Abstract
The development of highly productive, genetically stable manufacturing cell lines is on the critical path to IND filing for protein‐based biologic drugs. Here, we describe the Leap‐In Transposase® platform, a novel transposon‐based mammalian (e.g., Chinese hamster ovary) cell line development system that produces high‐titer stable pools with productivity and product quality attributes that are highly comparable to clones that are subsequently derived therefrom. The productivity distributions of clones are strongly biased toward high producers, and genetic and expression stability is consistently high. By avoiding the poor integration rates, concatemer formation, detrimental transgene recombination, low average expression level, unpredictable product quality, and inconsistent genetic stability characteristic of nonhomologous recombination methods, Leap‐In provides several opportunities to de‐risk programs early and reduce timelines and resources.
Keywords: cell line development, integration, Leap‐In transposase, stable pools, transposase, transposon
Assessment of integrated copy numbers by ddPCR and productivity assessments from representative production cultures demonstrate that >90% of the clones established by Leap–In transposons maintain the To productivity and copy number levels

1. INTRODUCTION
The development of robust, stable, high‐producer cell lines is critical for commercial manufacturing of therapeutic proteins, vaccines, and for gene therapy modalities, yet this step is often rate‐limiting. The labor‐intensive process of isolating stable, high‐producer cell lines is compounded, as next‐generation biologics frequently require the expression of multiple subunits at optimal ratios (Klein et al., 2012, Spiess et al., 2015). The limitations of traditional stable cell line generation approach using viral or plasmid‐based vectors include low integration rate, limited cargo size, genetic instability (Kim et al., 2011), and diminishing expression due to gene silencing (Moritz et al., 2015). These all hamper the standardization of efficient stable cell line development workflows.
Stable genomic integration mechanisms are frequently categorized into two groups: nonhomologous recombination‐based random integration and site‐specific recombinase‐mediated integration processes (Carver et al., 2020). However, there is a third category of stable integration mechanisms, which combines the high copy numbers obtained through random integration, with the intact transgene structure and open chromatin target sites characteristic of site‐specific landing pads. This third category is a transposon‐based mechanism, which has emerged as a DNA transfer tool for gene discovery and gene delivery applications in vertebrates.
Several transposon‐based systems have been characterized, including the natural medaka fish hAT gene family element Tol2 (Kawakami et al., 2000), the engineered Tc1/mariner transposons, named Sleeping Beauty (SB; Mikkelsen et al., 2003) and Frog Prince (Miskey et al., 2003), and the insect‐derived natural element PiggyBac (Yusa, 2015).
More recently, a novel transposon–transposase system, Leap‐In Transposase®, has been engineered from a frog transposon (Balasubramanian et al., 2018; Boldog, 2019). Here, we describe the characteristic structural and functional features of Leap‐In‐mediated integrations and illustrate how those unique features enable a more efficient, robust approach to manufacturing stable cell lines.
2. MATERIALS AND METHODS
2.1. Recombinant DNA methods
2.1.1. Vector construction
The recombinant genes were synthesized, and the expression constructs were assembled in ATUM's laboratories using proprietary technologies. The sequence of the assembled constructs was confirmed using Sanger sequencing.
The expression of codon‐optimized human IgG1 heavy and light chains was driven by the human and the murine EF1α promoters, respectively. The variable expression levels of the codon‐optimized glutamine synthetase gene were achieved by combinations of 5ʹ and 3ʹ regulatory elements, and coding sequence attenuation. The transcription units were flanked by the HS4 and the D4Z4 insulator elements. In the constructs, the segment containing the bacterial selection marker and replication origin was separated from the mammalian expression units by the left and right Leap‐In1 boundary elements. The selection in the GS experiments was performed only by glutamine deprivation (Fan et al., 2012). No methionine sulfoximine (MSX) was used.
The constructs designed for the DG44 experiments were essentially the same with the exception of the selection cassette. The murine dihydrofolate reductase (DHFR) gene was linked to a puromycin N‐acetyl transferase gene by various IRES sequences of different strengths. The transfected DG44 cells were selected in HT‐media supplemented with different amounts of methotrexate (MTX).
Leap‐In mRNA was manufactured using an AOF process by TriLink. The mRNA lots were released per ATUM's specifications.
2.2. Cell culture
HD‐BIOP3 (Horizon Discovery) and DG44 (Prof. Lawrence Chasin) cell‐derived stable pools and clones were used in the study. All cell culture procedures were performed in chemically defined media formulations. The cells were maintained by routine passages two to three times a week. Cells were counted by ViCell (Beckman). The cells were transfected using the Neon electroporation system (Thermo/Invitrogen) equipped with 100‐µl tips. Expression construct (transposon) DNA and Leap‐In transposase mRNA were co‐transfected. Stable pools were selected under glutamine‐free conditions for the BIOP3 lines and with HT‐formulation supplemented with various MTX concentrations for DG44. Single‐cell cloning was performed by the VISP system (Solentim). Monoclonality was confirmed by the VISP and Cell Metric (Solentim) instruments. Volumetric productivities were assessed in fed‐batch production runs following ATUM's standard feeding protocols in 24 deep well plates, tube spins, or shake flasks.
2.3. Protein purification
The product from model antibody cultures grown for product quality characterization was purified using protein A capture. The concentration of the purified protein was determined by A280 absorbance and using molecular extinction coefficients.
2.4. Analytical methods
2.4.1. Protein
Productivities were estimated by an Octet HTX (Pall) using protein A sensors. Protein charge variants were separated on Caliper chips (PerkinElmer) following the manufacturer's recommendation. PNGAse F‐released N‐linked glycan structures were identified and quantified by hydrophobic interaction chromatography and mass spectrometry.
2.4.2. Molecular biology
Droplet digital PCR (ddPCR) was performed using a BioRad QX200 system (BioRad) following the manufacturer protocol and using transposon‐specific primers. The primers and probes were synthesized by IDT. Transgene structure integrity characterization and genomic integration site identification were performed using targeted locus amplification (TLA) by Cergentis.
3. RESULTS AND DISCUSSION
3.1. The Leap‐In transposase® system
The Leap‐In transposase® system utilizes transposon‐based expression vectors and a cognate transposase enzyme. As is typical of transposase‐based gene transfer, the Leap‐In transposon vectors contain two inverted terminal repeat (ITR) sequences recognized by the transposase enzyme. The genetic elements between these ITRs are completely customizable, with no size or sequence limitations. For manufacturing cell line development applications, transposons are typically configured to contain open reading frames to express the target biomolecule, a gene encoding a selectable marker, and all the associated regulatory elements needed for highly efficient expression in the host cell. These elements are flanked by left and right ITRs and the target site duplication TTAW (TTAT or TTAA; Figure 1).
Figure 1.
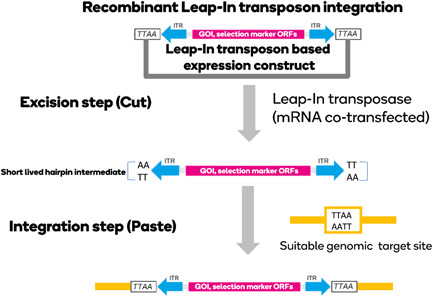
A schematic representation of transposition, the Leap‐In transposase‐mediated cut and paste transgene integration mechanism (explained in the text). For simplicity, only the TTAA recognition signature example is shown [Color figure can be viewed at wileyonlinelibrary.com]
The Leap‐In enzymes belong to the DDE/D integrase family (Nesmelova & Hackett, 2010). The DDE/D integrases mediate stable integration by a two‐step cut and paste mechanism (Mitra et al., 2008). To set a time limit for their intracellular presence, the transposases are introduced to the host cells by co‐transfecting Leap In mRNA with the transposon‐based expression construct. First, upon binding to the specific left and right ITRs, the enzyme initiates double‐stranded breaks in the flanking TTAW integration site signatures, and short‐lived TT/AA hairpin intermediates are formed at the ends of the released transposon. The ITR‐bound transposase enzyme then recognizes a suitable genomic target site characterized with signatures associated with open chromatin and the presence of a TTAW sequence. Once an appropriate genomic site is found, the enzyme makes a double‐stranded break at the TTAW site, resolves the hairpins, and integrates the transposon (Figure 1).
Characteristics of transposon‐mediated integration of expression constructs into a target cell genome include the following: (i) all sequences between the ITRs are faithfully integrated without deletions, insertions, or structural rearrangement, and (ii) when multiple copies of a transposon are introduced into a cell's genome, each insertion occurs at a separate location, so that no concatemeric structures are introduced that could be prone to rearrangement or silencing. To confirm that transposition by the Leap‐In system inserts multiple independent copies of structurally intact transposons, we analyzed clonal cell lines isolated from three uniquely different Leap‐In cell pools. Each cell pool was produced by co‐transfection of the glutamine synthetase knockout HD‐BIOP3 Chinese hamster ovary (CHO) host (provided by Horizon Discovery) with a different DNA plasmid‐borne transposon and Leap‐In transposase mRNA. Each transposon contained genes encoding an antibody heavy and light chain, regulatory elements directing their high‐level expression, and a glutamine synthetase selectable marker. Forty‐eight hours after transfection, the cells were placed into glutamine‐free media and incubated under static conditions until cell viability was >95%. To evaluate the genomic structure of the transposition events, one monoclonal line isolated from each pool was analyzed by TLA performed by Cergentis (Hottentot et al., 2017).
Transposition events were identified by looking for the characteristic sequences (TTAA and ITRs) at each end, as shown for two representative examples in Figure 2. Transposition of the transposon to the genome results in a duplication of the 5ʹ‐TTAA‐3ʹ target site (black letters in Figure 2) within the CHO genome (blue letters in Figure 2) on either side of the transposon. Between the target site duplications is the sequence of the two ITRs (pink letters in Figure 2) and between them are the entire contents of the transposon. Bacterial elements from the plasmid (the kanamycin gene and bacterial origin of replication) are not present in a transposon, and hence not present in the transposition‐mediated integration sites in the CHO genome. In contrast, if a transposon has been integrated by random fragmentation and nonhomologous integration, the ITRs will still be within their bacterial context, there will be a break at some location within the plasmid, and the two ends of that break will be adjacent to CHO genomic sequences. We identified 108 transposon integrations in the three clonal cell lines (Table 1). Out of the 108 integrations, only one integration in each cell line did not have the structure shown in Figure 2, indicating that under the applied transfection and selection conditions, >97% of the integrations were Leap‐In‐mediated transpositions.
Figure 2.
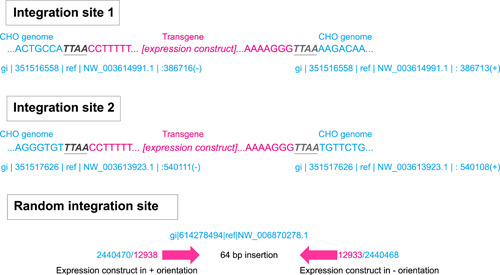
The nucleotide‐level structures of two Leap‐In‐mediated integration junctions from one recombinant cell. Blue: CHO genome sequence, black: target site duplication, pink: transposon. CHO, Chinese hamster ovary [Color figure can be viewed at wileyonlinelibrary.com]
Table 1.
The fraction of transposition‐based stable integrations in three Leap‐In‐mediated stable, mAb‐producing CHO clones
| Clone | Transposition | Nonhomologous recombination |
|---|---|---|
| 963‐0 | 24/25 | 1/25 |
| 964‐5 | 23/24 | 1/24 |
| AT‐G8 | 58/59 | 1/59 |
In addition, the TLA analysis revealed that the transposed Leap‐In transposons integrated as single‐copy transgenes, as shown in Figure 2. The integrated sequences included the entire segment of the expression construct located between the ITRs. The sequences, integrated via transposition, were without any deletion, truncations, or transgene–transgene fusions and maintained the original configuration of the expression construct. In contrast, all three sites where integration occurred by nonhomologous recombination contained transgene–transgene fusions.
The Leap‐In‐mediated integration mechanism results in multiple transposons in the genome. Each integration site contains only a single copy of the transposon and the transposons are structurally intact at the nucleotide level. This is in stark contrast to transgenes integrated by nonhomologous end joining, which are frequently rearranged (Lattenmayer et al., 2006; Tharmalingam et al., 2018). The selection marker attenuation levels used in the studies presented in this manuscript do not support the survival of recombinant cells established by random integration.
3.2. DHFR‐based selection of Leap‐In‐mediated stable DG44 pools demonstrates copy number‐dependent expression
CHO cells are commonly used for biological manufacturing. Two popular selection systems employ host cells that are incapable of synthesizing a critical metabolic intermediate. Hosts such as the DG44 cell line lack a functional gene for the DHFR enzyme, which is required for the synthesis of purine and thymidylate (Florin et al., 2011). When random integration methods are used with DHFR selection, an amplification step is frequently required to increase production titers to acceptable levels (Cacciatore et al., 2010). By using a transposon containing a DHFR gene driven by an attenuated promoter, we expect to be able to select pools with high transgene copy numbers in a single step.
A Leap‐In transposon construct was designed to contain an antibody heavy and light chain as well as associated expression regulatory elements together with a selection cassette. Stable pools were established by co‐transfecting transposon DNA and transposase mRNA into CHO DG44 DHFR−/− host cells and selecting cells for survival with different concentrations of the DHFR inhibitor MTX in a single step after transfection. Once cell pools had reached >95% viability (14–21 days depending on selection stringency), genomic DNA was prepared from samples and the average number of transposon integrations per genome was determined using ddPCR. Figure 3 shows that with increasing concentrations of MTX, a linear increase in the average transposon copy number was observed. Thus, the average number of transposons integrated per genome can be controlled by the stringency of selection.
Figure 3.
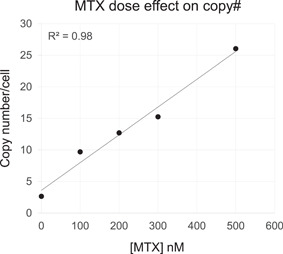
Correlation between selection stringency and integrated Leap‐In transposon copy number in stable DG44 pools selected in HT‐media. The stringencies were controlled by the indicated MTX concentrations during the entire duration of pool selections without any stepwise increase
Several additional pools were generated, using transposons with further attenuated DHFR expression. Average transposon copies per cell were measured for these pools also. The specific productivity of each pool was then determined in 25‐ml shake flask‐scale fed‐batch experiments. A strong linear correlation (R 2 = 0.84) was observed between the average number of integrated transposons per cell and the corresponding specific productivities (Figure 4).
Figure 4.
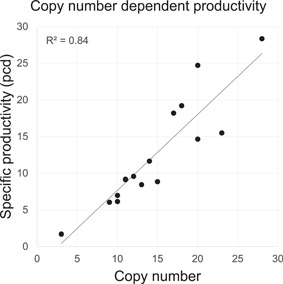
Correlation between transgene copy number and specific productivity in CHO DG44 stable pools established under different selection stringencies. CHO, Chinese hamster ovary
An increase in the number of transposons integrated into the CHO DG44 genome results in a proportional increase in the specific productivity of the pool, a feature of a copy number‐dependent expression system where the transgene expression cassettes are faithfully integrated into and insulated from the surrounding genome elements. It also demonstrates that, on average, each integration via transposition maintains the functional integrity of the transposon (LC, HC, regulatory elements, and selection cassette) without deleterious recombination or silencing.
3.3. Leap‐In transposase‐mediated stable pools established from GS−/− CHO cells
We designed Leap‐In transposons with five different glutamine synthetase cassettes with progressively decreasing glutamine synthetase expression levels, and thus progressively increasing selection stringencies, in the order h+, ht, ht+, hxt, and hxt+. Open reading frames encoding the same heavy and light chain of an antibody were synthesized and cloned into these five transposons. The heavy and light chains were under the control of identical EF1 promoter‐based regulatory elements and flanked by the same HS4 and D4Z4 insulators; hence, the five transposons differed only in the glutamine synthetase selection cassette. Transposons were co‐transfected with Leap‐In Transposase mRNA into GS knockout cells. Stable pools were subsequently established by transferring the cells into glutamine‐free media 48‐h post‐transfection. No additional selection pressure (e.g., MSX addition) was used during the selections. Figure 5a shows the viability selection curves for the five pools.
Figure 5.
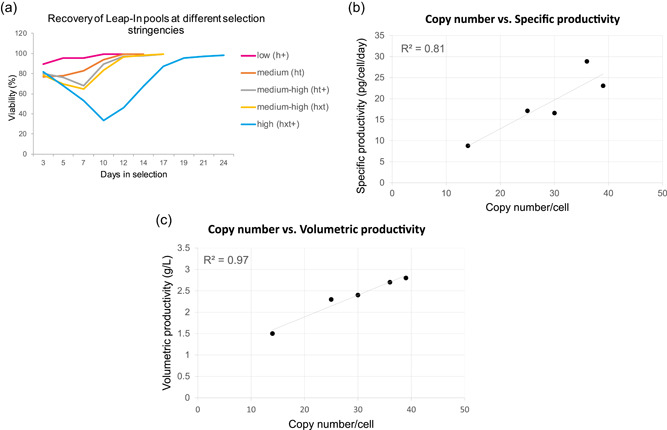
(a) Viability of GS KO cells transfected with different selection stringency transposons during stable pool selection in glutamine‐free media. (b, c) Correlation between the integrated transposon copy number and specific productivity (b) and volumetric productivity (c) in the five Leap‐In‐mediated stable pools established at different selection stringencies [Color figure can be viewed at wileyonlinelibrary.com]
After recovery, each pool was analyzed for average transposon copy number and grown in a 10‐day fed‐batch culture. Specific productivities and copy numbers are shown in Table 2.
Table 2.
Day 10 volumetric productivities, calculated specific productivities, and integrated transgene copy numbers
| Selection stringency | Copy/cell | Productivity on Day 10 | ||
|---|---|---|---|---|
| Designation | Strength | Volumetric (g/L) | Specific (pcd) | |
| h+ | Low | 14 | 1.5 | 8.8 |
| ht | Medium | 25 | 2.3 | 17.1 |
| hxt | Medium‐high | 30 | 2.4 | 16.55 |
| ht+ | Medium‐high | 39 | 2.8 | 23.05 |
| hxt+ | High | 36 | 2.7 | 28.85 |
The pool with the transposon conferring the least stringent (h+) selection recovered to >95% viability in less than a week. Pools with three intermediate stringencies (ht, hxt, and ht+) had reached 95% viability within about 12 days, but the most stringent (hxt+) selection took around 20 days to recover. Similar to the DHFR‐based selection (Figure 4), there is a correlation between the selection stringency, the integrated transgene copy numbers, and the specific productivities of the pools (Figure 5b).
These data differentiate the Leap‐In‐mediated stable GS pools from pools established by random integration where such correlation cannot be established (Noh et al., 2018). Strikingly, the volumetric productivity values correlated strongly (R 2 = 0.97) with the copy number (Figure 5c), further distinguishing the copy number‐dependent Leap‐In‐mediated expression from random integration systems.
3.4. Clonal distributions from Leap‐In pools are biased toward high‐expressing clones
Single cells were deposited into 96‐well plates (one plate for each pool) and monoclonal lines were derived from the four most productive pools presented in Table 2. We observed that under identical cloning conditions, we obtained gradually fewer viable clones from more stringently selected pools (Table 3). After expansion, the productivity of these clones was measured in 7‐day fed‐batch cultures in 24 deep well plates. The distribution of productivities is shown in Figure 6.
Table 3.
Data shown in Figure 6 were analyzed to show the distribution of productivity. For each pool, Q1 is the fraction of clones producing between 75% and 100% of the amount of antibody made by the most productive clone. Similarly, Q2 is the fraction of clones producing between 50% and 75%, Q3 is the fraction of clones producing between 25% and 50%, and Q4 is the fraction of clones producing between 0% and 25% of the amount of antibody made by the most productive clone
| Selection stringency | |||||
|---|---|---|---|---|---|
| Level | Low | Medium‐high | High | ||
| ht | ht+ | hxt | hxt+ | ||
| % of cells in | Q1 | 27.5 | 42.55 | 46.88 | 57 |
| Q2 | 49.3 | 36.17 | 34.38 | 29 | |
| Q3 | 18.8 | 17.02 | 15.63 | 14 | |
| Q4 | 4.3 | 4.26 | 3.13 | 0 | |
| Number of clones tested | 69 | 47 | 32 | 14 | |
| Outgrowth rate | 72% | 49% | 33% | 15% | |
Figure 6.
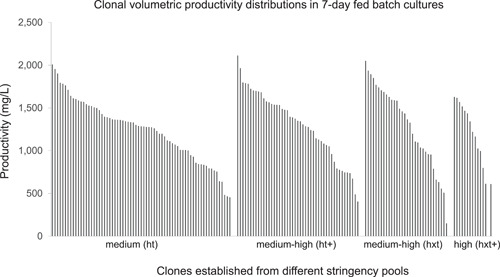
Productivity of clones isolated from four stable pools expressing the same monoclonal antibody but established under different selection stringencies. Productivities were measured by Octet at the end of 7‐day 24 deep well plates scale fed‐batch cultures
The productivity of the highest expressing clones from the first three pools was similar, ~2 g/L in the 7‐day fed‐batch cultures. Interestingly, the highest expressing clone from the most stringent (hxt+) selection produced just over 1.5 g/L, significantly less than the top clone from the other three pools. On the basis of these observations, in our current platform selection system, Leap‐In stable pools established by medium‐high stringency selections are the preferred choice to isolate clonal cell lines.
When the clonal productivity ranges of the four pools were divided into four equal quartiles and the clones sorted into these quartiles by their productivities, selection stringency‐dependent trends can be observed (Table 3). The selection stringency directly correlates with the Q1 fractions. The relatively small Q3 and Q4 fractions show a small inverse correlation with the selection stringency. The combined Q1 + Q2 fractions represent >75% of all clones, a remarkable characteristic bias toward high producer cells in Leap‐In‐generated stable pools.
The data in Table 3 and Figure 6 demonstrate that less than 100 monoclonal stable clones are sufficient to isolate high producer clones from Leap‐In‐mediated stable pools, even when established at medium stringency selection. In contrast, from random integration pools, many thousands of clones need to be screened in a successful cell line development campaign (Le et al., 2018). The unique clonal distributions of the Leap‐In‐mediated stable pools decrease clone screening requirements by one to two orders of magnitude, resulting in drastically reduced resource requirements.
3.5. The Leap‐In transposons are stable in the CHO genome
Genetic stability is a critical quality attribute (CQA) of commercial manufacturing cell lines, yet the current mainstream random integration mechanisms cannot control or even predict genetic stability. The frequently rearranged tandem transgene integrants, often exacerbated when gene amplification methods are used, result in genetically unstable recombinant loci.
We have analyzed the genetic stability of more than 80 clones derived from multiple external and internal R&D programs. To do so, the cells were passaged for between 60 and 90 population doublings, with and without selective pressure. Assessment of integrated copy numbers by ddPCR and productivity assessments from representative production cultures demonstrate that >90% of the clones established by Leap‐In transposons maintain the T0 productivity and copy number levels (Figure 7a,b). In the remaining <10% clones, productivity is decreased by less than 30% of the T0 value, a value generally considered as acceptable stability within the industry. In short, in Leap‐In‐mediated cell line development programs, instability during the ranking process further reduces the number of clones required to handle and triage during a development program.
Figure 7.
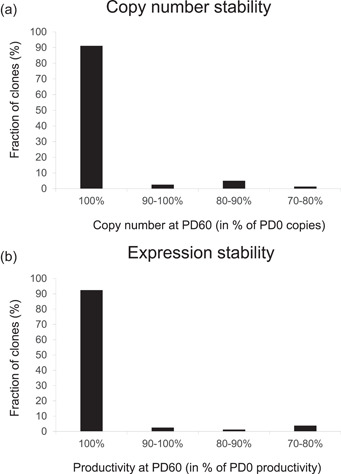
(a, b) Stability of copy number and productivity in Leap‐In stable clones
We also made a more in‐depth assessment of the genetic stability of one clone over 90 population doubling. Nucleotide sequence‐level data were derived from the recombinant transgene insertions using TLA technology performed by Cergentis (Hottentot et al., 2017). There were 58 transposase‐mediated integration sites detected in the cells analyzed at the T0 and the PD90 time points. More important, their flanking genomic sequences were identical at the two timepoints (Figure S1). The integration sites were mapped to the host genome scaffolds and were found at the exact same positions at both timepoints, demonstrating the consistent structural stability of Leap‐In transposase‐mediated stable integrations.
3.6. Products from Leap‐In pools and clones are highly comparable
The more homogeneous distribution of clonal productivities means that pool titers and clone titers are very comparable. In other words, the Leap‐In pools reliably predict their derivative clonal productivities. Data from nine different programs are shown in Table 4. This high degree of correlation means that pools can, in principle, be used for early process development work even before single‐cell cloning has begun. This also means that one can screen Leap‐In pools established using various vector elements, chain ratios, coding sequences, and so on, and be assured that the performance in the pools is predictive for the derivative clones.
Table 4.
Comparison of Leap‐In‐mediated stable pool and derivative clonal productivities in various cell line development (CLD) programs
| Productivity (mg/L) | Rel. stdev% | ||
|---|---|---|---|
| Pool | Clonal average | ||
| CLD program 1 | 3367 | 3824 | 6.24 |
| CLD program 2 | 3900 | 4480 | 7.21 |
| CLD program 3 | 3588 | 3621 | 17.73 |
| CLD program 4 | 4432 | 4483 | 4.75 |
| CLD program 5 | 3125 | 3039 | 20.17 |
| CLD program 6 | 3600 | 3891 | 5.38 |
| CLD program 7 | 4500 | 5333 | 4.72 |
| CLD program 8 | 3856 | 4024 | 3.98 |
| CLD program 9 | 4299 | 4970 | 9.23 |
The comparability of stable pools and clones is not limited to productivity. Three pools (ht, hxt, and ht+) described in Table 2 and clones from Figure 6 were used as models. We looked at two global physicochemical CQAs: charge profile (Figure 8a) and N‐linked glycan distribution (Figure 8b) in the same monoclonal antibody produced by the three Leap‐In‐mediated stable pools and randomly isolated high producer derivative clones from each.
Figure 8.
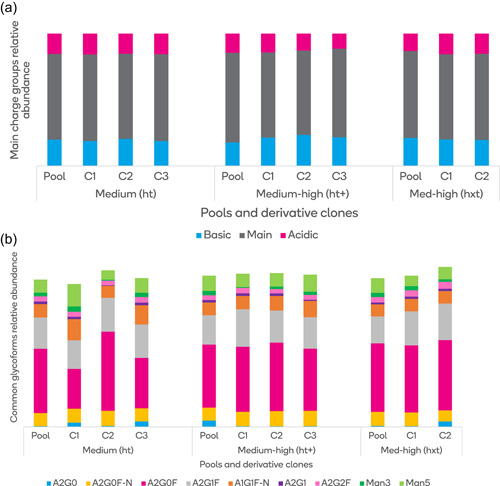
(a) The distribution of basic, main, and acidic charge groups produced by the three stable pools, established at different stringencies, and from their derivative clones. The data indicate that there is a strong charge profile comparability between the stable pools and the derivative clones. (b) The N‐linked glycan distribution in the three stable pools and derivative clones [Color figure can be viewed at wileyonlinelibrary.com]
The data in Figure 8b present the N‐linked glycan distributions in the three stable pools and derivative clones. Whereas the two higher stringency pools produce glycosylation profiles comparable to their derivative clones, the clones isolated from the lower stringency pool show more diversity in N glycan composition. This observation may guide selection stringency choice, depending on how homogeneous or diverse glycan structures are preferred.
3.7. Leap‐In pools are stable enough for process development and biological product DS manufacturing
On the basis of their productivity and product quality comparability, the Leap‐In‐mediated stable pools can be considered as representative cell substrates to the derivative final clones. This suggests that the pools may be used to support process development, analytical development, and IND‐enabling tox manufacturing. These activities can be initiated while the final clones are being identified, shortening the CMC development timelines.
As discussed, the individual Leap‐In clones demonstrate remarkable genetic stability. However, the inherent clonal growth differences driving population dynamics may change the clonal distribution of the stable pools over time. To evaluate whether changes in population dynamics permit the pools to be used for representative drug substance manufacturing, the same three stable pools presented in Figure 5 were subjected to a standard stability passage study in glutamine‐free formulation for 30 population doublings. At the end of the passages, fed‐batch production runs were performed using the Time 0 and the PD30 pools. The results are shown in Figure 9a–c.
Figure 9.
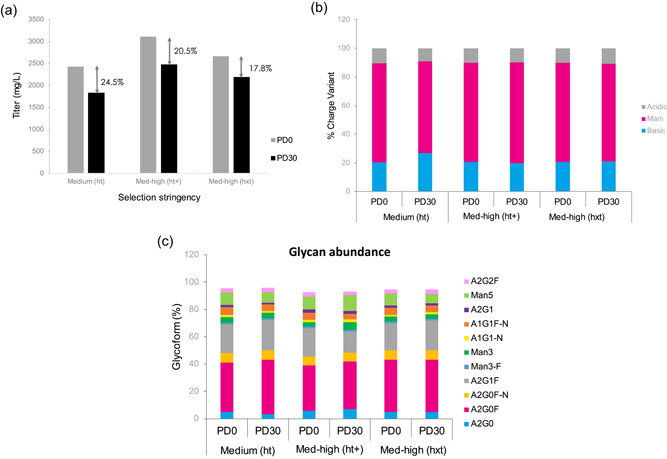
(a) The volumetric productivity in Leap‐In stable pools. Gray bars: Time 0, black bars: PD30. The arrows indicate the productivity change expressed in % of the T0 value. (b) Charge profile comparability between T0 and PD30 stable pools. (c) N‐linked glycosylation comparability between the T0 and PD30 stable pools [Color figure can be viewed at wileyonlinelibrary.com]
As expected, the volumetric productivities are lower in the PD30 pools as compared with their T0 counterparts. The degree of productivity decrease inversely correlates with the fraction of clones in Quartile 1 encompassing the top producers (R 2 = 0.99, data not shown). Nevertheless, the productivity change, observed in all three pools, is less than 25%. This is below the conventionally recognized ~30% productivity loss acceptance criteria for clonal stability.
Analytical comparability was established for charge profile and N‐linked glycosylation of the products made by the T0 and the PD30 pools (Figure 9b,c).
The data presented in Figure 9b,c demonstrate a high degree of product quality comparability between T0 and PD30 pools. In total, 30 population doublings, starting from a vial of 10E7 cells, are sufficient to seed a 5000‐L bioreactor that, at multi‐gram per liter, would produce kilogram quantities of bulk drug substance. The data endorse the Leap‐In system as a viable alternative to other approaches, aiming to utilize stable pools to shorten CMC development timelines (Hu et al., 2017; Rajendra et al., 2017; Scarcelli et al., 2017).
3.8. Summary
Compared with conventional random integration‐based technologies, the Leap‐In transposase‐mediated stable cell line development provides an array of valuable features, from genetically stable integrations through high expression levels of consistent ratios of multi‐cistronic units to robust pool‐to‐clone productivity and product quality comparability.
The structural and functional integrity of the Leap‐In‐mediated stable integrants leads to more homogeneous stable pools, where ~97% of the recombinant integration sites represent single and exact copies of the expression construct. As a consequence, there is a strong productivity and product quality comparability between Leap‐In‐mediated stable pools and their derivative clones.
This characteristic pool‐to‐clone comparability enables early de‐risking of the development programs. This is accomplished by triaging a number of representative predictive stable pools for optimal productivity and product quality, thus changing the traditional manufacturing cell line development paradigm. The productivity and product quality decisions and selections at pool ranking stage reliably predict the productivity and product quality spectrum of the final clones. Once the optimal stable pool is identified, only a small (<100) number of clones need to be screened and ranked to isolate genetically stable clones with the desired productivity and CQA profile. The observed high producer clone enrichment is more prominent in the Leap‐In‐mediated pools than by the alternative, industrially relevant epigenetic regulatory elements including UCOEs, insulators, and MARs that have been described elsewhere (Saunders et al., 2015).
Unlike the pools established by traditional expression technologies, the Leap‐In stable pools maintain genetic, productivity, and product quality stability. This unique stability of pools enables the early and efficient manufacturing of representative drug substances for analytical and formulation development studies as well as material for toxicology studies or even Phase I clinical trials. Also, based on their productivity and robust genetic stability, the Leap‐In‐mediated stable clones are attractive potential candidates to support perfusion‐based manufacturing processes, where in addition to solving engineering and logistical challenges, the development of production clones best suited for extended operation modes is also a mission‐critical task (Bielser et al., 2018).
In summary, the Leap‐In‐mediated manufacturing cell line development workflow results in high productivity, predictable product quality, robust genetic stability, and shortened CMC development timelines with significant resource reduction.
AUTHOR CONTRIBUTIONS
The first four authors, Sowmya Rajendran, Sowmya Balasubramanian, Lynn Webster, and Maggie Lee, contributed equally to this study.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors wish to thank Dr Claes Gustafsson and Dr Oren Beske for their critical comments and Medini Gore for the illustrations.
Rajendran, S. , Balasubramanian, S. , Webster, L. , Lee, M. , Vavilala, D. , Kulikov, N. , Choi, J. , Tang, C. , Hunter, M. , Wang, R. , Kaur, H. , Karunakaran, S. , Sitaraman, V. , Minshull, J. , & Boldog, F. (2021). Accelerating and de‐risking CMC development with transposon‐derived manufacturing cell lines. Biotechnology and Bioengineering. 118, 2301–2311. 10.1002/bit.27742
Sowmya Rajendran, Sowmya Balasubramanian, Lynn Webster and Maggie Lee contributed equally to this study.
Contributor Information
Jeremy Minshull, Email: jminshull@atum.bio.
Ferenc Boldog, Email: fboldog@atum.bio.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author on request.
REFERENCES
- Balasubramanian, S. , Peery, R. B. , Minshull, J. , Lee, M. , White, R. , Kelly, R. M. , & Barnard, G. C. (2018). Generation of high expressing Chinese hamster ovary cell pools using the Leap‐In transposon system. Biotechnology Journal, 13, 10. [DOI] [PubMed] [Google Scholar]
- Bielser, J.‐M. , Wolf, M. , Souquet, J. , Broly, H. , & Morbidelli, M. (2018). Perfusion mammalian cell culture for recombinant protein manufacturing—A critical review. Biotechnology Advances, 36, 1328–1340. [DOI] [PubMed] [Google Scholar]
- Boldog, F. (2019). A genomic case of structure‐function relationship: The Leap‐In transposase system. Proceedings of Cell Line Development and Engineering US.
- Cacciatore, J. J. , Chasin, L. A. , & Leonard, E. F. (2010). Gene amplification and vector engineering to achieve rapid and high‐level therapeutic protein production using the DHFR‐based CHO cell selection system. Biotechnology Advance, 28(6), 673–681. [DOI] [PubMed] [Google Scholar]
- Carver, J. , Ng, J. , Zhou, M. , Ko, P. , Zhan, D. , Yim, M. , Shaw, D. , Snedecor, B. , Laird, M. W. , Lang, S. , Shen, A. , & Hu, Z. (2020). Maximizing antibody production in targeted integration host by optimization of subunit gene dosage and position. Biotechnology Progress, 36(4):e2967. [DOI] [PubMed] [Google Scholar]
- Fan, L. , Kadura, I. , Krebs, L. E. , Hatfield, C. C. , Shaw, M. M. , & Frye, C. C. (2012). Improving the efficiency of CHO cell line generation using glutamine synthetase gene knockout cells. Biotechnology and Bioengineering, 109(4), 1007–1015. [DOI] [PubMed] [Google Scholar]
- Florin, L. , Lipske, C. , Becker, E. , & Kaufmann, H. (2011). Supplementation of serum free media with HT is not sufficient to restore growth properties of DHFR−/− cells in fed batch processes‐ Implications for designing novel CHO‐based expression platforms. Journal of Biotechnology, 152(4), 189–193. [DOI] [PubMed] [Google Scholar]
- Hottentot, Q. P. , van Min, M. , Splinter, E. , & White, S. J. (2017). Targeted locus amplification and next generation sequencing. In White S. J., & Cantsilieris S. (Eds.), Genotyping: Methods and protocols. Methods in molecular biology (Vol. 1492, pp. 185–196). Humana Press. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Hsu, W. , Pynn, A. , Ng, D. , Quicho, D. , Adem, Y. , Kwong, Z. , Mauger, B. , Joly, J. , Snedecor, B. , Laird, M. W. , Andersen, D. , & Shen, A. (2017). A strategy to accelerate protein production from a pool of clones in Chinese hamster ovary cells for toxicology studies. Biotechnology Progress, 33(6), 1449–1455. [DOI] [PubMed] [Google Scholar]
- Kawakami, K. , Shima, A. , & Kawakami, N. (2000). Identification of a functional transposase of the Tol2 element, an Ac‐like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proceedings of the National Academy of Sciences of the United States of America, 97(21), 11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. , O'Callaghan, M. O. , Droms, K. A. , & James, D. C. (2011). A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnology and Bioengineering, 108(10), 24342446. [DOI] [PubMed] [Google Scholar]
- Klein, C. , Sustmann, C. , Thomas, M. , Stubenrauch, K. , Croasdale, R. , Schanzer, J. , Brinkmann, U. , Kettenberger, H. , Regula, J. T. , & Schaefer, W. (2012). Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. mAbs, 4(6), 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattenmayer, C. , Loeschel, M. , Steinfellner, W. , Trummer, E. , Mueller, D. , Schriebl, K. , Vorauer‐Uhl, K. , Katinger, H. , & Kunert, R. (2006). Identification of transgene integration loci of different highly expressing recombinant CHO cell lines by FISH. Cytotechnology, 51, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, K. , Tan, C. , Gupta, S. , Guhan, T. , Barkhordarian, H. , Lull, J. , Stevens, J. , & Munro, T. (2018). A novel mammalian cell line development platform utilizing nanofluidics and optoelectro positioning technology. Biotechnology Progress, 34(6), 1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, J. G. , Yant, S. R. , Meuse, L. , Huang, Z. , Xu, H. , & Kay, M. A. (2003). Helper‐independent Sleeping Beauty transposon‐Transposase vectors for efficient non‐viral gene delivery and persistent gene expression in vivo. Molecular Therapy, 8(4), 654–665. [DOI] [PubMed] [Google Scholar]
- Miskey, C.s , Izsvak, Z.s , Plasterk, R. H. , & Ivics, Z. (2003). The Frog Prince: A reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Research, 31(23), 6873–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R. , Fain‐Thornton, J. , & Craig, N. L. (2008). PiggyBac can bypass DNA synthesis during cut and paste transposition. The EMBO Journal, 27(7), 1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, B. , Becker, P. B. , & Gopfert, U. (2015). CMV promoter mutants with a reduced propensity to productivity loss in CHO cells. Scientific Reports, 5, 16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesmelova, I. V. , & Hackett, P. B. (2010). DDE transposases: Structural similarity and diversity. Advanced Drug Delivery Reviews, 62(12), 1187–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, S. M. , Shin, S. , & Lee, G. M. (2018). Comprehensive characterization of glutamine synthetase‐mediated selection for the establishment of recombinant CHO cells producing monoclonal antibodies. Scientific Reports, 8, 5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra, Y. , Balasubramanian, S. , McCracken, N. A. , Norris, D. L. , Lian, Z. , Schmitt, M. G. T. , Frye, C. C. , & Barnard, G. C. (2017). Evaluation of piggyBac‐mediated CHO pools to enable material generation to support GLP toxicology studies. Biotechnology Progress, 33(6), 1436–1448. [DOI] [PubMed] [Google Scholar]
- Saunders, F. , Sweeney, B. , Antoniou, M. N. , Stephens, P. , & Cain, K. (2015). Chromatin function modifying elements in an industrial antibody production platform‐comparison of UCOE, MAR, STAR and cHS4 elements. PLOS One, 10(4):e0120096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli, J. J. , Shang, T. Q. , Iskr, T , Allen, M. J. , & Zhang, L. (2017). Strategic deployment of CHO expression platforms to deliver Pfizer's monoclonal antibody portfolio. Biotechnology Progress, 33(6), 1463–1467. [DOI] [PubMed] [Google Scholar]
- Spiess, C. , Zhai, Q. , & Carter, P. J. (2015). Alternative molecular formats and therapeutic applications for bispecific antibodies. Molecular Immunology, 67(2 part A), 95–106. [DOI] [PubMed] [Google Scholar]
- Tharmalingam, T. , Barkhordarian, H. , Tejeda, N. , Daris, K. , Yaghmour, S. , Yam, P. , Lu, F. , Goudar, C. , Munro, T. , & Stevens, J. (2018). Characterization of phenotypic and genotypic Diversity in subclones derived from a clonal cell line. Biotechnology Progress, 34, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa, K. (2015). piggyBac transposon. Microbiology Spectrum, 3(2):MDNA3‐0028‐2014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on request.


