Abstract
Appearance and taste are important factors in rice (Oryza sativa) grain quality. Here, we investigated the taste scores and related eating‐quality traits of 533 diverse cultivars to assess the relationships between—and genetic basis of—rice taste and eating‐quality. A genome‐wide association study highlighted the Wx gene as the major factor underlying variation in taste and eating quality. Notably, a novel waxy (Wx) allele, Wx la, which combined two mutations from Wx b and Wx in, exhibited a unique phenotype. Reduced GBSSI activity conferred Wx la rice with both a transparent appearance and good eating quality. Haplotype analysis revealed that Wx la was derived from intragenic recombination. In fact, the recombination rate at the Wx locus was estimated to be 3.34 kb/cM, which was about 75‐fold higher than the genome‐wide mean, indicating that intragenic recombination is a major force driving diversity at the Wx locus. Based on our results, we propose a new network for Wx evolution, noting that new Wx alleles could easily be generated by crossing genotypes with different Wx alleles. This study thus provides insights into the evolution of the Wx locus and facilitates molecular breeding for quality in rice.
Keywords: allelic variation, appearance and eating quality, intragenic recombination, Oryza sativa, waxy
Wxla is derived from intragenic recombination of the Wxb and Wxin alleles, and Wxla rice grains have a transparent appearance and excellent eating quality. The study of Wxla provides insight on the evolution of the Wx gene and suggests avenues for improvement of rice grain quality.
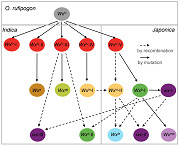
INTRODUCTION
Rice is the staple food for more than one‐third of the world's population. As a result of economic development and improvements in living standards, rice quality has become a major factor in rice production, due to its effect on market value and farmer incomes (Zhang, 2007; Zhang et al., 2008). Scientists in China have proposed the concept of Super Green Rice, with the objective of developing environmentally friendly rice varieties with high yield and good quality (Zhang, 2007). Processed white rice is the main form of rice as a commodity, and appearance and taste are the most important traits demanded by consumers (Zhou et al., 2019). Appearance is determined mainly by grain shape and transparency, whereas eating quality comprises physicochemical properties that include apparent amylose content (AAC), gel consistency (GC), gelatinization temperature (GT), and viscosity. Clarification of the relationships between quality traits and taste and elucidation of their genetic basis are main objectives in meeting the breeding goals for Super Green Rice.
Many studies in rice have aimed to dissect the genetic underpinnings of eating quality. The Waxy (Wx) gene, which encodes granule‐bound starch synthase I (GBSSI) and controls amylose content in the endosperm, is a major determinant of eating quality (Traore et al., 2011). In glutinous (waxy) rice, a 23‐bp repeat in the second exon of Wx causes the premature termination of translation and loss of GBSSI function (Wanchana et al., 2003). Several Wx alleles confer diverse GBSSI activities that affect amylose content (Ayres et al., 1997; Larkin and Park, 2003; Mikami et al., 2008; Zhang et al., 2019). Wx lv was reported to be the ancestral allele of the Wx gene in rice and the three main Wx alleles (Wx a , Wx b , Wx in) differ from Wx lv by substitution of functional residues (Zhang et al., 2019). Gelatinization temperature is mainly controlled by ALK, which encodes a putative soluble starch synthase II (SSIIa) and positively regulates GT (Gao et al., 2003). Most indica varieties have a high GT, caused by enhanced activity of ALK, which increases the contents of medium and long amylopectin chains that are more difficult to gelatinize. In addition to Wx and ALK, several genes that have been identified via mutants are thought to influence eating quality. For example, the flo5 mutant generated from a T‐DNA insertion in SSIIIa, which controls amylopectin chain elongation, shows a degree of polymerization ≥30 and a floury endosperm phenotype with decreased viscosity (Fujita et al., 2007). Moreover, mutation of starch‐branching enzyme I (SBEI) decreases GT, leading to improved eating quality (Satoh et al., 2003).
Despite these studies, the relationships between taste and grain‐quality traits, and the genetic basis of taste, remain unclear. Sensory testing is a direct way to evaluate taste in rice, but it is not practical for use in breeding or research, due to limitations in the numbers of professional tasters and the large amounts of cooked rice required for such tests (Lyon et al., 1999; Limpawattana and Shewfelt, 2010; Kwak et al., 2015). However, a commercially available taste analyzer can accurately predict taste and is suitable for scientific research (Champagne et al., 1996). In this study, we used taste scores (TSs) obtained from a rice taste analyzer (see Methods) to study the relationship between taste and eating‐quality traits in 533 different rice varieties. We performed genome‐wide studies on TS and eating‐quality traits to identify genes that are important for eating quality. Our results provide insights into the genetic bases of rice quality and have new genetic resource for rice breeding.
RESULTS
Phenotyping reveals eating‐quality trait relationships
To clarify the relationship between TS and eating‐quality traits, we investigated AAC, GC, GT, viscosity characteristics, and the TS in 533 different rice cultivars (Figure 1A). The viscosity characteristics obtained from a rapid visco‐analyzer (RVA) included six parameters (Figure S1); namely, peak viscosity (PKV), hot paste viscosity (HPV), breakdown viscosity (BDV), cold paste viscosity (CPV), setback viscosity (SBV), and consistency viscosity (CSV). The TS ranged from 30 (less preferred) to 87.8 (more preferred) in non‐glutinous varieties. The TS was positively correlated with GC, PKV and BDV and negatively correlated with AAC, SBV, and CSV (Figure 1). Among these quality traits, BDV and SBV contributed most to the TS.
Figure 1.
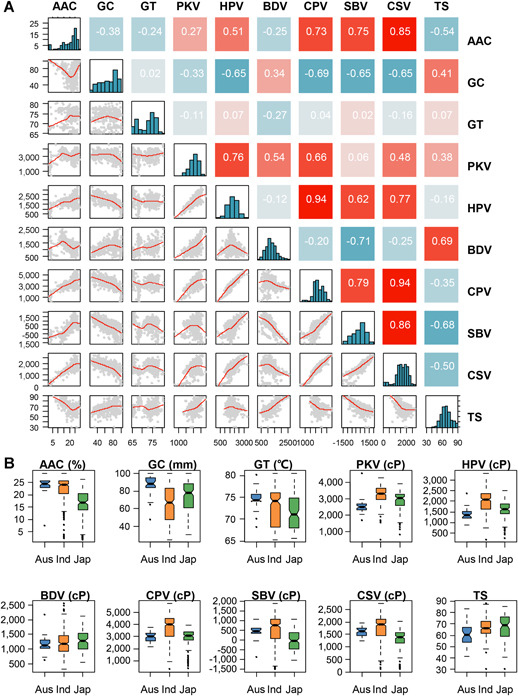
Phenotypic analysis reveals trait relationships and subpopulation characteristics
(A) Heatmap depicting Pearson's correlation coefficients between phenotype means for quality traits across all varieties within the study. Phenotypic distributions of quality traits are located within the diagonal. (B) Phenotypic distributions of quality traits divided by the aus (Aus), indica (Ind), and japonica (Jap) subpopulations. The number of varieties within each subpopulation was 50, 307, and 176, respectively. cP, centipoise.
Subpopulation differences are well known in O. sativa, especially between subspecies indica and japonica (Huang et al., 2012). Eating‐quality traits also showed distinct distributions within subpopulations (Figure 1B). In a previous study, a collection of 533 different cultivated rice accessions was divided into aus, indica and japonica groups, containing 50, 305, and 178 accessions, respectively (Zhou et al., 2017). The indica group had a higher AAC than the japonica group, as a result of different distributions of known Wx alleles (Ayres et al., 1997). The japonica group showed a higher TS than the indica group, and a correspondingly higher BDV and lower SBV. These results demonstrated the diversity among the accessions.
Wx is the main gene that confers eating quality
We performed a genome‐wide association study for nine eating‐quality traits and obtained TSs for the entire group of accessions. The Wx gene was identified as a major gene affecting AAC, GC, all RVA characteristics, and TS, whereas ALK was identified to influence GT (Figure S2; Table S1). Because no strong LD blocks were observed around the Wx locus, the peak SNP on chromosome 6 was identified to be the major functional site underlying the observed variation in Wx (Figure 2A, B). The G‐to‐T SNP in intron 1 divided cultivated rice into high AAC and intermediate AAC genotypes, and this variation explained 60.02% of the phenotypic variance in AAC and 33.63% of the variation in TS (Table S1). A stepwise regression for all Wx variants identified five known functional variants (Table S2). The four major variants, Int1‐1, Ex2‐112, Ex4‐77, and Ex6‐62, explained up to 80.64% of the phenotypic variance in AAC. We then divided the Wx locus into eight alleles according to these six variations (Figure 2C). Different Wx alleles conferred different AAC, GC, TS, appearance, and RVA characteristics (Figure 2C–E).
Figure 2.
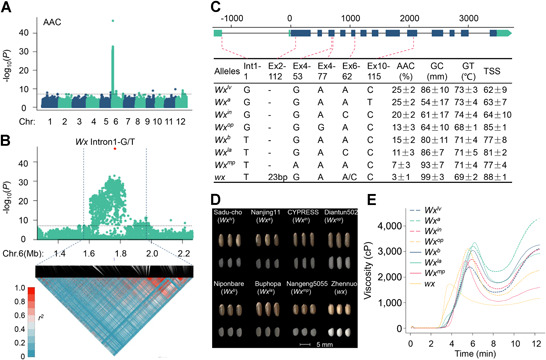
Association study identification of multiple Wx alleles conferring different qualities
(A) Manhattan plot for apparent amylose content (AAC) in the whole population. The dashed line represents the significance threshold (−log10(P) = 7.18). (B) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak on chromosome 6. The red dot indicates the position of nucleotide variation in the candidate gene. (C) Genotypes and phenotypes of different Wx alleles in 533 different rice varieties. (D) The appearance of brown rice and milled rice for representative cultivars carrying different Wx alleles. (E) Representative rapid visco‐analyzer (RVA) pasting properties for different Wx alleles. cP, centipoise.
A new Wx allele, Wx la
Seven out of the eight Wx alleles have been previously described, but one allele displayed a low AAC, a high GC, good RVA characteristics, and an attractive TS. Because this allele exhibited a low AAC and a transparent appearance, we named it Wx la (Figure 2C, D). The Wx b allele contained the Int1‐1 mutation, the Wx in allele contained the Ex6‐62 mutation, and Wx la contained a combination of both mutations. The Wx op and Wx mp alleles also exhibited a low AAC but an opaque appearance (Figure 2D). Single SNP differences existed between any two combinations of Wx b, Wx la and Wx mp, but quality assessments of varieties containing each allele differed. The AAC, GC and TS of Bophopa, which carried the Wx la allele, were intermediate between those of Nipponbare and Nangeng5055, which carried the Wx b and Wx mp alleles, respectively. Previous studies of diverse rice collections indicated that the Int1‐1 and Ex6‐62 variants at the Wx locus were associated with a low AAC (Larkin and Park, 2003; Hoai et al., 2014). However, the effect of this haplotype on rice grain quality was not analyzed.
Wx la exhibits an intermediate genetic effect between that of Wx b and Wx mp
To investigate further the relationship between DNA sequence variation at the Wx locus and eating quality and to understand how the Wx alleles regulate eating quality at the physiological level, the genetic effect of Wx alleles, including quality traits and GBSS activity, required evaluation in the same genetic background. We transformed alleles Wx b, Wx la and Wx mp into glutinous japonica variety Zhennuo (ZN), which carries the nonfunctional Wx allele, wx. These Wx alleles conferred apparently different appearances and eating qualities in the ZN genetic background (Figures 3A–D, 4A–E). ZN‐Wx la produced a transparent grain phenotype similar to that associated with ZN‐Wx b, whereas the grains of ZN‐Wx mp were less transparent (Figure 3A–C). The phenotypic values for quality traits and TS for the isoline with ZN‐Wx la were intermediate between those of isolines with ZN‐Wx b and ZN‐Wx mp (Figure 4A–E), suggesting that the Wx la allele indeed conferred genetic effects intermediate to those of Wx b and Wx mp.
Figure 3.
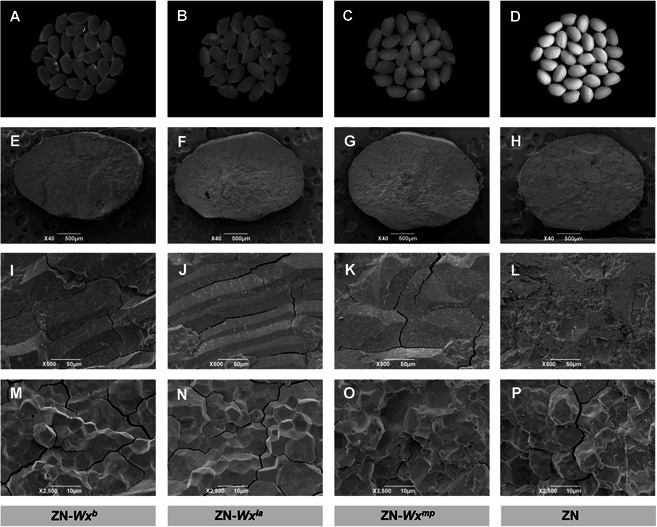
The appearance and morphology of starch granules for different Wx alleles of white rice
(A–D) Milled rice from genotypes ZN‐Wx b (A), ZN‐Wx la (B), ZN‐Wx mp (C) and ZN (D). (E–P) SEM images showing the morphology of the starch granules of ZN‐Wx b (E, I, M), ZN‐Wx la (F, J, N), ZN‐Wx mp (G, K, O) and ZN (H, L, P). Resolution, 40× (E–H), 500X (I–L) and 2,500X (M–P).
Figure 4.
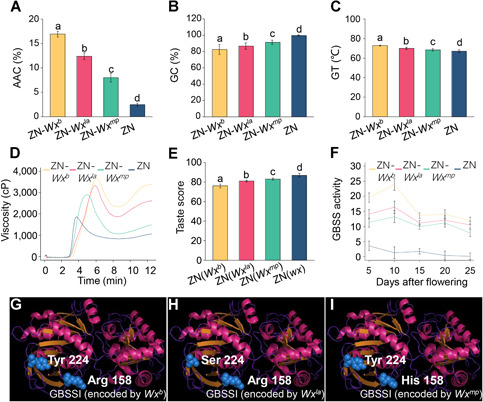
Different GBSSI enzyme activities lead to differences in qualities conferred by Wxb, Wxla and Wxmp
(A–E) The apparent amylose content (AAC) (A), gel consistency (GC) (B), gelatinization temperature (GT) (C), rapid visco‐analyzer (RVA) pasting properties (D) and taste score (E) of Wx b, Wx la and Wx mp in the ZN background. (F) The GBSSI activity of different Wx alleles during endosperm development. (G–I) Three‐dimensional homology modeling of GBSSI encoded by Wx b (G), Wx la (H) and Wx mp (I). ZN is a glutinous rice variety containing the wx allele. Error bars represent the SD of means (n = 5). Different letters above the bars indicate significant differences at P < 0.05, using Tukey's multiple‐comparison test. cP, centipoise.
Reduced GBSSI activity leads to the Wx la phenotype
The Wx b, Wx la and Wx mp alleles all have the Int1‐1 mutation; therefore, this mutation should not affect endosperm in the material in this study. Expression analysis revealed that isolines containing ZN‐Wx b, ZN‐Wx la, and ZN‐Wx mp had similar levels of mature Wx mRNA during seed development (Figure S3). However, the activity of Wx la‐encoded GBSSI was clearly lower than that of Wx b‐encoded GBSSI (Figure 4F), but higher than that of Wx mp‐encoded GBSSI. Wx la and Wx mp each contain a single SNP within the coding sequence compared with Wx b (Figure 2C). The structures of the GBSSI proteins translated from Wx la and Wx mp should be similar, because both possess enzymatic activity. The amino acids produced by the Arg to His mutation in Ex4‐53 did not affect polarity, whereas the Tyr to Ser amino acid substitution caused by the mutation in Ex6‐62 did (Figure 4G–I). Because the effect of the Ex4‐53 mutation was greater than that of the Ex6‐62 mutation, the variation generated by Ex4‐53 should be closer to the active site of the GBSSI protein.
Scanning electron microscopy (SEM) of transverse mature endosperm sections revealed many cavities in the endosperm of ZN‐Wx mp and ZN, compared with only a few cavities in that of ZN‐Wx b and ZN‐Wx la (Figure 3). Therefore, the cavities within the starch granules were probably responsible for the observed differences in the physicochemical properties and transparency between the different genotypes. A reduced AAC is therefore important for the formation of stable, transparent endosperm.
Wx la derives from intragenic recombination
The six mutations in Wx generated more than six alleles and some alleles contained two mutations (Figure 2C). Some alleles were formed by a second mutation within an existing allele, such as wx and Wx mp, which contain additional mutations to Wx b (Wanchana et al., 2003; Yang et al., 2013). However, Wx la contained two existing and widely distributed variations, Int1‐1 and Ex6‐62, and its origin prompted further analysis.
We performed an intensive haplotype analysis of the Wx locus in 4,726 rice accessions (Figures 5A, S4). The Wx alleles were divided into 16 haplotypes based on 52 variations; several alleles consisted of one or more haplotype: a single haplotype in Wx a, Wx op, Wx la, Wx mp, two haplotypes in Wx in Wx b, three haplotypes in wx, and five haplotypes in Wx lv. Among them, Wx lv‐I, Wx lv‐II, Wx lv‐III and Wx lv‐V, correspond to Wx lv‐III, Wx lv‐II, Wx lv‐IV and Wx lv‐I, respectively, in the study of Zhang et al. (2019). Clear indica–japonica differences existed among haplotypes and alleles. Wx la was a rare allele and was present only in japonica and potentially derived from Wx b, Wx in or wx. However, it was not possible to determine the origin of Wx la by analyzing variants within the Wx locus. Therefore, we studied 100‐kb sequences upstream and downstream of the Wx gene to investigate the origin of Wx la. Phylogenetic analysis of the 200‐kb sequences spanning Wx suggested that Wx la was most closely related to Wx in (Figure 5B).
Figure 5.
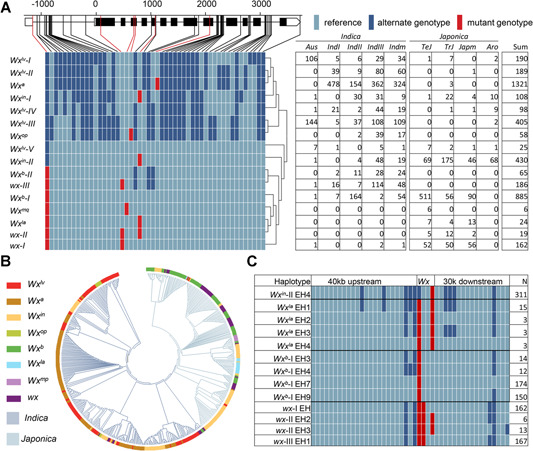
Genetic diversity and evolutionary relationship among multiple Wx alleles in rice
(A) Genotypes and distributions of haplotypes divided by 52 variants within the Wx gene. (B) Phylogenetic relationship of 200‐kb sequences flanking Wx generated from 500 cultivated rice accessions. (C) Extended haplotypes of Wx in, Wx la, Wx b, and wx.
To determine whether Wx la was derived from recombination involving Wx in, we examined the genomic regions flanking Wx in all accessions. In total, 50 extended haplotypes (EH) (Figure S5) were identified, including four extended haplotypes of Wx la (Figure 5C). Among these, Wx la EH1 differed only in Int1‐1 compared with Wx in‐II EH4, and Wx la EH2 differed only in Ex6‐62 compared with Wx b‐I EH3. However, the upstream and downstream regions of Wx la EH3 derived from different haplotypes. The downstream region of Wx la EH3 also derived from Wx in‐II EH4, whereas the upstream region of Wx la EH3 potentially derived from Wx b‐I EH3, Wx b‐II EH, wx‐I EH, wx‐II EH2, wx‐II EH3 or wx‐III EH1. We observed that existing alleles such as Wx op and Wx mp, which were generated by mutation, possessed a single extended haplotype, whereas alleles such as Wx in‐II and wx‐II, which were generated via recombination, possessed more haplotypes. These findings indicated that Wx la was generated from recombination of Wx in and Wx b or wx.
Intragenic recombination is a major driver of genetic diversity in Wx
On the basis of the extended haplotypes of Wx, we speculated on the potential origins of all Wx haplotypes (Figure 6). The Wx lv is the ancestral allele and the five haplotypes Wx lv ‐I to Wx lv ‐V all derived from O. rufipogon. The Wx a, Wx op, Wx in ‐I and Wx b ‐I alleles derived from mutations in Wx lv ‐II, Wx lv ‐III, Wx lv ‐IV and Wx lv ‐V, respectively. The alleles wx‐I and Wx mp were each derived from Wx b by a second mutation; however, the Wx in‐II, Wx b‐II, Wx la, wx‐II and wx‐III haplotypes were generated by intragenic recombination. These five haplotypes increased the genetic diversity of the Wx locus and indicated gene flow between indica and japonica.
Figure 6.
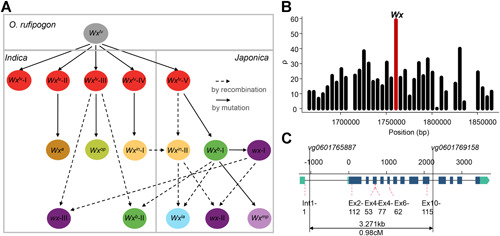
Proposed evolutionary relationships and estimation of the recombination rate at the Wx locus
(A) The evolutionary relationships among Wx alleles and haplotypes in rice. (B) Estimates of population recombination rate (ρ) around the Wx locus in 533 cultivars. (C) Estimates of the intragenic recombination rate at the Wx locus. vg0601765887 and vg0601769158 are flanking markers used to screen for recombinants. The location of six functional variations are marked with red dashed lines.
The Wx locus has a high recombination rate
A previous study reported a high intragenic recombination frequency (27.3 kb/cM) at the Wx locus (Inukai et al., 2000). We estimated the recombination rate around Wx using FastEPRR (Gao et al., 2016) and concluded that the Wx locus represented a recombination hotspot (Figure 6B), which greatly contributed to the genetic diversity of the Wx gene. The genetic distance between Int1‐1 and Ex6‐62 was estimated to be 0.082 cM, indicating that Wx la could easily have resulted from intragenic recombination (Figure S6).
To investigate whether new Wx alleles could be generated by intragenic recombination, we crossed Buphopa (allele Wx la) to Zhenshan97 (allele Wx a). The Wx region flanked by InDel variations vg0601765887 and vg0601769158 (http://ricevarmap.ncpgr.cn/) were screened in recombinants among 5,000 F2 plants from a cross between Buphopa and Zhenshan97. Forty‐nine recombinants were identified between the markers wx‐1765887 and wx‐1769158 (Figure S7; Table S3). Because vg0601765887 and vg0601769158 are only 3.271 kb apart, the recombination rate at the Wx locus was approximately 3.34 kb/cM, which is much higher than that reported in previous studies. All the recombinants could potentially generate new Wx alleles by self‐crossing; therefore, new alleles can be generated by crossing existing accessions containing different alleles.
DISCUSSION
In this study, we clarified the relationship between taste and traditional quality traits in 533 different cultivated rice varieties. An association study demonstrated that Wx was the most important gene that affected taste and eating‐quality traits and a new allele, Wx la, was identified, which conferred good eating quality and grain transparency. Enzyme activity leading to a low amylose content was the basis for the high quality of the cultivar containing Wx la and the Wx la allele derived from intragenic recombination. We concluded that the Wx gene has undergone complex selection during rice breeding, including intragenic recombination following hybridization, natural mutation, and artificial mutagenesis, and we propose a new evolutionary trajectory for the gene.
Preferences regarding rice quality differ among different cultures; consequently, taste and appearance vary tremendously throughout the world (Calingacion et al., 2014). Natural variation in several major genes accounts for much of the phenotypic variation in appearance and eating quality. The GS3 and GW5 genes define grain length and grain width, respectively, whereas ALK defines GT (Nakamura et al., 2005; Takano‐Kai et al., 2009; Liu et al., 2017; Zhou et al., 2017). These genes usually possess only two or three functional alleles; however, the Wx gene has at least 10 different functional alleles (currently, Wx lv, Wx a, Wx in, Wx op, Wx b, Wx la, Wx mq, Wx mp, Wx hp, and wx) (Ayres et al., 1997; Wanchana et al., 2003; Mikami et al., 2008; Liu et al., 2009; Yang et al., 2013; Zhang et al., 2019). The differing and increasing demands for quality has led to the selection of a wide range of mutants of the Wx gene, which generally contain a decreased level of amylose. The wild‐type Wx allele Wx lv first mutated to Wx a in indica and to Wx b in japonica (Zhang et al., 2019). Subsequently, Wx lv mutated to wx in in indica, and Wx b mutated to Wx mq in japonica by artificial mutagenesis. However, a low amylose content does not always lead to better taste; for example, indica varieties with a higher amylose content than japonica are more suitable for making fried rice.
Meiotic recombination is a major driver of genetic diversity, species evolution, and agricultural improvement. Intragenic recombination caused by both crossover and non‐crossover events involving chromatids during meiosis can lead to new alleles or new combinations of existing alleles (Hamant et al., 2006; Mézard et al., 2007). The amount of intragenic recombination in plant genomes varies among species; high recombination frequencies have been observed within genes in maize, but are seldomly observed in Arabidopsis (Yao and Schnable, 2005; Smagulova et al., 2011). Recombination events are not evenly distributed across chromosomes, but tend to occur in recombination hotspots. The mean recombination frequency in the rice genome was estimated at one in 250–300 kb/cM, whereas the recombination frequency within the Wx locus is about 10 times higher than the mean across the genome (Inukai et al., 2000). In this study, six Wx variants (five missense mutations and one nonsense mutation) were identified in a sample of 4,726 varieties. These variations are potentially capable of generating 25 + 1 = 33 different Wx alleles. In addition, it is possible to identify new variants that increase the diversity of Wx and rice taste. The crossing of varieties containing different Wx alleles can generate new alleles through intragenic recombination. After crossing the Wx a genotype (Ex10‐115) with the Wx la genotype (Int1‐1+Ex6‐62), we identified 49 recombinants (Figure S7) that could generate three types of new alleles: Int1‐1+Ex6‐62+Ex10‐115, Int1‐1+Ex10‐115, and Ex6‐62+Ex10‐115. Such new alleles enrich the genetic diversity of the Wx locus and could help to improve rice quality.
MATERIALS AND METHODS
Plant materials and phenotyping
Association mapping was performed on a sample of 533 Oryza sativa accessions. Information concerning the accessions, including names, countries of origin, geographical locations and subpopulation classification was reported in a previous study (Zhou et al., 2017). Approximately 36 seeds of each accession were germinated and transplanted in an experimental field at Wuhan (N30.49°, E114.36°) for 2 years. Harvested grains of the 533 accessions were air‐dried and stored at room temperature for at least 3 months before testing.
Phenotypic analyses of 10 quality traits were conducted over 2 years. The apparent amylose content (AAC) and gel consistency (GC) of milled rice flours were measured following the method of Bao et al. (2006). Gelatinization temperature (GT) was measured by degree of disintegration of milled rice soaked in KOH solution and evaluated using a 1–7 scale proportionate to the amount of disintegration (Gao et al., 2003). Flour pasting properties were assessed using a rapid visco‐analyzer (Perten, RVA4500) according to the method of Bao et al. (2006). For grain transparency analysis, mature seeds of different Wx genotypes were dried in a drying oven (37oC) for 24 h to ensure they had the same moisture content. Taste scores of cooked rice were evaluated using a taste analyzer kit (Satake, STA1B‐RHS1A‐RFDM1A, Japan) that included a taste analyzer, a freshness meter, and a hardness and viscosity analyzer. All procedures were carried out according to the manufacturer's protocol.
Genome‐wide association analyses
Genome‐wide association study (GWAS) analyses of quality traits were separately performed for indica and japonica accessions and on the entire population, using mixed linear models provided in the EMMAX program (Kang et al., 2010) that also accounted for population structure and relative kinship for statistical association purposes. In each panel, only SNPs with a MAF >5% and missing rates <15% were selected for association analyses. Finally, 5.2 million SNPs were used to estimate population structure and kinship coefficients (Zhao et al., 2015b), and for GWAS. Genome‐wide significance thresholds of the GWAS were determined using a modified Bonferroni correction as described (Li et al., 2012), in which the total number of SNPs (M) for threshold calculation was replaced by the effective number of SNPs (Me). The calculated genome‐wide significance thresholds, based on a nominal level of 0.05, were P = 6.6 × 10−8, 8.7 × 10−8, and 2.0 × 10−7 for the whole population, and indica and japonica populations, respectively (Table S1). The physical locations of the SNPs were identified using Rice Annotation version 7.0 of variety Nipponbare from Michigan State University (MSU).
Statistical analyses
Histograms, boxplots, correlations and GWAS analyses were constructed using phenotypic grand means for each variety. The Pearson correlation coefficient P‐values were calculated via a two‐sided t‐test using the cor.test() function in R (Ihaka and Gentleman, 1996). The phenotypic variation in quality traits explained by multiple SNPs was calculated using the lm() and step() functions in R.
Transgene analyses
The 7.1‐kb genomic fragments containing the Wx gene from Niponbare (Wx b), Buphopa (Wx la), Nangeng5055 (Wx mp) were cloned into the pCAMBIA1301S binary vector (Cambia) digested with KpnI and HindIII, to generate transgenic complementation constructs. Constructs were first introduced into E. coli strain Trans 5α and were sequenced to identify correct clones, which were then introduced into Agrobacterium tumefaciens strain EHA105 and transferred into relevant plant material via Agrobacterium‐mediated transformation (Toki, 1997). We used Zhennuo (ZN) as the recipient parent and positive lines were designated ZN‐Wx b, ZN‐Wx la, and ZN‐Wx mp.
Gene expression and measurement of GBSSI activity
Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer`s instructions. Total RNA was pre‐treated with DNaseI (Invitrogen) and about 2 μg total RNA was used to synthesize first‐strand cDNA using oligo (dT)18 primer (Promega). The first‐strand cDNA product was then diluted to a density of 10 ng/μL and 5 μL diluted cDNA was used as a template in a PCR reaction in a total volume of 11 μL. For quantitative real‐time PCR, 5.5 μL of SYBR Green I was added to the reaction mix and PCR was carried out in an ABI QuantStudio6 Flex machine according to the manufacturer's instructions. Melting curves and transcript data were calculated by QuantStudio Real‐Time PCR software. OsActin1 was used as an internal control and the relative expression level was calculated by 2−ΔΔC t. Each experiment was performed with at least three replicates.
Activity of GBSSI was measured using a plant Granule‐Bound Starch Synthase ELISA kit (ml076667, Shanghai Enzyme‐Linked Biotechnology Co., Ltd). Immature grains were freshly harvested at 5, 10, 15, 20, and 25 d after flowering. After removing the glumes, husks and embryos, the endosperms were ground into powder in liquid N2. The powder was placed in a 1.5‐mL centrifuge tube and analyzed according to the manufacturer's instructions.
Scanning electron microscopy
Cross‐sections of milled white rice grains were coated with gold under vacuum. Starch granule morphology was examined with a scanning electron microscope (JSM‐6390LV, JEOL, Japan) at an accelerating voltage of 10 kV, a spot size of 30 nm, and at magnifications of 40×, 400×, and 2,500×. Scanning electron microscopy (SEM) analysis was based on at least three biological replications of mounted specimens. All procedures were carried out according to the manufacturer's protocol.
Haplotype and population genetic analysis
The 4,726 O. sativa accessions used in this study consisted of three groups; the first group of 533 accessions was sequenced in our previous study (Zhao et al., 2015a); the second group containing 950 accessions was sequenced by Huang et al. (2011); and the third group consisting of 3,243 accessions was derived from the 3,000 Rice Genomes Project (3KRGP) (Li et al., 2014). The SNP and InDel variation data for Wx in all 4,726 accessions are available at RiceVarMap (http://ricevarmap.ncpgr.cn/). Subpopulation identities were inferred using ADMIXTURE (Alexander et al., 2009), and were also queried from RiceVarMap. In total, 101 SNPs and 22 InDels were identified in the 5,036 bp sequence of LOC_Os06g04200. Nucleotide diversity (π and θ) was calculated using the DnaSP program (Watterson, 1975; Tajima, 1983; Kumar et al., 2016). Haplotypes were extracted using the same program after removing low‐frequency variations and non‐informative InDels. Extended haplotypes (EH) spanning an 80.2‐kb region flanking Wx were used to distinguish the origins of Wx alleles on the basis of analysis of 4,726 accessions. Haplotypes were constructed from distinct SNPs (frequencies >0.05) identified within the 80‐kb region flanking Wx. The phylogenetic tree for 200‐kb sequences spanning Wx was constructed using the neighbor‐joining method in Mega 7.0 (Kumar et al., 2016).
Estimation of recombination rate
The recombination rates around the Wx locus were calculated using the R package FastEPRR (Gao et al., 2016). Genotypes of SNPs and InDels in 533 varieties were used to estimate the recombination rate with a 5‐kb sliding window. To estimate the intragenic recombination rate, indica accession Zhenshan97 was crossed to japonica accession Buphopa. Zhenshan97 and Buphopa carried the Wx a and Wx la alleles, respectively, at the Wx locus. Markers wx‐1765887 and wx‐1769158 (Table S3) designed from variations vg0601765887 and vg0601769158 (http://ricevarmap.ncpgr.cn) were used to screen for recombinants between vg0601765887 and vg0601769158 among 5,000 F2.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
H.Z. and D.X. conducted most of the experiments, including phenotyping, association analysis, transformation analysis, and population genetic analysis. D.Z., Y.L., P.L., and B.W. conducted parts of the phenotyping; G.W. and X.L. provided rice germplasm samples; G.G., Q.Z., J.X., S.Y., and X.L. participated in field management and logistics. Y.H. designed and supervised the study. Y.H., H.Z., and D.X. analyzed the data and wrote the paper. All authors reviewed the manuscript and approved the final version of the manuscript.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.13011/suppinfo
Figure S1. Rapid visco‐analyzer (RVA) pasting viscosity parameters (left) and RVA pasting properties of 533 different rice accessions (right)
BDV, breakdown viscosity; cP, centipoise; CPV, cold paste viscosity; CSV, consistency viscosity; HPV, hot paste viscosity; PKV, peak viscosity; SBV, setback viscosity.
Figure S2. Manhattan and QQ plots for 10 quality traits among the entire population of 533 rice accessions
Dashed lines represent the significance thresholds (‐log10(P) = 7.18).
Figure S3. Expression level of different Wx alleles in ZN isolines during endosperm development
ZN is a glutinous rice variety containing the wx allele. Plotted values are means ± SD (n = 5).
Figure S4. Distributions and genotypes of Wx haplotypes among 52 variants among different subpopulations
The reference genotype is depicted in light gray, alternative genotypes in dark gray, and mutant genotypes in red.
Figure S5. Table showing extended haplotypes (EH) corresponding to the 80‐kb region flanking the Wx gene (shown as a bar)
The reference genotype is depicted in light blue and alternative genotypes in dark blue.
Figure S6. Estimates of genetic distance among six functional variants of the Wx locus according to Inukai et al. (2000)
Figure S7. The genotypes of recombinants identified from the cross between two accessions carrying Wx a and Wx la alleles, respectively
Table S1. SNPs and candidate genes significantly associated with quality traits
Table S2. Genetic effects of six Wx variants on apparent amylose content (AAC)
Table S3. Primers used in this study
ACKNOWLEDGEMENTS
This work was supported by grants from the National Program on R&D of Transgenic Plants (2016ZX08009004), the Natural Science Foundation of China (91935303), the Ministry of Science and Technology (Grants 2016YFD0100501), the earmarked fund for the China Agriculture Research System (CARS‐01‐03) and the Postdoctoral Science Foundation of China (2017M622477).
Biographies


Zhou, H., Xia, D., Zhao, D., Li, Y., Li, P., Wu, B., Gao, G., Zhang, Q., Wang, G., Xiao, J., Li, X., Yu, S., Lian, X., and He, Y. (2021). The origin of Wx la provides new insights into the improvement of grain quality in rice. J. Integr. Plant Biol. 63: 878–888.
Edited by: Zhizhong Gong, China Agricultural University, China
REFERENCES
- Alexander, D.H. , Novembre, J. , and Lange, K. (2009). Fast model‐based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres, N.M. , McClung, A.M. , Larkin, P.D. , Bligh, H.F.J. , Jones, C.A. , and Park, W.D. (1997). Microsatellites and a single‐nucleotide polymorphism differentiate apparentamylose classes in an extended pedigree of US rice germ plasm. Theor. Appl. Genet. 94: 773–781. [Google Scholar]
- Bao, J. , Shen, S. , Sun, M. , and Corke, H. (2006). Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: Apparent amylose content, pasting viscosity and gel texture. Starch‐Stärke 58: 259–267. [Google Scholar]
- Calingacion, M. , Laborte, A. , Nelson, A. , Resurreccion, A. , Concepcion, J.C. , Daygon, V.D. , Mumm, R. , Reinke, R. , Dipti, S. , and Bassinello, P.Z. (2014). Diversity of global rice markets and the science required for consumer‐targeted rice breeding. PLoS One 9: e85106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne, E.T. , Richard, O.A. , Bett, K.L. , Grimm, C.C. , Vinyard, B.T. , Webb, B.D. , McClung, A.M. , Barton, F. , Lyon, B.G. , and Moldenhauer, K. (1996). Quality evaluation of U.S. medium‐grain rice using a Japanese taste analyzer. Cereal Chem. 73: 290–294. [Google Scholar]
- Fujita, N. , Yoshida, M. , Kondo, T. , Saito, K. , Utsumi, Y. , Tokunaga, T. , Nishi, A. , Satoh, H. , Park, J.H. , Jane, J.L. , Miyao, A. , Hirochika, H. , and Nakamura, Y. (2007). Characterization of SSIIIa‐deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144: 2009–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Ming, C. , Hu, W. , and Li, H. (2016). New software for the fast estimation of population recombination rates (FastEPRR) in the genomic era. G3 (Bethesda) 6: 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z. , Zeng, D. , Cui, X. , Zhou, Y. , Yan, M. , Huang, D. , Li, J. , and Qian, Q. (2003). Map‐based cloning of the ALK . gene, which controls the gelatinization temperature of rice. Sci. China Series C: Life Sci. 46: 661–668. [DOI] [PubMed] [Google Scholar]
- Hamant, O. , Ma, H. , and Cande, W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302. [DOI] [PubMed] [Google Scholar]
- Hoai, T.T.T. , Matsusaka, H. , Toyosawa, Y. , Suu, T.D. , Satoh, H. , and Kumamaru, T. (2014). Influence of single‐nucleotide polymorphisms in the gene encoding granule‐bound starch synthase I on amylose content in Vietnamese rice cultivars. Breed. Sci. 64: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Kurata, N. , Wang, Z.X. , Wang, A. , Zhao, Q. , Zhao, Y. , Liu, K. , Lu, H. , Li, W. , and Guo, Y. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Zhao, Y. , Wei, X. , Li, C. , Wang, A. , Zhao, Q. , Li, W. , Guo, Y. , Deng, L. , Zhu, C. , Fan, D. , Lu, Y. , Weng, Q. , Liu, K. , Zhou, T. , Jing, Y. , Si, L. , Dong, G. , Huang, T. , Lu, T. , Feng, Q. , Qian, Q. , Li, J. , and Han, B. (2011). Genome‐wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44: 32–39. [DOI] [PubMed] [Google Scholar]
- Ihaka, R. , and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5: 299–314. [Google Scholar]
- Inukai, T. , Sako, A. , Hirano, H.Y. , and Sano, Y. (2000). Analysis of intragenic recombination at wx in rice: Correlation between the molecular and genetic maps within the locus. Genome 43: 589–596. [DOI] [PubMed] [Google Scholar]
- Kang, H.M. , Sul, J.H. , Service, S.K. , Zaitlen, N.A. , Kong, S.Y. , Freimer, N.B. , Sabatti, C. , and Eskin, E. (2010). Variance component model to account for sample structure in genome‐wide association studies. Nat. Genet. 42: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, H.S. , Kim, M. , Lee, Y. , and Jeong, Y. (2015). Identification of key sensory attributes for consumer acceptance and instrumental quality of aseptic‐packaged cooked rice. Int. J. Food Sci. Technol. 50: 691–699. [Google Scholar]
- Larkin, P.D. , and Park, W.D. (2003). Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol. Breed. 12: 335–339. [Google Scholar]
- Li, J.Y. , Wang, J. , and Zeigler, R.S. (2014). The 3,000 rice genomes project: New opportunities and challenges for future rice research. GigaScience 3: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.X. , Yeung, J.M. , Cherny, S.S. , and Sham, P.C. (2012). Evaluating the effective numbers of independent tests and significant p‐value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpawattana, M. , and Shewfelt, R. (2010). Flavor lexicon for sensory descriptive profiling of different rice types. J. Food Sci. 75: S199–S205. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Chen, J. , Zheng, X. , Wu, F. , Lin, Q. , Heng, Y. , Tian, P. , Cheng, Z. , Yu, X. , and Zhou, K. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3: 17043. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Ma, X. , Liu, S. , Zhu, C. , Jiang, L. , Wang, Y. , Shen, Y. , Ren, Y. , Dong, H. , Chen, L. , Liu, X. , Zhao, Z. , Zhai, H. , and Wan, J. (2009). Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71: 609–626. [DOI] [PubMed] [Google Scholar]
- Lyon, B.G. , Champagne, E.T. , Vinyard, B.T. , Windham, W.R. , Barton, F.E. , Webb, B.D. , McClung, A.M. , Moldenhauer, K.A. , Linscombe, S. , and McKenzie, K.S. (1999). Effects of degree of milling, drying condition, and final moisture content on sensory texture of cooked rice. Cereal Chem. 76: 56–62. [Google Scholar]
- Mézard, C. , Vignard, J. , Drouaud, J. , and Mercier, R. (2007). The road to crossovers: Plants have their say. Trends Genet. 23: 91–99. [DOI] [PubMed] [Google Scholar]
- Mikami, I. , Uwatoko, N. , Ikeda, Y. , Yamaguchi, J. , Hirano, H.Y. , Suzuki, Y. , and Sano, Y. (2008). Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 116: 979–989. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y. , Francisco, P.B. , Hosaka, Y. , Sato, A. , Sawada, T. , Kubo, A. , and Fujita, N. (2005). Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 58: 213–227. [DOI] [PubMed] [Google Scholar]
- Satoh, H. , Nishi, A. , Yamashita, K. , Takemoto, Y. , Tanaka, Y. , Hosaka, Y. , Sakurai, A. , Fujita, N. , and Nakamura, Y. (2003). Starch‐branching enzyme I‐deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova, F. , Gregoretti, I.V. , Brick, K. , Khil, P. , Camerini‐Otero, R.D. , and Petukhova, G.V. (2011). Genome‐wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano‐Kai, N. , Jiang, H. , Kubo, T. , Sweeney, M. , Matsumoto, T. , Kanamori, H. , Padhukasahasram, B. , Bustamante, C. , Yoshimura, A. , and Doi, K. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 182: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki, S. (1997). Rapid and efficient Agrobacterium‐mediated transformation in rice. Plant Mol. Biol. Rep. 15: 16–21. [Google Scholar]
- Traore, K. , McClung, A.M. , Chen, M.H. , and Fjellstrom, R. (2011). Inheritance of flour paste viscosity is associated with a rice Waxy gene exon 10 SNP marker. J. Cereal Sci. 53: 37–44. [Google Scholar]
- Wanchana, S. , Toojinda, T. , Tragoonrung, S. , and Vanavichit, A. (2003). Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci. 165: 1193–1199. [Google Scholar]
- Watterson, G.A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Wang, J. , Fan, F.J. , Zhu, J.Y. , Chen, T. , Wang, C.L. , Zheng, T.Q. , Zhang, J. , Zhong, W.G. , and Xu, J.L. (2013). Development of AS‐PCR marker based on a key mutation confirmed by resequencing of Wx‐mp in Milky Princess and its application in japonica soft rice (Oryza sativa L.) breeding. Plant Breed. 132: 595–603. [Google Scholar]
- Yao, H. , and Schnable, P.S. (2005). Cis‐effects on meiotic recombination across distinct a1‐sh2 intervals in a common Zea genetic background. Genetics 170: 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Zhu, J. , Chen, S. , Fan, X. , Li, Q. , Lu, Y. , Wang, M. , Yu, H. , Yi, C. , Tang, S. , Gu, M. , and Liu, Q. (2019). Wxlv, the ancestral allele of rice waxy gene. Mol. Plant 12: 1157–1166. [DOI] [PubMed] [Google Scholar]
- Zhang, Q. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA 104: 16402–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Zhang, S. , Yang, J. , and Zhang, J. (2008). Yield, grain quality and water use efficiency of rice under non‐flooded mulching cultivation. Field Crops Res. 108: 71–81. [Google Scholar]
- Zhao, H. , Yao, W. , Ouyang, Y. , Yang, W. , Wang, G. , Lian, X. , Xing, Y. , Chen, L. , and Xie, W. (2015a). RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 43: D1018–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Yao, W. , Ouyang, Y. , Yang, W. , Wang, G. , Lian, X. , Xing, Y. , Chen, L. , and Xie, W. (2015b). RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 43: D1018–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Li, P. , Xie, W. , Hussain, S. , Li, Y. , Xia, D. , Zhao, H. , Sun, S. , Chen, J. , and Ye, H. (2017). Genome‐wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol. Plant 10: 634–644. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Xia, D. , and He, Y. (2019). Rice grain quality—traditional traits for high quality rice and health‐plus substances. Mol. Breed. 40: 1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article: http://onlinelibrary.wiley.com/doi/10.1111/jipb.13011/suppinfo
Figure S1. Rapid visco‐analyzer (RVA) pasting viscosity parameters (left) and RVA pasting properties of 533 different rice accessions (right)
BDV, breakdown viscosity; cP, centipoise; CPV, cold paste viscosity; CSV, consistency viscosity; HPV, hot paste viscosity; PKV, peak viscosity; SBV, setback viscosity.
Figure S2. Manhattan and QQ plots for 10 quality traits among the entire population of 533 rice accessions
Dashed lines represent the significance thresholds (‐log10(P) = 7.18).
Figure S3. Expression level of different Wx alleles in ZN isolines during endosperm development
ZN is a glutinous rice variety containing the wx allele. Plotted values are means ± SD (n = 5).
Figure S4. Distributions and genotypes of Wx haplotypes among 52 variants among different subpopulations
The reference genotype is depicted in light gray, alternative genotypes in dark gray, and mutant genotypes in red.
Figure S5. Table showing extended haplotypes (EH) corresponding to the 80‐kb region flanking the Wx gene (shown as a bar)
The reference genotype is depicted in light blue and alternative genotypes in dark blue.
Figure S6. Estimates of genetic distance among six functional variants of the Wx locus according to Inukai et al. (2000)
Figure S7. The genotypes of recombinants identified from the cross between two accessions carrying Wx a and Wx la alleles, respectively
Table S1. SNPs and candidate genes significantly associated with quality traits
Table S2. Genetic effects of six Wx variants on apparent amylose content (AAC)
Table S3. Primers used in this study


