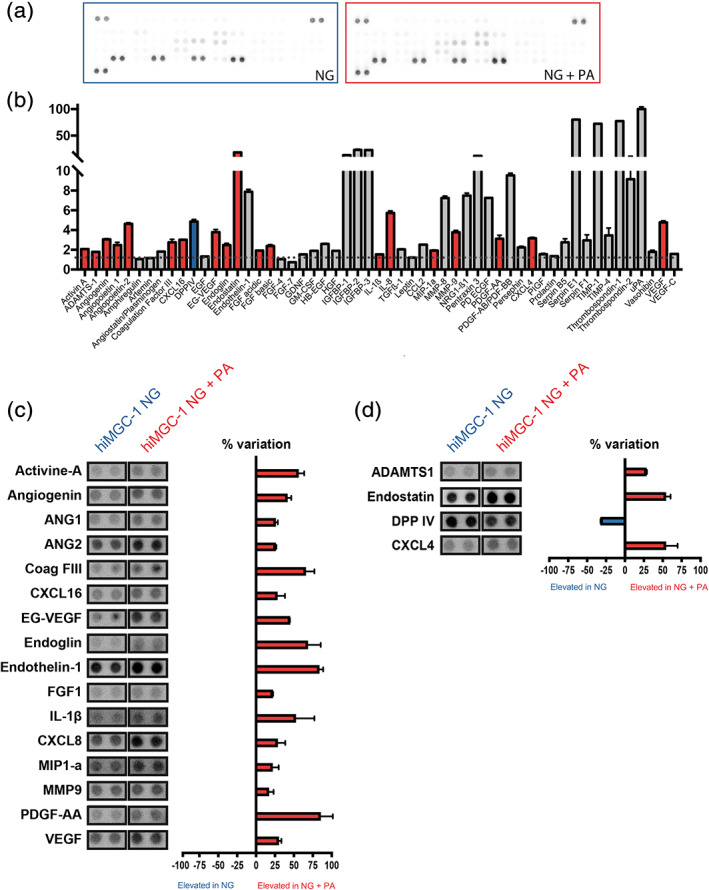
Elevated levels of potent angiogenic factors in the supernatant of hiMGCs treated with palmitate. Semi‐quantitative analysis of the expression level of 55 angiogenic proteins in the supernatant of hiMGCs (hiMGC‐1) treated for 24 hr using an angiogenic blot assay. (a) Representative photography of a blot using the supernatant of hiMGCs treated with NG (left) and NG + PA (right) as a probe. (b) Raw pixel intensity quantification of the 55 proteins found in the supernatant of hiMGCs treated with in NG + PA. Values are expressed in RDU versus membrane background (red bars represent upregulated proteins, blue bars represent downregulated proteins and grey bars represent unregulated proteins compared to the supernatant of hiMGCs cultured with NG). The supernatant of hiMGC‐1 treated for 24 hr with NG + PA contained detectable amounts of 41 angiogenic proteins. (c and d) Graph bar representation of the 20 regulated proteins. Values are expressed in RDU as a percent variation between hiMGCs treated with NG and NG + PA. (c) Regulated pro‐angiogenic proteins (red bars represent the proteins upregulated in the supernatant of hiMGCs treated with NG + PA). (d) Regulated anti‐angiogenic proteins (red bars represent the proteins upregulated in the supernatant of hiMGCs treated with NG + PA, the blue bar represents the protein upregulated in the supernatant of hiMGCs treated with NG). PA regulated 19 angiogenic proteins tested: 16 were proangiogenic and 3 were anti‐angiogenic. Error bars represent the SEM of the duplicate spots, each membrane was incubated with a pool of three independent supernatant. hiMGCs, human iPSC‐derived Müller glial cells; NG, normal glucose concentration (5 mM); PA, palmitate; RDU, relative density units [Color figure can be viewed at wileyonlinelibrary.com]
3. DISCUSSION
In this study, we developed for the first time an in vitro model of DR based on hiMGCs cultured under a defined condition in order to achieve the dyslipidemia features of the disease. We described an easy method to generate hiMGCs from ROs and culture conditions to expand and bank these glial cells. Thawed hiMGCs could be expanded up to P8 without losing their glial phenotype (Figure 1, S1 and S2) and transcriptome profiling revealed the high degree of similarity between hiMGCs and pMGCs (Figures 2 and 3). We showed that hiMGCs acquired functional features of MGCs and responded to a DR‐related stress by upregulating the expression of genes involved in inflammatory and angiogenic pathways (Figure 4) and by releasing DR‐related angiogenic factors such as VEGF, CXCL8, IL‐1β, and ANGPTL4 (Figure 5).
To thoroughly characterize these newly developed hiMGCs, we analyzed hiMGCs and pMGCs obtained from post‐mortem human samples by immunofluorescence and RNA sequencing. Our data indicated that hiMGCs were phenotypically close to pMGCs. First, hiMGCs expressed key markers of MGCs including VIM, GS, and SOX9. Using a combination of transcriptome analysis and data mining, we showed that hiMGCs had lost hiPSC features during the differentiation process and that their expression profile of 23 retinal markers unambiguously corresponded to a glial phenotype. Surprisingly, under our culture conditions, we found that hiMGCs did not express MGC‐specific proteins involved in transmembrane exchange such as APQ4, KCNJ 2 and 10, SLC1A3, and RLBP1. Nevertheless, these genes were similarly expressed in hiMGCs and pMGCs, suggesting that hiMGCs could be used as efficiently as pMGCs for in vitro models of MGCs. Furthermore, a single cell analysis has revealed that the expression of MGC markers could be restricted to subsets of glial cells with spatial differences. For instance, a subset of MGCs has been shown to express very low RLBP1 levels (Voigt et al., 2020) while differences in SLC1A3 and CD44 levels have been reported between the central retina and the periphery (Menon et al., 2019). These findings suggest that our protocol could favor the differentiation of a particular subset of MGCs at the expense of others. Further experiments are needed to determine if a global deregulation of these markers is due to a bias in the differentiation toward a specific subset of MGCs or to a global downregulation of these markers. Second, genes differentially expressed between hiMGCs and pMGCs (431 with a FC ranging from 0.33 to 7.65) were not limited to specific biological functions and only 2.3% of them were differentially expressed with a high FC (log2 FC ≥2). A GO annotation study of these 100 regulated transcripts revealed that the changes occurred in the organization of the extracellular matrix, and they could explain the small differences in cellular shapes observed in early passages (from P2 to P4). Interestingly, hiMGCs did not express GFAP. GFAP is not expressed by MGCs under physiological conditions but its expression is increased in Muller cells in response to injury (Dahl, 1979; Sarthy et al., 1991). GFAP expression has also been reported in pMGCs under basal culture conditions (Capozzi et al., 2018). Accordingly, we found a sustained expression of GFAP in pMGC‐2 and ‐3 but the expression of AIR specific transcripts remains surprisingly low in both pMGCs and hiMGCs indicating that both cell lines where closest to MGCs in physiological conditions rather than MGCs in pathological conditions. Thus, hiMGCs could be generated from donors and easily selected, expanded and stored. Our phenotypic analysis strongly suggested that hiMGCs were very close to pMGCs and that they could be an attractive alternative to the rare pMGCs.
To assess the potential use of hiMGCs in disease modeling, we determined a defined culture condition and developed a hiMGC‐based model of DR to test their response to stimuli involved in DR progression (HG and PA exposure). HiMGCs and pMGCs were cultured in the presence of HG and PA, a FFA with elevated plasma levels in T2D patients (Korani et al., 2012; P. S. Patel et al., 2010) and the expression of ATF3, CXCL8, CXCL2, and ANGPTL4, that play a key role in DR pathogenesis, was determined (Capozzi et al., 2018). We showed that PA, but not HG, similarly upregulated the expression of these genes in hiMGCs and pMGCs. Interestingly under our defined culture conditions, the response of hiMGCs and pMGCs to PA was independent of the glucose stimulation while a previous study has reported a slight potentiation in pMGCs (Capozzi et al., 2018). The absence of response of hiMGCs is in line with other in vitro studies on human retinal endothelial cells showing no effect of HG on the inflammatory and angiogenic responses (Chen et al., 2003; Mohamed et al., 2014). Similarly, RNAseq analysis has recently been performed on pMGCs by Capozzi et al, and they have concluded that HG alone did not significantly affect diabetes‐related pathways (Capozzi et al., 2018). Remarkably, hiMGC response to PA was very similar between hiMGC from the different donors, suggesting that hiMGCs could be reprogrammed from cells of different origins without affecting their potential response to injury.
Therefore, hiMGCs could be a valuable tool for future studies investigating DR‐related molecular pathways in human retinas or assessing new therapeutic drugs.
DR‐associated angiogenesis and edema are not solely related to VEGF (Semeraro et al., 2019). Cytokines released by activated glial cells are found in the vitreous of DR patients and are likely to participate in vascular remodeling (Abcouwer, 2017; McAuley et al., 2014). Since DR‐related transcripts involved in angiogenic and inflammatory pathways were regulated in response to PA, we assessed the use of hiMGCs as an adequate model to decipher VEGF‐independent vascular remodeling. We thus compared the expression of 55 angiogenic factors in the supernatant of control and PA‐treated hiMGCs. Among the proteins upregulated by PA, we only found two anti‐angiogenic factors while pro‐angiogenic factors strongly related to DR such as ANG2, endoglin, IL‐1β, CXCL8, MMP‐9, PDGF‐AA and VEGF were all upregulated. Among them, the expression levels of CXCL8 (IL‐8) and ANGPTL‐4 were high in hiMGCs and strongly induced by PA (228.9‐ and 5.0‐folds, respectively). CXCL8 induces vascular leakage and is involved in central nervous system edema (Semple, Kossmann, & Morganti‐Kossmann, 2010). Hypoxia upregulates its expression in glial cells (Yoshida, Yoshida, Khalil, Ishibashi, & Inomata, 1998) and its levels is increased in the vitreous of DR patients (Funatsu, Noma, Mimura, Eguchi, & Hori, 2009; Patel, Tombran‐Tink, Hykin, Gregor, & Cree, 2006; Yoshida et al., 1998; Yoshimura et al., 2009). Its upregulation has been shown to be independent of VEGF (Bromberg‐White et al., 2013). In patients resistant to anti‐VEGF therapy, CXCL8 levels correlate with therapy success (Jeon & Lee, 2014). Thus, under pathological conditions, CXCL8 could play a central role in the pathogenesis of DR by promoting vascular permeability and angiogenesis. ANGPTL4 is a well‐characterized transcriptional target of PPAR‐β/δ and it also plays an important role in angiogenesis in proliferative DR (Babapoor‐Farrokhran et al., 2015; Xin et al., 2013). Therapies directed against ANGPTL4 are now emerging to control endothelial cell proliferation in DR (Yang, Cheng, & Su, 2018). We showed that hiMGCs could be used as an appropriate model to test the effects of DR‐related pathological stimuli on MGCs. HiMGCs expressed canonical pro‐angiogenic molecules such as VEGF under pathological conditions as well as other valuable targets that have been associated with DR such as CXCL8 or ANGPTL4.
The role of dyslipidemia in the pathogenesis of DR is now widely accepted and supported by the results of the FIELD and ACCORD studies (Accord Study group et al., 2010; Keech et al., 2007). However, the relationship between dyslipidemia and DR is still poorly understood, and discrepancies in the association between the class of plasma lipids and DR progression suggest that subtle changes in lipid composition, that remains to be determined, may be responsible for DR worsening (Chang & Wu, 2013). Clinically, a poor response to anti‐VEGF treatment has been associated with the presence of lipoproteinaceous debris and cholesterol crystals that compose the fluid of hyperreflective cystoid spaces found in DME patients (Ahn, Han, Ahn, Kim, & Oh, 2020; Horii et al., 2012; Kashani et al., 2018). HiMGCs that strongly respond to plasma lipids such as PA will be a valuable tool to help to understand the biology of lipids associated with DR progression.
4. MATERIALS AND METHODS
4.1. Human iPSC maintenance
Human iPSC clone AHF1pi2 (Reichman, Terray, et al., 2014), FS2pi3 (Reichman et al., 2017) and 5f (Slembrouck‐Brec et al., 2019) were derived respectively from adult dermal fibroblasts, foreskin fibroblasts and post‐mortem MGCs (pMGC‐3). Briefly, cells were cultured on truncated recombinant human vitronectin (rhVTN‐N; ThermoFisher Scientific, Waltham, MA, Cat. #A14700) coated dishes with Essential 8™ medium (ThermoFisher Scientific, Cat. #SMC‐100B) as previously described (Slembrouck‐Brec et al., 2018) (Supplementary Table S4). HiPSCs were passaged with the enzyme‐free Gentle Cell Dissociation Reagent (StemCell Technologies, Vancouver, BC, Canada, Cat. #07174) every week. Detached cell aggregates were collected in Essential 8™ medium and carefully pipetted up and down to obtain uniform suspension of cell aggregates and re‐seeded at ratio of 1/10 to 1/60 depending on the confluence.
4.2. Retinal differentiation
For retinal differentiation, our xeno‐, feeder‐, and embryoid body‐free protocol was used as previously described (Slembrouck‐Brec et al., 2018). HiPSCs were expanded to 70–80% confluence in dishes coated with rhVTN‐N in Essential 8™ medium. At this time, defined as Day 0 (D0), hiPSCs were cultured in chemical defined Essential 6™ medium (ThermoFischer Scientific, Cat. #A1516401). After 2 days, the medium was replaced by E6N2 medium [Essential 6™ medium, N2 supplement (ThermoFischer Scientific, Cat. #17502048), 10 units/ml Penicillin and 10 μg/ml Streptomycin (ThermoFisher Scientific, Cat. #15140148]. The medium was changed every 2–3 days. On D28, identified self‐formed ROs were isolated, using a needle and cultured as floating structures in the ProB27 medium [DMEM/Nutrient Mixture F‐12 (DMEM/F12; ThermoFisher Scientific, Cat. # 11320074), 1X MEM non‐essential amino acids (ThermoFisher Scientific, Cat. #11140050), 2% B27™ supplement (Thermo Fischer Scientific, Cat. #17504044), 10 units/ml Penicillin and 10 μg/ml Streptomycin] supplemented with 10 ng/ml of recombinant human fibroblast growth factor‐2 (FGF2; PeproTech, Cranbury, NJ, Cat. #100‐18B). Half of the medium was changed every 2–3 days. At D35, FGF2 was removed of the ProB27 medium and ROs were cultured in floating condition for the next several weeks to follow retinal differentiation and maturation (Supplementary Table S5).
4.3. Human post‐mortem and hiPSC‐derived Müller glia cell isolation and amplification
Human post‐mortem retinal tissue or ROs were dissociated accordingly to the previously described protocol (Slembrouck‐Brec et al., 2018). All generated cell batches were found negative for Mycoplasma contamination. For hiMGCs, floating ROs were collected between D150 and D200. Distal RPE was discarded from the organoids under a stereomicroscope and neuro‐retinal structures were washed 3 times in Ringer solution (NaCl 155 mM; KCl 5 mM; CaCl2 2 mM; MgCl2 1 mM; NaH2PO4 2 mM; HEPES 10 mM; and Glucose 10 mM). RPE‐free ROs were dissociated with two units of pre‐activated papain at 28.7 μ/mg (Worthington Biochemical Corp., Lakewood, NJ, Cat. #LS003124) in Ringer solution during 25 min at 37°C. When a homogenous cell suspension is obtained, papain was inactivated by adding 1 ml of MGC medium [DMEM, high glucose −25 mM‐, pyruvate (ThermoFisher Scientific, Cat. #41966052), fetal bovine serum (FBS; ThermoFisher Scientific, Cat. #26140079) 10%, 100 units/ml Penicillin and 100 μg/ml Streptomycin). Cells were centrifuged at 110g and resuspended in pre‐warmed MGC medium. Retinal cells were plated at 50000 cell/cm2 in T‐25 cm2 flask previously coated with Geltrex matrix (ThermoFischer Scientific, Cat. #A1413302) and incubated at 37°C in a standard 5% CO2/95% air incubator (Supplementary Table S4). Medium was changed every 2–3 days.
4.4. Cryopreservation of MGCs
HiMGCs or pMGCs at passage 1 were cryopreserved in 10% DMSO, 90% FBS solution (4 × 105 Cells / 500 μl/cryotube) and placed in isopropanol‐based freezing container at −80°C for a minimum of 4 hr. Frozen tubes were kept in a −150°C freezer for long‐term storage. Frozen MGCs were thawed quickly at 37°C in a water bath and resuspended in prewarmed dedicated media for downstream investigations.
4.5. Immunostaining and imaging of MGCs
HiMGCs, pMGCs, and RO sections were fixed with 4% PFA in phosphate‐buffered saline solution (PBS; ThermoFisher Scientific, Cat. #10010023) for 5 min before immunostaining. After washes with PBS, nonspecific binding sites were blocked for 1 hr at room temperature with blocking buffer [PBS solution containing 0.2% gelatin (Merck, Darmstadt, Germany) and 0.25% Triton X‐100 (ThermoFisher Scientific, Cat. #28314)] and then overnight at 4°C with the primary antibody against human Vimentin (VIM, 1:100; Merck, clone VIM 3B4, Cat. #CBL 202), Glial fibrillary acidic protein (GFAP, 1:200, Cell Signaling Technology, Danvers, MA, clone D1F4Q, Cat. #12389), Glutamine synthetase (GS, 1:500, Merck, clone GS‐6, Cat. #MAB302), SRT‐BOX 9 (SOX9, 1:1000; Merck, Cat. #AB5535), SRT‐BOX 2 (SOX2, 1:400; Cell Signaling, clone D6D9, Cat. #3579) diluted in blocking buffer. Slides were washed three times in PBS with 0.1% Tween®20 (Merck, Cat. #P1379) and then incubated for 1 hr at room temperature with appropriate secondary antibodies conjugated with either AlexaFluor 488, 594 or 647 (Interchim) diluted at 1:600 in blocking buffer with 4′,6‐diamidino‐2‐phenylindole (DAPI) diluted at 1:1000 to counterstain nuclei. Fluorescent staining signals were captured with an Olympus FV1000 confocal microscope equipped with 405, 488, 543, and 633 nm lasers. Confocal images were acquired using a 1.55 or 0.46 μm step size and corresponded to the projection of 20–40 optical sections.
4.6. RNAseq analysis
Total RNA were extracted from P2 hiMGCs, pMGC‐1 and ‐2 or P3 pMGC‐3 using the Nucleospin RNA II kit (Macherey‐Nagel, Düren, Germany, Cat. #740955) according to the manufacturer's protocol, RNA quality and quantity were evaluated using BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA) with the RNA 6000 Nano Kit (Agilent Technologies, Cat. #5067‐1511). RNA sequencing libraries were constructed from 1 μg of total RNA using a modified TruSeq RNA Sample preparation kit protocol. Pass‐filtered reads (using Trimmomatic) were mapped using HiSAT2 and aligned to human reference genome GRCh38.95 (Kim, Langmead, & Salzberg, 2015). The count table of the gene features was obtained using HTSeq. Normalization and differential expression analysis values were computed with DESeq2 (Love et al., 2014). TPM were determined using Libinorm using htseq mode (Dyer, Shahrezaei, & Hebenstreit, 2019). All generated data that supports the findings of this study are available in the Data S1 of this article and all raw data have been deposited in NCBI’s Gene Expression Omnibus GSE166565.
Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847; Menon et al., 2019). Expression level in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406; Hoang et al., 2020).
4.7. Palmitate solubilization in culture medium
PA as other FFA is a hydrophobic molecule. To allow its solubilization in culture medium, PA (Merck, Cat. #P0500) was bound to bovine serum albumin FFA‐free (BSA, Merck, Cat. #A8806). To obtain the working concentration of PA‐culture medium (500 μM PA, 0.5% vol/vol EtOH), corresponding to a molar ratio PA/BSA = 3.8, we diluted at 1:200 an absolute EtOH solution of 0.1 M of PA in a BSA‐culture medium (0.88% wt/vol of BSA). As described in Supplementary Table S4, BSA‐culture medium is either prepared from 5.6 or 25 mM glucose DMEM.
4.8. Defined culture condition modeling DR
The hiMGCs and pMGCs were thawed and grow in MGC medium in a standard 5% CO2/95% air incubator until P3. Twenty‐four hours before passage, P3 MGCs were serum‐starved medium and changed in normo glucose (NG −5.6 mM‐) medium without BSA and EtOH [DMEM low glucose −5.6 mM‐, pyruvate (ThermoFisher Scientific, Cat. #31885) supplemented with 19.4 mM mannitol (Merck, Cat. #240184) and 50 units/ml Penicillin and 50 μg/ml Streptomycin] (Supplementary Table S4). Mannitol was added to keep osmotic condition similar to DMEM High glucose (25 mM glucose). P3 cells were then passed in Geltrex‐coated dishes (35,000 cells/cm2) in NG medium without BSA. After 4 hr, attached P4 hiMGCs and pMGCs were cultured for 18 hr (transcriptome analysis) or 24 hr (proteomic analysis) in different conditions using. First, a dose response to PA was performed using a NG medium [DMEM low glucose −5.6 mM‐, pyruvate supplemented with 19.4 mM mannitol, 50 units/ml Penicillin and 50 μg/ml Streptomycin, BSA 0.88 wt/vol, EtOH 0.5% vol/vol)] and increasing concentration of PA (0, 5, 250, and 500 μM). Then, four different media were used: NG [DMEM low glucose −5.6 mM‐, pyruvate supplemented with 19.4 mM mannitol, 50 units/ml Penicillin and 50 μg/ml Streptomycin, BSA 0.88 wt/vol, EtOH 0.5% vol/vol)], HG [DMEM high glucose −25 mM‐, pyruvate supplemented with 50 units/ml Penicillin and 50 μg/ml Streptomycin, BSA 0.88 wt/vol, EtOH 0.5% vol/vol], NG + PA (DMEM low glucose −5.6 mM‐, pyruvate supplemented with 19.4 mM mannitol, 50 units/ml Penicillin, and 50 μg/ml Streptomycin BSA 0.88 wt/vol, EtOH 0.5 vol/vol) or HG + PA (DMEM high glucose −25 mM‐, BSA 0.88 wt/vol, PA 500 μM, EtOH 0.5% vol/vol) (Table S4).
4.9. RNA isolation, reverse transcription and real‐time quantitative polymerase chain reaction (RT‐qPCR)
After 18 hr of treatment hiMGCs and pMGCs were lysed and RNA purified using the RNA XS kit (Macherey‐Nagel, Cat. #740902) according to the manufacturer's protocol. Total RNA was isolated and converted to cDNA using QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany Cat. #205314). Each reverse transcription assay was performed in a 20 μl reaction. Subsequent real‐time qPCR was performed using cDNA, Sybr Green PCR Master Mix (ThermoFisher Scientific, Cat. #4367659) in StepOne Plus real‐time PCR system (ThermoFisher Scientific, Applied Biosystems™) with the following profile: 10 min at 95°C, followed by a total of 40 two‐temperature cycles (15 s at 95°C and 1 min at 60°C). To verify the purity of the products, a melting curve was produced after each run according to the manufacturer's instructions. Results were expressed as fold induction after normalization by RPS26 gene expression. Primers for RT‐qPCR were purchased from IDT technology (Table S5).
4.10. Profiling of angiogenesis related protein in DR‐modeled hiMGC secretome
The relative expression profile of 55 human angiogenesis‐related proteins was performed using Proteome Profiler Human Angiogenesis Array kit (Bio‐Technee, Minneapolis, MNCat. #ARY007) following the manufacturer's instructions. Briefly, hiMGCs were cultured under NG or NG + PA conditions, 700 μl of 24 hr‐conditioned medium were mixed with a cocktail of biotinylated detection antibodies and incubated with the array membrane. Captured proteins are visualized using chemiluminescent detection reagents supplied and each duplicate spot represent the detection of one specific antigen. The signal detected is proportional to the amount of bound analyte and the images were acquired on a Fusion FX7 (Vilber, Marne‐la‐Vallée, France) with an exposure time of 10 min. Relative level of proteins was measured with the open source image processing program Fiji (Rueden et al., 2017) using pixel intensity after subtraction of local background level with MicroArray_Profile plugin (OptiNav, Bellevue, WA). Three duplicate reference spots (positive controls) placed at 3 corners are used to normalize pixel intensity for each membrane.
4.11. Statistical analysis
Graph Pad Prism 8 (GraphPad Software) was used for data analysis and graphic representation. All values are reported as a mean ± SEM. In RT‐qPCR experiments, statistical differences between groups were evaluated using a two‐tailed Kruskal–Wallis non‐parametric test, followed by Dunn's multiple comparisons.
CONFLICT OF INTEREST
OG, J‐AS, and SR are inventors on pending patents related to generation of retinal cells from human pluripotent stem cells.
AUTHOR CONTRIBUTIONS
Aude Couturier and Guillaume Blot contributed to the conception and design of the study, data collection, data analysis and interpretation and wrote the manuscript. Lucile Vignaud contributed to data collection, data analysis and interpretation. Céline Nanteau and Amélie Slembrouck‐Brec contributed to the collection of data. Valérie Fradot, Niyazi Acar, José‐Alain Sahel, and Ramin Tadayoni revised the manuscript for important intellectual content. Gilles Thuret provided human tissues and revised the manuscript. Jerome E. Roger performed the analysis and participated in the interpretation of bioinformatics data. Florian Sennlaub and Olivier Goureau contributed to the conception and design of the study and revised the manuscript. Xavier Guillonneau and Sacha Reichman contributed to the conception and design of the study, data analysis and interpretation and wrote the manuscript. Aude Couturier and Guillaume Blot contributed equally to this work as co‐first authors, and Xavier Guillonneau and Sacha Reichman contributed equally to this work as co‐senior authors. All the material can be requested to Xavier Guillonneau or Sacha Reichman.
Supporting information
FIGURE S1 Immunofluorescence comparison of fresh and thawed MGCs. Immunochemistry analysis of fresh P2 hiMGC‐1 (left panels), thawed hiMGC‐1 (center panels) and pMGC‐2 (right panels) for the MGC makers VIM and GFAP (top panels), GS and SOX9 (bottom panels). Nuclei from the top panels were stained with DAPI. Scale bar = 50 μm
FIGURE S2 Immunofluorescence comparison of hiMGCs and pMGCs from P2 to P8. Immunofluorescence analysis of hiMGC‐1 (A to G), and pMGC‐2 (H to N) for the MGC makers SOX9, VIM, GS, GFAP and nuclei stained with DAPI over the passages (a and h) P2, (b and i) P3, (c and j) P4, (d and k) P5, (e and l) P6, (f and m) P7 (g and n) P8. Scale bar = 50 μm
FIGURE S3 Comparison of the expression level of neuronal, MGC and microglial markers in hiMGCs, pMGCs, hMacroglia and mMGCs. Heatmap representation of the expression of neuronal, MGC and microglial markers in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). Expression in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847) {. Expression in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S4 Comparison of the expression level of gene with the highest hMacroglia AUC values in hiMGCs, pMGCs, hMacroglia and mMGCs. (a) Heatmap representation of the expression level of the 50 genes with the highest AUC in hMacroglia in the study by Menon et al. in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). (b) Heatmap representation of the expression of 7 classical MGC markers not found in the 50 genes with the highest AUC in hMacroglia. pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). Expression in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847). Expression level in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S5 Comparison of the expression level of gene with the lowest hMacroglia AUC values in hiMGCs, pMGCs, hMacroglia and mMGCs. (a) Heatmap representation of the expression level of the 50 genes with the lowest hMacroglia AUC score in the study by Menon et al. in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel).
Expression level in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847). Expression level in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S6 Pathway distribution of genes differentially expressed within hiMGCs and pMGCs. Schematic tree of cellular pathways repartition where genes are found differentially expressed (green) within pMGCs and hiMGCs, using Reactome database. (a) Pathway in which transcripts (431) are found differentially expressed. (b) Pathway in which transcripts (100) are found differentially expressed with a log2 FC ≥2
FIGURE S7 Expression levels of genes belonging to the extracellular matrix organization GO term in pMGCs and hiMGCs. Graph bar representation of the mean (± SEM) relative expression level of EMILIN1, collagens (COL9A3 and COL9A2), integrin (ITGB4) and cellular adhesion molecules (ICAM1) in P4 hiMGCs derived from three different iPS clones (right panels) and P4 pMGCs isolated from three different donors (left panel). For each of them, the expression expressed is the mean of n = 4 independent points).
MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs
FIGURE S8 hiMGCs and pMGCs responses to DR‐relevant stress. Human iPSC‐derived MGC‐(1 to 3) and pMGC‐(1 to 3) were treated for 18 hr as follow NG (DMEM 5.6 mM glucose, 19.4 mM mannitol, BSA 0.88 w/v, EtOH 0.5 v/v) or HG (DMEM 25 mM glucose, BSA 0.88 w/v, EtOH 0.5% v/v) or NG + PA (DMEM 5.6 mM glucose, 19.4 mM mannitol, BSA 0.88 w/v, EtOH 0,5 v/v) or HG + PA (DMEM 25 mM glucose, BSA 0.88 w/v, PA 500 μM, EtOH 0.5% v/v). Transcript relative expression were analyzed by RT‐qPCR. Graph bar representation of the mean (± SEM) relative expression of ATF3, CXCL8 (IL‐8), CXCL2 and ANGPTL4 in hiMGCs derived from three different iPS clones (left panels) and pMGCs isolated from three different donors (right panel) or iPS clones. For each of them, the expression expressed is the mean of n = 4 independent points)
TABLE S1 RNAseq data: list of non‐differentially expressed genes within hiMGCs and pMGCs
TABLE S2 RNAseq data: list of differentially expressed genes within hiMGCs and pMGCs
TABLE S3 RNAseq data: list of known MGC markers and their expression in hiMGCs and pMGCs
TABLE S4 RNAseq data: list of auto‐immune retinitis or healthy MGC markers and their expression in hiMGCs and pMGCs
TABLE S5 Culture media for hiMGC generation and defined culture conditions for DR modeling
TABLE S6 Primers sequences
ACKNOWLEDGMENTS
We wish to thank Frida Paulina Muniz Ruvalcaba for her critical reading, Stéphane Fouquet from the imaging facility of the Institut de la Vision and Wassila Carpentier for expert technical assistance. This work was supported by grants from INSERM, Agence Nationale pour la Recherche (Labex Lifesenses (ANR‐10‐LABEX‐65)) and IHU FOReSIGHT (ANR‐18‐IAHU‐01) supported by French state funds managed by the ANR within the Investissements d'Avenir program.
Couturier A, Blot G, Vignaud L, et al. Reproducing diabetic retinopathy features using newly developed human induced‐pluripotent stem cell‐derived retinal Müller glial cells. Glia. 2021;69:1679–1693. 10.1002/glia.23983
Aude Couturier and Guillaume Blot contributed equally as first authors.
Xavier Guillonneau and Sacha Reichman contributed equally as last authors.
Funding information IHU FOReSIGHT (ANR‐18‐IAHU‐01); Labex Lifesenses (ANR‐10‐LABEX‐65); INSERM
Contributor Information
Xavier Guillonneau, Email: xavier.guillonneau@inserm.fr.
Sacha Reichman, Email: sacha.reichman@inserm.fr.
DATA AVAILABILITY STATEMENT
All generated data that supports the findings of this study are available in the supplementary material of this article and avalaible for download from public database (GEO submission in process, for review purposes, fastaq files can be downloaded from the following private link: fastaq_download.)
REFERENCES
- Abcouwer, S. F. (2017). Muller cell‐microglia cross talk drives neuroinflammation in diabetic retinopathy. Diabetes, 66(2), 261–263. 10.2337/dbi16-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACCORD Study group , ACCORD Eye Study Group , Chew, E. Y. , Ambrosius, W. T. , Davis, M. D. , Danis, R. P. , … Fine, L. J. (2010). Effects of medical therapies on retinopathy progression in type 2 diabetes. The New England Journal of Medicine, 363(3), 233–244. 10.1056/NEJMoa1001288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, J. , Han, S. , Ahn, S. M. , Kim, S. W. , & Oh, J. (2020). Clinical implications of suspended scattering particles in motion observed by optical coherence tomography angiography. Scientific Reports, 10(1), 15. 10.1038/s41598-019-55606-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babapoor‐Farrokhran, S. , Jee, K. , Puchner, B. , Hassan, S. J. , Xin, X. , Rodrigues, M. , … Sodhi, A. (2015). Angiopoietin‐like 4 is a potent angiogenic factor and a novel therapeutic target for patients with proliferative diabetic retinopathy. Proceedings of the National Academy of Sciences of the United States of America, 112(23), E3030–E3039. 10.1073/pnas.1423765112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg‐White, J. L. , Glazer, L. , Downer, R. , Furge, K. , Boguslawski, E. , & Duesbery, N. S. (2013). Identification of VEGF‐independent cytokines in proliferative diabetic retinopathy vitreous. Investigative Ophthalmology & Visual Science, 54(10), 6472–6480. 10.1167/iovs.13-12518 [DOI] [PubMed] [Google Scholar]
- Capozzi, M. E. , Giblin, M. J. , & Penn, J. S. (2018). Palmitic acid induces Muller cell inflammation that is potentiated by co‐treatment with glucose. Scientific Reports, 8(1), 5459. 10.1038/s41598-018-23601-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi, M. E. , Hammer, S. S. , McCollum, G. W. , & Penn, J. S. (2016). Epoxygenated fatty acids inhibit retinal vascular inflammation. Scientific Reports, 6, 39211. 10.1038/srep39211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi, M. E. , McCollum, G. W. , Cousins, D. B. , & Penn, J. S. (2016). Linoleic acid is a diabetes‐relevant stimulator of retinal inflammation in human retinal Muller cells and microvascular endothelial cells. Journal of Diabetes and Metabolism, 7(12), 1–3. 10.4172/2155-6156.1000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. C. , & Wu, W. C. (2013). Dyslipidemia and diabetic retinopathy. The Review of Diabetic Studies, 10(2–3), 121–132. 10.1900/RDS.2013.10.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Jump, D. B. , Grant, M. B. , Esselman, W. J. , & Busik, J. V. (2003). Dyslipidemia, but not hyperglycemia, induces inflammatory adhesion molecules in human retinal vascular endothelial cells. Investigative Ophthalmology & Visual Science, 44(11), 5016–5022. 10.1167/iovs.03-0418 [DOI] [PubMed] [Google Scholar]
- Chung, S. H. , Shen, W. , Davidson, K. C. , Pébay, A. , Wong, R. C. B. , Yau, B. , & Gillies, M. (2019). Differentiation of retinal glial cells from human embryonic stem cells by promoting the notch signaling pathway. Frontiers in Cellular Neuroscience, 13, 527. 10.3389/fncel.2019.00527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, D. (1979). The radial glia of Müller in the rat retina and their response to injury. An immunofluorescence study with antibodies to the glial fibrillary acidic (GFA) protein. Experimental Eye Research, 28(1), 63–69. 10.1016/0014-4835(79)90106-4 [DOI] [PubMed] [Google Scholar]
- Dyer, N. P. , Shahrezaei, V. , & Hebenstreit, D. (2019). LiBiNorm: An htseq‐count analogue with improved normalisation of smart‐seq2 data and library preparation diagnostics. PeerJ, 7, e6222. 10.7717/peerj.6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastlake, K. , Wang, W. , Jayaram, H. , Murray‐Dunning, C. , Carr, A. J. F. , Ramsden, C. M. , … Limb, G. A. (2019). Phenotypic and functional characterization of Muller glia isolated from induced pluripotent stem cell‐derived retinal organoids: Improvement of retinal ganglion cell function upon transplantation. Stem Cells Translational Medicine, 8(8), 775–784. 10.1002/sctm.18-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid, S. , Sas, K. M. , Abcouwer, S. F. , Feldman, E. L. , Gardner, T. W. , Pennathur, S. , & Fort, P. E. (2019). New insights into the mechanisms of diabetic complications: Role of lipids and lipid metabolism. Diabetologia, 62(9), 1539–1549. 10.1007/s00125-019-4959-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat, A. , Sidiropoulos, K. , Viteri, G. , Forner, O. , Marin‐Garcia, P. , Arnau, V. , … Hermjakob, H. (2017). Reactome pathway analysis: A high‐performance in‐memory approach. BMC Bioinformatics, 18(1), 142. 10.1186/s12859-017-1559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu, H. , Noma, H. , Mimura, T. , Eguchi, S. , & Hori, S. (2009). Association of vitreous inflammatory factors with diabetic macular edema. Ophthalmology, 116(1), 73–79. 10.1016/j.ophtha.2008.09.037 [DOI] [PubMed] [Google Scholar]
- Grosche, A. , Hauser, A. , Lepper, M. F. , Mayo, R. , von Toerne, C. , Merl‐Pham, J. , & Hauck, S. M. (2016). The proteome of native adult Muller glial cells from murine retina. Molecular & Cellular Proteomics, 15(2), 462–480. 10.1074/mcp.M115.052183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, S. S. , & Busik, J. V. (2017). The role of dyslipidemia in diabetic retinopathy. Vision Research, 139, 228–236. 10.1016/j.visres.2017.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, C. , Simo‐Servat, A. , Bogdanov, P. , & Simo, R. (2017). Diabetic retinopathy: New therapeutic perspectives based on pathogenic mechanisms. Journal of Endocrinological Investigation, 40(9), 925–935. 10.1007/s40618-017-0648-4 [DOI] [PubMed] [Google Scholar]
- Hicks, D. , & Courtois, Y. (1990). The growth and behaviour of rat retinal Muller cells in vitro. 1. An improved method for isolation and culture. Experimental Eye Research, 51(2), 119–129. 10.1016/0014-4835(90)90063-z [DOI] [PubMed] [Google Scholar]
- Hoang, T. , Wang, J. , Boyd, P. , Wang, F. , Santiago, C. , Jiang, L. , … Blackshaw, S. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science, 370(6519), eabb8598. 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii, T. , Murakami, T. , Nishijima, K. , Akagi, T. , Uji, A. , Arakawa, N. , … Yoshimura, N. (2012). Relationship between fluorescein pooling and optical coherence tomographic reflectivity of cystoid spaces in diabetic macular edema. Ophthalmology, 119(5), 1047–1055. 10.1016/j.ophtha.2011.10.030 [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Wang, X. , Hu, B. , Mao, Y. , Chen, Y. , Yan, L. , … Tang, F. (2019). Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single‐cell RNA‐seq analysis. PLoS Biology, 17(7), e3000365. 10.1371/journal.pbio.3000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Da, W. , Sherman, B. T. , & Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jeon, S. , & Lee, W. K. (2014). Effect of intravitreal triamcinolone in diabetic macular edema unresponsive to intravitreal bevacizumab. Retina (Philadelphia, Pa.), 34(8), 1–6. [DOI] [PubMed] [Google Scholar]
- Kashani, A. H. , Green, K. M. , Kwon, J. , Chu, Z. , Zhang, Q. , Wang, R. K. , … Rosenfeld, P. J. (2018). Suspended scattering particles in motion: A novel feature of OCT angiography in exudative Maculopathies. Ophthalmol Retina, 2(7), 694–702. 10.1016/j.oret.2017.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech, A. C. , Mitchell, P. , Summanen, P. A. , O'Day, J. , Davis, T. M. , Moffitt, M. S. , Colman, P. G. , … & Field study investigators (2007). Effect of fenofibrate on the need for laser treatment for diabetic retinopathy (FIELD study): A randomised controlled trial. Lancet, 370(9600), 1687–1697. 10.1016/S0140-6736(07)61607-9 [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12(4), 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korani, M. , Firoozrai, M. , Maleki, J. , Ghahramanpour, F. , Heidari, I. , Fallah, S. , & Seifi, M. (2012). Fatty acid composition of serum lipids in patients with type 2 diabetes. Clinical Laboratory, 58(11–12), 1283–1291. [PubMed] [Google Scholar]
- Lawrence, J. M. , Singhal, S. , Bhatia, B. , Keegan, D. J. , Reh, T. A. , Luthert, P. J. , … Limb, G. A. (2007). MIO‐M1 cells and similar muller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells, 25(8), 2033–2043. 10.1634/stemcells.2006-0724 [DOI] [PubMed] [Google Scholar]
- Lei, Y. , Hu, L. , Yang, G. , Piao, L. , Jin, M. , & Cheng, X. (2017). Dipeptidyl peptidase‐IV inhibition for the treatment of cardiovascular disease ‐ recent insights focusing on angiogenesis and neovascularization. Circulation Journal, 81(6), 770–776. 10.1253/circj.CJ-16-1326 [DOI] [PubMed] [Google Scholar]
- Limb, G. A. , Salt, T. E. , Munro, P. M. , Moss, S. E. , & Khaw, P. T. (2002). In vitro characterization of a spontaneously immortalized human Muller cell line (MIO‐M1). Investigative Ophthalmology & Visual Science, 43(3), 864–869. [PubMed] [Google Scholar]
- Liu, L. , Li, Y. , Guan, C. , Li, K. , Wang, C. , Feng, R. , & Sun, C. (2010). Free fatty acid metabolic profile and biomarkers of isolated post‐challenge diabetes and type 2 diabetes mellitus based on GC‐MS and multivariate statistical analysis. Journal of Chromatography. B, Analytical Technologies in the Biomedical and Life Sciences, 878(28), 2817–2825. 10.1016/j.jchromb.2010.08.035 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski, S. W. , Lo, C. Y. , Sharov, A. A. , Nguyen, Q. , Fang, L. , Hung, S. S. , … Wong, R. C. (2019). A single‐cell transcriptome atlas of the adult human retina. EMBO Journal, 38(18), e100811. 10.15252/embj.2018100811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley, A. K. , Sanfilippo, P. G. , Hewitt, A. W. , Liang, H. , Lamoureux, E. , Wang, J. J. , & Connell, P. P. (2014). Vitreous biomarkers in diabetic retinopathy: A systematic review and meta‐analysis. Journal of Diabetes and its Complications, 28(3), 419–425. 10.1016/j.jdiacomp.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Menon, M. , Mohammadi, S. , Davila‐Velderrain, J. , Goods, B. A. , Cadwell, T. D. , Xing, Y. , … Hafler, B. P. (2019). Single‐cell transcriptomic atlas of the human retina identifies cell types associated with age‐related macular degeneration. Nature Communications, 10(1), 4902. 10.1038/s41467-019-12780-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed, I. N. , Hafez, S. S. , Fairaq, A. , Ergul, A. , Imig, J. D. , & El‐Remessy, A. B. (2014). Thioredoxin‐interacting protein is required for endothelial NLRP3 inflammasome activation and cell death in a rat model of high‐fat diet. Diabetologia, 57(2), 413–423. 10.1007/s00125-013-3101-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, J. I. , Tombran‐Tink, J. , Hykin, P. G. , Gregor, Z. J. , & Cree, I. A. (2006). Vitreous and aqueous concentrations of proangiogenic, antiangiogenic factors and other cytokines in diabetic retinopathy patients with macular edema: Implications for structural differences in macular profiles. Experimental Eye Research, 82(5), 798–806. [DOI] [PubMed] [Google Scholar]
- Patel, P. S. , Sharp, S. J. , Jansen, E. , Luben, R. N. , Khaw, K. T. , Wareham, N. J. , & Forouhi, N. G. (2010). Fatty acids measured in plasma and erythrocyte‐membrane phospholipids and derived by food‐frequency questionnaire and the risk of new‐onset type 2 diabetes: A pilot study in the European Prospective Investigation into Cancer and Nutrition (EPIC)‐Norfolk cohort. The American Journal of Clinical Nutrition, 92(5), 1214–1222. 10.3945/ajcn.2010.29182 [DOI] [PubMed] [Google Scholar]
- Peng, Y. R. , Shekhar, K. , Yan, W. , Herrmann, D. , Sappington, A. , Bryman, G. S. , … Sanes, J. R. (2019). Molecular classification and comparative Taxonomics of Foveal and peripheral cells in primate retina. Cell, 176(5), 1222–1237 e1222. 10.1016/j.cell.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman, S. , Sahel, J. A. , & Goureau, O. (2014). Production of in vitro retina from pluripotent human stem cells: A new therapeutic tool. Medical Science (Paris), 30(10), 845–848. 10.1051/medsci/20143010009 [DOI] [PubMed] [Google Scholar]
- Reichman, S. , Slembrouck, A. , Gagliardi, G. , Chaffiol, A. , Terray, A. , Nanteau, C. , … Goureau, O. (2017). Generation of storable retinal Organoids and retinal pigmented epithelium from adherent human iPS cells in Xeno‐free and feeder‐free conditions. Stem Cells, 35(5), 1176–1188. 10.1002/stem.2586 [DOI] [PubMed] [Google Scholar]
- Reichman, S. , Terray, A. , Slembrouck, A. , Nanteau, C. , Orieux, G. , Habeler, W. , … Goureau, O. (2014). From confluent human iPS cells to self‐forming neural retina and retinal pigmented epithelium. Proceedings of the National Academy of Sciences of the United States of America, 111(23), 8518–8523. 10.1073/pnas.1324212111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch, K. , Jadhav, A. P. , Trimarchi, J. M. , Stadler, M. B. , Roska, B. , Sun, B. B. , & Cepko, C. L. (2008). The transcriptome of retinal Müller glial cells. The Journal of Comparative Neurology, 509(2), 225–238. 10.1002/cne.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubsam, A. , Parikh, S. , & Fort, P. E. (2018). Role of inflammation in diabetic retinopathy. International Journal of Molecular Sciences, 19(4). 942. 10.3390/ijms19040942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden, C. T. , Schindelin, J. , Hiner, M. C. , DeZonia, B. E. , Walter, A. E. , Arena, E. T. , & Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics, 18(1), 529. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rungger‐Brandle, E. , Dosso, A. A. , & Leuenberger, P. M. (2000). Glial reactivity, an early feature of diabetic retinopathy. Investigative Ophthalmology & Visual Science, 41(7), 1971–1980. [PubMed] [Google Scholar]
- Sarthy, P. V. , Fu, M. , & Huang, J. (1991). Developmental expression of the glial fibrillary acidic protein (GFAP) gene in the mouse retina. Cellular and Molecular Neurobiology, 11(6), 623–637. 10.1007/BF00741450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeraro, F. , Morescalchi, F. , Cancarini, A. , Russo, A. , Rezzola, S. , & Costagliola, C. (2019). Diabetic retinopathy, a vascular and inflammatory disease: Therapeutic implications. Diabetes & Metabolism, 45(6), 517–527. 10.1016/j.diabet.2019.04.002 [DOI] [PubMed] [Google Scholar]
- Semple, B. D. , Kossmann, T. , & Morganti‐Kossmann, M. C. (2010). Role of chemokines in CNS health and pathology: A focus on the CCL2/CCR2 and CXCL8/CXCR2 networks. Journal of Cerebral Blood Flow and Metabolism, 30(3), 459–473. 10.1038/jcbfm.2009.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoe, T. , Kuribayashi, H. , Saya, H. , Seiki, M. , Aburatani, H. , & Watanabe, S. (2010). Identification of CD44 as a cell surface marker for Müller glia precursor cells. Journal of Neurochemistry, 115(6), 1633–1642. 10.1111/j.1471-4159.2010.07072.x [DOI] [PubMed] [Google Scholar]
- Slembrouck‐Brec, A. , Nanteau, C. , Sahel, J. A. , Goureau, O. , & Reichman, S. (2018). Defined Xeno‐free and Feeder‐free culture conditions for the generation of human iPSC‐derived retinal cell models. Journal of Visualized Experiments, 139. 10.3791/57795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slembrouck‐Brec, A. , Rodrigues, A. , Rabesandratana, O. , Gagliardi, G. , Nanteau, C. , Fouquet, S. , … Goureau, O. (2019). Reprogramming of adult retinal Muller glial cells into human‐induced pluripotent stem cells as an efficient source of retinal cells. Stem Cells International, 2019, 7858796. 10.1155/2019/7858796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout, S. , Chan‐Ling, T. , Holländer, H. , & Stone, J. (1993). The role of Müller cells in the formation of the blood‐retinal barrier. Neuroscience, 55(1), 291–301. 10.1016/0306-4522(93)90473-s [DOI] [PubMed] [Google Scholar]
- Turner, D. L. , & Cepko, C. L. (1987). A common progenitor for neurons and glia persists in rat retina late in development. Nature, 328(6126), 131–136. 10.1038/328131a0 [DOI] [PubMed] [Google Scholar]
- Voigt, A. P. , Binkley, E. , Flamme‐Wiese, M. J. , Zeng, S. , DeLuca, A. P. , Scheetz, T. E. , … Stone, E. M. (2020). Single‐cell RNA sequencing in human retinal degeneration reveals distinct glial cell populations. Cell, 9(2), 438. 10.3390/cells9020438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, A. P. , Whitmore, S. S. , Flamme‐Wiese, M. J. , Riker, M. J. , Wiley, L. A. , Tucker, B. A. , … Scheetz, T. E. (2019). Molecular characterization of foveal versus peripheral human retina by single‐cell RNA sequencing. Experimental Eye Research, 184, 234–242. 10.1016/j.exer.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X. , Rodrigues, M. , Umapathi, M. , Kashiwabuchi, F. , Ma, T. , Babapoor‐Farrokhran, S. , … Sodhi, A. (2013). Hypoxic retinal Muller cells promote vascular permeability by HIF‐1‐dependent up‐regulation of angiopoietin‐like 4. Proceedings of the National Academy of Sciences of the United States of America, 110(36), E3425–E3434. 10.1073/pnas.1217091110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W. , Peng, Y. R. , van Zyl, T. , Regev, A. , Shekhar, K. , Juric, D. , & Sanes, J. R. (2020). Cell atlas of the human fovea and peripheral retina. Scientific Reports, 10(1), 9802. 10.1038/s41598-020-66092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Cheng, Y. , & Su, G. (2018). A review of the multifunctionality of angiopoietin‐like 4 in eye disease. Bioscience Reports, 38(5). 10.1042/BSR20180557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, A. , Yoshida, S. , Khalil, A. K. , Ishibashi, T. , & Inomata, H. (1998). Role of NF‐kappaB‐mediated interleukin‐8 expression in intraocular neovascularization. Investigative Ophthalmology & Visual Science, 39(7), 1097–1106. [PubMed] [Google Scholar]
- Yoshimura, T. , Sonoda, K.‐H. , Sugahara, M. , Mochizuki, Y. , Enaida, H. , Oshima, Y. , … Ishibashi, T. (2009). Comprehensive analysis of inflammatory immune mediators in vitreoretinal diseases. PLoS One, 4(12), e8158–e8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Wang, C. , Shi, K. , & Yin, X. (2018). Relationship between dyslipidemia and diabetic retinopathy: A systematic review and meta‐analysis. Medicine (Baltimore), 97(36), e12283. 10.1097/md.0000000000012283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Immunofluorescence comparison of fresh and thawed MGCs. Immunochemistry analysis of fresh P2 hiMGC‐1 (left panels), thawed hiMGC‐1 (center panels) and pMGC‐2 (right panels) for the MGC makers VIM and GFAP (top panels), GS and SOX9 (bottom panels). Nuclei from the top panels were stained with DAPI. Scale bar = 50 μm
FIGURE S2 Immunofluorescence comparison of hiMGCs and pMGCs from P2 to P8. Immunofluorescence analysis of hiMGC‐1 (A to G), and pMGC‐2 (H to N) for the MGC makers SOX9, VIM, GS, GFAP and nuclei stained with DAPI over the passages (a and h) P2, (b and i) P3, (c and j) P4, (d and k) P5, (e and l) P6, (f and m) P7 (g and n) P8. Scale bar = 50 μm
FIGURE S3 Comparison of the expression level of neuronal, MGC and microglial markers in hiMGCs, pMGCs, hMacroglia and mMGCs. Heatmap representation of the expression of neuronal, MGC and microglial markers in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). Expression in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847) {. Expression in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S4 Comparison of the expression level of gene with the highest hMacroglia AUC values in hiMGCs, pMGCs, hMacroglia and mMGCs. (a) Heatmap representation of the expression level of the 50 genes with the highest AUC in hMacroglia in the study by Menon et al. in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). (b) Heatmap representation of the expression of 7 classical MGC markers not found in the 50 genes with the highest AUC in hMacroglia. pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel). Expression in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847). Expression level in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S5 Comparison of the expression level of gene with the lowest hMacroglia AUC values in hiMGCs, pMGCs, hMacroglia and mMGCs. (a) Heatmap representation of the expression level of the 50 genes with the lowest hMacroglia AUC score in the study by Menon et al. in pMGCs and hiMGCs (left), hMacroglia (middle), and mMGCs (right panel).
Expression level in pMGCs and hiMGCs are expressed in FPKM (to allow comparison with the Hoang et al study). Average expression in hMacroglia and AUC were determined by Menon et al for the macroglial cell cluster of a human retina and downloaded from public repository (GSE137537, GSE137847). Expression level in FPKM for cell‐sorted MGCs from control mice were determined by Hoang et al and downloaded from public repository (GSE 135406). FPKM: Fragment per Kilobase Million. Heatmap for hiMGCs, pMGCs and mMGCs represent the log2(FPKM+1) values for each transcript. MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs. mMGCS: mouse cell‐sorted MGCs. hMacroglia: macroglial cell cluster of a human retina
FIGURE S6 Pathway distribution of genes differentially expressed within hiMGCs and pMGCs. Schematic tree of cellular pathways repartition where genes are found differentially expressed (green) within pMGCs and hiMGCs, using Reactome database. (a) Pathway in which transcripts (431) are found differentially expressed. (b) Pathway in which transcripts (100) are found differentially expressed with a log2 FC ≥2
FIGURE S7 Expression levels of genes belonging to the extracellular matrix organization GO term in pMGCs and hiMGCs. Graph bar representation of the mean (± SEM) relative expression level of EMILIN1, collagens (COL9A3 and COL9A2), integrin (ITGB4) and cellular adhesion molecules (ICAM1) in P4 hiMGCs derived from three different iPS clones (right panels) and P4 pMGCs isolated from three different donors (left panel). For each of them, the expression expressed is the mean of n = 4 independent points).
MGCs: Muller Glial Cells, pMGCs: primary MGCs, hiMGCs human induced Pluripotent‐derived MGCs
FIGURE S8 hiMGCs and pMGCs responses to DR‐relevant stress. Human iPSC‐derived MGC‐(1 to 3) and pMGC‐(1 to 3) were treated for 18 hr as follow NG (DMEM 5.6 mM glucose, 19.4 mM mannitol, BSA 0.88 w/v, EtOH 0.5 v/v) or HG (DMEM 25 mM glucose, BSA 0.88 w/v, EtOH 0.5% v/v) or NG + PA (DMEM 5.6 mM glucose, 19.4 mM mannitol, BSA 0.88 w/v, EtOH 0,5 v/v) or HG + PA (DMEM 25 mM glucose, BSA 0.88 w/v, PA 500 μM, EtOH 0.5% v/v). Transcript relative expression were analyzed by RT‐qPCR. Graph bar representation of the mean (± SEM) relative expression of ATF3, CXCL8 (IL‐8), CXCL2 and ANGPTL4 in hiMGCs derived from three different iPS clones (left panels) and pMGCs isolated from three different donors (right panel) or iPS clones. For each of them, the expression expressed is the mean of n = 4 independent points)
TABLE S1 RNAseq data: list of non‐differentially expressed genes within hiMGCs and pMGCs
TABLE S2 RNAseq data: list of differentially expressed genes within hiMGCs and pMGCs
TABLE S3 RNAseq data: list of known MGC markers and their expression in hiMGCs and pMGCs
TABLE S4 RNAseq data: list of auto‐immune retinitis or healthy MGC markers and their expression in hiMGCs and pMGCs
TABLE S5 Culture media for hiMGC generation and defined culture conditions for DR modeling
TABLE S6 Primers sequences
Data Availability Statement
All generated data that supports the findings of this study are available in the supplementary material of this article and avalaible for download from public database (GEO submission in process, for review purposes, fastaq files can be downloaded from the following private link: fastaq_download.)


