Abstract
Tuberculosis is a significant health problem without an effective vaccine to combat it. A thorough understanding of the immune response and correlates of protection is needed to develop a more efficient vaccine. The immune response against Mycobacterium tuberculosis (Mtb) is complex and involves all aspects of the immune system, however, the optimal protective, non‐pathogenic T cell response against Mtb is still elusive. This review will focus on discussing CD4 T cell immunity against mycobacteria and its importance in Mtb infection with a primary focus on human studies. We will in particular discuss the large heterogeneity of immune cell subsets that have been revealed by recent immunological investigations at an unprecedented level of detail. These studies have identified specific classical CD4 T cell subsets important for immune responses against Mtb in various states of infection. We further discuss the functional attributes that have been linked to the various subsets such as upregulation of activation markers and cytokine production. Another important topic to be considered is the antigenic targets of Mtb‐specific immune responses, and how antigen reactivity is influenced by both disease state and environmental exposure(s). These are key points for both vaccines and immune diagnostics development. Ultimately, these factors are holistically considered in the definition and investigations of what are the correlates on protection and resolution of disease.
Keywords: CD4 T cells, Mtb‐specific T cells, mycobacteria, TB
1. INTRODUCTION
The goal of the End TB Strategy is to reduce TB deaths by 90% and TB incidence of new cases per year by 80% by year 2030, compared with 2015. 1 The ambitious goal of a fast deceleration of disease incidence could be achieved by a multipronged approach including increases in access to TB medical care, addressing socioeconomic factors, as well as research and technological breakthroughs especially in diagnostics, therapeutics, and vaccines.
Despite significant worldwide control efforts over the last 20 years, the progress toward elimination of tuberculosis has slowed. The World Health Organization (WHO) estimates that approximately one‐quarter of the world's population (1.7 billion total) is infected with Mtb. Mtb is responsible for 1.3 million deaths among HIV‐negative individuals annually and ~ 10 million new infections are reported each year. 2 In 2019, tuberculosis was the ninth leading cause of death worldwide and the leading cause from a single infectious agent, ranking above HIV/AIDS. 2 The severity of this situation is compounded by the fact that many cases in low‐income and middle‐income countries go undiagnosed and thus untreated. Additionally, with the COVID‐19 pandemic in 2020/21, tuberculosis management has been neglected; patients have discontinued their treatment due to lockdowns, new cases are not visiting clinics despite symptoms, and co‐infection with SARS‐CoV‐2 may lead to increased mortality. 3 Thus, it is expected that the number of tuberculosis cases will rise further in the coming years.
2. THE SPECTRUM OF MTB INFECTIONS
Mtb infections are traditionally classified into active TB (ATB) infection or a quiescent/latent state (LTBI). ATB is typically defined as the presence of symptoms and/or Mtb smear/culture positivity. However, it is now well accepted that Mtb infection should be seen as a continuous spectrum with high heterogeneity and no clear segregation between the LTBI and ATB group. 4 , 5 , 6
The distinction between ATB and LTBI is often made based on presence of symptoms and culture positivity for simplicity in clinical and research settings. Typically, LTBI will have a positive tuberculin skin test (TST) and/or Interferon Gamma Release assay (IGRA), but this may also be true for individuals who have eliminated their infection. Therefore, the IGRA+ group contains a spectrum of individuals from those who have cleared their infection to individuals with subclinical TB disease, and not all within this group have the same likelihood of developing active disease. 4 Moreover, the TST and IGRA tests have limited usefulness in areas with high TB burden and TB endemic areas. LTBI individuals with a more recent infection, or with presence of co‐morbidity factors such as HIV, or diabetes are at higher risk of developing active disease. In addition to co‐morbidities and time since infection, severity of ATB is also dependent on additional factors, such as the infecting Mtb strain and how far the infection has progressed. In this review, we compare Mtb‐specific classical CD4 T cell immune responses in LTBI (usually defined as IGRA+) vs ATB (individuals with symptoms) as this is commonplace in the scientific literature. However, we realize that improved diagnostics and molecular tools to allow more granularity on the disease spectrum will allow more in‐depth studies in the future and the characterization of immune correlates of protection that can more closely cover this spectrum.
3. THE NEED FOR IMPROVED VACCINATION AND IMMUNODIAGNOSTICS
The majority of infected individuals control the pathogen by mounting a successful, long‐lived, and protective immune response, leading to either elimination of the bacteria or a persistent latent infection which is not associated with significant clinical symptoms. However, approximately 10% of latently infected individuals eventually develop active disease. 7 , 8 The risk of developing ATB is higher in individuals that are immunocompromised (due to age, corticosteroid use, malnutrition, and HIV infection). The lengthy treatment is expensive and requires a combination of multiple antibiotics.
In many parts of the world, access to these drugs is limited and compliance with the drug regime is often poor, thus favoring the development of drug resistant strains. The development of drug resistant strains of Mtb pose a threat to global health and the success of the End TB Strategy. Worldwide, 4.1% of new cases and 19% of previously treated cases are infected with rifampicin‐resistant (RR‐TB) or multidrug‐resistant TB (MDR‐TB). 2 About 6.2% of these MDR‐TB cases are classified as extensively drug‐resistant TB (XDR‐TB), which has been identified in 123 WHO member states. 2 The prevalence of MDR cases both complicates the schedule and increases cost of treatment. Most importantly, the existence of antibiotic resistant strains emphasizes the need for development of a vaccine solution to curb their spread.
The vaccination of children with M. bovis BCG results in a 60%‐80% decrease in the incidence of active TB. However, in most developed countries BCG vaccination is not recommended due to the relatively low incidence of disease and variable effectiveness in preventing first time pulmonary TB in adults, a large fraction of active disease cases. 2 A new vaccine against TB is required, preferably targeting adolescents and adults who represent the vast majority of new cases 1 , 2 , 9 and are responsible for spreading Mtb infection. Ideally, a vaccine should be protective irrespective of Mtb infection status, that is, both in individuals with and without evidence of latent infection, and prevent progression to active disease, reinfection, and reactivation. A recent prevention of infection trial using BCG and a subunit TB vaccine candidate provided encouraging results showing reduced rates of sustained QuantiFERON conversions. 10
As a complimentary approach to developing a new vaccine, advanced diagnostic tools could theoretically identify individuals at high risk of developing active TB disease through systematic screening. The high‐risk individuals could then be treated before they become infectious, which would also contribute to a reduction of TB cases. Immunodiagnostic tests for TB rely on the detection of an immune response against mycobacterial antigens, either by delayed hypersensitivity reaction (Tuberculin skin test; TST), or by detection of IFNγ following in vitro stimulation (IGRA). A TST can produce a false‐positive result due to prior BCG vaccination, and may produce a false‐negative result due to other factors such as immunosuppression or malnutrition. 11
In summary, new vaccines and immunodiagnostics could provide a quantum leap in the fight against TB. However, to accomplish these goals a precise understanding of the characteristics of immune responses and their impact on disease progression and susceptibility is required. The rest of this review will focus on these issues.
4. CD4 T CELLS AND THEIR IMPORTANCE IN MTB INFECTION
Human T cell responses to Mtb involve classically restricted CD4 and CD8 αβ T cells, 12 , 13 and non‐classically restricted T cells such as NKT (CD1), MAIT (MR1) and γδ T cells. 14 , 15 , 16 Depletion of CD4 T cells demonstrated that while CD8+ T cells and other immune cells also play a protective role against Mtb, they alone cannot compensate for the lack of a dominant CD4 T cell response. 17 The importance of CD4 T cells in the defense against Mtb is also supported by the fact that patients with HIV infection (which leads to reduced CD4 T cell counts) are more susceptible to primary Mtb infection, reinfection, and reactivation. 18
CD4 T cells primarily act by secreting a variety of cytokines that attract other immune cells to the site of infection and initiate the differentiation of different CD4 T cell subsets capable of performing effector functions. The ability of T cells to recognize Mtb‐infected antigen presenting cells is a key step in containing the infection. Srivastava et al was able to demonstrate using mouse models that direct recognition of Mtb‐infected cells by CD4 T cells is required for control of the infection. 19
IFNγ +production by CD4 T cells is commonly associated with control of Mtb infection. 8 , 20 , 21 , 22 , 23 The essential role of IFNγin the protective immunity to mycobacteria is made apparent in individuals with genetic defects in the IFNγ receptor, who have an increased susceptibility to infection with mycobacteria. 24 However, several reports demonstrated other anti‐tuberculosis CD4 T cell effector functions not accounted for by IFNγ production. The likelihood of developing ATB does not correlate with either the amount of produced IFNγ or the pattern of co‐production with other cytokines. 25 Accordingly, the focus of our review is on classical CD4 T cells, since they represent a major component of the T cell response against Mtb. We consider both IFNγ production, as well as other effector and phenotypic functions associated withCD4T cells and control of Mtb infection (Figure 1, Table 1).
FIGURE 1.
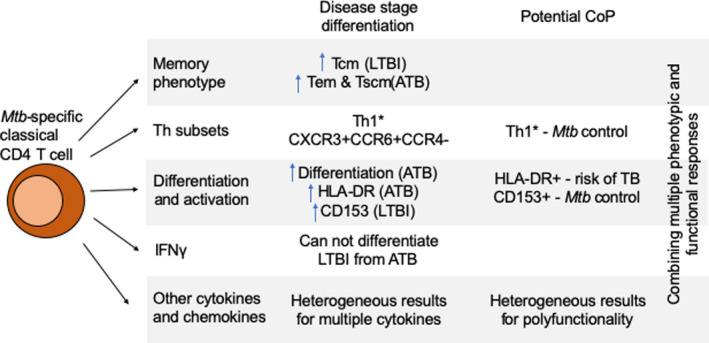
Summary of characteristics of Mtb‐specific classical CD4 T cells for differentiation of disease stages and potential correlates of protection. Mtb‐specific classical CD4 T cells express different characteristics depending on the disease stage of the individual. There is a higher frequency of Tcm, Th1* and CD153 in LTBI. ATB has higher frequency of Tem and Tscm, HLA‐DR expression and increased differentiation. Th1* and CD153 are important for control of Mtb infection. Other cytokines and chemokines include: IL‐2, IL‐10, IL‐17, TNFα, and CXCL9/10/11/12/13
TABLE 1.
Differential CD4 T cell phenotypes observed in major stages of Mtb infection (LTBI, ATB, and severe ATB), and following BCG vaccination
| Mtb‐specific CD4 T cell phenotype | Infection stage | |||
|---|---|---|---|---|
| Vaccinated | LTBI | ATB | Severe ATB | |
| Memory phenotype | ||||
| Tcm (CCR7+CD45RA−) | Humans ↑ Tcm phenotype in vaccinated newborns 53 |
Humans ↑ Tcm phenotype compared to ATB 52 |
||
| Tem (CCR7‐CD45RA‐) |
Humans |
|||
| Tscm (CD45RA+CCR7+CD27+) |
Humans ↑ Tscm compared to LTBI 55 |
|||
| Th subset | ||||
|
Th1*‐Th1/Th17 cells (CXCR3+CCR6+CCR4−) TFs: RORC, Tbet |
Humans Re‐vaccination with BCG boosted reactivation of “polyfunctional Th1/Th17 lymphocytes” 69 NHPs |
Humans |
NHPs ↑ CD4 T cells expressing a “hybrid Th1/Th17 immune response” detected using single‐cell RNA‐sequencing in TB granulomas of NHPs infected with Mtb 75 |
|
| Differentiation markers | ||||
| Markers Associated With Less Differentiated Phenotype | ||||
| CD27 |
Humans |
|||
| CD127 |
Humans ↑ CD127 in Mtb‐specific CD4 T‐cells compared to ATB 52 |
|||
| Activation markers | ||||
| CD153 (CD30L) |
Humans ↑ CD153 Mtb‐specific CD4+ T cells compared to ATB 39 , 41 Mice ↑ CD153 expression on Mtb‐specific Th1 cells in the lung parenchyma of Mtb‐infected mice 41 , 96 NHPs ↑ CD153 expression on Ag‐specific CD4 T cells in the airways compared to blood in Mtb‐infected NHPs that inversely correlates with granuloma bacterial load 38 |
Mice CD153 deficient mice develop high pulmonary bacterial loads and die early after Mtb infection 41 |
||
| HLA‐DR |
Humans ↑ HLA‐DR in Mtb‐specific CD4 T cells compared to LTBI 88 , 92 , 93 , 94 , 95 |
|||
| CD38 |
Humans ↑ CD38in Mtb‐specific CD4 T cells compared to LTBI 84 , 88 , 89 , 97 , 98 |
|||
| Ki67 |
Humans ↑ Ki67in Mtb‐specific CD4 T cells compared to LTBI 88 , 89 , 97 , 98 |
|||
| PD‐1 |
Humans ↑ PD‐1 in Mtb‐specific CD4 T cells compared to LTBI 87 |
|||
| Cell adhesion molecules | ||||
| CD62L (L‐selectin) |
Humans Mtb‐specific CD4T cells were confined to the GPA33‐CD62L‐ Th1* subset in IGRA+ individuals 42 |
Humans Mtb‐specific CD4+ T cells from tubercular pleural fluid were effector/memory cells with high CD45RO expression, but low CD62L, CCR7, and CD27 expression 74 |
||
| Chemokines and their receptors | ||||
| CXCR3 |
Mice ↑ CXCR3 on KLGR1‐ Mtb‐specific T cells derived from the lung parenchyma (compared to lung vasculature) of vaccinated mice challenged with Mtb. Adoptive transfer of these parenchymal T cells resulted in greater control of infection compared to more terminally differentiated KLGR1+ T cells localized to the lung vasculature 200 , 201 , 202 , 203 , 204 |
Humans ↑ CXCR3 on Mtb‐tetramer+ CD4 T cells in LTBI compared to ATB 68 |
||
| IP‐10 |
Humans ↑ IP‐10 levels in Mtb‐infected individuals (both LTBI and ATB) compared to uninfected individuals 136 . IP‐10 expression may be increased in ATB compared to LTBI 137 , but its ability to distinguish ATB from LTBI is controversial 139 . |
|||
| Cytokines | ||||
| IFNγ |
Humans ↑ IFNγ response in Mtb‐infected individuals compared to uninfected individuals, but IFNγ expression cannot differentiate LTBI from ATB |
|||
| IL‐17 |
Humans ↑ IFNγ+IL‐17+ CD4 T cells in severe ATB compared to less severe disease 119 |
|||
| TNFɑ | Complex role of TNFα in the immune response against Mtb | |||
|
Humans ↑ single‐positive TNFɑ Mtb‐specific CD4 T cells in ATB compared to LTBI 122 |
||||
| IL‐10 |
Humans ↑ IL‐10 in lungs and serum of individuals with active pulmonary TB 120 , 129 , 132 |
|||
|
Polyfunctional cytokine responses Dual or Triple‐producing (INFγ,IL‐2,TNFɑ) cells |
Reports on polyfunctional responses conflicting 17 | |||
|
Humans ↑ of double and triple producing T cells in both children and adults post BCG‐vaccination, but mainly children 41 NHPs ↑ in CD4 T cells dually producing IFNγ and TNFα following intravenous administration of BCG that were associated with reduced disease pathology 194 |
Humans ↑ IL‐2/IFNγ ratio after long‐term stimulation with PPD differentiates LTBI from ATB 106 , 107 , 142 |
Humans ↑ frequency of triple producing T cells compared to LTBI 57 , 120 , 122 , 142 , 143 , 196 , 197 , 198 |
||
Generally, a marker is included under a specific stage of Mtb infection if it has been described as increased in that stage relative to the other disease stages. Differentially expressed markers that may serve as potential correlate of protection are italicized in red; Dx markers that can distinguish LTBI from ATB are italicized in blue.
5. STRATEGIES TO DEFINE THE PHENOTYPE OF MTB‐SPECIFIC CD4 T CELLS
Flow cytometry is by far the most commonly used technique to interrogate the phenotype of Mtb‐specific CD4 T cells. There are several strategies to identify Mtb‐specific CD4 T cells from bulk CD4 T cells. The most common strategy is to stimulate cells in vitro with Mtb‐derived reagents: either whole preparations (eg, Mtb lysate, PPD), or peptides (such as the ones used for the IGRA assay targeted against the two proteins ESAT‐6 and CFP10, or “megapools”, see below). The advantage of peptides is that they are defined synthetic reagents with little variation across batches and thus generate results highly consistent across experiments. They can also be selected based on their MHC binding to target either CD4 or CD8 T cells. 26 , 27 However, they are usually limited to only a few proteins or epitopes of interest. To overcome this limitation, our group has designed a peptide pool that combine 300 primarily MHC class II restricted epitopes that represent more than 80 Mtb proteins (see below).
Regardless of the stimuli used, surrogate markers of antigen‐specificity are needed to identify the cells with antigen‐specific reactivity following stimulation. For Mtb‐specific T cells, the most commonly used measurement is IFNγ. However, as discussed more below, not all Mtb‐specific CD4 T cells express IFNγ. Many more cytokines are produced such as IL‐2, TNF, and IP‐10, as well as other cellular changes occurring in response to Mtb stimuli. To overcome the hurdle of having to specifically select which cytokines to measure, there are assays that measure the expression of surface proteins that are specifically induced upon T cell activation. 28 These surface proteins, typically called activation‐induced markers, encompasses TNF family receptors OX40, CD137, and CD154, as well as CD69 and PD‐L1.
Another strategy to characterize the phenotype of antigen‐specific T cells is to use multimeric staining reagents (eg, MHC tetramers), which require precise knowledge of the epitopes’ HLA restriction and HLA expression of the subjects. Multimers captures all T cells capable of binding a given epitope:MHC combination, thus only work in subjects that express the specific MHC allele, and recognize the specific epitope. 29 Therefore, multimers are ideally suited for in‐depth characterization of a small representative set of epitope specificities. In contrast, stimulation with peptide pools and measurement of cytokines or upregulation of activation markers are possible in most subjects, albeit with some limitations. Mtb‐specific T cells that do not express the chosen markers escape detection. In addition, bystander activation can lead to T cells being captured that do not directly react to the epitopes.
6. EPITOPE MEGAPOOLS AS A UNIVERSAL TOOL FOR MEASURING CD4 T CELL RESPONSES
It is often possible to assess T cell responses directly ex vivo by using pools of different epitopes or peptides, so that the overall frequency of responding cells is enhanced. 30 , 31 , 32 , 33 , 34 This approach is particularly key to analyze small sample volumes. This “megapool” approach is based on large numbers of peptides pooled and formulated using sequential lyophilization. 35 Specifically, for detection of Mtb‐specific responses we have described and validated a comprehensive megapool of 300 Mtb epitopes representing more than 80 Mtb proteins, 29 derived from a proteome‐wide screen for epitopes and antigens recognized by IGRA+ individuals. 36 This original pool, named “MTB300”, and versions thereof have been used by a number of studies to measure and phenotype Mtb‐specific responses. 28 , 50 Due to the overlap of epitopes recognized by different species MHC this pool has also been shown to capture T cell reactivity in mice and non‐human primates. 38 , 41 , 44 , 49 , 50
These studies have contributed to our understanding of immunity against Mtb and other mycobacteria. For example, peptide megapools contributed to the identification of peptide MHC ligands for TCR groups 37 and the phenotyping of Mtb‐specific CD4 T cells. 42 MTB300 was used when CD153 on CD4 T cells was identified as a major mediator of host protections against pulmonary Mtb infection 41 and the subsequent evaluation of the role of CD153 in Mtb infection in humans. 39 , 43 This approach has also been useful in providing evidence for mutations that are detrimental to host immunity against Mtb and other mycobacteria. 45 , 47 , 48
7. MEMORY PHENOTYPE OF MTB‐SPECIFIC CD4 T CELLS
As mentioned above, understanding the complexity of CD4 T cells responses to Mtb and their functional attributes is key to developing correlates of protection and informing the design and testing of vaccines and immunodiagnostics. In this and the following sections, we address these issues.
CD4 T cells can be divided into naïve and memory populations based on the expression of CCR7 and CD45RA. Naive cells have a CD45RA+CCR7+ phenotype. Memory cells can be further partitioned into three different phenotypes: central memory (CCR7+CD45RA−, Tcm), effector memory (CCR7−CD45RA−, Tem), and effector memory re‐expressing CD45RA (CCR7−CD45RA+, Temra). 51
In LTBI, Mtb‐specific CD4 T cells have been shown to predominantly express the CCR7+CD45RA− Tcm phenotype, 52 similarly to BCG‐specific CD4 T cells after BCG vaccination in newborns. 53 In contrast, Mtb‐specific CD4 T cells in ATB have the CCR7−CD45RA− Tem phenotype. 52 , 54 , 55 These cells might also represent effector T cells (Teff) since Teff are expected to downregulate CCR7. 56 Individuals with ATB have a higher proportion of effector memory cells, likely with less tissue homing capacity but higher effector functions, compared to latently infected individuals. 57 , 58
Whereas the vast majority of Mtb‐specific CD4 T cells falls into the memory compartment, a small but not negligible fraction of Mtb‐specific CD4 T cells have a naive CD45RA+CCR7+CD27+ phenotype. 36 , 53 , 54 These cells, initially named naive‐like cells, were subsequently identified as stem cell memory T cells, or Tscm. Tscm are a subset of long‐lived memory CD4 T cells that can hold specificity to multiple pathogenic or self‐derived antigens in humans and hold enhanced ability for self‐renewal and multipotency. 59 Transcriptomic analysis of tetramer sorted cells showed that Mtb‐specific Tscm cells have a transcriptomic profile highly similar to bulk Tscm but also share phenotypic and functional properties with both central memory and effector T cells. 60 Based on these results, it was suggested that Mtb‐specific Tscm might therefore represent a less differentiated subset of Mtb‐specific T cells.
Mtb‐specific T cells with a Tscm phenotype are induced after primary Mtb infection 60 and BCG vaccination, 61 and their blood frequency is increased in ATB compared to LTBI. 55 The function of antigen‐specific Tscm in the context of TB remain unclear. Adoptive transfer of Mtb‐specific memory T cells with a naive‐like phenotype in mice showed a higher degree of protection compared to Mtb‐specific Tem transfer, 55 suggesting they might hold an important protective role in TB.
Taken together, these data suggest a heterogeneity within Mtb‐specific memory CD4 T cell subsets that varies depending on an individual's position on the spectrum of infection. This is important to consider in progression studies and vaccine efficacy trials.
8. T‐HELPER SUBSETS OF MTB‐SPECIFIC CD4 T CELLS
Different T‐helper (Th) subsets can be defined based on their surface expression of chemokine receptors and/or specific transcription factors. The classical Th subsets express different combinations of CXCR3, CCR6, and CCR4, Th1 (CXCR3+CCR6−CCR4−), Th17 (CXCR3−CCR6+CCR4+), and Th2 (CXCR3+CCR6−CCR4+). 62
IFNγ, IL‐2, and TNFα producing CD4 T cells (classically called Th1) and IL‐17 producing CD4 T cells (Th17) cells are considered to be the main T cell subsets responding to Mtb infection. Th2 and regulatory T (Treg) cells are also another subset of CD4 T cells that have been found at the site of infection, 63 but they play a different role in immunity. 64 Previous studies have reported Th2 and Tregs working to impair Th1/Th17 and CD8 cytotoxic T cells (CTLs), 65 indicating a suppressive action.
Another Th subset involved in the immune responses against Mtb infection is Th1* (also called Th1 co‐expressing CCR6, Th17.1, Th1Th17, Th17/Th1, and Th1/Th17 cells), which form their own distinct population of CXCR3+CCR6+CCR4− cells co‐expressing the transcription factors Tbet and RORC. 62
9. TH1* AS A CRUCIAL SUBSET OF MTB‐SPECIFIC T CELLS
Our work and others have shown that Th1* contain the majority of Mtb‐specific T cells in IGRA+ individuals. 36 , 62 , 66 We have also found that mycobacteria‐specific (including non‐tuberculous mycobacteria; NTM) epitopes are also recognized by Th1* cells, in both Mtb‐infected and uninfected individuals. 67 Moreover, this specific Th population is present at a higher frequency in IGRA+ individuals compared to Mtb‐uninfected controls, 66 unlike other Th subsets that were present at similar frequencies in both cohorts. These primary observations suggested a role for these Th1* cells in the containment of Mtb infection. In a follow‐up study, using a DRB5*01:01 tetramer loaded with CFP10 52 , 53 , 54 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , we found that more than 90% of tetramer+ T cells were Th1*. 66 This finding was confirmed in a study by Strickland et al, who also reported that Mtb‐specific CD4 T cells are predominantly Th1* in HIV‐negative IGRA+ subjects. 68 Interestingly, they also found that the Th1* subset is contracted in individuals with active TB. 68
Transcriptomic profiling highlighted that Th1* cells have a specific gene signature which is distinct from other Th subsets, and associated with TB susceptibility (CCR2, IL12RB2), augmented cell survival and proliferation (BAFF, MDR1, KIT), and CTL‐like cytotoxic cell killing (transcription factor EOMES, granzyme A, granzyme K, perforin), suggesting a role in disease control in LTBI individuals (ie, preventing transition to active disease). 66
Other studies have confirmed an important role for Th1* cells in Mtb infection. Re‐vaccination with BCG was shown to boost the reactivation of “polyfunctional Th1/Th17 lymphocytes” (likely Th1*, although not confirmed with cell surface staining) in a cohort of IGRA+young adults in the absence of isoniazid treatment (INH). 69 Furthermore, Th1* cells are found in more than one stage of Mtb infection as these cells have also been identified in blood from individuals with active TB, 70 albeit at lower frequency compared to LTBI. In a non‐human primate model of latent Mtb infection, both CCR6 and CXCR3 were upregulated on Mtb‐specific CD4 T cells in the airways, and CXCR3+CD4 T cells accumulated in the granulomas and their frequency correlated with bacterial burden. 71 CXCR3 is known to be important for T cell migration from the blood to the lung in the context of TB 72 and other respiratory infections. 73 Thus, CXCR3+CCR6+ CD4 T cells might encompass most antigenic reactivity in TB due to their unique ability to circulate between the blood and the site of infection.
10. FURTHER HETEROGENEITY WITHIN THE TH1* SUBSET
Th1* are capable of producing many different cytokines and effector molecules upon polyclonal stimulation, including IFNγ, IL‐2, TNFɑ, IL‐17, CCL3, GZMB, IL‐22, and CCL4. However, Mtb‐specific Th1* cells produce IFNγ, IL‐2, and TNFɑ, but not IL‐17 in IGRA+ individuals. 66 The difference between polyclonal and Mtb‐specific stimuli indicated that it is possible to identify cellular heterogeneity within the overall Th1* subset. Applying bulk transcriptomics on sorted memory CD4 T cells in IGRA+ individuals, we revealed 74 differentially expressed genes that were able to distinguish IGRA+ individuals and Mtb‐uninfected controls. 42 In this study, we further refined the Mtb‐reactivity was restricted to the GPA33−CD62L− compartment within Th1*. 42 CD62L had also previously been found to be downregulated in Mtb‐specific CD4T cells. 74
A recent study that used single‐cell RNA sequencing to define cellular responses associated with control of Mtb infection (ie, bacterial killing in granulomas), identified higher proportions of CD4 T cells expressing a “hybrid Th1/Th17 immune response”, 75 thus further strengthening the hypothesis that Th1* have a role in containment of Mtb infection. This may be mediated in part by their expression of CCR6, which mediates cell homing to inflamed tissues, 76 and thus allows peripheral localization. Tissue resident memory cells are also known to co‐express CXCR3 and CCR6. 77
In ATB, several studies also reported that Mtb reactivity within circulating CD4 T cells maps to the Th1* subset. 70 , 78 However, another report studying Mtb‐tetramer+ CD4 T cells showed a more diverse expression of chemokine receptors, with downregulation of CXCR3 in ATB compared to LTBI. 68
A caveat in using chemokine receptors for phenotyping Mtb‐specific CD4 T cells reside in the fact that most assays identifying Mtb‐specific CD4 T cells rely on in vitro stimulation, which impacts the surface expression of chemokine receptors. 68 , 70 For such studies, the use of tetramers for identifying Mtb‐specific CD4 T cells is thus preferred over in vitro stimulation. 66 , 68 Alternatively, pre‐sorting of CD4 T cell subsets based on chemokine receptors followed by antigen‐specific in vitro stimulation can be utilized. 42 The low stability of chemokine receptor expression on the cell surface might explain the variability observed between some studies, especially in the case of active TB where cells might be already pre‐activated in vivo.
Taken together, these studies point toward an important role of Th1* cells in Mtb infection. Future studies using in‐depth phenotyping of Mtb‐specific T cells in different disease stages will improve our understanding of this particular Th subset, as well as other cells involved in the immune response against Mtb.
11. THE IMPACT OF T CELL DIFFERENTIATION AND ACTIVATION
Mtb‐specific CD4 T cells differ between TB disease states in terms of their differentiation phenotype. Numerous studies have reported a reduction in CD27 expression in Mtb‐specific CD4 T cells of ATB infected individuals compared to LTBI. 79 , 80 , 81 , 82 , 83 , 84 , 85 CD27 expression has been further assessed as a potential diagnostic marker for improving active TB diagnosis in children. 86 CD127 was shown to be downregulated in Mtb‐specific CD4 T cells of ATB infected individuals compared to LTBI, 52 whereas PD1 was upregulated. 87
The CD27−CD127−PD1+ phenotype is typically associated with more differentiated T cells (compared to CD27+CD127+PD1− cells), including effector T cells. 56 Thus, in active disease, Mtb‐specific CD4 T cells bear a phenotype that reflect enrichment for highly differentiated effector T cells compared to latent infection.
In addition to their differentiation phenotype, another major distinction between Mtb‐specific CD4 T cells in ATB vs LTBI lies in the expression of activation markers. In ATB, HLA‐DR, CD38 and Ki67 are strongly upregulated in Mtb‐specific CD4 T cells compared to LTBI. 68 , 84 , 88 , 89 Upregulation of these three markers in antigen‐specific T cells was also reported during acute infection with HIV, EBV, or CMV compared to chronic infection. 90 , 91 In particular, HLA‐DR expression on Mtb‐specific CD4 T cells has shown promising sensitivity and specificity to discriminate between ATB and LTBI, 88 , 92 and it was also proposed as a potential prognostic marker for progression to active disease. 93 , 94 , 95 More recently, our work suggest that HLA‐DR marks a subset of Mtb‐specific CD4 T cells with an effector phenotype that have recently proliferated upon infection (Tippalagama et al manuscript in preparation).
CD153 (CD30 ligand, encoded by TNFSF8) expressed by CD4 T cells is another cell surface molecule that has been shown to be differentially expressed between LTBI and ATB individuals. In LTBI, the frequency of CD153+ Mtb‐specific CD4 T cells is increased compared to ATB. 39 , 41 CD153 expression on CD4 T cells was shown to be critical for mounting protective immune responses against Mtb in mice 41 , 96 and was associated with lower bacterial load in humans. 39 Recently it was shown that the phenotypic profile of Mtb‐specific CD4 T cells, using HLA‐DR, CD27, and CD153, can be used to assess severity of TB disease and monitor treatment. 43
Overall, these studies provide strong evidence that the measurement of activation markers is a powerful tool for capturing CD4 T cells that are important to the control of Mtb infection.
12. EFFECT OF TB TREATMENT AND HIV CO‐INFECTION
Following treatment, Mtb‐specific CD4 T cells of individuals with ATB have reduced expression of HLA‐DR, CD38, Ki67, PD1 and increased expression of CD27 and CD153 compared to prior to therapy initiation. 39 , 87 , 88 , 97 Thus, upon ATB treatment Mtb‐specific CD4 T cells shift towards a phenotype similar to LTBI. Moreover, HLA‐DR, CD38 and Ki67 expression on Mtb‐specific CD4 T cells positively correlated with mycobacterial load upon treatment. 88 More recently, Vickers et al 98 showed that the pre‐treatment frequency of CD27+CD38+HLADR+ CD4 T cells after in vitro stimulation with PPD can discriminate between slow and fast responders during treatment. Thus, measuring differences in T cell populations pre‐treatment can give insights on treatment status of a patient and potentially predict treatment failures. With the increased availability of compact flow cytometers, these findings have the potential tobe translated into tools to help diagnose and classify Mtb infections and monitor treatment in less developed and rural endemic areas.
Co‐infection with HIV is a major risk factor for developing active TB, and it strongly affects the CD4 T cell compartment. Several studies have focused on determining the effect of HIV co‐infection on the phenotype of Mtb‐specific CD4 T cells. Mostly, they have found no major differences between HIV seronegative and seropositive TB states. In ATB‐HIV co‐infected individuals, Mtb‐specific CD4 T cells express high levels of HLA‐DR and low levels of CD27, compared to LTBI‐HIV co‐infected subjects. 81 , 92 The frequency of CD153+ Mtb‐specific CD4 T cells is also reduced in ATB compared to LTBI, regardless of HIV seropositive status. 39
13. MTB‐SPECIFIC CD4 T CELL RESPONSES ASSOCIATED WITH IL‐2 PRODUCTION
As mentioned above, the IGRA tests, which measures Mtb‐specific IFNγ in responses, fails to accurately differentiate between LTBI and ATB. In particular, these tests are often considered ineffective when used to determine a child's state of infection. 99 , 100 Since Mtb infection is associated with a spectrum of disease manifestations, it is important to develop tests reflecting more closely the spectrum of Mtb infections so that an intervention tailored to the individual patient can be selected. 101 , 102 Hence, parameters other than IFNγ alone, may be key for future TB diagnostics. This is also relevant for individuals who are highly exposed to Mtb (household contacts of patients with TB) but consistently test negative for both TST and IGRA, that is, so called “resistors”. 103
Early within the immune response mounted by CD4 T cells, rapid short‐lived IL‐2 secretion is genetically controlled and is key in signaling proliferation and differentiation of other immune cells. 104 IL‐2 is important for extracellular killing of mycobacteria, as well as granuloma formation. IL‐2 has been found to be an accurate indicator for differentiating between ATB and LTBI. 105 Particularly, IL‐2 in combination with other cytokines makes for useful ratios that accurately distinguish ATBI and LTBI. 106 Specific ratios of IL‐2 to IFNγ in long‐term stimulation are suggestive of LTBI. 107
Besides being a strong indicator of ATB, IL‐2 has also been shown to induce Foxp3+ Treg growth and response, without impairment of macaque anti‐TB immunity. 108 CD4+CD25+Foxp3+ Treg cells may be essential to a healthy TB response in humans 109 and may actually function to guide Treg cell differentiation of certain CD4 T cells. 110 Because CD4+CD25+Foxp3+ Treg cells likely have a relevant role within severe Mtb infection, IL‐2’s involvement may be important for therapy. 111
14. IL‐17 AND TNF INVOLVEMENT IN MTB‐SPECIFIC CD4 T CELL RESPONSES
One frequently studied cytokine in Mtb infections is IL‐17, which works synergistically and cross‐regulatorily with IFNγ. 112 Stimulation with IL‐23 triggers Th17 cells to produce and secrete IL‐21, IL‐22, and IL‐17, the latter of which plays a multifaceted role in TB. 113 IL‐17 has been shown to induce non‐hematopoietic cells to produce CXCL13, an important chemokine required for localization that will be discussed later. 114 A further unique role for IL‐17 may be the regulation of hypoxic TB granulomas, 115 which may be linked to an inverse relationship with IL‐17 and TB disease in individuals with active TB. Furthermore, via IL‐1β, and IL‐6/STAT3 pathways, IL‐17 has been shown to directly—without the involvement of APC or CD8+ T cells—increase CD4 T cell‐resistance to immune regulation. 116 These functions contribute to the maintenance of Mtb within the lungs.
While IL‐17 does have a strong supporting role in control of Mtb infection, it also has a drawback: IL‐17 has been shown to increase antigen load leading to tissue damage. 117 This is particularly relevant considering the evidence that multidrug resistant strains of TB induce higher levels of IL‐17 producing cells. 118 Furthermore, IFNγ +IL‐17+CD4 T cells tend to be more pronounced during more severe episodes of ATB. 119
In addition to IL‐17, other cytokines that are members of the tumor necrosis factor family also correlate with ATB. As with inflammatory cytokines, TNFα works synergistically with IFNγ inMtb infection and may be more frequently produced during an active Mtbinfection. 120 In an analysis of multiple cytokines after Mtb‐antigen stimulation it was found that only TNFα was not significantly more abundant in LTBI. 121 Furthermore, increased single‐positive TNFα Mtb‐specific CD4 T cells were also found to be highly predictive of active TB. 122
TNF‐α is primarily produced by macrophages, but can also be secreted by CD4 T cells, and plays a managerial role. TNFα activates macrophages via an autocrine/paracrine mechanism, recruits lymphocytes and monocytes to the infection cite via chemokine signaling, restricts mycobacterial growth in granulomas and promotes tissue inflammation and apoptosis, and promotes DC maturation via TNFR1 and DC survival via TNFR2. 123 , 124 , 125 , 126 Furthermore, it may also be involved in controlling Treg responses. 127 Due to the complex role of TNFα in the immune response against Mtb, it is no surprise individuals who receive TNFα‐neutralizing medication also have an increased likelihood of developing active TB. 128
15. ADDITIONAL PLAYERS IN CD4 FUNCTIONAL RESPONSES; IL‐10, CHEMOKINES, AND POLYFUNCTIONALITY
There is also evidence for Mtb‐specific IL‐10 production in humans with active TB, where IL‐10 mediates inhibition of antigen presentation to T cells, and therefore mediates a decreased ability to clear infection contributing to TB pathogenesis. 129 Furthermore, IL‐10 is produced after BCG vaccination, and is responsible for a subsequent reduction of Mtb‐specific Th immune responses. 130 , 131 IL‐10 has also been shown to be elevated in serum from active pulmonary TB patients. 132
Chemokines are significant for their role in signaling the location of infection. For example, Th17 cells are known to express CXCL9, CXCL10, and CXCL11 upon Mtb challenge, which in turn recruit IFNγ producing CD4 T cells to the lung. 133 Interferon gamma‐induced protein 10 (CXCL10 or IP‐10) in particular has been shown to be at least as effective as IFNγin diagnostic assays for Mtb infection. 134 , 135 When testing for IP‐10 levels, the QuantiFERON‐TB Gold Plus (QFT‐Plus) test revealed that in both individuals with LTBI and ATB IP‐10 levels were elevated compared to uninfected subjects, and IP‐10 was found to be partially increased in IGRA+ individuals. 136 IP‐10 is a promising biomarker for ATB 137 , 138 but there is still controversy surrounding IP‐10’s ability to accurately distinguish between ATB and LTBI. 139
Lastly, polyfunctional CD4 T cells are also scrutinized for their role within Mtb infection. 140 In this review, polyfunctional CD4 T cells are defined as dual‐ or triple‐ producing CD4 T cells that secrete pro‐inflammatory cytokines in response to Mtb. 141 Pertaining to Mtb infection, higher frequency of triple producing T cells (IFNγ, IL‐2, and TNFα) was reported during ATB as compared to LTBI; and IL‐2/IFNγ are increased during LTBI. 142 During treatment of TB, triple producers are increased in numbers, 143 and after completion of treatment and the clearance of TB, polyfunctional producers decrease. 144 children, 145 and correlations between polyfunctional T cell response to BCG and inhibition of BCG induces polyfunctional T cell activity in both adults and children—though primarily in Mtb 146 have been reported.
16. THE BREADTH OF TARGETS OF CD4 T CELL RESPONSES
Mtb has a large genome which encompasses approximately 4000 open reading frames (ORFs). A relatively small fraction of these ORFs have been described as antigens. Knowledge about the breadth of responses and the specific T cells responding could aid in the design of new diagnostics, therapies, and vaccination strategies. These may include the discovery of antigens recognized both during Mtb infection and following BCG vaccination. Antigens that are exclusively recognized following BCG vaccination, and not after natural Mtb infection, could potentially be removed to improve vaccine efficacy.
Our group performed a proteome‐wide screen to detect HLA class II restricted epitopes and antigens recognized in IGRA+ individuals without any signs of active TB. 36 This involved the approximately 4000 ORFs of the Mtb genome and 20 000 predicted HLA class II binders. T cell reactivity against these peptides were measured using ELISPOT to detect ex vivo PBMC production of IFNγ. A total of 82 antigens, accounting for approximately 80% of the total IFNγ response, were recognized by more than 10% of IGRA+ subjects, and each subject recognized 24 epitopes on average. These results underline the breadth of immune responses to Mtb in IGRA+ individuals. Our proteome‐wide screen for Mtb‐reactivity was performed in an IGRA+ cohort from San Diego (CA, USA). 36 Subsequent studies of T cell reactivity against the most frequently recognized antigens in cohorts from nine different geographical locations revealed similar response magnitudes and significant correlation between the original cohort and other worldwide locations. 147 Comparison of the reactivity patterns between IGRA+ samples from other locations and the USA cohort revealed a significant correlation both in terms of immunogenicity (magnitude of responses), and immunodominance (eg, relative frequency of recognition). In side‐by‐side comparisons of antigen‐specific T cell reactivity a specific hierarchy of reactivity can be seen. 29 , 36 , 147
17. DISEASE STAGE SPECIFIC ANTIGENS
While IGRA can distinguish prior or current Mtb infection from BCG vaccination and some of the NTM exposure, 102 both the classical TST and IGRA tests are unable to reliably discriminate between active TB and LTBI. Thus, there is a need for a more extensive search for a panel of antigens that can distinguish LTBI and active disease, and better reflect the spectrum of Mtb infections.
Disease stage specific antigens have been described by multiple studies (reviewed in 148). Mtb expresses distinct proteins in the different stages of infection that are expected to give rise to stage‐specific immune responses and antigen recognition, which have indeed been reported. 149 , 150 In granulomas, Mtb is believed to be in a dormant state, triggered by a range of stress factors including hypoxia, low pH, nitric oxide, nutrient deprivation, and host immune pressure. 151 Under these conditions, genes encoded by the DosR regulon are upregulated 152 , 153 and several antigens encoded by this regulon have been described as preferentially recognized by individuals with LTBI 149 , 154 , 155 , 156 (reviewed in 148). In addition, some proteins have been described and referred to as “resuscitation antigens”. 157 , 158 These are small bacterial proteins that promote proliferation of dormant mycobacteria and are therefore believed to be involved in the reactivation of Mtb. 159 However, these antigens have not been studied in the context of being preferentially expressed or recognized by a certain stage of Mtb infection.
Additionally, a proteome‐wide screen for Mtb‐reactivity in disease stages other than healthy IGRA+, such as individuals with active TB or BCG vaccinated individuals, has not yet been performed. It would address an important gap in knowledge; as to date most investigations have targeted either the antigens known to be recognized in LTBI, or specific antigen subgroups selected on the basis of a particular hypothesis.
A proteome‐wide screen would be of interest for both diagnostic and vaccine development considerations. Boosting immune responses against antigens expressed during the active phase of infection might translate into reduced incidence or reactivation of infection. Conversely, boosting responses against antigens exclusively recognized in the latent phase might be of limited value toward the prevention of infection or reactivation. Interestingly, immunization with Mtb‐specific antigens improved BCG‐induced protection in mice, whereas boosting with antigens that are also present in BCG did not. 160 Furthermore, since the spectrum of antigenic specificities associated with different disease stages is incompletely defined, current immunologic tests do not distinguish active disease from LTBI, nor do they quantify the risk of a latently infected individual for progression to active TB. More efficacious predictive diagnostic tests could lead to targeted therapy prior to progression to ATB.
It is also possible that antigens and epitopes recognized during the different stages of Mtb infection might be identical but functionally different in terms of T cell subset, memory phenotype, and cytokine profile elicited. More investigations will establish whether the inclusion of additional or novel antigens in diagnostics and measurement of specific biomarkers can help distinguish individuals with active TB from Mtb‐infected healthy individuals. There is a huge diversity between studies not only in subjects and locations, but also regarding which antigens are investigated, the type of antigenic stimuli, the cell types investigated, and how the antigen‐specific response is measured, 148 which contributes to the difficulty to correlate between different studies.
18. MTB‐SPECIFIC IMMUNE RESPONSES CROSS‐REACT WITH NON‐TUBERCULOUS MYCOBACTERIA (NTM)
Mtb‐specific reactivity can also be influenced by exposure to non‐tuberculous mycobacteria (NTM) and other environmental microbes. Significant reactivity exists in Mtb uninfected individuals that is directed against epitopes conserved among bacteria in the mycobacteria genus, a factor to be considered in diagnostic applications, but also potentially offering an avenue to boost general and widespread reactivity. 40 , 67
Non‐tuberculous mycobacteria vary in their ability and the extent to which they cause clinically significant symptoms or disease in humans. 161 They also vary in other factors such as in vitro growth characteristics and ecological niche, living and multiplying in a variety of human and environmental reservoirs. 161 , 162 , 163 The majority of NTM are present ubiquitously in the environment including soil, seawater, treated/untreated freshwater and a variety of organic and inorganic surfaces. 161 , 162 , 163 Exposure to NTM may result in colonization, infection, and/or pathology that is detectable in the skin or respiratory and gastrointestinal tracts of healthy humans. 161 , 164
Mtb‐uninfected individuals who have not received BCG are capable of responding to Mtb‐derived antigens. 67 NTM are capable of inducing cross‐reactive T‐cell responses to Mtb‐derived epitopes. 165 , 166 , 167 , 168 Higher baseline positive responses to PPD in Mtb‐uninfected adults compared to children might reflect the increased likelihood of NTM exposure with age. 169
In our studies we found that control subjects, who were both TB‐negative and not immunized with BCG, also responded, albeit to a lower extent to Mtb‐derived T cell epitopes. 36 This led to a follow up study focusing on Mtb/NTM cross‐reactivity at the level of the specific epitopes. 67 This analysis revealed that their reactivity was likely due to previous NTM exposure since the epitopes they recognize were also conserved in NTM species. 67
19. POTENTIAL FUNCTIONAL CONSEQUENCE OF ENVIRONMENTAL EXPOSURES ON MTB‐SPECIFIC IMMUNE RESPONSES
It is possible that T cell epitopes conserved between Mtb, NTM, and BCG that elicit cross‐reactive responses offer protection in the form of heterologous immunity or, to the complete contrary, act deleteriously by preventing the institution of BCG‐induced protective responses, creating diagnostic challenges, 163 , 170 and/or confounding evaluation of investigative vaccination strategies. 171 It is hypothesized that exposure to environmental mycobacteria contributes to variable BCG efficacy, 172 , 173 with increasing NTM exposure negatively correlated with efficacy. Several intriguing hypotheses have been proposed to explain how environmental NTM exposure may or may not provide protection against Mtb and contribute to variable BCG efficacy. The “masking” and “blocking” hypotheses propose very different mechanisms to explain how NTM cross‐reactivity contributes to this variability (recently reviewed in 174).
In addition to being influenced by NTM, we have found evidence that Mtb‐specific epitope reactivity is influenced by the microbiome. 40 We observed differential recognition of Mtb‐derived epitopes that was associated with the time period when individuals with active TB undergo treatment. These “treatment sensitive” epitopes are more conserved in the microbiome than “persistent” epitopes. Thus, the strong antibiotic regimen against TB results in the loss of reactivity against a subset of Mtb epitopes, broadly conserved across the microbiome. The influence of epitope conservation in the microbiome in active TB using longitudinal samples and subject‐specific microbiome sequences remains to be determined.
20. IMMUNE CORRELATES OF PROTECTION
An immune correlate of protection (CoP) is a statistical correlation between a clinical endpoint associated with protection, such as protection against infection, disease, severe disease, or reinfection, and an immune marker, which can or cannot itself play a causative role in the protective response following vaccination or natural infection. 175 A validated CoP, indicative of the ideal immune response an effective vaccine should elicit, could be utilized in the vaccine development pipeline to ascertain or prioritize prospective antigens for inclusion in vaccines or to optimize various aspects of vaccine administration including route, dose, delivery method, adjuvant, and schedule. 175 , 176
Efforts to improve TB vaccination efficacy are obstructed by incomplete understanding of the immune responses affording protection against Mtb. No confirmed CoPs, with ultimate validation determined in a phase III efficacy trial with disease and infection endpoints, 176 for a TB vaccine exist as no TB vaccine trials have been conducted demonstrating both efficacy and sufficient sample number for definitive CoP identification. 177 Live Mtb challenge in humans would be the ideal model for CoP identification, but is an impossibility for obvious ethical and technical reasons. 178 Thus, it is important to mine other sample sets from both human and animals, particularly non‐human primates (NHPs), in order to identify potential CoPs that warrant further evaluation. 179 Although other immune cell types (ie, NK cells, CD8 T cells), as well as general features of the broad immunologic landscape (ie, monocyte/T lymphocyte ratio; reviewed in 178) have been suggested as potential correlates of protective immunity, here, we focus on the functional signatures of CD4 T cells as CoPs (Figure 1, Table 1), as this cell subset alone is capable of mediating potent anti‐mycobacterial immunity. 17
21. CD4 T CELLS SECRETING IFNγ
The long‐held paradigm, based largely on animal, but also human studies, is that BCG mediates its protective effects via secretion of IFNγ by Th1 polarized CD4 T cells. 17 , 178 , 180 Thus, IFNγ is the gold standard biomarker by which to assess protection provided by BCG or other candidate TB vaccines. 181 Defective IFNγ signaling, such as in individuals who develop neutralizing antibodies against IFNγ, 182 increases susceptibility to mycobacterial infection and disease. 45 , 47 , 48 In addition, in the MVA85A vaccine efficacy trial, while boosting with MVA85A did not improve protection upon primary inoculation with BCG, low BCG‐specific IFNγ production by PBMCs from BCG‐vaccinated infants associated with increased risk of developing TB disease over the next three years of life. 183
Despite evidence that IFNγ is needed for host resistance to Mtb, the correlation between IFNγ and protection against TB is notoriously inconsistent between studies. 120 , 184 , 185 In contrast to the MVA85A efficacy trial, the only other infant CoP study using vaccine samples conducted by Kagina et al found no association between IFNγ‐secreting CD4 T cells from BCG‐vaccinated South African infants and protection against culture‐positive TB two years after vaccination. 25 Moreover, IFNγ positively correlated with symptoms of active pulmonary disease such as fever and weight loss in Mtb‐infected individuals. 186 Thus, it is critical to look beyond Th1 at other T cell subpopulations—such as polyfunctional T cells producing multiple cytokines, subsets expressing specific activation markers, and memory subsets at different stages of differentiation and thus equipped with different tissue homing and effector capabilities, for their suitability as CoPs.
22. POLYFUNCTIONAL CD4 T CELLS AS A POTENTIAL CORRELATE OF PROTECTION
As discussed above, CD4 T cells can be further characterized based on their ability to produce multiple cytokines. Multifunctional/polyfunctional cells had first been associated with protection in other infectious diseases, namely Leishmania 187 and HIV, particularly when antigen load is low. 188 , 189 Moreover, studies in mice found an association between IFNγ/TNFα/IL‐2 triple‐producing or IFNγ/IL‐2 double‐producing T cells and protection against TB. 190 , 191 , 192 , 193 Intravenous administration of BCG induced polyfunctional CD4 T cells dually producing IFNγ and TNFα that were associated with reduced disease pathology in NHPs. 194
However, the relationship between polyfunctionality and protection against human Mtb infection is less clear. 17 In support of a protective role, some studies report increased polyfunctional T cells in patients with LTBI compared to ATB. Moreover, reduced polyfunctional T cell responses in patients with active disease could be recovered with antibiotic treatment for TB. 122 , 143 , 191 , 195 However, on the opposite spectrum, others report an association between increased polyfunctional T cell responses and ATB. 57 , 120 , 122 , 143 , 196 , 197 , 198 Adding to the controversy, the CoP infant vaccine efficacy trial conducted by Kagina et al 25 reported no correlation between polyfunctional BCG‐specific CD4 T cells and protection against developing active TB.
Thus, polyfunctional T cells are induced by Mtb infection, but whether these cells are also a correlate of protection is not fully determined. It is likely that the disease stage and bacterial load plays a role in the polyfunctional response 57 , 198 and perhaps not protection per se. 199 Alternatively, expression of certain combinations of cytokines may be relevant to protection, but only within the context of a particular subset of Mtb‐specific CD4 T cells. This highlights the importance of looking at additional cell surface activation, migration, and memory markers in order to combine a differentiation phenotype with a functional secretory profile.
23. MEMORY CD4 T CELLS WITH LUNG HOMING CAPACITY
A more discriminative approach combining polyfunctional responses with phenotypic characterization of surface markers, indicative of a T cell's memory differentiation status and thus effector capabilities, may be critical to separate immunopathology from protective antigen‐specific T cell responses that could be used as CoPs. 56 , 120 , 184 , 185 Post‐vaccination measurement of multifunctional responses in Mtb‐specific, relatively undifferentiated, memory T cell subsets retaining the capacity to traffic to the lung may be more indicative of protective immunity against TB. 19 Specifically, vaccinated mice challenged with Mtb have increased frequencies of KLRG1− Mtb‐specific CD4 T cells, derived from activated, replicating PD‐1 high cells, 200 that produce IL‐2 in the lungs compared to unvaccinated naive mice. 201 , 202 KLRG1 is frequently associated with terminal differentiation while IL‐2 production is often associated with a central memory phenotype. 180 Intravascular staining of Mtb challenged mice revealed that Mtb‐specific CD4 T cells localized to the lung parenchyma highly express the activation marker PD‐1 and the chemokine receptor CXCR3, involved in CD4 T cell homing to inflamed tissues, while intravascular Mtb‐specific CD4 T cells highly expressed the terminal differentiation marker KLRG1. 203 , 204 In support of a role in host protection, adoptive transfer of the Mtb‐specific parenchymal CD4 T cells induced much greater control of Mtb infection compared to the intravascular subset. Thus, based on murine studies, migration markers associated with the ability of a CD4 T cells to exit the circulation and enter the lung to interact with Mtb‐infected APCs are promising CoP candidates. 180
24. OTHER T CELL SUBSETS AND BIOMARKERS
In line with the putative protective role for memory cells with lung tissue homing capacity, antigen‐specific Th1* CD4 T cells, expressing the tissue memory marker CCR6 and tissue homing chemokine receptor CXCR3, are a promising CoP candidate. Mtb challenge in BCG‐vaccinated macaques, a particularly useful model for CoP identification, also reported an association between protection and Th1*, with high levels of antigen‐specific Th1/Th17 CD4T cells found in the bronchoalveolar lavage fluid of macaques that received BCG. 205
Other cell subsets and cell surface markers involved in activation are being explored as potential CoPs. Activated HLA‐DR+CD4 T cells were associated with increased risk of TB in BCG vaccinated infants, 93 implicating a role for T cell activation status in determining the individual response to BCG inoculation.
CD153 is a promising CoP candidate that has been positively correlated with Mtb control. It was recently identified through RNA‐sequencing of lung parenchyma residing CD4 T cells that have been shown to be protective in mice. 41 CD153+CD4 T cells increase in the lung tissue of Mtb‐infected mice and CD153−/− mice exhibit higher bacterial loads. 41 Mtb‐specific CD153+CD4 T cells correlated with control of lung granulomas in NHP and, as mentioned above, individuals with active TB had lower expression of CD153 on their CD4 T cells. 41 TNFSF8 (the gene that encodes CD153) was also shown to be differentially expressed in a population of BAL CD4 T cells from IV‐BCG vaccinated NHPs who were protected from Mtb challenge. 205 Furthermore, CD153 has been shown to be inversely correlated to bacterial load, suggesting either enhancement of CD4 survivability and/or enhancement of NK cell proliferation. 39
Taken together, these studies highlight the promising value of functional signatures of CD4 T cells as immune CoPs in Mtb infection. The recent encouraging result that IV BCG vaccination prevents or substantially limits Mtb infection in NHP models 205 has important implications for the identification of immune CoPs. However, given the high diversity in results and outcomes across studies and models, these various studies also highlight that a single immune marker is unlikely to predict candidate vaccine‐induced protection. Thus, it is necessary to employ a systems biology approach to identify different T cell subsets, cytokine secretory profiles, and specific markers involved in adhesion, migration, activation, and co‐simulation that could yield insight into potential new CoPs. Additionally, identified correlates may be highly vaccine specific (eg, only useful to evaluate BCG efficacy) and differ from protective mechanisms at play in natural infection.
25. MTB INFECTION AND THE COVID‐19 PANDEMIC
25.1. Direct effect: Relationship between ATB & LTBI and the susceptibility to COVID‐19 and disease severity
Several studies have attempted to investigate the risk factors associated with COVID‐19 and Mtb infection in an attempt to prioritize treatments for the most vulnerable. A case control study of 36 COVID‐19 confirmed patients in China were followed up and categorized based on disease severity. 206 The results suggested a positive correlation between Mtb infection, susceptibility to SARS‐CoV‐2 and COVID‐19 disease severity. 206 Similar studies have emerged from India, Italy, the Philippines, and South Africa, 207 , 208 , 209 , 210 , 211 albeit they are all limited by a lack of social determinants and comorbidities that could influence co‐infections. Over time as more and larger studies describing COVID‐19 and Mtb infections are published there will be more evidence to support the relationship between Mtb infection and COVID‐19.
25.2. Indirect effects of the COVID‐19 pandemic on TB care
A CDC report from 2004, followed up on healthcare workers in a TB moderate to high incidence are in Taiwan. 212 The screening led to the discovery of 60 suspected ATB cases amongst the healthcare workers. Investigations into the origin of the cluster revealed that an elderly patient was admitted for 12 weeks to the general ward before he was diagnosed with ATB. Between 1998 and 2002, all specialized TB hospitals in Taiwan were closed due to the SARS outbreak and as a result more cases were being managed in a general hospital setting, increasing the nosocomial transmission of Mtb. 212 This meant that Mtb infections were either overlooked or misdiagnosed during the outbreak.
Similarly, during the current COVID‐19 pandemic, medical and human resources were being re‐directed to the care of COVID‐19 patients, while the care of most other diseases was left on the backburner. TB patients were suddenly confronted with the lack of access to diagnosis and treatment facilities. They were also less likely to leave the safety of their homes to obtain necessary treatment. The Hinduja Hospital, a tertiary care hospital in India, observed a drop of 85% out‐patient visits in April 2020, following the lockdown. 213 The indirect effects of COVID‐19 are not always obvious. For India, the lockdown may result in an around an additional 40 000 TB cases annually for the next five years, and a 5.7% increase in TB deaths. 213
TB has been around far longer than COVID‐19 and the pandemic has caused TB to be left on the sideline, while the world focuses their attention and resources on resolving the current crisis. However, the surge in interest towards COVID‐19 can also be beneficial toward developing newer and better diagnostics and therapeutics against TB. For instance, many studies have found that COVID‐19 disease and severity correlate with an increased frequency in circulating HLA‐DR+CD4 T cells. 214 , 215 , 216 As discussed previously in this review, HLA‐DR expression on Mtb‐specific CD4 T cells has also been repeatedly associated with ATB compared to LTBI y. 68 , 84 , 88 , 89 Thus, there seem to be similarities in the immune cell subsets and immune pathways that correlates with protection and/or disease severity against SARS‐CoV‐2 and Mtb. Understanding the complex relationship between these two pathogens and elucidating the molecular mechanisms behind susceptibility and disease severity in single and co‐infections will be fundamental to the development of preventive and treatment strategies for Mtb and Mtb/SARS‐CoV‐2 infections.
26. CONCLUSION AND OUTLOOK
Pathogen‐specific T cell immunity is a key host mechanism to control Mtb infection. Understanding the complexity of T cell responses is crucial to help with the fight against Mtb. A thorough understanding of the nature of responding classical CD4 T cells and the specific epitopes and antigens they recognize in different Mtb infection states will aid in immunodiagnostics, treatment monitoring and vaccine efficacy trials. For instance, unraveling the heterogeneity and biology of Th1*, the major CD4 T cell subset that contain antigen‐specific CD4 T cells in latent Mtb infection might help design better vaccines. Understanding the role of non‐IFNγ producing CD4 T cells and identify their alternative secreted cytokines, chemokines and surface markers might improve the sensitivity and specificity of immunodiagnostic tests. The discovery of CD4 T cell antigens that can be recognized in LTBI but not ATB will help better discriminate between Mtb infection states and more closely reflect the spectrum of infection.
There is a large heterogeneity between different studies aiming to identify which immune parameters will be of most use for correlates of protection. The need to have more control for technical variability is crucial, which would allow for comparison between studies. A limitation in comparing results between different studies is the variability in the definition of the different study cohorts included, in particularly for LTBI where there is often considerable heterogeneity. There is also a need for further research defining and characterizing Mtb‐specific T cell responses in cohorts representing the entire spectrum of infection, as well as ages and co‐morbidities. The so far elusive CoPs are likely not one single immune marker, but instead a combination of secreted and expressed functional molecules acting together.
In addition, it is important to consider the diversity between populations, and the environment they live in, as well as the complex host‐pathogen interactions between humans and Mtb, as well as other environmental and commensal bacteria.
Systems biology approaches that combines several molecular levels of information (proteome, genome, transcriptome) and environment (such as microbiomes) and other large‐scale endeavors have been and will continue to be very successful in identifying and characterizing the immune response against Mtb.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health grant number 75N93019C00067.
Morgan J, Muskat K, Tippalagama R, Sette A, Burel J, Lindestam Arlehamn CS. Classical CD4 T cells as the cornerstone of antimycobacterial immunity. Immunol Rev. 2021;301:10–29. 10.1111/imr.12963
This article is part of a series of reviews covering Immunity to Mycobacteria appearing in Volume 301 of Immunological Reviews.
Jeffrey Morgan, Kaylin Muskat and Rashmi Tippalagama contributed equally to the writing of this article.
DATA AVAILABILITY STATEMENT
Data discussed were all retrieved from published literature as specified in the reference list.
REFERENCES
- 1. WHO . End TB strategy. 2015. https://www.who.int/tb/strategy/End_TB_Strategy.pdf?ua=1
- 2. WHO . Global tuberculosis report 2019. Geneva: World Health Organization; 2017. Licence: CC BY‐NCSA3.0 IGO. [Google Scholar]
- 3. Crisan‐Dabija R, Grigorescu C, Pavel CA, et al. Tuberculosis and COVID‐19: lessons from the past viral outbreaks and possible future outcomes. Can Respir J. 2020;2020:1401053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pai M, Behr MA, Dowdy D, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. [DOI] [PubMed] [Google Scholar]
- 5. Modlin RL, Bloom BR. TB or not TB: that is no longer the question. Sci Transl Med. 2013;5(213):213sr6 [DOI] [PubMed] [Google Scholar]
- 6. Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends Microbiol. 2009;17(5):183‐188. [DOI] [PubMed] [Google Scholar]
- 7. Comstock GW. Epidemiology of tuberculosis. Am Rev Respir Dis. 1982;125(3 Pt 2):8‐15. [DOI] [PubMed] [Google Scholar]
- 8. Kaufmann SH. Is the development of a new tuberculosis vaccine possible? Nat Med. 2000;6(9):955‐960. [DOI] [PubMed] [Google Scholar]
- 9. Schrager LK, Chandrasekaran P, Fritzell BH, et al. WHO preferred product characteristics for new vaccines against tuberculosis. Lancet Infect Dis. 2018;18(8):828‐829. [DOI] [PubMed] [Google Scholar]
- 10. Nemes E, Geldenhuys H, Rozot V, et al. Prevention of M. tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N Engl J Med. 2018;379(2):138‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang H, Kruh‐Garcia NA, Dobos KM. Purified protein derivatives of tuberculin–past, present, and future. FEMS Immunol Med Microbiol. 2012;66(3):273‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin PL, Flynn JL. CD8 T cells and Mycobacterium tuberculosis infection. Semin Immunopathol. 2015;37(3):239‐249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayer‐Barber KD, Barber DL. Innate and adaptive cellular immune responses to mycobacterium tuberculosis infection. Cold Spring Harb Perspect Med. 2015;5(12):a018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boom WH. Gammadelta T cells and Mycobacterium tuberculosis . Microbes Infect. 1999;1(3):187‐195. [DOI] [PubMed] [Google Scholar]
- 15. Huang S. Targeting innate‐like T cells in tuberculosis. Front Immunol. 2016;7:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Rhijn I, Moody DB. Donor unrestricted T cells: a shared human T cell response. J Immunol. 2015;195(5):1927‐1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional signatures of human CD4 and CD8 T cell responses to Mycobacterium tuberculosis . Front Immunol. 2014;5:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barnes PF, Bloch AB, Davidson PT, Snider DE Jr. Tuberculosis in patients with human immunodeficiency virus infection. N Engl J Med. 1991;324(23):1644‐1650. [DOI] [PubMed] [Google Scholar]
- 19. Srivastava S, Ernst JD. Cutting edge: direct recognition of infected cells by CD4 T cells is required for control of intracellular Mycobacterium tuberculosis in vivo. J Immunol. 2013;191(3):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boom WH. The role of T‐cell subsets in Mycobacterium tuberculosis infection. Infect Agents Dis. 1996;5(2):73‐81. [PubMed] [Google Scholar]
- 21. Kaufmann SHE. How can immunology contribute to the control of tuberculosis? Nat Rev Immunol. 2001;1(1):20‐30. [DOI] [PubMed] [Google Scholar]
- 22. Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93‐129. [DOI] [PubMed] [Google Scholar]
- 23. Jung YJ, Ryan L, LaCourse R, North RJ. Properties and protective value of the secondary versus primary T helper type 1 response to airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2005;201(12):1915‐1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newport MJ, Huxley CM, Huston S, et al. A mutation in the interferon‐gamma‐receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335(26):1941‐1949. [DOI] [PubMed] [Google Scholar]
- 25. Kagina BM, Abel B, Scriba TJ, et al. Specific T cell frequency and cytokine expression profile do not correlate with protection against tuberculosis after bacillus Calmette‐Guerin vaccination of newborns. Am J Respir Crit Care Med. 2010;182(8):1073‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paul S, Lindestam Arlehamn CS, Scriba TJ, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods. 2015;422:28‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paul S, Croft NP, Purcell AW, et al. Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system. PLoS Comput Biol. 2020;16(5):e1007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dan JM, Lindestam Arlehamn CS, Weiskopf D, et al. A cytokine‐independent approach to identify antigen‐specific human germinal center T follicular helper cells and rare antigen‐specific CD4+ T cells in blood. J Immunol. 2016;197(3):983‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindestam Arlehamn CS, McKinney DM, Carpenter C, et al. A Quantitative analysis of complexity of human pathogen‐specific CD4 T cell responses in healthy m. tuberculosis infected South Africans. PLoS Pathog. 2016;12(7):e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bancroft T, Dillon MB, da Silva AR, et al. Th1 versus Th2 T cell polarization by whole‐cell and acellular childhood pertussis vaccines persists upon re‐immunization in adolescence and adulthood. Cell Immunol. 2016;304–305:35‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hinz D, Seumois G, Gholami AM, et al. Lack of allergy to timothy grass pollen is not a passive phenomenon but associated with the allergen‐specific modulation of immune reactivity. Clinical Exp Allergy. 2016;46(5):705‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. da Silva AR, Paul S, Sidney J, et al. Definition of human epitopes recognized in tetanus toxoid and development of an assay strategy to detect Ex vivo Tetanus CD4+ T cell responses. PLoS One. 2017;12(1):e0169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grifoni A, Angelo MA, Lopez B, et al. Global assessment of dengue virus‐specific CD4(+) T cell responses in dengue‐endemic areas. Front Immunol. 2017;8:1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weiskopf D, Cerpas C, Angelo MA, et al. Human CD8+ T‐cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis. 2015. 212(11):1743‐1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carrasco Pro S, Sidney J, Paul S, et al. Automatic generation of validated specific epitope sets. J Immunol Res. 2015;2015:763461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindestam Arlehamn CS, Gerasimova A, Mele F, et al. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 2013;9(1):e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Glanville J, Huang H, Nau A, et al. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547(7661):94‐98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kauffman KD, Sallin MA, Hoft SG, et al. Limited pulmonary mucosal‐associated invariant T Cell accumulation and activation during Mycobacterium tuberculosis infection in Rhesus Macaques. Infect Immun. 2018;86(12):e00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Du Bruyn E, Ruzive S, Lindestam Arlehamn CS, et al. Mycobacterium tuberculosis‐specific CD4 T cells expressing CD153 inversely associate with bacterial load and disease severity in human tuberculosis. Mucosal Immunol. 2020. 10.1038/s41385-020-0322-6 (online ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scriba TJ, Carpenter C, Pro SC, et al. Differential recognition of Mycobacterium tuberculosis‐specific epitopes as a function of tuberculosis disease history. Am J Respir Crit Care Med. 2017;196(6):772‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sallin MA, Kauffman KD, Riou C, et al. Host resistance to pulmonary Mycobacterium tuberculosis infection requires CD153 expression. Nat Microbiol. 2018;3(11):1198‐1205. [DOI] [PubMed] [Google Scholar]
- 42. Burel JG, Lindestam Arlehamn CS, Khan N, et al. Transcriptomic analysis of CD4(+) T cells reveals novel immune signatures of latent tuberculosis. J Immunol. 2018;200(9):3283‐3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Riou C, Du Bruyn E, Ruzive S, et al. Disease extent and anti‐tubercular treatment response correlates with Mycobacterium tuberculosis‐specific CD4 T‐cell phenotype regardless of HIV‐1 status. Clin Transl Immunol. 2020;9(9):e1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patankar YR, Sutiwisesak R, Boyce S, et al. Limited recognition of Mycobacterium tuberculosis‐infected macrophages by polyclonal CD4 and CD8 T cells from the lungs of infected mice. Mucosal Immunol. 2020;13(1):140‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kong XF, Martinez‐Barricarte R, Kennedy J, et al. Disruption of an antimycobacterial circuit between dendritic and helper T cells in human SPPL2a deficiency. Nat Immunol. 2018;19(9):973‐985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moguche AO, Musvosvi M, Penn‐Nicholson A, et al. Antigen availability shapes T cell differentiation and function during tuberculosis. Cell Host Microbe. 2017;21(6):695‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okada S, Markle JG, Deenick EK, et al. Immunodeficiencies. Impairment of immunity to Candida and Mycobacterium in humans with bi‐allelic RORC mutations. Science. 2015;349(6248):606‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martinez‐Barricarte R, Markle JG, Ma CS, et al. Human IFN‐gamma immunity to mycobacteria is governed by both IL‐12 and IL‐23. Sci Immunol. 2018;3(30):eaau6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wood MP, Wood LF, Templeton M, et al. Transient immune activation in BCG‐vaccinated infant Rhesus Macaques is not sufficient to influence oral simian immunodeficiency virus infection. J Infect Dis. 2020;222(1):44‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mothe BR, Lindestam Arlehamn CS, Dow C, et al. The TB‐specific CD4(+) T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis. 2015;95(6):722‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708‐712. [DOI] [PubMed] [Google Scholar]
- 52. Pollock KM, Whitworth HS, Montamat‐Sicotte DJ, et al. T‐cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis. 2013;208(6):952‐968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soares AP, Kwong Chung CK, Choice T, et al. Longitudinal changes in CD4(+) T‐cell memory responses induced by BCG vaccination of newborns. J Infect Dis. 2013;207(7):1084‐1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tena‐Coki NG, Scriba TJ, Peteni N, et al. CD4 and CD8 T‐cell responses to mycobacterial antigens in African children. Am J Respir Crit Care Med. 2010;182(1):120‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Orlando V, La Manna MP, Goletti D, et al. Human CD4 T‐cells with a naive phenotype produce multiple cytokines during mycobacterium tuberculosis infection and correlate with active disease. Front Immunol. 2018;9:1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who's who of T‐cell differentiation: human memory T‐cell subsets. Eur J Immunol. 2013;43(11):2797‐2809. [DOI] [PubMed] [Google Scholar]
- 57. Petruccioli E, Petrone L, Vanini V, et al. IFNgamma/TNFalpha specific‐cells and effector memory phenotype associate with active tuberculosis. J Infect. 2013;66(6):475‐486. [DOI] [PubMed] [Google Scholar]
- 58. El Fenniri L, Toossi Z, Aung H, et al. Polyfunctional Mycobacterium tuberculosis‐specific effector memory CD4+ T cells at sites of pleural TB. Tuberculosis. 2011;91(3):224‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gattinoni L, Lugli E, Ji Y, et al. A human memory T cell subset with stem cell‐like properties. Nat Med. 2011;17(10):1290‐1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mpande CAM, Dintwe OB, Musvosvi M, et al. functional, antigen‐specific stem cell memory (TSCM) CD4(+) T cells are induced by human Mycobacterium tuberculosis infection. Front Immunol. 2018;9:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan J, Tenant J, Pacatte T, et al. A subset of mycobacteria‐specific CD4(+) IFN‐gamma(+) T Cell expressing naive phenotype confers protection against tuberculosis infection in the lung. J Immunol. 2019;203(4):972‐980. [DOI] [PubMed] [Google Scholar]
- 62. Acosta‐Rodriguez EV, Rivino L, Geginat J, et al. Surface phenotype and antigenic specificity of human interleukin 17‐producing T helper memory cells. Nat Immunol. 2007;8(6):639‐646. [DOI] [PubMed] [Google Scholar]
- 63. da Silva MV, Tiburcio MG, Machado JR, et al. Complexity and controversies over the cytokine profiles of T helper cell subpopulations in tuberculosis. J Immunol Res. 2015;2015:639107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cardona P, Cardona PJ. Regulatory T cells in Mycobacterium tuberculosis infection. Front Immunol. 2019;10:2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rahman S, Gudetta B, Fink J, et al. Compartmentalization of immune responses in human tuberculosis: few CD8+ effector T cells but elevated levels of FoxP3+ regulatory t cells in the granulomatous lesions. Am J Pathol. 2009;174(6):2211‐2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Arlehamn CL, Seumois G, Gerasimova A, et al. transcriptional profile of tuberculosis antigen‐specific T cells reveals novel multifunctional features. J Immunol. 2014;193(6):2931‐2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lindestam Arlehamn CS, Paul S, Mele F, et al. Immunological consequences of intragenus conservation of Mycobacterium tuberculosis T‐cell epitopes. Proc Natl Acad Sci USA. 2015;112(2):E147‐155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Strickland N, Muller TL, Berkowitz N, et al. Characterization of Mycobacterium tuberculosis‐specific cells using MHC class II tetramers reveals phenotypic differences related to HIV infection and tuberculosis disease. J Immunol. 2017;199(7):2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rakshit S, Ahmed A, Adiga V, et al. BCG revaccination boosts adaptive polyfunctional Th1/Th17 and innate effectors in IGRA+ and IGRA‐ Indian adults. JCI Insight. 2019;4(24):e130540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nikitina IY, Panteleev AV, Kosmiadi GA, et al. Th1, Th17, and Th1Th17 lymphocytes during tuberculosis: Th1 lymphocytes predominate and appear as low‐differentiated CXCR3(+)CCR6(+) cells in the blood and highly differentiated CXCR3(+/‐)CCR6(‐) Cells in the lungs. J Immunol. 2018;200(6):2090‐2103. [DOI] [PubMed] [Google Scholar]
- 71. Shanmugasundaram U, Bucsan AN, Ganatra SR, et al. Pulmonary Mycobacterium tuberculosis control associates with CXCR3‐ and CCR6‐expressing antigen‐specific Th1 and Th17 cell recruitment. JCI Insight. 2020;5(14):e137858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hoft SG, Sallin MA, Kauffman KD, Sakai S, Ganusov VV, Barber DL. The rate of CD4 T cell entry into the lungs during Mycobacterium tuberculosis infection is determined by partial and opposing effects of multiple chemokine receptors. Infect Immun. 2019;87(6):e00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kohlmeier JE, Cookenham T, Miller SC, et al. CXCR3 directs antigen‐specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J Immunol. 2009;183(7):4378‐4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qiao D, Li L, Guo J, et al. Mycobacterium tuberculosis culture filtrate protein 10‐specific effector/memory CD4(+) and CD8(+) T cells in tubercular pleural fluid, with biased usage of T cell receptor Vbeta chains. Infect Immun. 2011;79(8):3358‐3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gideon HP, Hughes TK, Wadsworth MH, et al. Single‐cell profiling of tuberculosis lung granulomas reveals functional lymphocyte signatures of bacterial control. bioRxiv. 2010. https://www.biorxiv.org/content/10.1101/2020.10.24.352492v1 [Google Scholar]
- 76. Thomas SY, Banerji A, Medoff BD, Lilly CM, Luster AD. Multiple chemokine receptors, including CCR6 and CXCR3, regulate antigen‐induced T cell homing to the human asthmatic airway. J Immunol. 2007;179(3):1901‐1912. [DOI] [PubMed] [Google Scholar]
- 77. Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: diversity, function, and antigen specificity. PLoS One. 2011;6(1):e16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Arrigucci R, Lakehal K, Vir P, et al. Active tuberculosis is characterized by highly differentiated effector memory Th1 cells. Front Immunol. 2018;9:2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Streitz M, Tesfa L, Yildirim V, et al. Loss of receptor on tuberculin‐reactive T‐cells marks active pulmonary tuberculosis. PLoS One. 2007;2(8):e735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jiang J, Wang X, Wang X, et al. Reduced CD27 expression on antigen‐specific CD4+ T cells correlates with persistent active tuberculosis. J Clin Immunol. 2010;30(4):566‐573. [DOI] [PubMed] [Google Scholar]
- 81. Schuetz A, Haule A, Reither K, et al. Monitoring CD27 expression to evaluate Mycobacterium tuberculosis activity in HIV‐1 infected individuals in vivo. PLoS One. 2011;6(11):e27284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nikitina IY, Kondratuk NA, Kosmiadi GA, et al. Mtb‐specific CD27low CD4 T cells as markers of lung tissue destruction during pulmonary tuberculosis in humans. PLoS One. 2012;7(8):e43733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Petruccioli E, Petrone L, Vanini V, et al. Assessment of CD27 expression as a tool for active and latent tuberculosis diagnosis. J Infect. 2015;71(5):526‐533. [DOI] [PubMed] [Google Scholar]
- 84. Ahmed MIM, Ntinginya NE, Kibiki G, et al. Phenotypic changes on Mycobacterium tuberculosis‐specific CD4 T cells as surrogate markers for tuberculosis treatment efficacy. Front Immunol. 2018;9:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Latorre I, Fernandez‐Sanmartin MA, Muriel‐Moreno B, et al. Study of CD27 and CCR4 markers on specific CD4(+) T‐cells as immune tools for active and latent tuberculosis management. Front Immunol. 2018;9:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Portevin D, Moukambi F, Clowes P, et al. Assessment of the novel T‐cell activation marker‐tuberculosis assay for diagnosis of active tuberculosis in children: a prospective proof‐of‐concept study. Lancet Infect Dis. 2014;14(10):931‐938. [DOI] [PubMed] [Google Scholar]
- 87. Day CL, Abrahams DA, Bunjun R, et al. PD‐1 expression on Mycobacterium tuberculosis‐specific CD4 T Cells is associated with bacterial load in human tuberculosis. Front Immunol. 2018;9:1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Adekambi T, Ibegbu CC, Cagle S, et al. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J Clin Invest. 2015;125(5):1827‐1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ahmed A, Adiga V, Nayak S, et al. Circulating HLA‐DR+CD4+ effector memory T cells resistant to CCR5 and PD‐L1 mediated suppression compromise regulatory T cell function in tuberculosis. PLoS Pathog. 2018;14(9):e1007289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Callan MF, Tan L, Annels N, et al. Direct visualization of antigen‐specific CD8+ T cells during the primary immune response to Epstein‐Barr virus In vivo. J Exp Med. 1998;187(9):1395‐1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8(4):379‐385. [DOI] [PubMed] [Google Scholar]
- 92. Riou C, Berkowitz N, Goliath R, Burgers WA, Wilkinson RJ. Analysis of the phenotype of Mycobacterium tuberculosis‐specific CD4+ T cells to discriminate latent from active tuberculosis in HIV‐uninfected and HIV‐infected individuals. Front Immunol. 2017;8:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fletcher HA, Snowden MA, Landry B, et al. T‐cell activation is an immune correlate of risk in BCG vaccinated infants. Nat Commun. 2016;7:11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Riou C, Gray CM, Lugongolo M, et al. A subset of circulating blood mycobacteria‐specific CD4 T cells can predict the time to Mycobacterium tuberculosis sputum culture conversion. PLoS One. 2014;9(7):e102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mpande CA, Musvosvi M, Rozot V, et al. Mycobacterium tuberculosis‐specific T cell activation identifies individuals at high risk of tuberculosis disease. medRxiv. 2006. https://www.medrxiv.org/content/10.1101/2020.06.26.20135665v1 [Google Scholar]
- 96. Tang C, Yamada H, Shibata K, et al. A novel role of CD30L/CD30 signaling by T‐T cell interaction in Th1 response against mycobacterial infection. J Immunol. 2008;181(9):6316‐6327. [DOI] [PubMed] [Google Scholar]
- 97. Ahmed MIM, Ziegler C, Held K, et al. The TAM‐TB assay‐a promising TB immune‐diagnostic test with a potential for treatment monitoring. Front Pediatr. 2019;7:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Vickers MA, Darboe F, Muefong CN, et al. Monitoring anti‐tuberculosis treatment response using analysis of whole blood Mycobacterium tuberculosis specific T cell activation and functional markers. Front Immunol. 2020;11(2236). https://www.frontiersin.org/articles/10.3389/fimmu.2020.572620/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Machingaidze S, Wiysonge CS, Gonzalez‐Angulo Y, et al. The utility of an interferon gamma release assay for diagnosis of latent tuberculosis infection and disease in children: a systematic review and meta‐analysis. Pediatr Infect Dis J. 2011;30(8):694‐700. [DOI] [PubMed] [Google Scholar]
- 100. Nenadic N, Kirin BK, Letoja IZ, Plavec D, Topic RZ, Dodig S. Serial interferon‐gamma release assay in children with latent tuberculosis infection and children with tuberculosis. Pediatr Pulmonol. 2012;47(4):401‐408. [DOI] [PubMed] [Google Scholar]
- 101. Lin PL, Flynn JL. The end of the binary era: revisiting the spectrum of tuberculosis. J Immunol. 2018;201(9):2541‐2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Barry CE 3rd, Boshoff HI, Dartois V, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Micro. 2009;7(12):845‐855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lu LL, Smith MT, Yu KKQ, et al. IFN‐gamma‐independent immune markers of Mycobacterium tuberculosis exposure. Nat Med. 2019;25(6):977‐987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL‐2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR‐specific competition. J Immunol. 2004;172(10):6136‐6143. [DOI] [PubMed] [Google Scholar]
- 105. Qiu X, Wang H, Tang Y, et al. Is interleukin‐2 an optimal marker for diagnosing tuberculosis infection? A systematic review and meta‐analysis. Ann Med. 2020;52(7):376‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Suzukawa M, Akashi S, Nagai H, et al. Combined analysis of IFN‐gamma, IL‐2, IL‐5, IL‐10, IL‐1RA and MCP‐1 in QFT supernatant is useful for distinguishing active tuberculosis from latent infection. PLoS One. 2016;11(4):e0152483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sun Q, Wei W, Sha W. Potential role for Mycobacterium tuberculosis specific IL‐2 and IFN‐gamma responses in discriminating between latent infection and active disease after long‐term stimulation. PLoS One. 2016;11(12):e0166501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chen CY, Huang D, Yao S, et al. IL‐2 simultaneously expands Foxp3+ T regulatory and T effector cells and confers resistance to severe tuberculosis (TB): implicative Treg‐T effector cooperation in immunity to TB. J Immunol. 2012;188(9):4278‐4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Churina EG, Urazova OI, Novitskiy VV. The role of foxp3‐expressing regulatory T cells and T helpers in immunopathogenesis of multidrug resistant pulmonary tuberculosis. Tuberc Res Treat. 2012;2012:931291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Thompson LJ, Lai JF, Valladao AC, Thelen TD, Urry ZL, Ziegler SF. Conditioning of naive CD4(+) T cells for enhanced peripheral Foxp3 induction by nonspecific bystander inflammation. Nat Immunol. 2016;17(3):297‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Shevach EM. Application of IL‐2 therapy to target T regulatory cell function. Trends Immunol. 2012;33(12):626‐632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Khader SA, Cooper AM. IL‐23 and IL‐17 in tuberculosis. Cytokine. 2008;41(2):79‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lyadova IV, Panteleev AV. Th1 and Th17 cells in tuberculosis: protection, pathology, and biomarkers. Mediators Inflamm. 2015;2015:854507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gopal R, Monin L, Slight S, et al. Unexpected role for IL‐17 in protective immunity against hypervirulent Mycobacterium tuberculosis HN878 infection. PLoS Pathog. 2014;10(5):e1004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Domingo‐Gonzalez R, Das S, Griffiths KL, et al. Interleukin‐17 limits hypoxia‐inducible factor 1alpha and development of hypoxic granulomas during tuberculosis. JCI Insight. 2017;2(19):e92973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Crawford MP, Sinha S, Renavikar PS, Borcherding N, Karandikar NJ. CD4 T cell‐intrinsic role for the T helper 17 signature cytokine IL‐17: effector resistance to immune suppression. Proc Natl Acad Sci USA. 2020;117(32):19408‐19414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Torrado E, Cooper AM IL‐17 and Th17 cells in tuberculosis. Cytokine Growth Factor Rev. 2010;21(6):455‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Basile JI, Geffner LJ, Romero MM, et al. Outbreaks of mycobacterium tuberculosis MDR strains induce high IL‐17 T‐cell response in patients with MDR tuberculosis that is closely associated with high antigen load. J Infect Dis. 2011;204(7):1054‐1064. [DOI] [PubMed] [Google Scholar]
- 119. Jurado JO, Pasquinelli V, Alvarez IB, et al. IL‐17 and IFN‐γ expression in lymphocytes from patients with active tuberculosis correlates with the severity of the disease. J Leukoc Biol. 2012;91(6):991‐1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sutherland JS, Adetifa IM, Hill PC, Adegbola RA, Ota MOC. Pattern and diversity of cytokine production differentiates between Mycobacterium tuberculosis infection and disease. Eur J Immunol. 2009;39(3):723‐729. [DOI] [PubMed] [Google Scholar]
- 121. Coppola M, Villar‐Hernandez R, van Meijgaarden KE, et al. Cell‐mediated immune responses to in vivo‐expressed and stage‐specific Mycobacterium tuberculosis antigens in latent and active tuberculosis across different age groups. Front Immunol. 2020;11:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Harari A, Rozot V, Enders FB, et al. Dominant TNF‐alpha+ Mycobacterium tuberculosis‐specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med. 2011;17(3):372‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002;168(9):4620‐4627. [DOI] [PubMed] [Google Scholar]
- 124. Fallahi‐Sichani M, El‐Kebir M, Marino S, Kirschner DE, Linderman JJ. Multiscale computational modeling reveals a critical role for TNF‐alpha receptor 1 dynamics in tuberculosis granuloma formation. J Immunol. 2011;186(6):3472‐3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Stenger S. Immunological control of tuberculosis: role of tumour necrosis factor and more. Ann Rheum Dis. 2005;64(suppl_4):iv24‐iv28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Maney NJ, Reynolds G, Krippner‐Heidenreich A, Hilkens CMU. Dendritic cell maturation and survival are differentially regulated by TNFR1 and TNFR2. J Immunol. 2014;193(10):4914‐4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chen X, Oppenheim JJ. The phenotypic and functional consequences of tumour necrosis factor receptor type 2 expression on CD4(+) FoxP3(+) regulatory T cells. Immunology. 2011;133(4):426‐433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Keane J, Gershon S, Wise RP, et al. Tuberculosis associated with infliximab, a tumor necrosis factor α–neutralizing agent. N Eng J Med. 2001;345(15):1098‐1104. [DOI] [PubMed] [Google Scholar]
- 129. Redford PS, Murray PJ, O'Garra A. The role of IL‐10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011;4(3):261‐270. [DOI] [PubMed] [Google Scholar]
- 130. Xu H, Jia Y, Li Y, et al. IL‐10 dampens the Th1 and Tc activation through modulating DC functions in BCG vaccination. Mediators Inflamm. 2019;2019:8616154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pitt JM, Stavropoulos E, Redford PS, et al. Blockade of IL‐10 signaling during bacillus Calmette‐Guerin vaccination enhances and sustains Th1, Th17, and innate lymphoid IFN‐gamma and IL‐17 responses and increases protection to Mycobacterium tuberculosis infection. J Immunol. 2012;189(8):4079‐4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Verbon A, Juffermans N, Van Deventer SJ, Speelman P, Van Deutekom H, Van Der Poll T. Serum concentrations of cytokines in patients with active tuberculosis (TB) and after treatment. Clin Exp Immunol. 1999;115(1):110‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Khader SA, Bell GK, Pearl JE, et al. IL‐23 and IL‐17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8(4):369‐377. [DOI] [PubMed] [Google Scholar]
- 134. Ruhwald M, Aabye MG, Ravn P. IP‐10 release assays in the diagnosis of tuberculosis infection: current status and future directions. Expert Rev Mol Diagn. 2012;12(2):175‐187. [DOI] [PubMed] [Google Scholar]
- 135. Sutherland JS, Mendy J, Gindeh A, et al. Use of lateral flow assays to determine IP‐10 and CCL4 levels in pleural effusions and whole blood for TB diagnosis. Tuberculosis. 2016;96:31‐36. [DOI] [PubMed] [Google Scholar]
- 136. Petrone L, Vanini V, Chiacchio T, et al. Evaluation of IP‐10 in Quantiferon‐Plus as biomarker for the diagnosis of latent tuberculosis infection. Tuberculosis. 2018;111:147‐153. [DOI] [PubMed] [Google Scholar]
- 137. Qiu X, Tang Y, Zou R, et al. Diagnostic accuracy of interferon‐gamma‐induced protein 10 for differentiating active tuberculosis from latent tuberculosis: a meta‐analysis. Sci Rep. 2019;9(1):11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chegou NN, Sutherland JS, Malherbe S, et al. Diagnostic performance of a seven‐marker serum protein biosignature for the diagnosis of active TB disease in African primary healthcare clinic attendees with signs and symptoms suggestive of TB. Thorax. 2016;71(9):785‐794. [DOI] [PubMed] [Google Scholar]
- 139. Goletti D, Raja A, Ahamed Kabeer BS, et al. IFN‐[gamma], but not IP‐10, MCP‐2 or IL‐2 response to RD1 selected peptides associates to active tuberculosis. J Infect. 2010;61(2):133‐143. [DOI] [PubMed] [Google Scholar]
- 140. Lewinsohn DA, Lewinsohn DM, Scriba TJ. Polyfunctional CD4(+) T cells as targets for tuberculosis vaccination. Front Immunol. 2017;8:1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Bhatt K, Verma S, Ellner JJ, Salgame P. Quest for correlates of protection against tuberculosis. Clin Vaccine Immunol. 2015;22(3):258‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Caccamo N, Guggino G, Joosten SA, et al. Multifunctional CD4(+) T cells correlate with active Mycobacterium tuberculosis infection. Eur J Immunol. 2010;40(8):2211‐2220. [DOI] [PubMed] [Google Scholar]
- 143. Day CL, Abrahams DA, Lerumo L, et al. Functional capacity of Mycobacterium tuberculosis‐specific T cell responses in Humans is associated with mycobacterial load. J Immunol. 2011. 187(5):2222‐2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Young JM, Adetifa IM, Ota MO, Sutherland JS. Expanded polyfunctional T cell response to mycobacterial antigens in TB disease and contraction post‐treatment. PLoS One. 2010;5(6):e11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Ritz N, Strach M, Yau C, et al. A comparative analysis of polyfunctional T cells and secreted cytokines induced by Bacille Calmette‐Guerin immunisation in children and adults. PLoS One. 2012;7(7):e37535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Smith SG, Zelmer A, Blitz R, Fletcher HA, Dockrell HM. Polyfunctional CD4 T‐cells correlate with in vitro mycobacterial growth inhibition following Mycobacterium bovis BCG‐vaccination of infants. Vaccine. 2016;34(44):5298‐5305. [DOI] [PubMed] [Google Scholar]
- 147. Carpenter C, Sidney J, Kolla R, et al. A side‐by‐side comparison of T cell reactivity to fifty‐nine Mycobacterium tuberculosis antigens in diverse populations from five continents. Tuberculosis. 2015;95(6):713‐721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Meier NR, Jacobsen M, Ottenhoff THM, Ritz N. A systematic review on novel Mycobacterium tuberculosis antigens and their discriminatory potential for the diagnosis of latent and active tuberculosis. Front Immunol. 2018;9:2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Leyten EMS, Lin MY, Franken KLMC, et al. Human T‐cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis . Microbes Infect. 2006;8(8):2052‐2060. [DOI] [PubMed] [Google Scholar]
- 150. Wilkinson S, Haslov D, et al. Human T‐ and B‐cell reactivity to the 16 kDa α‐crystallin protein of Mycobacterium tuberculosis . Scand J Immunol. 1998;48(4):403‐409. [DOI] [PubMed] [Google Scholar]
- 151. Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis . Annu Rev Microbiol. 2001;55:139‐163. [DOI] [PubMed] [Google Scholar]
- 152. Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis . J Biol Chem. 2004;279(22):23082‐23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha ‐crystallin. Proc Natl Acad Sci USA. 2001;98(13):7534‐7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Demissie A, Leyten EMS, Abebe M, et al. Recognition of stage‐specific mycobacterial antigens differentiates between acute and latent infections with Mycobacterium tuberculosis . Clin Vaccine Immunol. 2006;13(2):179‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Geluk A, Lin MY, van Meijgaarden KE, et al. T‐Cell recognition of the HspX protein of Mycobacterium tuberculosis correlates with latent M. tuberculosis infection but not with M. bovis BCG vaccination. Infect Immun. 2007;75(6):2914‐2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Roupie V, Romano M, Zhang L, et al. Immunogenicity of eight dormancy regulon‐encoded proteins of Mycobacterium tuberculosis in DNA‐vaccinated and tuberculosis‐infected mice. Infect Immun. 2007;75(2):941‐949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Commandeur S, van Meijgaarden KE, Lin MY, et al. Identification of human T‐cell responses to Mycobacterium tuberculosis resuscitation promoting factors in long‐term latently infected individuals. Clin Vaccine Immunol. 2011;18(4):676‐683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Schuck SD, Mueller H, Kunitz F, et al. Identification of T‐cell antigens specific for latent Mycobacterium tuberculosis infection. PLoS One. 2009;4(5):e5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kell DB, Young M. Bacterial dormancy and culturability: the role of autocrine growth factors. Curr Opin Microbiol. 2000;3(3):238‐243. [DOI] [PubMed] [Google Scholar]
- 160. Aagaard C, Knudsen NPH, Sohn I, et al. Immunization with Mycobacterium tuberculosis‐specific antigens bypasses T cell differentiation from prior bacillus Calmette‐Guerin vaccination and improves protection in mice. J Immunol. 2020;205(8):2146‐2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Forbes BA, Hall GS, Miller MB, et al. Practice guidelines for clinical microbiology laboratories: mycobacteria. Clin Microbiol Rev. 2018;31(2):e00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Tortoli E. Phylogeny of the genus Mycobacterium: many doubts, few certainties. Infect Genet Evol. 2012;12(4):827‐831. [DOI] [PubMed] [Google Scholar]
- 163. Griffith DE, Aksamit T, Brown‐Elliott BA, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367‐416. [DOI] [PubMed] [Google Scholar]
- 164. Somoskovi A, Salfinger M. Nontuberculous mycobacteria in respiratory infections: advances in diagnosis and identification. Clinics Lab Med. 2014;34(2):271‐295. [DOI] [PubMed] [Google Scholar]
- 165. Fine PE. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. 1995;346(8986):1339‐1345. [DOI] [PubMed] [Google Scholar]
- 166. Wilson ME, Fineberg HV, Colditz GA. Geographic latitude and the efficacy of bacillus Calmette‐Guerin vaccine. Clinical Infect Dis. 1995;20(4):982‐991. [DOI] [PubMed] [Google Scholar]
- 167. Collins FM. Immunogenicity of various mycobacteria and the corresponding levels of cross‐protection developed between species. Infect Immun. 1971;4(6):688‐696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Orme IM, Collins FM. Adoptive protection of the Mycobacterium tuberculosis‐infected lung. Dissociation between cells that passively transfer protective immunity and those that transfer delayed‐type hypersensitivity to tuberculin. Cell Immunol. 1984;84(1):113‐120. [DOI] [PubMed] [Google Scholar]
- 169. Scriba TJ, Tameris M, Mansoor N, et al. Modified vaccinia Ankara‐expressing Ag85A, a novel tuberculosis vaccine, is safe in adolescents and children, and induces polyfunctional CD4+ T cells. Eur J Immunol. 2010;40(1):279‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Falkinham JO 3rd. Environmental sources of nontuberculous mycobacteria. Clin Chest Med. 2015;36(1):35‐41. [DOI] [PubMed] [Google Scholar]
- 171. Ivanyi J. Function and potentials of M. tuberculosis epitopes. Front Immunol. 2014;5:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Mangtani P, Abubakar I, Ariti C, et al. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis. 2014;58(4):470‐480. [DOI] [PubMed] [Google Scholar]
- 173. Shah JA, Lindestam Arlehamn CS, Horne DJ, Sette A, Hawn TR. Nontuberculous Mycobacteria and heterologous immunity to tuberculosis. J Infect Dis. 2019;220(7):1091‐1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Kuan R, Muskat K, Peters B, Lindestam Arlehamn CS. Is mapping the BCG vaccine‐induced immune responses the key to improving the efficacy against tuberculosis? J Intern Med. 2020;288(6):651‐660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Plotkin SA, Gilbert PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. 2012;54(11):1615‐1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Ellner JJ, Hirsch CS, Whalen CC. Correlates of protective immunity to Mycobacterium tuberculosis in humans. Clin Infect Dis. 2000;30(Suppl 3):S279‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Fletcher HA. Systems approaches to correlates of protection and progression to TB disease. Semin Immunol. 2018;39:81‐87. [DOI] [PubMed] [Google Scholar]
- 178. Satti I, McShane H. Current approaches toward identifying a correlate of immune protection from tuberculosis. Expert Rev Vaccines. 2019;18(1):43‐59. [DOI] [PubMed] [Google Scholar]
- 179. Barker L, Hessel L, Walker B. Rational approach to selection and clinical development of TB vaccine candidates. Tuberculosis. 2012;92(Suppl 1):S25‐S29. [DOI] [PubMed] [Google Scholar]
- 180. Sakai S, Mayer‐Barber KD, Barber DL. Defining features of protective CD4 T cell responses to Mycobacterium tuberculosis . Curr Opin Immunol. 2014;29:137‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Abebe F. Is interferon‐gamma the right marker for bacille Calmette–Guérin‐induced immune protection? The missing link in our understanding of tuberculosis immunology. Clin Exp Immunol. 2012;169(3):213‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Browne SK. Anticytokine autoantibody‐associated immunodeficiency. Annu Rev Immunol. 2014;32:635‐657. [DOI] [PubMed] [Google Scholar]
- 183. Tameris MD, Hatherill M, Landry BS, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo‐controlled phase 2b trial. Lancet. 2013;381(9871):1021‐1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Mittrucker HW, Steinhoff U, Kohler A, et al. Poor correlation between BCG vaccination‐induced T cell responses and protection against tuberculosis. Proc Natl Acad Sci USA. 2007;104(30):12434‐12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Vordermeier HM, Chambers MA, Cockle PJ, Whelan AO, Simmons J, Hewinson RG. Correlation of ESAT‐6‐specific gamma interferon production with pathology in cattle following Mycobacterium bovis BCG vaccination against experimental bovine tuberculosis. Infect Immun. 2002;70(6):3026‐3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Tsao TC, Huang CC, Chiou WK, Yang PY, Hsieh MJ, Tsao KC. Levels of interferon‐gamma and interleukin‐2 receptor‐alpha for bronchoalveolar lavage fluid and serum were correlated with clinical grade and treatment of pulmonary tuberculosis. Int J Tuberc Lung Dis. 2002;6(8):720‐727. [PubMed] [Google Scholar]
- 187. Darrah P, Patel D, de Luca P, et al. Multifunctional TH1 cells define a correlate of vaccine‐mediated protection against Leishmania major. Nat Med. 2007;13:843‐850. [DOI] [PubMed] [Google Scholar]
- 188. Flynn JL, Chan J. Tuberculosis: latency and reactivation. Infect Immun. 2001;69(7):4195‐4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Almeida J, Price D, Papagno L, et al. Superior control of HIV‐1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473‐2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Forbes EK, Sander C, Ronan EO, et al. Multifunctional, high‐level cytokine‐producing Th1 cells in the lung, but not spleen, correlate with protection against Mycobacterium tuberculosis aerosol challenge in mice. J Immunol. 2008;181(7):4955‐4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Aagaard C, Hoang TT, Izzo A, et al. Protection and polyfunctional T cells induced by Ag85B‐TB10.4/IC31 against Mycobacterium tuberculosis is highly dependent on the antigen dose. PLoS One. 2009;4(6):e5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lindenstrom T, Agger EM, Korsholm KS, et al. Tuberculosis subunit vaccination provides long‐term protective immunity characterized by multifunctional CD4 memory T cells. J Immunol. 2009;182(12):8047‐8055. [DOI] [PubMed] [Google Scholar]
- 193. Derrick SC, Yabe IM, Yang A, Morris SL. Vaccine‐induced anti‐tuberculosis protective immunity in mice correlates with the magnitude and quality of multifunctional CD4 T cells. Vaccine. 2011;29(16):2902‐2909. [DOI] [PubMed] [Google Scholar]
- 194. Sharpe S, White A, Sarfas C, et al. Alternative BCG delivery strategies improve protection against Mycobacterium tuberculosis in non‐human primates: protection associated with mycobacterial antigen‐specific CD4 effector memory T‐cell populations. Tuberculosis. 2016;101:174‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Marin ND, Paris SC, Rojas M, Garcia LF. Functional profile of CD4+ and CD8+ T cells in latently infected individuals and patients with active TB. Tuberculosis. 2013;93(2):155‐166. [DOI] [PubMed] [Google Scholar]
- 196. Qiu Z, Zhang M, Zhu Y, et al. Multifunctional CD4 T cell responses in patients with active tuberculosis. Sci Rep. 2012;2:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Mueller H, Detjen AK, Schuck SD, et al. Mycobacterium tuberculosis‐specific CD4+, IFN[gamma]+, and TNF[alpha]+ multifunctional memory T cells coexpress GM‐CSF. Cytokine. 2008;43(2):143‐148. [DOI] [PubMed] [Google Scholar]
- 198. Caccamo N, Guggino G, Joosten SA, et al. Multifunctional CD4+T cells correlate with active Mycobacterium tuberculosis infection. Eur J Immunol. 2010;40(8):2211‐2220. [DOI] [PubMed] [Google Scholar]
- 199. Urdahl KB, Shafiani S, Ernst JD. Initiation and regulation of T‐cell responses in tuberculosis. Mucosal Immunol. 2011;4(3):288‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Reiley WW, Shafiani S, Wittmer ST, et al. Distinct functions of antigen‐specific CD4 T cells during murine Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA. 2010;107(45):19408‐19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Knudsen NP, Agger EM, Andersen P Control of chronic mycobacterium tuberculosis infection by CD4 KLRG1‐ IL‐2‐secreting central memory cells. J Immunol. 2013;190(12):6311‐6319. [DOI] [PubMed] [Google Scholar]
- 202. Woodworth JS, Aagaard CS, Hansen PR, Cassidy JP, Agger EM, Andersen P. Protective CD4 T cells targeting cryptic epitopes of Mycobacterium tuberculosis resist infection‐driven terminal differentiation. J Immunol. 2014;192(7):3247‐3258. [DOI] [PubMed] [Google Scholar]
- 203. Anderson KG, Mayer‐Barber K, Sung H, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9(1):209‐222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Sakai S, Kauffman KD, Schenkel JM, et al. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma‐homing CD4 T cells. J Immunol. 2014;192(7):2965‐2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Darrah PA, Zeppa JJ, Maiello P, et al. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature. 2020;577(7788):95‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Chen Y, Wang Y, Fleming J, et al. Active or latent tuberculosis increases susceptibility to COVID‐19 and disease severity. medRxiv. 2020. [Google Scholar]
- 207. Tadolini M, Codecasa LR, Garcia‐Garcia JM, et al. Active tuberculosis, sequelae and COVID‐19 co‐infection: first cohort of 49 cases. Eur Respir J. 2020;56(1). https://www.medrxiv.org/content/10.1101/2020.03.10.20033795v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Stochino C, Villa S, Zucchi P, Parravicini P, Gori A, Raviglione MC. Clinical characteristics of COVID‐19 and active tuberculosis co‐infection in an Italian reference hospital. Eur Respir J. 2020;56(1):2001708. https://erj.ersjournals.com/content/56/1/2001398.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Motta I, Centis R, D'Ambrosio L, et al. Tuberculosis, COVID‐19 and migrants: preliminary analysis of deaths occurring in 69 patients from two cohorts. Pulmonology. 2020;26(4):233‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Sy KTL, Haw NJL, Uy J. Previous and active tuberculosis increases risk of death and prolongs recovery in patients with COVID‐19. Infect Dis (Lond). 2020;52(12):902‐907. [DOI] [PubMed] [Google Scholar]
- 211. Davies M‐A. HIV and risk of COVID‐19 death: a population cohort study from the Western Cape Province, South Africa. medRxiv. 2020. [Google Scholar]
- 212. Chou M‐Y, Yeh P‐F, Liu J‐H, et al. Nosocomial Transmission of Mycobacterium tuberculosis found through screening for severe acute respiratory syndrome—Taipei, Taiwan, 2003. MMWR Morb Mortal Wkly Rep. 2004;53(15):321‐322. https://www.medrxiv.org/content/10.1101/2020.07.02.20145185v2 [PubMed] [Google Scholar]
- 213. Udwadia ZF, Vora A, Tripathi AR, Malu KN, Lange C, Sara RR. COVID‐19 ‐Tuberculosis interactions: when dark forces collide. Indian J Tuberc. 2020;67(4S):S155‐S162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Mathew D, Giles JR, Baxter AE, et al. Deep immune profiling of COVID‐19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369(6508):eabc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Kuri‐Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID‐19. Sci Immunol. 2020;5(49):eabd7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Meckiff BJ, Ramirez‐Suastegui C, Fajardo V, et al. Imbalance of regulatory and cytotoxic SARS‐CoV‐2‐reactive CD4(+) T cells in COVID‐19. Cell. 2020;183(5):1340‐1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data discussed were all retrieved from published literature as specified in the reference list.


