Abstract
Caenorhabditis elegans is the first and only metazoan model that enables whole-body gene knockdown by simply feeding their standard laboratory diet, E. coli, carrying RNA interference (RNAi)-expressing constructs. The simplicity of the RNAi treatment, small size, and fast reproduction rate of C. elegans allow us to perform whole-animal high-throughput genetic screens in wild-type, mutant, or otherwise genetically modified C. elegans. In addition, more than 65% of C. elegans genes are conserved in mammals including human. In particular, C. elegans metabolic pathways are highly conserved, which supports the study of complex diseases such as obesity in this genetically tractable model system. In this chapter, we present a detailed protocol for automated high-throughput whole-animal RNAi screening to identify the pathways promoting obesity in diet-induced and genetically driven obese C. elegans. We describe an optimized high-content screening protocol to score fat mass and body fat distribution in whole animals at large scale. We provide optimized pipelines to automatically score phenotypes using the open-source CellProfiler platform within the context of supercomputer clusters. Further, we present a guideline to optimize information workflow from the automated microscope to a searchable database. The approaches described here enable unveiling the whole network of gene-gene and gene-environment interactions that define metabolic health or disease status in this proven model of human disease, but similar principles can be applied to other disease models.
Keywords: Whole-animal screening, C. elegans, Genetics of obesity, Automated microscopy, Live screening, Fat screening
1. Introduction
Caenorhabditis elegans has become a leading model organism to tackle important biomedical questions. It is a powerful system due to its short life span (less than 30 days), high reproduction rate (300–500 eggs per animal in total with peak around 150 eggs/day), fast development (90 h from hatching egg to adult at 15 °C), transparency of the organism, and fully sequenced and annotated genome [1]. Furthermore, C. elegans has a stereotypical distribution of cells identical from worm to worm and its neuronal connectome has been completely traced, providing great advantages for studying biological processes that rely on proper cell-to-cell, tissue-to-tissue, or whole-body communication [2].
The most basic biological processes and their effectors (genes) are conserved from C. elegans to human, which makes research in C. elegans translatable to mammals and even humans. Although there are important differences in terms of tissue and organ function, and some key mammalian metabolic players are missing (i.e., leptin) most of the core lipid, sugar, and protein metabolism pathways are highly conserved between worms and mammals. For example, adipose triglyceride lipase, hormone-sensitive lipase, and lysosomal lipases play essential roles in fat storage and mobilization in C. elegans and mammals [3, 4]. TOR kinase, AMPK, sterol response element-binding protein, and many other transcription factors similarly control metabolism-gene regulation and cell responses to nutrients in C. elegans and mammals. Loss of function of such regulators causes severe metabolic defects such as obesity in worms [5-7]. In addition, several pathways systemically regulating metabolism are also conserved. Importantly, the major components of the insulin-signaling pathway are conserved and chronically defective insulin signaling leads to symptoms associated with insulin resistance including obesity in worms [8]. These and other examples suggest that unwinding C. elegans metabolic players and how they are modulated by changing environmental factors, including food quantity and quality or genetic defects, would help us better understand metabolic disease in humans.
C. elegans is also the first model system enabling whole-genome systemic RNA interference in vivo. This capability is particularly critical to understand metabolic disease since metabolic status is the product of coordinated action of multiple cells and organ systems. In C. elegans, RNAi can be delivered by simply feeding worms with bacteria that overproduces double-stranded RNA against a specific worm gene upon isopropylthiogalactoside (IPTG) induction [9]. RNAi libraries developed by Julie Ahringer’s group cover close to 87% of the worm genome and these feeding constructs are readily available from Source BioScience [10].
The fast development and reproduction rate imply that C. elegans can be used as an in vivo model to perform high-throughput (HT) screening. Automated and quantitative high-throughput screening methods have been developed and improved throughout the past years, and these HT assays allow us to identify lead compounds in in vivo HT chemical screens and genes in RNAibased screens [11].
Previously we described an improved oil red O staining technique to provide an image-based quantitative measurement of fat mass that correlated to biochemically measured triglyceride mass [12]. This method uses isopropanol to preserve worm-tissue structure, which is otherwise fragile (i.e., by paraformaldehyde-based fixation). This method can be used as a powerful quantitative technique to identify genes that alter body fat mass in C. elegans [12]. Here we present several advances to the processing and imaging of the worm populations, and we delineate considerations and solutions to the handling of the massive amount of data generated by image-based HT screening of C. elegans.
Comparing to our previous method, we optimized conditions in preparation, RNAi treatment, and data analysis. In this chapter, we describe detailed step-by-step procedures with improvement on reducing non-RNAi variants such as position of the wells, growth conditions, and precipitation of ORO dyes, and moreover we incorporate the data processing power from supercomputer to enable analyses of high-resolution composite images from high-content screening experiments.
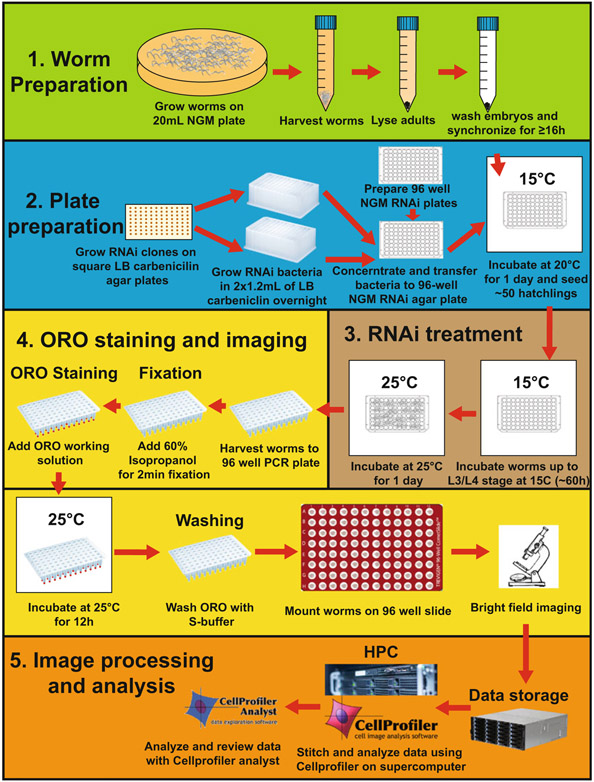
Studying metabolic diseases such as obesity is a challenge due to the complex genetic interactions between multiple pathways throughout development, between cells and organs, and the modifier effect of the environment, in particular the diet. Our whole-animal automated high-throughput genetic screening method provides an approach to unveil the genetic pathways underlying genetic- and diet-induced obesity in intact animals (Fig. 1).
Fig. 1.
RNAi screening workflow. Worm preparation is shown in green (see Subheading 3.3); bacterial RNAi preparation is shown in blue (see Subheading 3.2); worm RNAi treatment is shown in brown (see Subheading 3.4); ORO staining and imaging procedures are shown in yellow (see Subheadings 3.5 and 3.6); and image analysis and data transfer and processing are shown in orange (see Subheading 4)
2. Materials
2.1. Bacteria and Worm Strains
RNAi Library from Ahringer laboratory can be acquired from Source BioScience: http://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ (see Note 1).
C. elegans strains NL2099 (rrf-3(pk1426)II (see Note 2) and GMW004 (rrf-3(pk1426) II; daf-2(e1368) III) can be acquired from Caenorhabditis Genetics Center (CGC) [13]: https://cbs.umn.edu/cgc/home (see Note 3).
L4440 empty vector transformed bacteria can be obtained from Addgene: https://www.addgene.org/1654/ (see Note 4).
daf-16 RNAi bacteria can be found in Ahringer RNAi library, number 1717, plate 18, well H1 (see Note 5).
2.2. Reagents
Potassium phosphate buffer: 108.3 g KH2PO4, 35.6 g K2HPO4, and water to 1 L, pH 6.0. Sterilize by autoclaving.
NGM RNAi agar: 3 g NaCl, 2.5 g peptone, 17 g agar, and water to 1 L. Sterilize by autoclave and cool to 55 °C. After cooling to 55 °C add in the following order: 1 mL of 5 mg/mL cholesterol dissolved in ethanol, 1 mL of 1 M CaCl2, 1 mL of 1 M MgSO4, 25 mL of 1 M potassium phosphate, pH 6.0, IPTG (1 mM final concentration), and 1 mL of 50 mg/mL carbenicillin.
LB broth: 10 g Bacto-tryptone, 5 g yeast extract, 10 g NaCl, and water to 1 L, pH to 7.5 with NaOH, and sterilize by autoclaving. Before use, add 1 mL of 50 mg/mL carbenicillin.
LB agar: 10 g Bacto-tryptone, 5 g yeast extract, 10 g NaCl, 17 g agar, and water to 1 L, pH to 7.5 with NaOH, and sterilize by autoclaving. Cool to around 55 °C and add 1 mL of 50 mg/mL carbenicillin.
S-basal without cholesterol buffer (S-buffer): 5.85 g NaCl, 1 g K2HPO4, 6 g KH2PO4, water to 1 L. Sterilize by autoclaving.
Lysis buffer: 195 μL 10 N NaOH, 600 μL sodium hypochlorite, and water to 3 mL
S-buffer with 0.01% Triton-X: Add 100 μL of Triton-X 100% to 1 L of S-buffer (resuspend with stirring bar for ≥1 h).
60% Isopropanol: Mix 30 mL of 100% isopropanol with 20 mL of water in a 50 mL conical tube.
Oil red O stock solution (0.5%): Resuspend 0.25 g of oil red O in 50 mL of 100% isopropanol by overnight shaking at room temperature. Prepare at least 2 days before use. Can be stored for ≥4 weeks at room temperature.
Diluted S-buffer (20%): 20 mL S-buffer and 80 mL of distilled water.
2.3. Supplies and Equipment
Omnitray single-well square plates.
96-pin replicator.
96-Well clear V-bottom 2 mL polypropylene deep-well plate.
96-Well cell culture plates.
Breathe-Easy sealing membrane.
HT115 E. coli strain.
AlumaSeal 96™ sealing film.
Sealing mats for 96-well PCR plates.
96-Well channel vacuum manifold.
96-Well PCR plates.
Petri dish (100 mm × 15 mm).
Sterile reservoirs.
37 °C Shaking incubator with holders for 96-well plates.
Benchtop centrifuge with 96-well plate adaptors.
Vertical flow biological hood.
Comet assay 96-well slides.
Upright or inverted microscope with an automated stage capable of doing high-throughput tile capture and stitching of images of 96-well slides (see Note 6).
Digital Triple Heat Block.
Multichannel pipettes and regular pipettes.
3. Methods
3.1. Reagent Preparation
3.1.1. Preparing Agar Plates (1 Week Before Experiment)
Make and autoclave 1 L LB agar as described in Subheading 2.
Place LB agar in a 60 °C water bath for 5–10 min to cool down.
Add 1 mL of 50 mg/mL carbenicillin.
Pour LB agar into plates in a hood. 20 mL of LB agar can be poured into 100 mm Petri dish or omnitray single-well square plates.
Place the plates in the hood for 30 min to solidify.
Store the plates in a sealed box at 4 °C. LB agar plates can be stored at 4 °C for up to 1 month (see Note 7).
3.1.2. Preparing NGM RNAi Plates (1 Week Before Experiment)
Make and autoclave 1 L NGM agar as described in Subheading 2.
Place NGM agar in a 60 °C water bath for 5–10 min to cool down.
In a biological hood, add in the following order and thoroughly mix after each ingredient: cholesterol, CaCl2, MgSO4, potassium phosphate, IPTG, and carbenicillin (volumes and stock concentrations listed in materials). Keep molten medium in water bath until pouring plates in the hood.
For a 100 mm petri dish, pour 20 mL of NGM agar in the hood and go to step 9. For pouring 96-well cell culture plates, go to step 5.
Right after step 3, take a heat block pre-warmed to 70 °C into the hood.
Bed clean aluminum foil in the heat block, and place a sterile reservoir and a 96-well cell culture plate on top of the foil.
Pour NGM agar into the sterile reservoir.
Use multichannel pipettes to transfer 150 μL of NGM agar to every well of the 96-well plate (see Note 8).
Place the plates in the hood for 30 min to solidify.
Store the plates in a sealed box in 4 °C. NGM RNAi agar plates can be stored at 4 °C for up to 1 month.
3.1.3. Preparing Oil Red O Working Solution (First Day of Experiment)
Prepare 0.5% oil red O stock solution several days beforehand.
Filter the oil red O stock solution with a 0.45 μm pore size filter.
Prepare a 60% working solution with filtered sterile water and shake for ≥2 h at room temperature.
Filter the solution with a 0.45 μm filter and shake for another 2 days at room temperature prior to usage.
3.2. Bacteria RNAi Strain Preparation
3.2.1. Growing Bacteria Colonies on Square LB Agar Plate (3 Days Before Experiment)
Clean the bench area with Cavicide™ and turn on the gas and fire on the bench (this step can also be performed in a hood).
Prepare sterilized water, 95% ethanol, and 10% bleach in three separate containers (e.g., empty 200 μL pipette boxes) next to the fire.
Briefly dip the 96-pin replicator into 10% bleach, rinse thoroughly with water, and wet in 95% ethanol. Flame every side of the replicator to make sure that it is sterile.
Place the replicator back into 95% ethanol.
Transport RNAi library to the bench with dry ice from −80 °C.
Remove the seal of the RNAi library plates next to the flame.
Flame the replicator quickly to evaporate the excess ethanol and wait for 5–15 s for it to cool down (see Note 9).
Dip the tips of the replicator into the library plate and make sure that they touch the bacteria in every single well.
Quickly transfer and stamp the tips of replicator on the LB square plate, and make smooth rounded movements to increase bacterial growth area without overlapping RNAi clones (see Note 10).
Briefly dip the replicator into 10% bleach and repeat steps 3 and 4 for each successive plate.
Re-seal the library plates with AlumaSeal 96™ Sealing Film and store back at −80 °C (do not ever let the library thaw!).
Incubate the RNAi clones seeded in the LB square plate at 37 °C overnight.
For control RNAi clones, use sterile toothpicks or wire loops to streak bacteria on the 10 cm LB agar carbenicillin 50 μg/mL plates from the glycerol stock and incubate at 37 °C overnight.
After incubation, record the RNAi clones that have no colonies.
The LB agar plates can be parafilmed and stored at 4 °C for up to 2 weeks.
3.2.2. Growing Bacteria in 96 Deep-Well Plates (2 Days Before Experiment)
In the biological hood, fill the 96 deep-well plate with 1200 μL/well of LB broth with 50 μg/mL carbenicillin using a multichannel pipette.
Sterilize the bench with Cavicide™ and turn on the gas and fire on the bench (this step can also be performed in hood).
Prepare sterilized water, 95% ethanol, and 10% bleach in three separate containers (e.g., empty 200 μL pipette boxes) next to the fire.
Dip the 96-pin replicator into 10% bleach, thoroughly rinse with water, then briefly dip in 95% ethanol, and flame every side of the replicator to make sure that it is sterile.
Dip replicator back in 95% ethanol, and quickly flame again to evaporate excess ethanol. Wait for 5–15 s for replicator to cool down.
Touch the replicator pins to the mini RNAi bacterial lawns.
Dip the tips of the replicator into the 1.2 mL LB broth carbenicillin in two 96 deep-well plates (see Note 11).
Re-sterilize replicator by briefly dipping it into 10% bleach and repeat steps 4 and 5 sequentially for the next plates.
Use sterile toothpicks or wire loops to pick and transfer colonies from positive and negative control 10 cm LB agar carbenicillin 50 μg/mL plates to all empty wells of a 96 deep-well plate and record these wells (see Note 12).
Seal the 96 deep-well plates with Breathe-Easy sealing membrane and shake at 1000 rpm at 37 °C overnight.
3.2.3. Seeding RNAi Bacteria in 96-Well NGM RNAi Plates (1 Day Before Experiment)
Take out 96 deep-well plates from the shaker and centrifuge for 10 min at 4415 × g.
Thoroughly clean the sink area with Cavicide™. Next to the sink, remove the seal, and discard the supernatant into a bucket containing bleach by quick inversion of the plate. Place plates upside down on a stack of clean paper towels and move to the hood.
Using a 12-channel pipette, add 100 μL of diluted S-buffer (20%) to each well of just one of the plates with RNAi bacterial pellets, and resuspend the bacterial pellets by vigorously pipetting.
Transfer bacterial suspension from plate replicate 1 to plate replicate 2 (we had grown two 96 deep-well plates of culture for each RNAi clone set). Use the transferred suspension to resuspend pellets of replicate plate 2. Do not add S buffer to the second plate (see Note 13).
Add 20 μL of distilled water to the wells located in the four edges of the plate. Then, transfer 100 μL of the bacterial suspension into the wells of the 96-well NGM RNAi agar plate using multichannel pipette (see Note 14).
Dry the bacterial suspensions in the hood for around 5 h, until no liquid remains (do not overdry the plates, covering faster drying wells with Breath-Easy if necessary).
Place the lids on the NGM plates and incubate the plates overnight in a 20 °C incubator for induction of expression of double-stranded RNAs.
3.3. Worm Preparation
3.3.1. Egg Preparation and Synchronization (1 Day Before Experiment)
Sterilize the bench with Cavicide™ and carry out the next steps next to flame.
Add 10–15 mL of S-buffer to a worm plate with ≥2000 gravid adult worms.
Using S-buffer, transfer worm/egg suspension to a 15 mL conical tube.
Centrifuge at ≤2000 × g for 30 s.
Discard supernatant above 3 mL using sterilized glass Pasteur pipettes connected to a vacuum line.
Add 3 mL of lysis buffer.
Shake vigorously for 1 min and then add 9 mL of S-buffer.
Centrifuge at 2000 × g for 30 s.
Discard supernatant above 3 mL using sterilized glass Pasteur pipettes connected to a vacuum line.
Add 3 mL of lysis buffer.
Shake vigorously for 1 min and add 9 mL of S-buffer.
Centrifuge at 2000 × g for 30 s.
Using sterilized glass Pasteur pipettes connected to the vacuum line, discard as much supernatant as you feel comfortable with, without disturbing the pellet. The volume remaining should be ≤500 μL; otherwise two additional washing steps are necessary.
Add 14 mL of S-buffer.
Centrifuge at 2000 × g for 30 s.
Wash by repeating steps 13–15 three times.
Discard as much supernatant as you feel comfortable with, without disturbing the pellet using sterilized glass Pasteur pipettes connected to the vacuum line, and fill up to the 10 mL mark with S-buffer.
Rotate embryos in a 20 °C incubator for ≥16 h.
The next day, estimate worm density (see Note 15).
Dilute the worm suspension with S-buffer, or concentrate by centrifugation, to bring the density to 10 worms/μL.
3.3.2. Seeding Worms (First Day of Experiment)
Take out the NGM plates with RNAi bacteria from the 20 °C incubator and move them to the hood.
In the hood, pour the worm suspension into a 50 mL sterile reservoir.
Use a multichannel pipette to seed 5 μL of worms from the reservoir to each well (constantly move the reservoir to keep the worms suspended).
Dry the plates in the hood for around 15 min.
3.4. RNAi Incubation (First Day of Experiment)
Move the plates to a 15 °C incubator and incubate for 60 h (see Note 16).
Transfer the plates to a 25 °C incubator for 24 h.
Check the worms growing in the wells to make sure that they are at the gravid stage.
3.5. Oil Red O (ORO) Staining
Filter the ORO working solution with a 0.45 μm filter.
Pour S-buffer into a reservoir (see Note 17).
Using a 12-channel pipette, add 100 μL S-buffer to each well.
Using a 12-channel pipette, transfer the whole-worm suspension to a 96-well PCR plate.
Connect the 96-channel vacuum manifold to a vacuum source and calibrate the height for aspiration using a mock PCR plate with water. Calibrate to leave ~25 μL of water behind (see Note 18).
Once aspirator is calibrated, aspirate the S-buffer from the 96-well PCR plates containing worms using the 96-channel vacuum manifold.
Wash twice by adding and aspirating 100 μL of S-buffer using a multichannel pipette and 96-channel vacuum manifold.
Using a 12-channel pipette, add 200 μL of 60% isopropanol to each well, intentionally disrupting the worm pellet while adding the isopropanol.
Let worms sink for about 2 min.
Aspirate 60% isopropanol using 96-channel vacuum manifold.
Using a 12-channel pipette, add 200 μL of ORO working solution to each well. Intentionally disrupt the worm pellet while adding the ORO solution or resuspend by pipetting but do not vortex, invert, or flick the plates. Avoid ORO droplets from reaching the walls of the tubes to prevent precipitation of the dye.
Seal the 96-well PCR plates using sealing mats, and put plates into a sealed box with a wet paper towel (see Note 19).
Incubate the box at 25 °C for 12–16 h (see Note 20).
Remove the seal and aspirate the ORO working solution using the 96-channel vacuum manifold.
Wash the wells twice with 100 μL of S-buffer.
Add 100 μL of S-buffer with 0.01% Triton.
Use a 12-channel pipette to mount 8 μL of worms on the Comet assay 96-well slides and carefully place the coverslips for imaging (see Note 21).
3.6. Image Acquisition and Processing
To capture images from the 96-well slide, we recommend using an automated microscope or high-content screening platform. In our case, we use a Nikon Eclipse Ti microscope with an automated stage.
The optics include a 10× 0.45NA objective and a DsRi2 camera. Initial storage into local SSD drives (≥10 TB) renders fastest capturing rates (see Note 22).
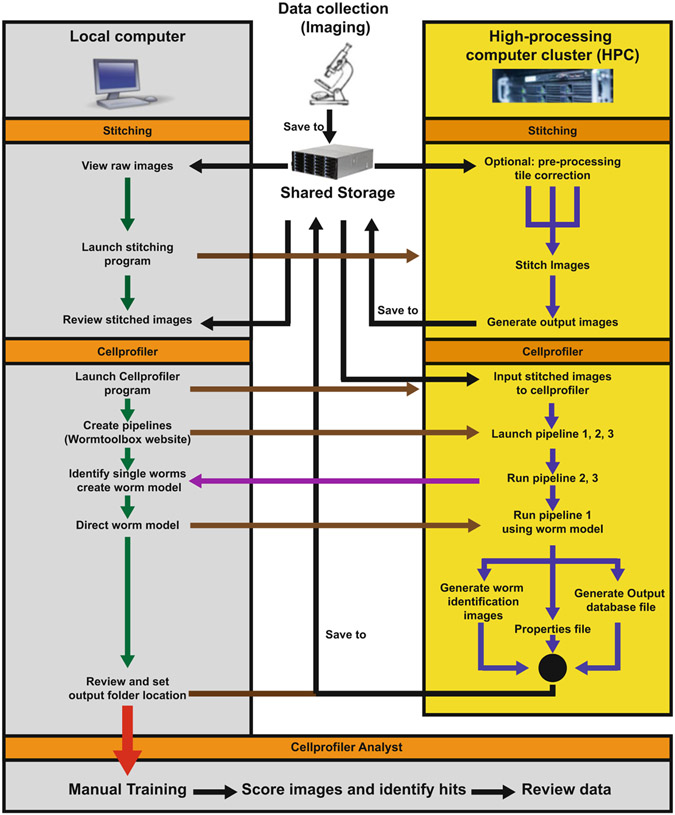
It is recommended that all data processing is completed on a high-processing computer (HPC) cluster. In the HPC context, image processing and analysis can be done in parallel (see Notes 23-31, Fig. 2).
Fig. 2.
Post-HCS image processing workflow. Imaging data may be saved in a shared storage positioned between local computers and a local or cloud-based HPC cluster. Both homemade encoded image stitching (available at http:/orourkeeyleenlab.wix.com/obesity-aging-lab/outreach) and CellProfiler data analysis can be run on an HPC cluster. In this diagram, black arrows indicate saving and loading data to and from shared storage; green arrows indicate data processing steps within a local computer; brown arrows indicate control of programs on the HPC cluster; blue arrows indicate automated (and often parallelized) programs running in the HPC cluster; purple arrows indicate direct communication from the HPC cluster to a local computer in the pipeline 2 and 3 to optimize worm models manually; and red arrow indicates that the output of CellProfiler will input into CellProfiler Analyst on the local computer for final data analysis and hit detection
Acknowledgments
We want to thank Carolina Wählby, Anne Carpenter, Jonah Larkins-For, and Annie Lee-Conery for their contributions to development of the earlier versions of these protocols. We would like to thank Wei Ma for practical advice on developing the assays. We also thank the University of Virginia Advanced Research Computing Services for support on Rivanna HPC cluster usage. This work would not have been possible without the generous support of the W. M. Keck Foundation.
Footnotes
Two major sets of RNAi feeding libraries are currently available, the initial Ahringer library and the supplementary library. The initial Ahringer library covers 72% of the worm genome, and the supplementary library covers another 15% of all genes, so the combination of both libraries covers 87% of the worm genome. All the feeding library sets are delivered as glycerol stocks and should be stored at −80 °C.
NL2099 strain carries a 3015 base-pair deletion between exons 4 and 11 of rrf-3 gene, and this allele(pk1426) can be detected by PCR using an internal forward primer: AATTGGAAGAATGAGTCACG, an external forward primer: AAATCATACGTCATCGATGC, and an external reverse primer: GCCACGAAATACCATTGCC. Amplification of genomic preps from wild-type N2 strain with internal forward primer and external reverse primer results in a 0.7 kb band, while amplification of genomic prep from NL2099 results in a 85 kb band. In addition, no product can be amplified on N2 strain with external forward primer and external reverse primer due to the size-limiting condition of this PCR. No product can be amplified on NL2099 strain using internal forward primer and external reverse primer because the internal forward primer sits in the deleted sequence. We use NL2099 strain for screen because rrf-3 is an RNA-dependent RNA polymerase and the rrf-3(pk1426) allele makes this strain hypersensitive to RNAi. Also, worms with this allele become sterile at 25 °C, which allows us to image and quantitate fat in adult worms without the interfering signal of the progeny.
GMW004 is a double-rrf-3(pk1426);daf-2 (e1368) mutant. DAF-2 is the worm insulin receptor and the e1368 allele of daf-2 is a hypomorphic thermosensitive mutation that leads to an obesity phenotype at 25 °C. daf-2(e1368) is a singlenucleotide substitution from G to A, causing a change of protein sequence from S573 to L573. This allele can be detected by PCR amplification of genomic DNA using forward primer CTCACCATTTGTCCCTTC and reverse primer CAATCGTCACCGTTTATCTC, followed by incubation with restriction enzyme TSP45I, which only cut wild-type daf-2 allele on the mutation site.
L4440 is the 2790 bp empty plasmidic vector of the RNAi constructs. It is shipped from Addgene as transformed bacteria and generally used as negative control for C. elegans RNAi screens.
DAF-16 is the C. elegans homolog of human FOXO, which is a transcription factor inhibited by insulin signaling. Reduced insulin signaling causes activation of DAF-16. Loss of DAF-16 is sufficient to rescue the obesity phenotype of daf-2 mutant worms. Hence, daf-16 RNAi can be used as a positive control in a daf-2 fat-suppressor screen.
The Nikon Eclipse Ti microscope with 10 × 0.45 NA objective and DsRi2 camera is suitable for this purpose.
These LB carbenicillin agar square plates need to be prepared well ahead or thoroughly dried in the hood. Otherwise, moisture will cross-contaminate stamped RNAi clones.
Pipette tips need to be prewarmed by pipetting up and down molten NGM RNAi agar several times before transferring. While aspirating the 96-well plates, place the tips on the side and the bottom of the well, and aspirate slowly to prevent bubbles from forming. It is very important to avoid bubbles in the wells because worms will crawl into the agar through bubbles, leading to irregular feeding, and will make it difficult to harvest worms for further processing.
It is important to make sure that the replicator is not too hot or it will kill the bacteria. There are two extra tips on the replicator that allow the user to get a sense of the pins’ temperature; simply touch the two extra tips with your hand while making sure not to touch any other tip of the replicator.
While stamping the replicator, it is important to be gentle and not poke into the agar. Also, every tip of the replicator has to touch the LB agar to secure the successful transfer of all RNAi clones.
One 96 deep-well plate worth of bacterial culture is insufficient food to sustain the growth of ~50 worms into gravid adulthood. Two or three 96 deep-well plates are preferred per RNAi library plate. In our experimental setup, two strains of worms (GMW004 and NL2099) are tested, so 4–6 96 deep-well plates are prepared for each RNAi library plate.
There are some RNAi clones that would not grow on the LB agar omnitray (not represented in the original library or experimental error). Take advantage of these empty wells and seed positive and negative control RNAi clones in these wells.
As referred in Note 11, two or three 96 deep-well plates of samples are grown for each RNAi library plate to be tested. These replicates need to be combined to support the growth of ~50 worms into gravid adulthood. If there are two deep-well plates per original plate, add 100 μL of 20% S-buffer to the first deep-well plate and resuspend the bacteria by pipetting. Then transfer the sample to the second plate and resuspend the bacteria of the second plate.
While transferring bacteria to the 96-well NGM RNAi agar plate, it is important to be careful not to poke into the agar. After carefully transferring 100 μL of bacteria, it is necessary to add 20 μL water to the edge wells because these wells dry much faster than the wells at the center of the plate. If these wells become too dry, the agar may crack, worms will crawl into the cracks, and that well will not be scorable.
To estimate the density of the worms, thoroughly resuspend the hatchlings by inverting 15 mL conical several times, take three or more 2 μL aliquots of worms, place them in a clean microscope slide, count the number of live hatchlings under the dissecting microscope, and average the hatchlings/microliter.
The 15 °C incubation can take longer or shorter than 60 h. The development of worms needs to be checked every 2 h starting from 50 h to 70 h, to ensure that the worms are at late L3 or early L4 stage for transferring to 25 °C incubator. It is critical not to transfer daf-2 mutant worms earlier than L3 because they will enter the dauer program and arrest, and also no later than L4 stage to observe full penetrance of the rrf-3 sterility phenotype.
0.01% of Triton in the S-buffer helps prevent the worms from sticking to the pipette tips, and reduce worm loss.
The amount of liquid remaining in the 96-well PCR plates is determined by the height of the 96-channel manifold. This can be adjusted and calibrated by filling water in a 96-well PCR plates and testing the water level after aspiration. It is important to keep around 25 μL of solution to minimize the chances of accidental loss of worms by aspiration.
Keeping the plate completely sealed and incubating in a humid box are necessary because the oil red O solution is a saturated solution, and loss of water causes precipitation of black crystals that prevent quantification of ORO levels.
It is important to restrict oil red O staining to ≤16 h at 25 ° C because longer incubation may change the tone of the oil red O staining and affect the quality of the images.
It requires some practice to be able to pipette up most of the worms in such a small amount of liquid. To achieve this, it is important to place the tip at the very bottom of the well and aspirate as fast as possible (i.e., use a “bad” pipetting procedure). Check the slide and make sure that you have worms in every well. If the worms in some wells are not transferred, use a single-channel pipette to transfer again.
The images are most likely to be stored on the local computer connected to microscope. To analyze them on an HPC cluster you will need to transfer them to a fast-transfer storage unit locally, remotely, or in the cloud. Your local administrator should advise you on the best available option.
When performing a high-throughput image-based genetic screening, capturing, storing, and processing approximately thirty 25 MB images per channel per well become bottlenecks due to the limited storage and processing power of regular lab computers or even workstations. For example, for the screening setup described here, each stitched RGB image of a whole well is 9001 × 8928 pixels, with each whole-well tiff image sized at around 250 MB, so one 96-well plate represents approximately 25 GB of data. Analyzing these data is an even bigger challenge than storage. Our single-worm identification algorithms, although highly accurate, are computationally demanding. Therefore, it can take from a day to a week to run a 0.25–1 TB dataset on a conventional desktop. A previous paper from our lab used compressed images to achieve analysis (5 Mb *.bmp file/well). However, this is not suboptimal due to the reduction in image resolution and consequent loss of information [14].
When naming folders and images avoid spaces. This will be useful when using a command line. If you are unable to change the names to avoid spaces you will need to use quotation marks at the beginning and at the end of the path to the file while using the command line.
As introduced in Subheading 3.6, data acquisition begins with automated imaging of the experimental plates. These data will be written directly to a mounted PC, often the one used to set up and begin microscopic data collection. A few considerations help ensure fast data collection and transfer, including local writing space, naming conventions, and an automated transfer framework. For local storage, it is recommended to have at least 10 TB of solid-state drive (SSD) space. SSD is much faster than its alternative, hard drive disk (HDD), and should be used to prevent bottlenecks. Data acquisition is also the optimal step to enforce naming conventions. Since there are no built-in frameworks for naming, it is up to the screening lab to provide and enforce easy-to-read naming conventions that will ensure smooth data processing. Finally, to prevent any impedance to data collection, it is recommended to have a second mounted PC that can separately upload data to the desired server. This allows for a slower upload speed, which is often unavoidable, while still maintaining scope operation. A detailed list of our setup and examples of file naming can be found at http://orourkeeyleenlab.wix.com/obesity-aging-lab/outreach.
While some of the processing steps can be executed on a personal computer, utilization of an HPC cluster will dramatically increase computational speeds and allow for use of fullresolution raw screening images. Interactive processing steps (i.e., refining worm identification) will require extra considerations, and a rudimentary knowledge of command-line language may be necessary to operate some of the programs described herein within the context of an HPC cluster. First, several systems and software tools must be installed, created, or optimized. These include CellProfiler (CP), CellProfiler Analyst (CPA), a MySQL database (or an alternative, but CP/CPA-compatible, database of your choice), and appropriate CellProfiler pipelines [15]. It is important to confer with a local information technologist to ensure proper installation and setup of these programs and communication with HPC cluster.
The setup and optimization of CellProfiler pipelines for new screens may be done outside of the HPC cluster environment, which facilitates the use of CP interactive optimization tools, as long as special attention is paid to any and all naming and PATH conventions required for CP and CPA to generate and retrieve data. CellProfiler’s export and import tools work within a HPC cluster environment as long as the final modifications to the pipeline—including image folder location and PATH names—are correctly assigned in the HPC environment. At this stage pay special attention to the MySQL database used to ensure that files are sent to the correct locations. Depending on the preferred CP analysis output, CSV data sheets can be generated instead of databases with CellProfiler, which can then be used to manually analyze data. This approach is less robust and may result in errors due to CellProfiler Analyst’s particular file location requirements.
A comprehensive analysis of the images is possible after the initial steps are completed, and can be easily completed using the scripts provided at http://orourkeeyleenlab.wix.com/obesity-aging-lab/outreach.
This data analysis process will require work on the HPC cluster through programs such as FastX or similar. FastX provides a graphical user interface (GUI)-enabled program for HPC cluster operation, and is an easy solution for users unfamiliar with linux or remote systems. If your institution does not have access to this program, or prefers a different one, alternatives should work just as well. Note that most alternatives may require basic command-line knowledge.
After initial data have been collected, post-processing can be done. This may include image review, and further image analysis if necessary. It is recommended the user conducts analysis within the HPC cluster to ensure that all image data can be retrieved without permission issues. Due to the preestablished database, it should be easy to locate and retrieve any files for further use. Another consideration is kinds of storage, in particular, the differences between private user storage and shared storage. It is recommended that both be set up in all cases: the shared storage for the organization and maintenance of all raw image data sets (terabytes of information), and the private user storage for processing images, outputting data, and linking of relevant experimental metadata or information fro databases. In this way, private data folders would beoptimized for processing, and commonly used and large image files are not copied to all users, but instead located in a central location. Nevertheless, naming conventions for files, subfolders, and folders should be established and strictly followed.
For users that want to compile an easy-to-use image viewer, programs such as Omero exist. Omero is an open-source program that allows for broad image organization and utilization. It may be possible to set the HPC cluster to send data outputs directly to Omero. Alternatively, integration of the MySQL database into Omero may provide the most success. These types of programs are not strictly necessary for post-processing, but may allow for additional organization and integration of screen results with data banks (i.e., wormbase, ensemble). Optimal data processing and organization solutions would vary with type and scale of the screens, as well as with lab resources.
References
- 1.Muschiol D, Schroeder F, Traunspurger W (2009) Life cycle and population growth rate of Caenorhabditis elegans studied by a new method. BMC Ecol 9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulston JE, Horvitz HR (1977) Postembryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56:110–156 [DOI] [PubMed] [Google Scholar]
- 3.Wang MC, O’Rourke EJ, Ruvkun G (2008) Fat metabolism links germline stem cells and longevity in C. elegans. Science 322:957–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narbonne P, Roy R (2009) Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457:210–214 [DOI] [PubMed] [Google Scholar]
- 5.Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J (2002) TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNx translation. Curr Biol 12:1448–1461 [DOI] [PubMed] [Google Scholar]
- 6.McKay RM, McKay JP, Avery L, Graff JM (2003) C. elegans: a model for exploring the genetics of fat storage. Dev Cell 4:131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sze JY, Victor M, Loer C, Shi Y, Ruvkun G (2000) Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403:560–564 [DOI] [PubMed] [Google Scholar]
- 8.Garcia AM, Ladage ML, Dumesnil DR, Zaman K, Shulaev V, Azad RK et al. (2015) Glucose induces sensitivity to oxygen deprivation and modulates insulin/IGF-1 signaling and lipid biosynthesis in Caenorhabditis elegans. Genetics 200:167–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabara H, Grishok A, Mello CC (1998) RNAi in C. elegans: soaking in the genome sequence. Science 282:430–431 [DOI] [PubMed] [Google Scholar]
- 10.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–237 [DOI] [PubMed] [Google Scholar]
- 11.O’Rourke EJ, Conery AL, Moy TI (2009) Whole-animal high-throughput screens: the C. elegans model. Methods Mol Biol 486:57–75 [DOI] [PubMed] [Google Scholar]
- 12.O’Rourke EJ, Soukas AA, Carr CE, Ruvkun G (2009) C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab 10:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12:1317–1319 [DOI] [PubMed] [Google Scholar]
- 14.Wählby C, Conery AL, Bray MA, Kamentsky L, Larkins-Ford J, Sokolnicki KL et al. (2014) High- and low-throughput scoring of fat mass and body fat distribution in C. elegans. Methods 68:492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamentsky L, Jones TR, Fraser A, Bray M, Logan D, Madden K et al. (2011) Improved structure, function, and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27: 1179–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]