Abstract
Purpose
Amplified MRI (aMRI) has been introduced as a new method of detecting and visualizing pulsatile brain motion in 2D. Here, we improve aMRI by introducing a novel 3D aMRI approach.
Methods
3D aMRI was developed and tested for its ability to amplify sub‐voxel motion in all three directions. In addition, 3D aMRI was qualitatively compared to 2D aMRI on multi‐slice and 3D (volumetric) balanced steady‐state free precession cine data and phase contrast (PC‐MRI) acquired on healthy volunteers at 3T. Optical flow maps and 4D animations were produced from volumetric 3D aMRI data.
Results
3D aMRI exhibits better image quality and fewer motion artifacts compared to 2D aMRI. The tissue motion was seen to match that of PC‐MRI, with the predominant brain tissue displacement occurring in the cranial‐caudal direction. Optical flow maps capture the brain tissue motion and display the physical change in shape of the ventricles by the relative movement of the surrounding tissues. The 4D animations show the complete brain tissue and cerebrospinal fluid (CSF) motion, helping to highlight the “piston‐like” motion of the ventricles.
Conclusions
Here, we introduce a novel 3D aMRI approach that enables one to visualize amplified cardiac‐ and CSF‐induced brain motion in striking detail. 3D aMRI captures brain motion with better image quality than 2D aMRI and supports a larger amplification factor. The optical flow maps and 4D animations of 3D aMRI may open up exciting applications for neurological diseases that affect the biomechanics of the brain and brain fluids.
Keywords: 3D steerable pyramid, amplified MRI, cardiac‐gated cine MRI, optical flow
1. INTRODUCTION
The human brain undergoes constant motion and deformation due to a range of physiological dynamics. Blood vessel pulsation, together with cerebrospinal fluid (CSF) motion, apply changing pressure on brain tissue, which in turn, results in small motions and deformations. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 The physiological and biomechanical response of the human brain in vivo is thought to be altered in various neurological disorders, such as hydrocephalus, 9 , 10 , 11 , 12 Chiari I malformation, 13 , 14 idiopathic intracranial hypertension, 15 and age‐related diseases in small cerebral vessels. 16 , 17 , 18 , 19 , 20 Thus, the ability to observe the manifestation of these disorders in the form of altered brain motion is thought to be of great interest.
Several methods, such as tagged MRI, 21 , 22 , 23 phase‐contrast MRI (PC‐MRI), 24 , 25 and cine displacement encoding with stimulated echoes (DENSE) MRI 26 , 27 , 28 have been introduced to quantify and explore this pulsatile motion. Recently, amplified MRI (aMRI) has been introduced as a new brain motion detection and visualization method, 29 , 30 which enables one to dramatically amplify the brain tissue response due to blood pulsation and CSF motion. Together with the high spatial and temporal resolution underlying the raw data acquisition, and conspicuous brain tissue and CSF contrast, aMRI allows one to observe the biomechanical response of the brain in exquisite detail.
aMRI is based on the use of a phase‐based motion magnification algorithm 31 applied to 2D multi‐slice cardiac gated (cine MRI) data, which results in an amplified “movie” of brain motion. Preliminary data have shown that aMRI may be a promising tool for differentiating abnormal from normal motion in Chiari I malformation patients 30 ; in this study, a patient exhibited increased caudal midbrain tissue displacement and downward displacement at the level of the brainstem and craniocervical junction consistent with previous studies using phase contrast (PC‐MRI). 1 , 32 , 33 In addition, aMRI has shown promise for visualizing cerebrovascular motion using amplified flow imaging (aFlow). 34 aFlow can capture the characteristics of transient events present in brain tissue (ie, blood flow interaction with arterial walls) when applied to 3D PC‐MRA. By incorporating dynamic mode decomposition into the aMRI data processing pipeline, it has potential application in the assessment of evolving intracranial aneurysms. 34 In addition, by generating amplified strain maps, aMRI has also been shown to have relevance in studying the effects of head impacts on brain health, as a tool for tracking tissue strain. 35 In their study on subconcussive impacts, Champagne et al used aMRI together with diffusion tensor imaging and helmet accelerometer data to gather insight on the region‐specific vulnerability of the corpus callosum to microstructural changes in white‐matter integrity. 35
In light of the promise of aMRI for various clinical applications, this work sets out improvements to the aMRI algorithm and acquisition. Notably, the original aMRI approach employs a 2D post‐processing algorithm which only amplifies motion occurring in the in‐plane direction, while ignoring motion occurring in the out‐of‐plane (ie, third) direction, which gives rise to motion artifacts in the amplified movie. Furthermore, the 2D aMRI algorithm has thus far only been applied to multi‐slice data. Such data typically only support thick slices, which increase motion artifacts due to partial volume effects. In addition, each slice is captured with a slightly different heart rate, which can result in an asynchronized data acquisition that can lead to more motion artifacts in the amplified movie.
In this study, we introduce a novel 3D aMRI post‐processing algorithm, which we apply to 3D cardiac gated balanced steady‐state free precession (bSSFP) cine data to improve the image quality and visualization of the original 2D aMRI output. By capturing motion in all three directions, the 3D aMRI approach improves the image quality and visualization of amplified cardiac‐ and CSF‐induced brain motion in all three directions. The new 3D aMRI algorithm incorporates an extended version of the 2D steerable pyramid filters 36 , 37 used in the 2D aMRI algorithm, allowing one to capture both in‐plane and out‐of‐plane brain motion. To further increase the spatial resolution and improve the performance of the 3D aMRI algorithm, we also applied it to 3D volumetric cine in vivo data. In addition, in an effort to assist clinicians’ interpretation of the 3D movies in the form of 2D maps, we generated optical flow vector maps 38 as a qualitative visual tool for capturing the brain’s motion. These maps were visually compared alongside PC‐MR for their ability to capture the predominant brain tissue displacement, which typically occurs in the cranial‐caudal direction around the midbrain region.
Last, the 3D aMRI algorithm was tested on 2D multi‐slice cine fast low angle shot (FLASH) in vivo data to demonstrate its applicability to different MRI contrasts.
The combination of the 3D aMRI post‐processing algorithm and the 3D volumetric acquisition provides a novel and promising imaging tool that can assist clinicians in the noninvasive assessment of brain pathologies that alter the brain’s biomechanical responses. In addition, by producing 4D animations from 3D aMRI when applied to a volumetric dataset, the complete brain tissue and CSF motion is captured to highlight the mechanism that is thought to drive the passage of CSF throughout the brain.
2. METHODS
The following convention (Supporting Information Figure S1) will be used throughout: “volumetric 3D aMRI” and “multi‐slice 3D aMRI” refer to the 3D algorithm applied to 3D (volumetric) and multi‐slice data, respectively. By the same token, “volumetric 2D aMRI” and “multi‐slice 2D aMRI” refer to the 2D aMRI algorithm applied to volumetric and multi‐slice data, respectively.
2.1. Human subjects
With ethical approval granted by The University of Auckland Human Participants Ethics Committee (ref: 018466), and informed consent, experiments were conducted on seven healthy adult volunteers. (The volunteers were a mix of both male and female with an age range between 25 and 65 y old.)
2.2. MRI acquisition
Scans were performed on volunteers on a 3T MAGNETOM Skyra system (Siemens Healthcare, Erlangen, Germany), XQ Gradients (45 mT/m @ 200 T/m/s), and a 32‐channel head coil. Both 2D multi‐slice and 3D volumetric (true 3D) cardiac‐gated (cine) MRI datasets were acquired. A bSSFP cine sequence was run in the sagittal plane with peripheral pulse gating to target a common scan time of 2:40 min and a field of view (FOV) of 23 cm2. The parameters for the 2D multi‐slice cine sequence were as follows: sagittal plane, matrix size = 192 × 192, repetition time (TR)/echo time (TE)/flip‐angle = 35 ms/1.5 ms/43°, acceleration factor = 3, 30 slices, minimum achievable slice‐thickness of 3 mm (resolution = 1.2 × 1.2 × 3 mm), zero slice gap and slice interleaving, 25 cardiac phases, scan time = 2:40 min. The 3D volumetric bSSFP cine sequence used a matrix size = 194 × 240, TR/TE/flip‐angle = 46 ms/1.7 ms/26°, acceleration factor = 3, partition‐thickness of 1.2 mm (resolution of 1.2 × 1.2 × 1.2 mm). To achieve whole brain coverage, 104 slices and prospective binning to 18 cardiac phases were acquired on the 24‐y‐old male, and 112 slices and prospective binning to 13 cardiac phases for the 44‐y‐old female. The scan time for the 3D volumetric sequence to cover the whole adult brain was approximately 10‐12 min, with the variation dependent on the brain size and cardiac input stability.
To test the ability of the 3D aMRI algorithm to amplify cine images of a different contrast, a 2D multi‐slice cine fast low angle shot (FLASH) sequence was acquired with a FOV = 23 cm2, matrix size = 192 × 192, TR/TE/flip‐angle = 70 ms/3.4 ms/12°, acceleration factor = 2, and 30 slices.
For reproducibility, we also collected five more datasets from volunteers using a 3T SIGNA Premier system (GE, USA), SuperG Gradients (80 mT/m @ 200 T/m/s), and a 46‐channel head coil. The parameters for the 3D volumetric cine FIESTA sequence were as follows: FOV = 23 cm2, matrix size = 256 × 256, TR/TE/flip‐angle = 2.9 ms/1 ms/25°, Hyperkat eight acceleration, partition‐thickness of 1.2 mm (resolution of 1.2 × 1.2 × 1.2 mm), peripheral pulse gating. A total of 116 slices were used for whole brain coverage and retrospectively binning to 20 cardiac phases was applied. The 3D volumetric cine FIESTA sequence took approximately 2.5 min.
For comparison to past studies using the common phase contrast MRI (PC‐MRI) approach, we also collected one set of cine PC‐MRI data and 3D volumetric aMRI data on a same volunteer using the 3T GE Signa system, with the following parameters: FOV = 23 cm2, velocity encoding = 1 cm/s, matrix size = 256 × 256, TR/TE/flip‐angle = 50 ms/12 ms/10°. Phase‐encoding was performed in the right‐left direction in the axial plane, and in the superior‐inferior direction for the sagittal and coronal planes. The resulting images were masked to remove the background noise and highlight the brain motion in these directions of interest.
2.3. Motion amplification
Both 2D and 3D aMRI are based on the Eulerian perspective for the flow field, where the properties of a voxel of fluid, such as pressure and velocity, evolve over time. This differs from the Lagrangian perspective, where the trajectory of particles is tracked over time. In the Eulerian approach to motion magnification, the motion is not explicitly estimated, but rather magnified by amplifying temporal intensity changes at fixed voxel, 30 assuming that the motion is subtle (sub‐voxel).
While 2D aMRI was limited at capturing only in‐plane motion, 3D aMRI captures and amplifies motion in all three directions. The 3D motion amplification is achieved by extending the linear complex steerable pyramid filters from 2D to 3D.
The 3D steerable pyramid is a linear decomposition in which steerable filters are used in a multi‐scale recursive scheme (low‐pass filtering and down sampling), resulting in a decomposition in terms of scale and orientation. The scale tuning of the filters is constrained by a recursive system, and the orientation tuning is constrained by the property of steerability. 37
3D aMRI starts by decomposing the data into scales and orientations using the 3D steerable pyramid. Initially, the image is separated into low‐ and high‐pass subbands. The low‐pass image is then divided into six oriented bandpass subbands and a lower‐pass subband. This last one is then subsampled by a factor of 2, both in the x, y, and z directions. The recursivity is achieved by inserting another level of decomposition in the lower branch. 39
The scales (levels) basis functions are band pass filters in the frequency domain. They are calculated in polar coordinates by multiplying a low‐pass filter Ls −1 of the previous scale with a high pass filter Hs of the current scale. The low‐pass and high‐pass filters for each scale are given by the following equations:
| (1) |
| (2) |
where s is the scaling factor of the level, and the band pass filter for the level is given by,
| (3) |
The angular filters are the 3D cones oriented along the six vertices of cuboctahedron (Table 1) and satisfy the following equation in the frequency domain:
| (4) |
where are the direction of the axes of symmetry of the six basis filters Bj.
TABLE 1.
Directions cosines for the axes of symmetry of 6 basis filters Bj with geometry based on the vertices of the cuboctahedron
| j | |||
|---|---|---|---|
| 0 | 0 | ||
| 1 | 0 | ||
| 2 | 0 | ||
| 3 | 0 | ||
| 4 | 0 | ||
| 5 | 0 |
The resulting filter in the frequency domain (Figure 1) for each level and orientation is given by:
| (5) |
where and every scale and orientation in the decomposition is constructed as follows:
| (6) |
where s and j are the scaling factor and orientation direction, respectively, is the Fourier transform of the image, and F −1 is the inverse Fourier transform.
FIGURE 1.
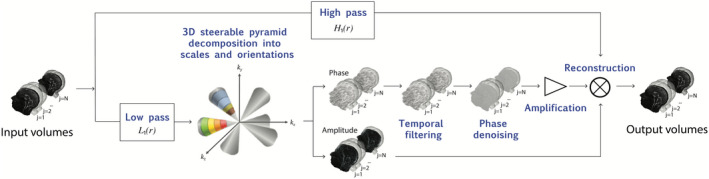
The 3D amplified MRI (aMRI) algorithm pipeline. The volumetric/multi‐slice cine MRI is decomposed by the 3D complex steerable pyramid into scales and orientations. The colors represent the frequency response of the different filters (scales and orientations). Each filter is a bandpass with specific orientation and satisfies Equation (5). Small amplitudes have been zeroed in order to emphasize the filter's conic shape. The phases of the decomposition are separated from the amplitude component, and independently temporally band pass filtered at each spatial location, orientation, and scale. The filtered phases are then “spatially” filtered again to increase the phases signal‐to‐noise ratio using amplitude‐weighted Gaussian spatial smoothing, and then multiplied by an amplification parameter and added to the original amplitude component. The 4D data are then reconstructed to produce an amplified 4D movie
For both the 2D and 3D algorithms, the steerable pyramid decomposition outputs a complex number (amplitude and phase) at each scale and orientation. The phases of the decomposition contain information about sub‐voxel motion, and we exploit this in order to achieve motion magnification; this is similar to the Fourier theorem, where phase variation (rotation) in the frequency domain corresponds to translation in the spatial domain. The phases are temporarily band‐passed in order to isolate the cardiac temporal frequency and to remove any DC component. In addition, in order to increase the performance of the algorithm, and to enable motion magnification with minimal noise artifacts, the band‐passed phases are spatially filtered with an amplitude‐weighted Gaussian smoothing filter. Next, the band‐passed phases are multiplied by a user‐defined amplification factor, α, and added to the original phase component. Attenuation of motion of the other temporal frequencies can be achieved by adding the band‐passed phases to a reference phase image/volume, which in our case was chosen to be the first volume. The volume is then reconstructed to synthesize an amplified 4D movie with the desirable range of temporal frequencies.
The 3D aMRI algorithm was used to amplify the motion of the brain during the cardiac cycle in all three directions simultaneously in both multi‐slice and volumetric data. In addition, both datasets were also amplified using the original 2D aMRI algorithm. The following parameters were used for both 2D and 3D aMRI: amplification factor α = 25, band‐pass filter of the heart rate frequency (±0.1) in addition to attenuating the motion related to all other temporal frequencies, and amplitude‐weighted Gaussian smoothing with . The amplification factor α was chosen according to the original 2D aMRI 30 study as , where is a displacement function, and is the spatial wavelength. By assuming a maximum displacement of the brain stem (midbrain, pons, and medulla) of approximately μm, 14 and minimum resolve wavelength 4 × 2.4 mm (due to amplitude‐weighted Gaussian smoothing), we chose an α within the boundary , that supports sufficient amplification and with minimum artifacts and distortions. Note that different input parameters will result in a different amplification factor boundary. For example, the selection of larger value of the variance in the Gaussian smoothing parameter will result in a bigger amplification boundary, so one should choose it carefully according to the desired application.
2.4. In vivo data comparative maps and visualization tools
For all aMRI datasets, anatomical snapshots of the cardiac cycle and maximum differences maps were produced to compare the brain tissue motion response and motion artifacts. Each successive frame was subtracted from the first frame of the cardiac cycle, and a “difference map” was chosen based on the maximum difference in overall signal intensity.
A built‐in MATLAB Farneback optical flow algorithm 38 was applied to the aMRI data, to aid the visualization of the amplified motion, and to capture the predominantly midbrain tissue displacement which typically occurs in the cranial‐caudal direction (as seen in PC‐MRI). The Farneback method is based on signal decomposition into polynomial basis and analytical solution of the optic flow problem, 38 and assesses the change within a small selected neighborhood size of each vector. It aligns a floating image to the reference image in the optimal position and orientation by modeling the signal with polynomial basis functions, weighted by an applicability function that determines the neighborhood’s importance, and using orientation tensors to compute disparity between the image pair. The optical flow algorithm has several advantages in that it is fast and accurate for both small and large motion. Additionally, since it follows the tissue boundaries (such as between CSF and brain parenchyma) temporally across the cardiac frames, optical flow is reliable in the presence of global signal intensity changes.
In addition, to show that 3D aMRI can be used with other contrast mechanisms, we also applied it to cine FLASH in vivo data. Finally, 4D animations of volumetric 3D aMRI were produced in order to visualize the whole brain tissue, and CSF motion using IMARIS 6.4.3.
3. RESULTS
Several important observations were made upon applying the 3D and 2D aMRI algorithms to multi‐slice and volumetric cine data. Improvements in the image quality came in the form of reduced motion artifacts. Here we define two types of motion artifacts:
Category 1—Motion artifacts occurring due to partial voluming arising from thick slices. These artifacts give rise to coherent (and unrealistic) brain motion in localized regions, as well as an additional erroneous intensity variation across the entire brain.
Category 2—Motion artifacts occurring due to the amplification algorithm’s inability to capture out‐of‐plane motion. These artifacts are more incoherent, manifesting as imaging blurring and ringing artifacts that obscure the anatomy.
As shown in Figures 2, 3, 4, 5 and Supporting Information Videos [Link], [Link], [Link], [Link], both the 3D aMRI algorithm and volumetric cine data together contribute to the overall reduction in motion artifacts (in both categories), with the algorithm itself having the more significant effect on artifact reduction. 3D aMRI reduces motion artifacts (category 2) by capturing and amplifying out‐of‐plane motion in addition to in‐plane motion. Volumetric cine data decrease motion artifacts (category 1) by reducing partial volume effects due to the thinner slices obtainable by the 3D acquisition. As such, the higher resolution in the axial and coronal planes dramatically improves the performance of both 2D and 3D aMRI. It was also observed that 3D aMRI supports a larger amplification factor compared to 2D aMRI.
FIGURE 2.
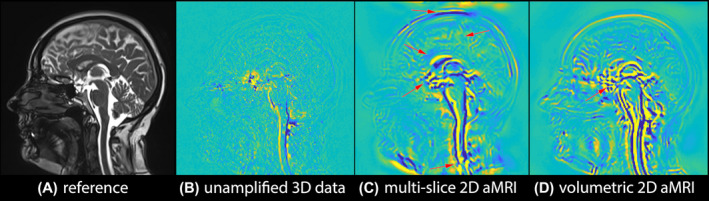
Multi‐slice 2D aMRI vs volumetric 2D aMRI. Anatomical reference (A), and maximum difference maps of the original (unamplified) 3D cine data (B), multi‐slice 2D aMRI (C), and volumetric 2D aMRI (D). Multi‐slice 2D aMRI contains more motion artifacts (red arrows) than volumetric 2D aMRI. Motion artifacts can still be seen in volumetric 2D aMRI (red arrow), because 2D aMRI does not amplify in‐ and out‐of‐plane motion. Here, the heart rate temporal frequency (±0.1) motion was amplified with an amplification parameter of 25 and an amplitude‐weighted Gaussian smoothing with , while attenuating the motion related to all other temporal frequencies
FIGURE 3.
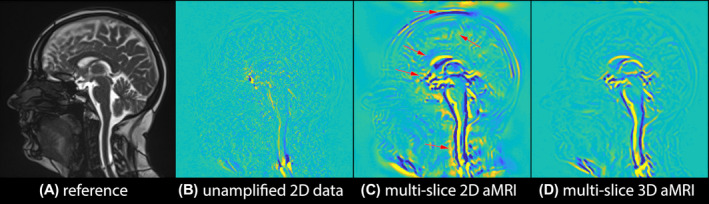
Multi‐slice 2D aMRI vs multi‐slice 3D aMRI. Anatomical reference (A), and maximum difference maps calculated from the original (unamplified) multi‐slice cine data (B), multi‐slice 2D aMRI (C), and multi‐slice 3D aMRI (D). Multi‐slice 3D aMRI contains fewer motion artifacts than multi‐slice 2D aMRI, especially around the cortex, corpus callosum, spinal cord, optic chiasm, and scalp (red arrows)
FIGURE 4.
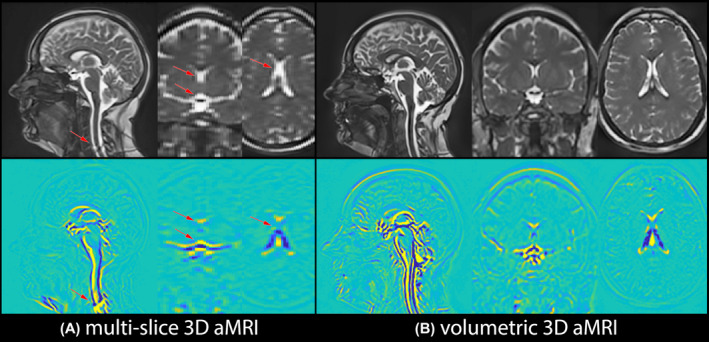
Multi‐slice 3D aMRI vs volumetric 3D aMRI. Maximum difference maps calculated from multi‐slice 3D aMRI and volumetric 3D aMRI together with their corresponding anatomical images. The finer resolution of the volumetric cine data enables one to observe motion in the axial and coronal plane in greater detail, and with fewer artifacts
FIGURE 5.
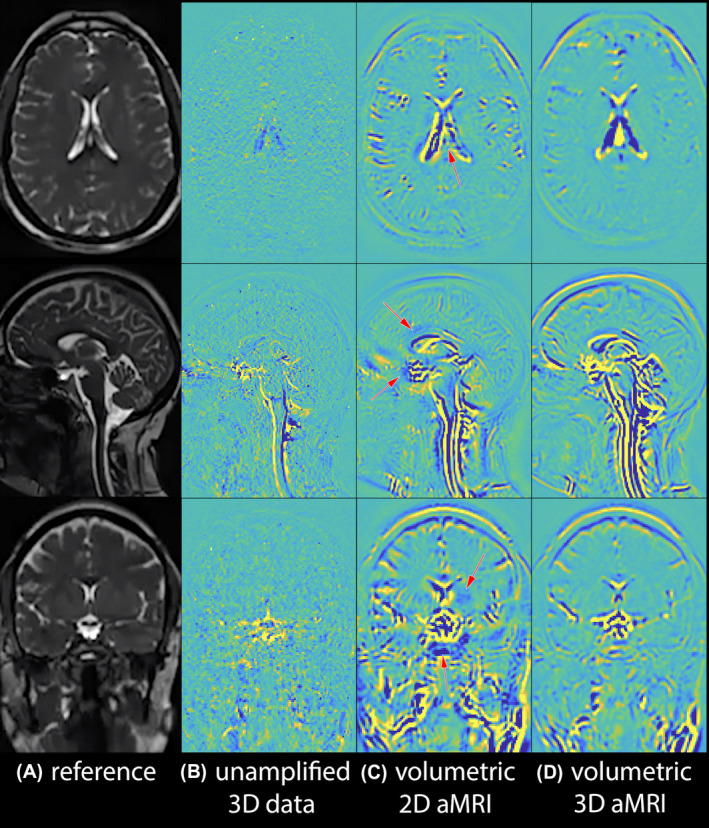
Volumetric 2D aMRI vs volumetric 3D aMRI. Anatomical reference (A), and maximum difference maps calculated from the original (unamplified) 3D cine data (B), volumetric 2D aMRI (C), and volumetric 3D aMRI (D). Volumetric 3D aMRI succeeded in capturing in‐ and out‐of‐plane motion, while significantly decreasing motion artifacts compared to volumetric 2D aMRI. This is well demonstrated in the region of the optic chiasm on the coronal image where brain motion is concentric as opposed to inferior/superior
Figure 2 and Supporting Information Video S1 depict multi‐slice 2D aMRI and volumetric 2D aMRI results. The expected characteristic brain motion amplification can be seen in both datasets, but fewer and less extensive motion artifacts (category 1) are seen in volumetric 2D aMRI, particularly in the cortex, spinal cord, and scalp. This is most evident in Figure 2B, where multi‐slice 2D aMRI produced significant coherent‐appearing scalp motion, induced by larger differences in geometry that exist between slices. However, artifacts (in category 2) remain in the volumetric 2D aMRI dataset since 2D aMRI is unable to capture and amplify in‐ and out‐of‐plane motion. These artifacts, which manifest as blurring accompanied by ringing features that move over time, are particularly prominent in the spinal cord region and around the optic chiasm.
Figure 3 and Supporting Information Video S2 depict multi‐slice 2D aMRI and multi‐slice 3D aMRI results. Motion artifacts (category 2) due to in‐ and out‐of‐plane motion are drastically reduced in multi‐slice 3D aMRI compared to multi‐slice 2D aMRI. A significant reduction in motion artifacts was also observed compared to volumetric 2D aMRI, especially in the spinal cord region and around the optic chiasm. This suggests that a large portion of the motion artifacts are caused by the 2D aMRI algorithm—which ignores in‐ and out‐of‐plane motion.
Figure 4 and Supporting Information Video S3 depict multi‐slice 3D aMRI and volumetric 3D aMRI results. Both outputs succeeded at capturing in‐ and out‐of‐plane motion, but the thinner slices of the volumetric cine data enabled one to observe the motion in the axial and coronal plane in greater detail, and with fewer artifacts.
Figure 5 and Supporting Information Video S4 depict volumetric 2D aMRI and volumetric 3D aMRI results. Here, it can be seen that compared with 2D aMRI, the 3D aMRI algorithm supports a larger amplification factor, and captures motion in all three planes while also suppressing motion artifacts.
3.1. Qualitative comparison of motion between 3D aMRI and PC‐MRI
In both 3D volumetric aMRI and cine PC‐MRI, the general characteristic of brain motion was found to be similar (Figure 6, Supporting Information Video S5). In both sequences, the predominant tissue displacement was in the cranial‐caudal direction in the sagittal and coronal planes, and expanding/contracting motion in the axial plane, with the largest brain tissue displacement occurring around the midbrain, cerebellar tonsils, brainstem, and hypothalamus. Minimal displacement occurred in the frontal lobe, parietal lobe, occipital lobe, temporal lobe, and posterior cerebellum. On 3D aMRI, this motion was clearly captured in the form of vectors by the optical flow map, which visually represented the velocity fields seen on PC‐MR.
FIGURE 6.
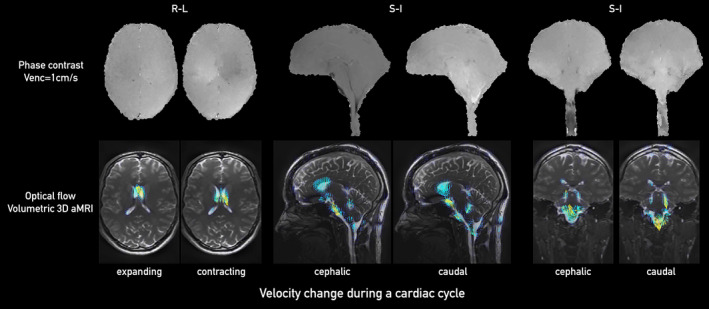
Comparison between PC‐MRI (top) and optical flow maps calculated from volumetric 3D aMRI (bottom) for sagittal (left two panels), coronal (middle two panels), and axial (right two panels) planes. Here, we show the pair of 12th and 19th phases of a cardiac cycle (the same total number of 20 phases collected for each of the PC‐MRI and volumetric 3D aMRI methods). The optical flow maps capture the relative brain tissue deformation over time and the physical change in shape of the ventricles by the relative movement of the surrounding tissues
3.2. 3D aMRI applied to in vivo cine FLASH
We applied our newly developed algorithm on a 2D cine FLASH in vivo data in order to demonstrate the robustness and ability of the new method in amplifying other types of MRI contrasts. In both volumetric 3D aMRI and multi‐slice cine FLASH 3D aMRI, brain motion was found to be very similar, as can be observed in the maximum difference maps (Figure 7 and Supporting Information Video S6). Taking the maximum intensity projection (MIP) of the amplified FLASH dataset allows one to observe the pulsation of some of the major blood vessels of the brain.
FIGURE 7.
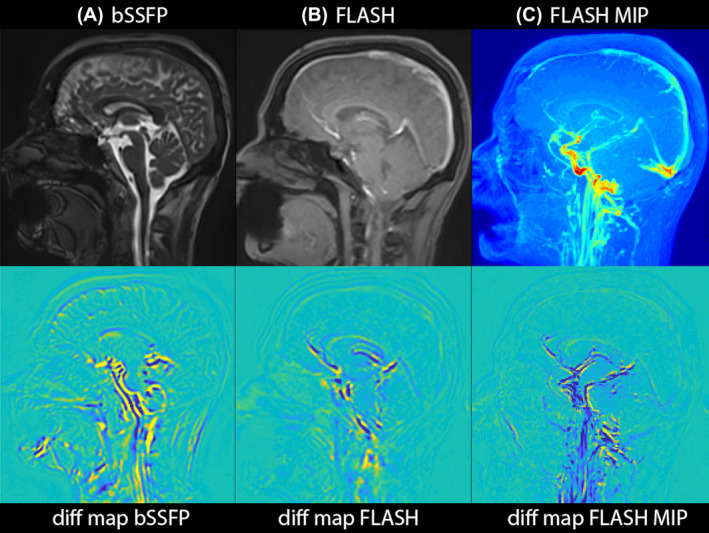
3D aMRI applied to FLASH, demonstrating the applicability of the 3D aMRI algorithm to other MR contrasts. Maximum difference maps calculated from 3D aMRI are shown for volumetric (bSSFP) data (A) and 2D FLASH cine data (B). C, An MIP of the amplified FLASH data is shown together with its corresponding difference map, allowing one to visualize the pulsation of the major blood vessels
3.3. 4D animation
Optical flow maps calculated from volumetric 3D aMRI, shown in Figure 6 and Supporting Information Video S7, show the ability of our method to capture the brain tissue motion over time and display the physical change in shape of the ventricles by the relative movement of the surrounding tissues. In addition, a 4D animation of volumetric 3D aMRI, shown in Figure 8 and Supporting Information Videos [Link], [Link], shows that the novel 3D algorithm can capture the complete brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain.
FIGURE 8.
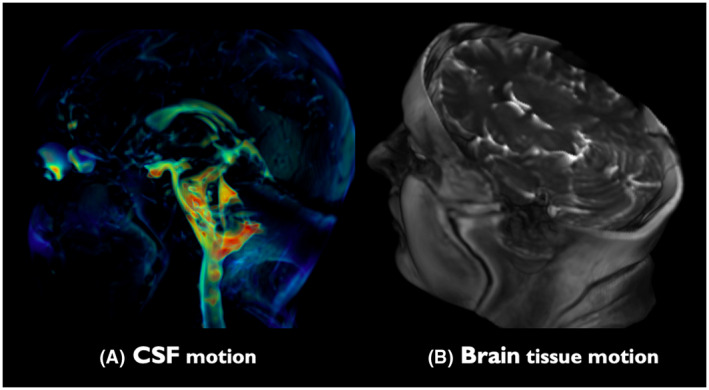
4D animation of volumetric 3D aMRI (Supporting Information Videos [Link], [Link]). The novel 3D algorithm exquisitely captures the brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain. The heart rate temporal frequency (±0.1) motion was amplified with an amplification parameter of 50 and amplitude‐weighted Gaussian smoothing with , while attenuating the motion related to all other temporal frequencies
4. DISCUSSION
This work introduces a novel 3D aMRI post‐processing approach that enables one to visualize the amplified cardiac‐ and CSF‐induced sub‐voxel brain motion in all three directions with striking detail. By extending the original 2D aMRI to 3D aMRI, applying it to volumetric in vivo data, and using a combination of advanced image processing visualization tools, this method can exquisitely highlight the 3D piston‐like motion of the brain and change in ventricular shape as the brain moves through the cardiac cycle.
This study shows the progressive benefit of moving from (the original) multi‐slice 2D aMRI approach to a volumetric 3D aMRI approach. Multi‐slice datasets limit the achievable out‐of‐plane resolution. Conversely, volumetric data yield higher resolution images that allow better delineation of the brain‐CSF interface, and fewer contaminating partial volume effects that can give rise to motion artifacts. This effect was first highlighted by applying the 2D aMRI algorithm to multi‐slice and volumetric data (Figure 2, Supporting Information Video S1). While the 2D aMRI algorithm considerably enhanced the visualization of the pulsatile motion of the brain (as expected 29 , 30 ), volumetric 2D aMRI notably reduced motion artifacts in the cortex, spinal cord, and scalp. However, artifacts remain in the volumetric 2D aMRI dataset since 2D aMRI is unable to capture and amplify in‐ and out‐of‐plane motion. These artifacts, which manifest as blurring accompanied by ringing features that move over time, are particularly prominent in the spinal cord region and around the optic chiasm.
3D aMRI introduces two main advantages compared to 2D aMRI. First, it can capture motion in all three directions, which results in fewer motion artifacts. Second, 3D aMRI can support larger amplification factor and as a result can reveal smaller brain motions compared to 2D aMRI. The ability of 3D aMRI to support a larger amplification factor is achieved by the extension of the steerable pyramid from 2D to 3D. This is because the level of amplification is directly related to the spatial support of the 3D filters (Equation 5), which are smaller compared to 2D filters. The smaller the filters are in the frequency domain, the larger their support is in the spatial domain, thus the window by which the motion can be shifted is larger. 31 Here, the ratio of the 3D filter size and the image (volume) dimensions in the frequency domain is smaller by a factor of (which is less than 1) compared to the 2D filter, where c is a constant and z is the length (number of pixels) of the volume in the slice direction.
A direct comparison between 3D and 2D aMRI could be seen in multi‐slice 2D aMRI and multi‐slice 3D aMRI (Figure 3, Supporting Information Video S2). Since 3D aMRI encapsulates motion in all three planes, the resulting images have considerably reduced artifacts compared to both multi‐slice and volumetric 2D aMRI. This implies that a larger portion of motion artifacts originate from the inability of 2D aMRI to capture in‐ and out‐of‐plane motion (rather than originating from partial volume effects). This finding is consistent with previous DENSE studies, 3 , 5 , 26 whereby these regions have been shown to have higher amplitudes of motion. In addition, this suggests that even if the underlying raw data acquisition is multi‐slice or non‐isotropic, there is a clear benefit of using the 3D aMRI algorithm, and as such this algorithm should be the method of choice over the 2D aMRI algorithm.
Nevertheless, it was found that the most benefit to be gained is through volumetric 3D aMRI (Figures 4, 5, Supporting Information Videos S3 and S4), where both motion artifacts categories are reduced significantly. The ability of 3D aMRI to support larger amplification factor and to capture and amplify in‐ and out‐of‐plane motion together with the thinner slices of the volumetric cine data enabled the observation of the piston‐like motion of the ventricles in the axial and coronal plane in greater detail.
Previous studies have demonstrated the complex characteristics of the brain’s biomechanics, with multiple factors in play. 1 , 32 , 33 However, the predominant tissue and fluid displacement can be seen in this work to be in the cranial‐caudal direction in the sagittal and coronal planes and with an expansion/contraction motion in the axial plane, with the largest brain tissue displacement occurring around the midbrain, brainstem, cerebellar tonsil, and hypothalamus regions. Minimal displacement occurs in the frontal lobe, parietal lobe, occipital lobe, temporal lobe, and posterior cerebellum due to the decreased compliance of brain tissue toward the cranium boundary. These findings can be seen in both cine PC‐MRI and aMRI (Figure 6, Supporting Information Video S5) and are consistent with previous work on PC‐MRI by Pelc et al. 1
Furthermore our optical flow maps applied to 3D aMRI (Figure 6, Supporting Information Video S7) can assist with the visualization of the complex motion characteristics of the pulsating brain during the cardiac cycle. Since the optical flow algorithm follows the tissue boundaries (such as CSF and brain parenchyma) throughout the temporal frames, it is reliable in the presence of global signal intensity changes across the temporal frames. The underlying tissue contrast provided by the bSSFP sequence used by aMRI also provides the ability to visualize this motion with reference to the brain anatomy. Note that these data were acquired on a small number of participants, and that intracranial pulsatility may have variations due to the transfer of pulsation out of the cranium through either venous or CSF outflow pathways, which can be subject dependent. 27 , 32
This work also demonstrates the versatility of 3D aMRI to other underlying MR contrast mechanisms (Figure 7, Supporting Information Video S6). The algorithm amplifies temporal intensity changes at fixed positions (or voxels) under the main assumptions that the underlying motions are sub‐voxel and that the data are relatively smooth. As a result, we postulate that it can be applied to different types of 3D bio‐imaging modalities, 40 such as CT, fluorescent microscopy, ultrasound, and OCT, although one should be careful with the last two, due to high speckle noise that exists in coherence imaging techniques. Applying 3D aMRI to FLASH datasets revealed similar brain motion characteristics to that observed with bSSFP, with the additional advantage of being able to amplify major blood vessel motion which was shown to have a potential application in the assessment of evolving intracranial aneurysms. 34 It should be noted that the aMRI algorithm amplifies subtle intensity variations within each voxel regardless of the source that causes these changes. Some of these changes are induced by motion, and some of them can be due to noise and local B0 field gradients interacting with the motion. Compared to natural scene videos, which capture the intensity changes due to optical flow (which will be predominantly due to visual motion), MR signals contain more underlying information that rely on a range of different physiological processes in addition to motion. A future interesting approach could be to amplify the signal from the MR phase and amplitude data separately, to explore which of these most contributes to the apparent motion seen after amplification. This could perhaps reveal which signal is related specifically to apparent motion.
The intertwinement of blood vessel pulsation, brain tissue, and CSF motion that form the biomechanical response of brain tissue seen with 3D aMRI, may provide us a lens to investigate the underlying conditions and characteristics of the human brain. The optical flow maps applied to volumetric 3D aMRI show the method’s ability to capture the brain tissue motion over time. The direction of pulsatile motion can be visualized and may assist radiologists with the interpretation of the complex brain motion and in particular help to highlight pathological brain movement. In addition, the great reduction in motion artifact together with high spatial resolution obtained with volumetric 3D aMRI may enable an accurate quantification of the 3D displacement field by using custom registration algorithms, and with that the extraction of in vivo biomechanical properties of the brain (such as stiffness), as was demonstrated by Weaver et al. 41 , 42 , 43 In their work, the intrinsic motion of the brain captured by a standard cine phase‐contrast MRI sequence was shown to be a viable alternative to traditional MR elastography (MRE) for measuring the biomechanical properties of brain tissue (obviating the need for specialized MRE hardware and software).
Finally, for the first time, 4D animations of volumetric 3D aMRI are shown to highlight the brain tissue and CSF motion, thus assisting in the observation of the physical change in shape of the ventricles by the relative movement of the surrounding tissues, and in particular the region of the thalamus and basal ganglia. This movement may be instrumental in propelling CSF, which fulfills an important role in the drainage of cerebral waste, 44 , 45 , 46 from the lateral ventricles where it is formed, to the subarachnoid space. The motion within the tissue itself is seen to change direction during each cardiac cycle and may be instrumental in the process of driving extracellular fluid through the extracellular spaces.
5. CONCLUSIONS
Here, we introduce a novel 3D aMRI post processing and image visualization approach that enables one to visualize the amplified cardiac‐ and CSF‐induced brain motion in all three directions with striking detail. Compared with the traditional multi‐slice 2D aMRI approach, 3D aMRI coupled with isotropic volumetric data exquisitely captures 3D brain motion, with better image quality, fewer artifacts, and supports larger amplification factor. The motion seen on volumetric 3D aMRI within the brain tissue may be instrumental in the process of driving extracellular fluid through the extracellular spaces.
In an effort to assist radiologists’ interpretation of the 4D movies in the form of 2D maps, we employ an optical flow approach to highlight the brain’s piston‐like motion. Used together with the 4D animation, our novel method may help understand the dynamics of what drives the passage of CSF through the ventricular system, and the extracellular fluid within the brain tissue, and open up exciting applications for a range of diseases and disorders that affect the biomechanics of the brain and brain fluids.
Supporting information
FIGURE S1 Data acquisition and processing schematic and nomenclature
VIDEO S1 Multi‐slice 2D aMRI contains more motion artifacts (red arrows) than volumetric 2D aMRI. Motion artifacts can still be seen in volumetric 2D aMRI (red arrow), because 2D aMRI does not amplify in‐ and out‐of‐plane motion
VIDEO S2 Multi‐slice 3D aMRI contains fewer motion artifacts than multi‐slice 2D aMRI, especially around the cortex, corpus callosum, spinal cord, optic chiasm, and scalp (red arrows)
VIDEO S3 The finer resolution of the volumetric cine data enables the observation of cardiac induce motions in the axial and coronal plane in greater detail, and with fewer artifacts compared to multi‐slice 3D aMRI
VIDEO S4 Volumetric 3D aMRI succeeded in capturing in‐ and out‐of‐plane motion, while significantly decreasing motion artifacts compared to volumetric 2D aMRI. This is well demonstrated in the region of the optic chiasm on the coronal image where brain motion is concentric as opposed to inferior/superior
VIDEO S5 Volumetric 3D aMRI compared with phase contrast. Motion seen in the amplified videos capture the relative brain tissue deformation over time and the physical change in shape of the ventricles by the relative movement of the surrounding tissues and show similar characteristic motion as seen in phase contrast
VIDEO S6 3D aMRI applied to FLASH demonstrating the applicability of the 3D aMRI algorithm to other MR contrasts. In both volumetric 3D aMRI and multi‐slice cine FLASH 3D aMRI, brain motion was found to be very similar. Taking the maximum intensity projection (MIP) of the amplified FLASH dataset enables the observation of the pulsation of some of the major blood vessels of the brain
VIDEO S7 Optical flow maps calculated from volumetric 3D aMRI. The optical flow maps capture the brain tissue motion over time and display the physical change in shape of the ventricles by the relative movement of the surrounding tissues
VIDEO S8 4D animation of volumetric 3D aMRI of CSF and brain tissue motion. The novel 3D algorithm exquisitely captures the brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain
VIDEO S9 4D animation of volumetric 3D aMRI of CSF and brain tissue motion. The novel 3D algorithm exquisitely captures the brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain
ACKNOWLEDGMENTS
Grant support and other assistance: NSF graduate fellowship, National Institutes of Health (R21NS111415), University of Auckland FDRF strategic initiatives grant, the New Zealand Provincial Growth Fund, and Trust Tairāwhiti. We acknowledge Siemens Healthineers and GE Healthcare for their assistance with the 3D cine (a.k.a. 3D aMRI) acquisition. We are also grateful to Dame Bronwen Holdsworth for proof‐reading our manuscript, to Prof David Dubowitz for helpful discussions, and to the Centre for Advanced MRI (CAMRI) and Mātai Medical Research technologists for their assistance with the scanning.
Terem I, Dang L, Champagne A, et al. 3D amplified MRI (aMRI). Magn Reson Med. 2021;86:1674–1686. 10.1002/mrm.28797
Itamar Terem and Leo Dang contributed equally to this work.
REFERENCES
- 1. Enzmann DR, Pelc NJ. Brain motion: measurement with phase‐contrast MR imaging. Radiology. 1992;185:653‐660. [DOI] [PubMed] [Google Scholar]
- 2. Alperin N, Vikingstad EM, Gomez‐Anson B, Levin DN. Hemodynamically independent analysis of cerebrospinal fluid and brain motion observed with dynamic phase contrast MRI. Magn Reson Med. 1996;35:741‐754. [DOI] [PubMed] [Google Scholar]
- 3. Sloots JJ, Biessels GJ, Zwanenburg JJM. Cardiac and respiration‐induced brain deformations in humans quantified with high‐field MRI. Neuroimage. 2020;210:116581. [DOI] [PubMed] [Google Scholar]
- 4. Mestre H, Tithof J, Du T, et al. Flow of cerebrospinal fluid is driven by arterial pulsations and is reduced in hypertension. Nat Commun. 2018;9:4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adams AL, Kuijf HJ, Viergever MA, Luijten PR, Zwanenburg JJM. Quantifying cardiac‐induced brain tissue expansion using DENSE. NMR Biomed. 2019;32:e4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirsch S, Klatt D, Freimann F, Scheel M, Braun J, Sack I. In vivo measurement of volumetric strain in the human brain induced by arterial pulsation and harmonic waves. Magn Reson Med. 2013;70:671‐683. [DOI] [PubMed] [Google Scholar]
- 7. Poncelet BP, Wedeen VJ, Weisskoff RM, Cohen MS. Brain parenchyma motion: measurement with cine echo‐planar MR imaging. Radiology. 1992;185:645‐651. [DOI] [PubMed] [Google Scholar]
- 8. Wagshul ME, Eide PK, Madsen JR. The pulsating brain: a review of experimental and clinical studies of intracranial pulsatility. Fluids Barriers CNS. 2011;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamada S, Kelly E. Cerebrospinal fluid dynamics and the pathophysiology of hydrocephalus: new concepts. Semin Ultrasound CT MR. 2016;37:84‐91. [DOI] [PubMed] [Google Scholar]
- 10. Ringstad G, Emblem KE, Eide PK. Phase‐contrast magnetic resonance imaging reveals net retrograde aqueductal flow in idiopathic normal pressure hydrocephalus. J Neurosurg. 2016;124:1850‐1857. [DOI] [PubMed] [Google Scholar]
- 11. Bradley WG. Magnetic resonance imaging of normal pressure hydrocephalus. Semin Ultrasound CT MRI. 2016;37:120‐128. [DOI] [PubMed] [Google Scholar]
- 12. Bradley WG, Bahl G, Alksne JF. Idiopathic normal pressure hydrocephalus may be a “two hit” disease: Benign external hydrocephalus in infancy followed by deep white matter ischemia in late adulthood. J Magn Reson Imaging. 2006;24:747‐755. [DOI] [PubMed] [Google Scholar]
- 13. Dawes BH, Lloyd RA, Rogers JM, Magnussen JS, Bilston LE, Stoodley MA. Cerebellar tissue strain in chiari malformation with headache. World Neurosurg. 2019;130:e74‐e81. [DOI] [PubMed] [Google Scholar]
- 14. Pahlavian SH, Oshinski J, Zhong X, Loth F, Amini R. Regional quantification of brain tissue strain using displacement‐encoding with stimulated echoes magnetic resonance imaging. ASME J Biomech Eng. 2018;140:081010. [DOI] [PubMed] [Google Scholar]
- 15. Alperin N, Ranganathan S, Bagci AM, et al. MRI evidence of impaired CSF homeostasis in obesity‐associated idiopathic intracranial hypertension. AJNR Am J Neuroradiol. 2013;34:29‐34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi Y, Wardlaw JM. Update on cerebral small vessel disease: a dynamic whole‐brain disease. Stroke Vasc Neurol. 2016;1:83‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sack I, Streitberger K‐J, Krefting D, Paul F, Braun J. The influence of physiological aging and atrophy on brain viscoelastic properties in humans. PLoS One. 2011;6:e23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsao CW, Seshadri S, Beiser AS, et al. Relations of arterial stiffness and endothelial function to brain aging in the community. Neurology. 2013;81:984‐991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bradley WG Jr. CSF flow in the brain in the context of normal pressure hydrocephalus. AJNR Am J Neuroradiol. 2015;36:831‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lv H, Kurt M, Zeng NA, et al. MR elastography frequency–dependent and independent parameters demonstrate accelerated decrease of brain stiffness in elder subjects. Eur Radiol. 2020;30:6614‐6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xing F, Woo J, Gomez AD, et al. Phase vector incompressible registration algorithm for motion estimation from tagged magnetic resonance images. IEEE Trans Med Imaging. 2017;36:2116‐2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knutsen AK, Wang WT, McEntee JE, et al. Using tagged MRI to quantify the 3D deformation of a cadaver brain in response to angular acceleration. Computational Biomechanics for Medicine. 2013;169‐183. [Google Scholar]
- 23. Knutsen AK, Magrath E, McEntee JE, et al. Improved measurement of brain deformation during mild head acceleration using a novel tagged MRI sequence. J Biomech. 2014;47:3475‐3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bouillot P, Brina O, Delattre BMA, et al. Neurovascular stent artifacts in 3D‐TOF and 3D‐PCMRI: influence of stent design on flow measurement. Magn Reson Med. 2019;81:560‐572. [DOI] [PubMed] [Google Scholar]
- 25. Yamada S, Tsuchiya K, Bradley WG, et al. Current and emerging MR imaging techniques for the diagnosis and management of CSF flow disorders: a review of phase‐contrast and time‐spatial labeling inversion pulse. Am J Neuroradiol. 2015;36:623‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adams AL, Viergever MA, Luijten PR, Zwanenburg JJM. Validating faster DENSE measurements of cardiac‐induced brain tissue expansion as a potential tool for investigating cerebral microvascular pulsations. Neuroimage. 2020;208:116466. [DOI] [PubMed] [Google Scholar]
- 27. Soellinger M, Rutz AK, Kozerke S, Boesiger P. 3D cine displacement‐encoded MRI of pulsatile brain motion. Magn Reson Med. 2009;61:153‐162. [DOI] [PubMed] [Google Scholar]
- 28. Zhong X, Meyer CH, Schlesinger DJ, et al. Tracking brain motion during the cardiac cycle using spiral cine‐DENSE MRI. Med Phys. 2009;36:3413‐3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holdsworth SJ, Rahimi MS, Ni WW, Zaharchuk G, Moseley ME. Amplified magnetic resonance imaging (aMRI). Magn Reson Med. 2016;75:2245‐2254. [DOI] [PubMed] [Google Scholar]
- 30. Terem I, Ni WW, Goubran M, et al. Revealing sub‐voxel motions of brain tissue using phase‐based amplified MRI (aMRI). Magn Reson Med. 2018;80:2549‐2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wadhwa N, Rubinstein M, Durand F, Freeman WT. Phase‐based video motion processing. ACM Trans Graph. 2013;32:1. [Google Scholar]
- 32. Wagshul ME, Eide PK, Madsen JR. The pulsating brain: a review of experimental and clinical studies of intracranial pulsatility. Fluids Barriers CNS. 2011;8. 10.1186/2045-8118-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hofmann E, Warmuth‐Metz M, Bendszus M, Solymosi L. Phase‐contrast MR imaging of the cervical CSF and spinal cord: volumetric motion analysis in patients with Chiari I malformation. AJNR Am J Neuroradiol. 2000;21:151‐158. [PMC free article] [PubMed] [Google Scholar]
- 34. Abderezaei J, Martinez J, Terem I, et al. Amplified flow imaging (aFlow): a novel MRI‐based tool to unravel the coupled dynamics between the human brain and cerebrovasculature. IEEE Trans Med Imaging. 2020;39:4113‐4123. 10.1109/TMI.2020.3012932. [DOI] [PubMed] [Google Scholar]
- 35. Champagne AA, Peponoulas E, Terem I, et al. Novel strain analysis informs about injury susceptibility of the corpus callosum to repeated impacts. Brain Commun. 2019;1:fcz021. 10.1093/braincomms/fcz021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mathewson J, Hale D. Detection of channels in seismic images using the steerable pyramid. SEG Technical Program Expanded Abstracts. 2008:859‐863. [Google Scholar]
- 37. Luche CAD, Delle Luche CA, Denis F, Baskurt A. 3D steerable pyramid based on conic filters. Wavelet Applications in Industrial Processing. 2004. 10.1117/12.516184. [DOI] [Google Scholar]
- 38. Farnebäck G. Two‐frame motion estimation based on polynomial expansion. Image Anal. 2003;363‐370. [Google Scholar]
- 39. Simoncelli EP, Freeman WT. The steerable pyramid: a flexible architecture for multi‐scale derivative computation. In 2nd IEEE International Conference on Image Processing, Washington, DC, vol III, 1995. pp. 444‐447. [Google Scholar]
- 40. Perrot V, Salles S, Vray D, Liebgott H. Video magnification applied in ultrasound. IEEE Trans Biomed Eng. 2019;66:283‐288. [DOI] [PubMed] [Google Scholar]
- 41. Weaver JB, Pattison AJ, McGarry MD, et al. Brain mechanical property measurement using MRE with intrinsic activation. Phys Med Biol. 2012;57:7275‐7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gordon‐Wylie SW, Solamen LM, McGarry MDJ, et al. MR elastography at 1 Hz of gelatin phantoms using 3D or 4D acquisition. J Magn Reson. 2018;296:112‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Solamen LM, McGarry MDJ, Fried J, Weaver JB, Lollis SS, Paulsen KD. Poroelastic mechanical properties of the brain tissue of normal pressure hydrocephalus patients during lumbar drain treatment using intrinsic actuation MR elastography. Acad Radiol. 2020;28:457‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mestre H, Kostrikov S, Mehta RI, Nedergaard M. Perivascular spaces, glymphatic dysfunction, and small vessel disease. Clin Sci. 2017;131:2257‐2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spector R, Robert Snodgrass S, Johanson CE. A balanced view of the cerebrospinal fluid composition and functions: focus on adult humans. Exp Neurol. 2015;273:57‐68. [DOI] [PubMed] [Google Scholar]
- 46. Tarasoff‐Conway JM, Carare RO, Osorio RS, et al. Clearance systems in the brain—implications for Alzheimer disease. Nat Rev Neurol. 2015;11:457‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Data acquisition and processing schematic and nomenclature
VIDEO S1 Multi‐slice 2D aMRI contains more motion artifacts (red arrows) than volumetric 2D aMRI. Motion artifacts can still be seen in volumetric 2D aMRI (red arrow), because 2D aMRI does not amplify in‐ and out‐of‐plane motion
VIDEO S2 Multi‐slice 3D aMRI contains fewer motion artifacts than multi‐slice 2D aMRI, especially around the cortex, corpus callosum, spinal cord, optic chiasm, and scalp (red arrows)
VIDEO S3 The finer resolution of the volumetric cine data enables the observation of cardiac induce motions in the axial and coronal plane in greater detail, and with fewer artifacts compared to multi‐slice 3D aMRI
VIDEO S4 Volumetric 3D aMRI succeeded in capturing in‐ and out‐of‐plane motion, while significantly decreasing motion artifacts compared to volumetric 2D aMRI. This is well demonstrated in the region of the optic chiasm on the coronal image where brain motion is concentric as opposed to inferior/superior
VIDEO S5 Volumetric 3D aMRI compared with phase contrast. Motion seen in the amplified videos capture the relative brain tissue deformation over time and the physical change in shape of the ventricles by the relative movement of the surrounding tissues and show similar characteristic motion as seen in phase contrast
VIDEO S6 3D aMRI applied to FLASH demonstrating the applicability of the 3D aMRI algorithm to other MR contrasts. In both volumetric 3D aMRI and multi‐slice cine FLASH 3D aMRI, brain motion was found to be very similar. Taking the maximum intensity projection (MIP) of the amplified FLASH dataset enables the observation of the pulsation of some of the major blood vessels of the brain
VIDEO S7 Optical flow maps calculated from volumetric 3D aMRI. The optical flow maps capture the brain tissue motion over time and display the physical change in shape of the ventricles by the relative movement of the surrounding tissues
VIDEO S8 4D animation of volumetric 3D aMRI of CSF and brain tissue motion. The novel 3D algorithm exquisitely captures the brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain
VIDEO S9 4D animation of volumetric 3D aMRI of CSF and brain tissue motion. The novel 3D algorithm exquisitely captures the brain tissue and CSF motion, helping to highlight the “piston‐like” motion of the ventricles that is thought to drive the passage of CSF throughout the brain


