Graphical abstract
Keywords: Coronavirus Infectious Disease Ontology, Ontology summarization, Ontology visualization, Ontology orientation, Comprehension of an ontology, Summarization networks
Abstract
The current intensive research on potential remedies and vaccinations for COVID-19 would greatly benefit from an ontology of standardized COVID terms. The Coronavirus Infectious Disease Ontology (CIDO) is the largest among several COVID ontologies, and it keeps growing, but it is still a medium sized ontology. Sophisticated CIDO users, who need more than searching for a specific concept, require orientation and comprehension of CIDO.
In previous research, we designed a summarization network called “partial-area taxonomy” to support comprehension of ontologies. The partial-area taxonomy for CIDO is of smaller magnitude than CIDO, but is still too large for comprehension. We present here the “weighted aggregate taxonomy” of CIDO, designed to provide compact views at various granularities of our partial-area taxonomy (and the CIDO ontology). Such a compact view provides a “big picture” of the content of an ontology. In previous work, in the visualization patterns used for partial-area taxonomies, the nodes were arranged in levels according to the numbers of relationships of their concepts. Applying this visualization pattern to CIDO's weighted aggregate taxonomy resulted in an overly long and narrow layout that does not support orientation and comprehension since the names of nodes are barely readable. Thus, we introduce in this paper an innovative visualization of the weighted aggregate taxonomy for better orientation and comprehension of CIDO (and other ontologies). A measure for the efficiency of a layout is introduced and is used to demonstrate the advantage of the new layout over the previous one. With this new visualization, the user can “see the forest for the trees” of the ontology. Benefits of this visualization in highlighting insights into CIDO’s content are provided. Generality of the new layout is demonstrated.
1. Introduction
The frantic worldwide search for mediations and vaccines for the COVID-19 infection would greatly benefit from a COVID-19 standard reference ontology for this extensive research. Several COVID-19 ontologies exist already. Most of them are accessible through the NCBO BioPortal [1]. The largest is the Coronavirus Infectious Disease Ontology (CIDO) [2] (6,938 concepts in February 2021), which was created to provide a standardized representation of various coronavirus infectious diseases [2], [3].
CIDO follows the OBO Foundry [4] principles and extensively reuses concepts from about 20 other ontologies [5] including the Chemical Entities of Biological Interest (ChEBI) [6] and the Human Phenotype Ontology (HPO) [7]. For example, drug concepts are reused from ChEBI, the National Drug File - Reference Terminology (NDF-RT) [8], and the Drug Ontology (DrON) [9]. CIDO contains 244 original COVID-19-specific concepts. Concepts are interconnected by 201 lateral relationship types such as caused by, infection with and treatment for. Various areas such as etiology, diagnosis, and treatment of coronavirus diseases are covered.
Other smaller COVID-19 ontologies emphasize different aspects of the disease. The COVID-19 ontology (COVID-19) (2,268 concepts) [10] covers predominantly concepts related to cell types, genes, and proteins involved in virus-host-interactions, as well as relevant epidemiological and medical concepts. It is similar to CIDO with more emphasis on presentations affecting various body systems. The COVID-19 Infectious Disease Ontology (IDO-COVID-19) (486 concepts) [11] extends the Infectious Disease Ontology (IDO) [12] and the Virus Infectious Disease Ontology (VIDO) [13] to solely represent concepts of relevant viral diseases. The WHO COVID-19 Rapid Version CRF semantic data model (COVIDCRFRAPID) (398 concepts) [14], captures the semantic references relevant to the WHO case report form (CRF). Two other relevant small ontologies are the Ontology for Collection and Analysis of COviD-19 Data (CODO) (90 concepts) [15], and the COVID-19 Surveillance Ontology (COVID19) (52 concepts) [16].
The ACT COVID Ontology v3.0 (2,446 concepts) is available on GitHub [17] as a set of SQL files loadable into a database. It supports cohort identification by reusing concepts of ICD [18], LOINC [19], CPT® (Current Procedural Terminology) [20] and the National Drug Code (NDC) [21].
Before users adapt a COVID-19 ontology as a reference ontology for research, they should acquire an understanding of its structure and content. In this paper, we address this need for the CIDO ontology, which is not only the largest COVID ontology, but is dynamically updated and enriched with new concepts. Another advantage of CIDO is its rich collection of lateral semantic relationships. The runner up in terms of size, the COVID-19 ontology, has no lateral relationships, which inhibits the utilization of our most commonly used summarization network.
In our previous research, we designed several kinds of general purpose summarization networks [22]. For ontologies with lateral semantic relationships we created the “area taxonomy” and its refinement, the “partial-area taxonomy” [23], [24], [25]. These two summarization networks were effective in summarizing small ontologies with a few hundred concepts. For example, in [26], [27], [28], we summarized the Ontology of Clinical Research (OCRe) [29], the Sleep Domain Ontology (SDO) [30], the Cancer Chemoprevention Ontology (CanCO) [31] and the Drug Discovery Investigations ontology (DDI) [32]. We also successfully summarized small hierarchies of large terminologies, e.g., the Biological Process hierarchy of the NCI Thesaurus (NCIt) [23], and the Specimen hierarchy of SNOMED CT [25], both of a magnitude of 1000 ~ 2000 concepts. (We will use the names “ontology” and “terminology” interchangeably when this is correct for a specific example.)
However, for larger ontologies and larger hierarchies of terminologies such as SNOMED CT, NCIt, Gene Ontology (GO) [33], ChEBI [6], or Uberon [34], the partial-area taxonomy has too many nodes to enable their layout on a computer screen in a readable way. E.g., the “Clinical finding hierarchy” in the January 2021 release of SNOMED CT with 114,493 concepts is summarized by a partial-area taxonomy with 14,956 nodes. One can use zooming-in in a browsing tool, but then only a section of the partial-area taxonomy is visible. However, Ochs et al. [35] have derived a partial-area taxonomy for the small 'bleeding' subhierarchy of about 1000 concepts of SNOMED CT’s 'Clinical finding' hierarchy. The result was utilized in [36].
For ontologies without lateral relationships, but with multiple parents, we designed the “tribal abstraction network” [37]. We also demonstrated summarization networks for several specialized terminologies such as one [38] for the Medical Entity Dictionary [39], and another one [40] for the National Drug File - Reference Terminology (NDF-RT) [8].
Other researchers have published relevant work on ontology summarization in the context of the semantic web. For example, Peroni et al. [41] described an ontology summary derived using topology-based criteria to identify key concepts in an ontology. Our summarization network is not limited to the concept level, but takes the overall structure into account. Many approaches [42] have been developed for ontology modularization which is to partition an ontology into modules for ontology reuse. There is extensive research regarding various techniques of ontology visualization. See [43], [44] for interesting review. We note that all these ontology visualization techniques are applicable for only small ontologies, in contrast, our work is visualizing large ontologies by first obtaining a summarization network and then visualizing it.
Out of the above choices, the summarization network that is most appropriate for CIDO is the “partial-area taxonomy” (from this point on called “taxonomy” for short.) A taxonomy is a network of nodes representing partial-areas connected via hierarchical child-of relationships. However, the taxonomy for the CIDO release (1.0.108) used in this research has 519 nodes and is therefore still too large for display on a single screen to achieve comprehension, even though it is less than 10% of the size of CIDO.
In this paper, we present the “weighted aggregate partial-area taxonomy” of CIDO (weighted aggregate taxonomy or WAT, for short) to provide a compact summarization network capable of summarizing it on one single screen. The number of nodes in the WAT of an ontology is adjustable and is controlled by a parameter defining at what size a partial-area is considered large. By the choice of this parameter we can control the granularity of the summarization. However, size is not the only problem. The shape of the summarization network is also an issue.
The WAT summary of CIDO is long and narrow and does not lend itself to provide a complete, easily readable display on one screen. Thus, we introduce an innovative layout for the WAT of CIDO that better fits onto a screen enabling a readable display. To make the improvement objective, we introduce a measure for assessing the efficiency of a layout and show that the new layout is indeed more efficient. This visualization provides better support for orientation and comprehension of CIDO. The benefit of this new layout is demonstrated in identifying parts of CIDO that are relevant for research on medications and vaccines for COVID-19. Generality of the use of the new layout is also demonstrated.
2. Background
2.1. The Coronavirus Infectious Disease Ontology (CIDO)
As an OBO library ontology [4], CIDO is a community-based biomedical ontology in the area of coronavirus infectious diseases. CIDO provides standardized human- and computer-interpretable annotation and representation of various infectious coronavirus diseases, including their etiology, transmission, epidemiology, pathogenesis, diagnosis, prevention, and treatment. Its development follows the OBO Foundry Principles [2].
CIDO has the aim to support a fundamental understanding of the host-coronavirus interaction mechanisms, to support the rational development of vaccines and drugs [2], [3], [45], [46]. To achieve these goals, CIDO semantically classifies related entities and interlinks these entities in an ontological framework. CIDO is released in OWL format and is freely available on the GitHub repository [47]. CIDO has also been deposited on BioPortal [48] and Ontobee [49]. Currently, CIDO is at version 1.0.184 with a total of 6,938 concepts and 371 properties of which 201 are relationships.
2.2. Area taxonomy and Partial-area taxonomy
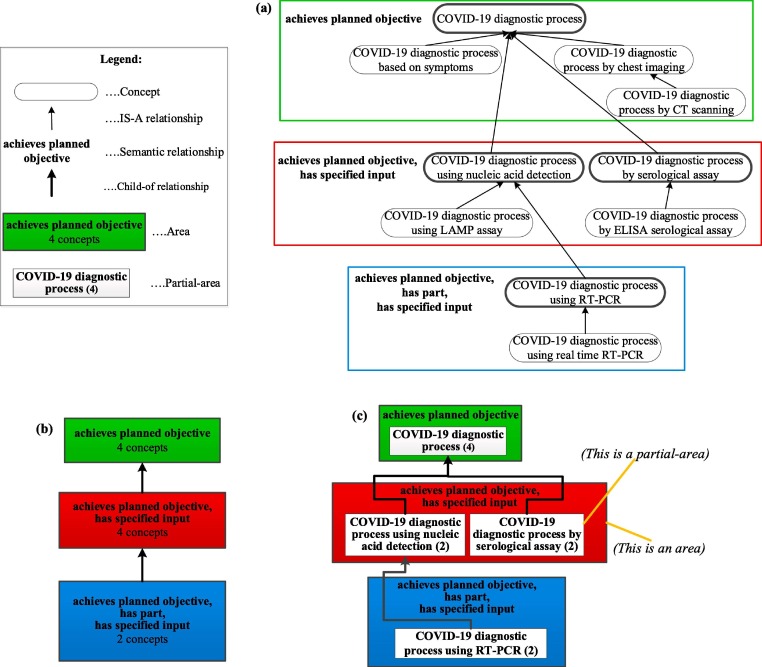
Biomedical ontologies represent complex domain knowledge and are not easy to comprehend. We have previously designed various summarization networks that provide “big picture” views of ontologies to achieve better ontology comprehension [22], [37], [40], [50]. A summarization network is composed of nodes, summarizing sets of similar concepts and connected by hierarchical child-of relationships. Fig. 1 demonstrates the derivation of two kinds of summarization networks for a hierarchy excerpt of 10 concepts from the CIDO ontology.
Fig. 1.
Two summarization networks for an excerpt from CIDO: (a) An excerpt of 10 COVID-19 diagnostic process concepts from CIDO. Concepts are represented by boxes with rounded corners and are connected by hierarchical IS-A relationships shown as thin black arrows. Nonhierarchical semantic (lateral) relationships are written in bold inside colored rectangular boxes. The boxes indicate sets of concepts with the same sets of lateral relationship types. Boxes with a different numbers of lateral relationship types have different colors. (b) The area taxonomy summarization network for the excerpt in (a), where the nodes, shown as boxes filled with solid colors, represent areas and the links are the hierarchical child-of relationships (shown as thick black upward arrows) connecting area nodes. Area nodes are color-coded by cardinalities of their sets of lateral relationship types. (c) The partial-area taxonomy summarization network for (a). The nodes are partial-areas represented as white boxes within area nodes. Numbers in () indicate how many concepts are summarized by partial-areas. As in (b), hierarchical child-of links connecting partial-area nodes are displayed as thick upward arrows.
Summarization is achieved as follows. Instead of showing the lateral relationships separately for each concept, we have overlaid rectangular boxes to group together sets of concepts that have exactly the same set of lateral relationship types listed inside those boxes. For example, there are four concepts with only one lateral relationship type achieves planned objective at the top of Fig. 1(a). The green rectangle around them indicates their structural similarity, i.e., having the same set of lateral relationship types.
As concepts become more specific when moving down in the hierarchy, additional lateral relationship types are introduced. The two child concepts of 'COVID-19 diagnostic process,' namely 'COVID-19 diagnostic process using nucleic acid detection' and 'COVID-19 diagnostic process by serological assay,' inherit its relationship type achieves planned objective and have one additional lateral relationship type has specified input. These two concepts with their respective child concepts, having the same two lateral relationship types, are enclosed in the red box.
A set of concepts with exactly the same set of lateral relationship types, is called an area . Every colored rectangle in Fig. 1(a) becomes one area node in Fig. 1(b). Thus, an area node summarizes the concepts of an area. An area node is labeled by the set of names of lateral relationship types of its concepts and the number of concepts it summarizes. For example, the top four concepts enclosed in the green box in Fig. 1(a) are summarized as the green area node labeled as 'achieves planned objective − 4 concepts' in Fig. 1(b).
There are four concepts with a bold border in Fig. 1(a). These concepts are called roots of their areas, because they have no IS-A links pointing to any concept inside of their “own” area. Areas may have multiple roots. For example, the red area has two roots, 'COVID-19 diagnostic process using nucleic acid detection' and 'COVID-19 diagnostic process by serological assay.' The child-of relationships hierarchically connecting areas are based on the configurations of area roots in the underlying ontology. More specifically, an area A is child-of an area B if a root in A has a parent in B. For example, the red area is child-of the green area in Fig. 1(b), since both roots in the red area are child concepts of the root in the green area named 'COVID-19 diagnostic process.' Area nodes and child-of links together comprise the area taxonomy [23] .
Clearly, Fig. 1(b), the area taxonomy with three areas, provides a compact summary of Fig. 1(a). However, Fig. 1(b) omits too many details, because it is based solely on structure. Consider the two root concepts in the red box in Fig. 1(a): 'COVID-19 diagnostic process using nucleic acid detection' and 'COVID-19 diagnostic process by serological assay' have the same set of two lateral relationship types (i.e., the same structure). However, each has a different semantics as reflected by its name. Hence, the red area node in Fig. 1(b) contains concepts with substantially different semantics. Thus, we refine an area into smaller units such that a unit contains a group of concepts with similar semantics. For example, the root 'COVID-19 diagnostic process using nucleic acid detection' and its child 'COVID-19 diagnostic process using LAMP assay' have similar semantics. Such a smaller unit is called a partial-area.
A partial-area within an area consists of one root concept of the area and all its descendant concepts in this area, represented as a white partial-area node within an area node in Fig. 1(c). A partial-area node is labeled by the name of the root concept representing the semantics of the partial-area, and the number of all concepts in this partial-area inside (). For example, the root concept 'COVID-19 diagnostic process using nucleic acid detection' and its child concept are summarized by the partial-area node 'COVID-19 diagnostic process using nucleic acid detection (2).' Partial-area nodes are similarly connected by child-of links, forming the partial-area taxonomy (PAT) [24]. The partial-area taxonomy in Fig. 1(c) provides a better summary of Fig. 1(a) with more details than Fig. 1(b), preserving important structural and semantic features of Fig. 1(a).
3. Methods
3.1. Comprehension and orientation of large ontologies
In this paper, we address orientation and comprehension of an ontology. In the Cambridge Dictionary [51] one of the definitions of comprehension is the ability to understand completely and be familiar with. One of the definitions of orientation is the position of something in relation to its surroundings. In this section we refer to these definitions. What is the interpretation of these terms in the context of an ontology?
Comprehension of an ontology involves obtaining knowledge about the various aspects of the ontology including its purpose, the identity of the curator(s), information about its use, as well as structural information such as format and numerical dimensions displayed in BioPortal [52] as metrics of the ontology. For example, the format of CIDO is OWL. As of February 20, 2021, Release 1.0.184 contains 6,938 concepts and 371 properties. Its maximum depth is 38, its maximum and average numbers of children are 403 and 3, respectively, etc. Another aspect relates to the content. What are the major subjects of the concepts in the ontology? How many concepts are there for each subject? (Note: The CIDO version analyzed in this paper is version 1.0.108, which was released on June 14, 2020, and contains 5,138 concepts.)
We interpret orientation in an ontology as the position of a concept, relative to its parent(s) and children. This corresponds to the immediate hierarchical neighborhood [53]. Such orientation can refer either to a concept or to a node representing a major subject or a secondary subject.
In our previous research we used visualizations of taxonomies for quality assurance of the corresponding ontologies. The OAF software tool [54], designed at the SABOC Center at NJIT, computes the taxonomy of an ontology and its partial-areas. The OAF tool can also generate a readable layout of taxonomies of small ontologies. Our many quality assurance papers rely on the OAF tool for generating publication figures. When the task at hand is to select random samples of study and control concepts from partial-areas, one does not need to visualize or comprehend the taxonomy.
It is important to note that the number of nodes in the taxonomy of an ontology depends not only on the number of concepts, but by the definition of “taxonomy,” on the number of relationship types. In the worst case, adding a relationship type to an ontology, may double the number of partial-areas, because the concepts of each existing area may be split into two kinds, those that have the new relationship type and those that don't. This splitting could also affect the content of the partial-areas. For example, if the root of a partial-area does not acquire the new relationship type, but some of its concepts do, those concepts have to be removed from the area, thus creating a different partial-area, in another area.
Previously [55], we presented a figure of one large hierarchy of the Gene Ontology (GO), for the purpose of auditing it. This was possible due to the hierarchy having only a very small number of relationships. The figure included areas with hundreds of partial-areas. In order to display all of the partial-areas, the algorithm had to truncate the names of many of them. Therefore, this figure did not allow a sufficient degree of comprehension of GO.
In this paper, we present a new approach to make the 5,138 concepts in the version 1.0.108 of CIDO comprehensible by summarization. The version 1.0.108 of CIDO has 113 relationship types. As a result, the number of partial-areas in its taxonomy is 519, which is considerably larger than what can be displayed on a computer screen in a readable way. The problem is even more severe when considering, for example, the large hierarchies of SNOMED CT (January 2021 Release) with 114,493 concepts in the 'Clinical finding' hierarchy and 58,445 concepts in the 'Procedure' hierarchy. The corresponding numbers of relationship types are 17 and 29.
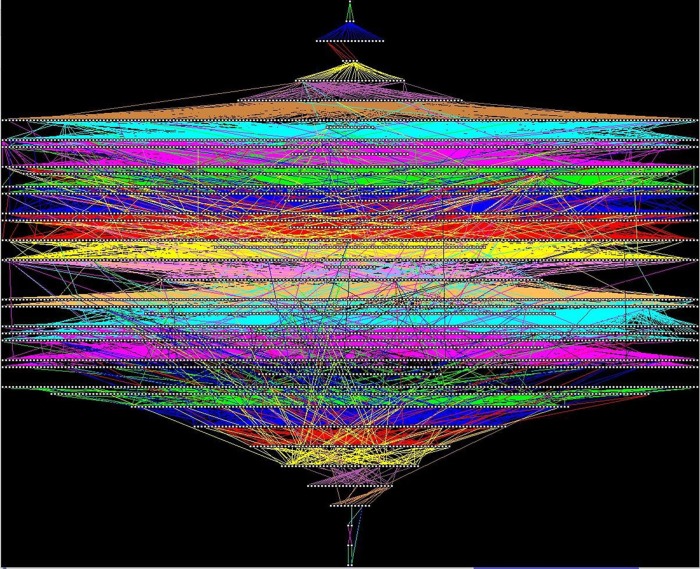
To illustrate the need for visual comprehension of CIDO, Fig. 2 , shows a color picture of the layout of its hierarchy. Small white boxes represent concepts, placed in levels according to their longest hierarchical distances from the root concept 'Thing' (calculated with Topological Sort). 'Thing' is visible (as white dot) at the upper edge of the diagram. All the IS-A links emanating from a given level are drawn with the same color. In the figure we see that some IS-A links from a concept in one level point to concepts in higher levels (closer to the root) rather than in the level immediately above, as most IS-A links do. This figure with its overwhelming appearance illustrates the challenges of visual comprehension of CIDO.
Fig. 2.
Colorful layout of CIDO concepts in layers according to their longest distance from the root. All IS-A links emanating from a level are in the same color.
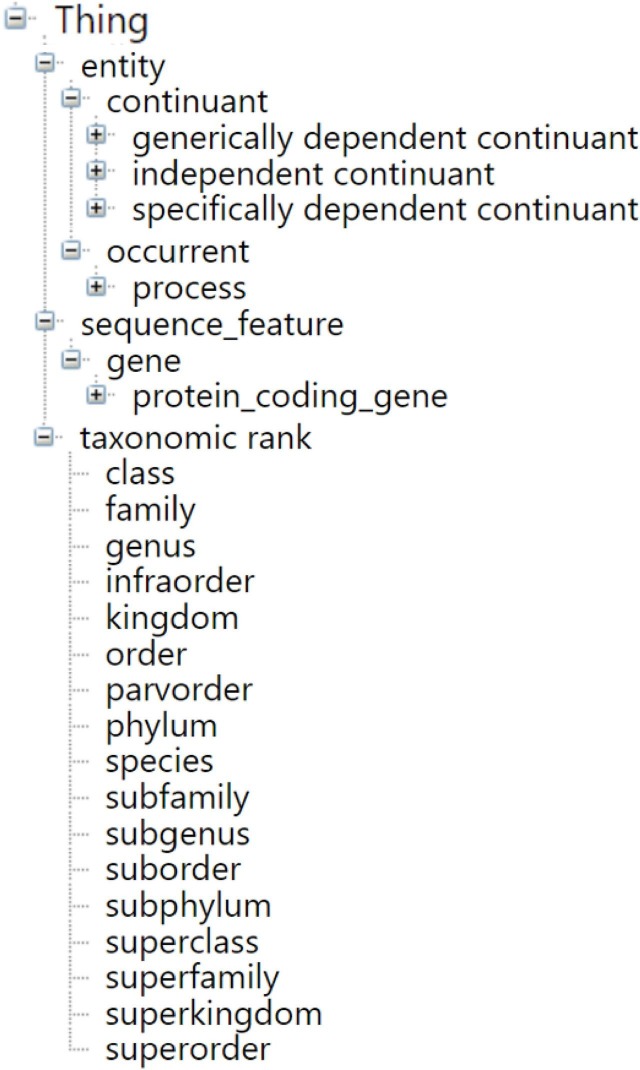
To use a metaphor, many readers start reading a book by first looking at the table of content to obtain a comprehension to the themes in the book and how many pages are devoted to each of them. One may suggest a similar approach for an ontology where the children of the root serve as chapters and the grandchildren represent sections in chapters. Contrary to a book, there is no natural order among the children, but a list may still provide an overview of the content and the major subjects of the ontology. Fig. 3 shows the four top levels of CIDO concepts in an indented format. Only four concepts, namely 'process,' 'sequence_feature,' 'gene,' and 'protein_coding_gene' have a “common English” name. We exclude from this counting the 'taxonomic rank' concept, which has many children but no grandchildren and thus provides no insight about the CIDO content. All the other names stem from the OBO approach of assigning abstract, “philosophy-inspired” names to top concepts. Hence this approach, which can provide some insights into the content of hierarchies of SNOMED CT and NCIt, does not provide sufficient comprehension support for CIDO.
Fig. 3.
The four top levels of CIDO concepts in an indented format.
In [2], the designers of CIDO describe some important concepts in CIDO and the relationship structure among them. As explained there, this “schema” was guiding the CIDO curators in designing and populating CIDO. Such a schema is important for the disciplined curation of an ontology. However, the role of a summarization network is different. It captures the state of the ontology post-curation, to examine which are the major subjects in the resulting ontology and how many concepts belong to each such subject. Moreover, the number of concepts is used to determine which subjects are considered major subjects, depending on the desired granularity.
3.2. Aggregate taxonomies
Our partial-area taxonomy may provide a useful summarized view of an ontology. However, this summarization network may still be too large for an easily comprehensible display on a screen. For example, the partial-area taxonomy of CIDO has 177 areas and 519 partial-areas. To obtain a more compact summary of an ontology, we defined the weighted aggregate taxonomy (WAT) [56], [57]. The idea is to differentiate between major partial-areas summarizing many concepts and minor ones, by using an integer bound b. The weight of a partial-area is defined as the number of the descendants of its root 'r' in the whole ontology. This definition intentionally counts not only the concepts of the partial-area 'r' itself, but also the concepts of all its descendants in other partial-areas, since they are also semantic refinements of 'r.'
In a WAT, only partial-areas with weights ≥ b are represented as nodes. Each such node represents a major subject in the topic modeled by the ontology, since the root 'r' of this partial-area has at least b descendants, some in its own partial-area 'r,' and others in descendant partial-areas. All these concepts are refinements of 'r', hence 'r' deserves to be designated a major subject. However, the partial-areas with a weight less than the bound b are not deleted, they are just hidden. Their contribution is aggregated into the closest ancestor partial-area with weight ≥ b. To control the size of the WAT, we can vary the bound b.
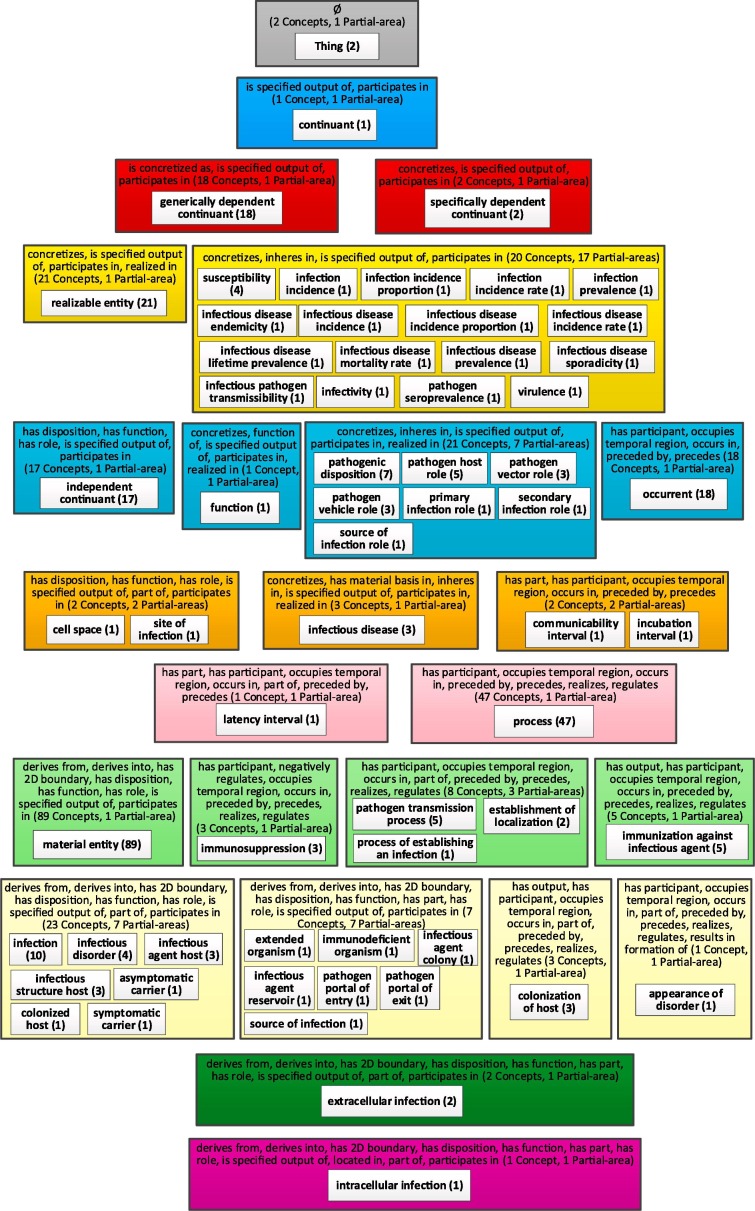
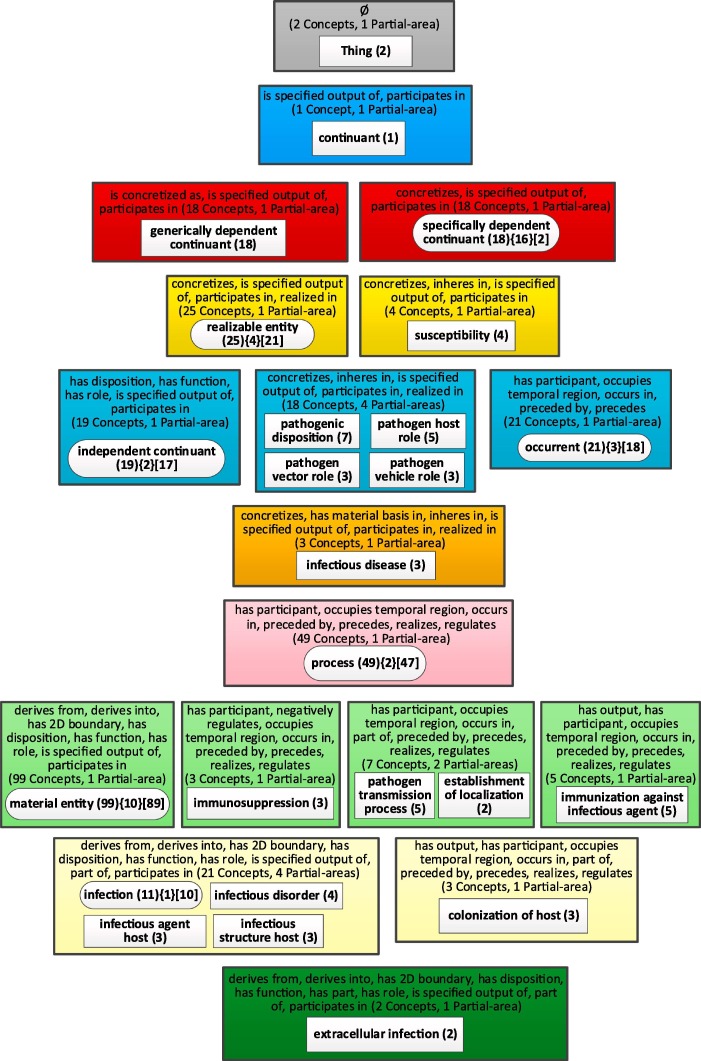
To illustrate the aggregation process and expansion process of a node, we will use the Infectious Disease Ontology (IDO) [12], which is also listed on BioPortal [58]. The choice of IDO for this purpose is influenced by the fact that CIDO was designed on top of IDO. The advantage of illustrating the aggregate taxonomy with IDO rather than CIDO is that IDO is much smaller, including 362 concepts and 43 properties, 27 of them are relationships and the maximum depth is eight. Fig. 4 shows the partial-area taxonomy of the IDO, which contains 63 partial-areas in 25 areas. As explained in the Background, the areas are arranged in color-coded levels according to the number of relationship types. The taxonomy of IDO consists of 11 levels. Hence, this taxonomy is visually long and narrow with at most 18 and an average of 5.27 partial areas in a level. Hence, the complete figure of the taxonomy is barely readable on a portrait layout page (which Fig. 4 demonstrates). Almost all screens have landscape layout, and on such a screen the figure is not readable. The user needs to zoom in for readability and can thus view only part of the taxonomy at one time, which increases the cognitive load. In addition, such a large figure with so many details is overwhelming for any user wishing to comprehend it.
Fig. 4.
The partial-area taxonomy of IDO.
The taxonomies in Fig. 4 and the following figures were generated by the OAF tool, designed by Ochs et al. [54] at the SABOC Center of NJIT, but they are indeed not readable without zooming in. For readability, we redrew Fig. 4 using Visio. Fig. 4 does not show the child-of relationships between partial-areas, since adding them would complicate the figure to an unacceptable degree. The reason is that while many child-of relationships are between partial-areas in consecutive levels, some of those relationships are between partial-areas in areas that are several levels apart. For example, the partial-area 'occurrent (18)' at level 5 is a child of the partial-area 'Thing (2)' at level 1, and a parent of the partial-area 'process (47)' at level 7. By clicking on different focus concepts, the user can dynamically explore the child-of relationships in the taxonomy using a feature of the OAF tool.
A network of 63 nodes (63 partial-areas in Fig. 4) is overwhelming for comprehension for most users even if it was readable. To obtain a more compact summary of IDO, we derived its WAT. There are 39 partial-areas of one concept in Fig. 4. By selecting a bound b = 2, all of them will be aggregated into their larger closest ancestor partial-areas. Fig. 5 shows the WAT of IDO obtained for b = 2. It contains 25 aggregate partial-areas in 18 areas. This WAT is also arranged in color-coded levels according to the numbers of relationship types. This figure uses the same graphical conventions as in the previous taxonomy figure and is more readable than Fig. 4.
Fig. 5.
The WAT of IDO obtained for b = 2.
In Fig. 5, some of the white boxes are rectangles containing one number, while others have rounded corners and list three numbers. A rectangle represents a partial-area and the number is its size (number of concepts summarized). A box with rounded corners represents an aggregate partial-area and the three numbers are the number of summarized concepts including concepts aggregated from small partial-areas, the number of small partial-areas aggregated into it, and the original size of the partial-area itself before the aggregation. For example, in the third (red) level on the left is the partial-area 'generically dependent continuant' with 18 concepts. To its right is the aggregate partial-area 'specifically dependent continuant.' The original size of this partial-area in the partial-area taxonomy is 2 (as can be seen in the IDO taxonomy in Fig. 4), and the number of concepts summarized by this aggregate partial-area in the WAT (b = 2) is 18, since 16 descendants of the partial-area, each with only one concept, were aggregated into it. We note that some of the nodes at the top levels have names such as 'continuant', 'occurrent', and 'specifically dependent continuant.' Those names are imported from the BFO [59] which is an OBO Foundry ontology [4].
The nodes of the WAT represent major subjects in IDO modeling. The WAT itself is therefore called a major subject network. Examples of major subjects beyond those already mentioned are 'pathogenic disposition (7)', 'infectious disease (3)', 'process (49){2}[47],' 'infection (11){1}[10],' and 'immunization against infectious agent (5).' This illustrates that the WAT provides a good summary of the content of IDO by listing major subjects and the numbers of concepts each of them summarizes.
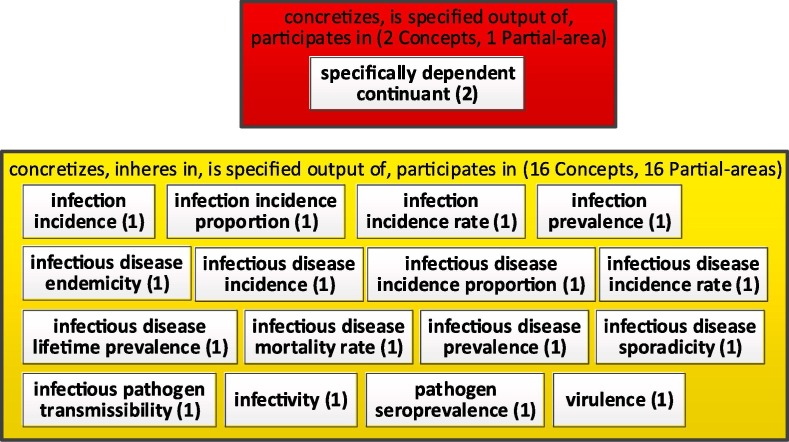
Furthermore, when more details are desired by the users, they can drill down (with the OAF software tool) into a major subject to display the hidden small partial-areas that were aggregated into this major subject partial-area. Suppose we want to concentrate on the 'specifically dependent continuant' major subject, in Fig. 5. We can “drill down,” into its 16 + 1 = 17 partial-areas and generate a new taxonomy to create a secondary subject network, where each node represents a secondary subject under this major subject. We show the taxonomy network in Fig. 6 .
Fig. 6.
Secondary subject network under ‘specifically dependent continuant’.
The network of secondary subjects exposes what concepts are summarized under the title 'specifically dependent continuant.' Example concepts include 'infection incidence,' 'infection prevalence,' and 'infectious disease mortality rate.' The major subject network shown by the aggregate taxonomy of Fig. 5 provides a top-level summary of the content of IDO. Once a user identifies a major subject of their interest, it is possible to drill down using OAF and obtain the secondary subject network with more details. By limiting the size of each figure to at most 25 nodes, they are relatively easy to comprehend.
How does the WAT support orientation? The major subject network supports top level orientation with regards to the subject, where the diagram shows its parent(s) and children among major subjects. For example, the parent of 'occurrent' is an 'entity' and its child is 'process'. The secondary subject network offers orientation on the level of partial-areas. That is, for a given partial-area, the network shows its parent and child partial-areas. A user can look at the concept network in a partial-area to obtain orientation on the concept level. If one is interested in a more far reaching orientation, the diagram enables access to grandparent(s) and grandchildren in each of the above display modes.
3.3. Child-of-based layout of an aggregate taxonomy
The IDO aggregate taxonomy of Fig. 5 is still deficient in terms of supporting comprehension and orientation. Due to the large number of relationship types, the figure is long and narrow. Even though the number of nodes is limited to 25, which is considered as a comprehensible size, the landscape layout of a typical screen prevents a readable display on a screen. Note that we present it here in a portrait layout so the reader can follow the examples of the aggregation process. Furthermore, it is not easy to identify the parent(s) or children of a node of interest, although OAF enables their dynamic highlighting as mentioned above. Another issue is that a parent may appear several levels above a child, which is counterintuitive. These problems stem from the fact that the partial-area taxonomy is embedded in the area taxonomy. Since each area is labeled by its own list of relationship types, layout of areas by the numbers of relationship types is one sensible option.
Recall, however, that the name of a partial-area is equal to the name of the root of the partial-area, which provides its semantics to the whole set of concepts. This is much more informative than the list of relationship types that labels an area. As a matter of fact, in Fig. 5, most of the space in any area with a few partial-areas, is devoted to the list of relationship types. Thus, the name of an (aggregate) partial-area, which corresponds to a major subject of the ontology, is written in small letters, requiring zooming in on a screen for readability. If the name is long, it may be truncated, eliminating essential information.
To overcome the above issues of the relationship-based layout, we introduce in this paper a new, alternative layout for an aggregate taxonomy, which we call the child-of-based layout. The child-of relationship hierarchy, which is not displayed in the relationship-based layout, is the backbone of the child-of-based layout of the aggregate taxonomy. As explained in the Background Section, a child-of link between partial-areas follows the pattern of the IS-A relationship from the root of the child partial-area. Hence, there is a well-defined hierarchy connecting the partial-areas that forms an acyclic network. Thus, in this new layout, we arrange the partial-areas (and the areas containing them, which follow a similar child-of pattern) in layers according to their hierarchical distance to the root partial-area of the aggregate taxonomy (named 'Thing' after the root of the ontology). (The distance is measured by the number of child-of links in a path consisting of partial-areas and child-of links connecting them. In case of multiple paths to the root partial-area with different lengths, the partial-area is placed in a layer according to the longest path.)
The topological sort algorithm [60] is used to divide the partial-areas into the proper layers. In this way, a large majority of the child-of relationships will appear between partial-areas in consecutive layers. This will simplify the layout algorithm for the diagram and will also make it easy for users to follow child-of paths in the resulting diagram, simplifying orientation.
The reason for the large number of levels in the (previous) relationship-based layout of the aggregate taxonomy of IDO is the large number of relationship types of IDO. The bottom area in Fig. 5 has 10 relationship types. Furthermore, for nine integers between 1 and 10, there exists an area with this number of relationship types. In the (new) child-of-based layout, the number of layers does not depend on the number of relationship types.
As a further improvement, in contrast to the relationship-based layout, we list in the child-of-based layout only the relationship types that are newly introduced in an area, but not the inherited relationship types. This frees up space in area boxes. The newly introduced relationship types typically embody the most pertinent information for users. If a user wants to know all relationship types defined for a partial-area, it is possible to obtain them by traversing the path(s) to the root partial-area, collecting the introduced relationship types along the way.
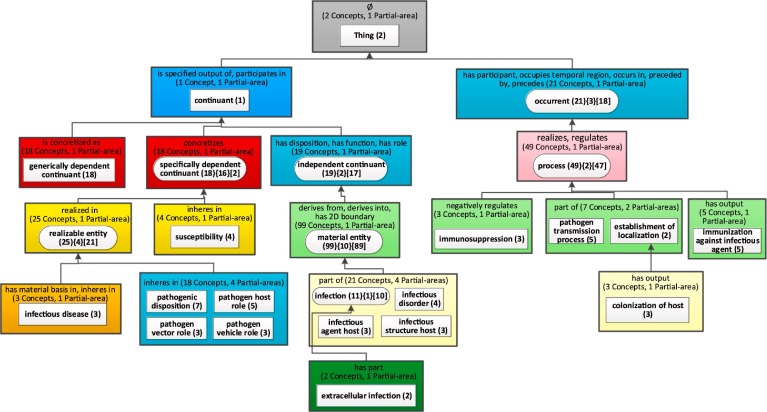
In OAF, when displaying a child-of-based layout, a user can obtain the complete list of relationship types from the system. In cases where an area does not introduce a relationship type, but inherits relationship types (from multiple parent partial-areas), no relationship type will be displayed for the area. This convention prevents the clutter that filled the relationship-based layout of an aggregate taxonomy with a long list of relationship types (see Fig. 5) that a user is typically not interested in. Fig. 7 presents the improved child-of-based layout for the above weighted aggregate taxonomy of the IDO ontology, which constitutes a great improvement over Fig. 5. Note that although the levels of the nodes are organized by ‘child-of’ distance from the root node, the nodes are still preserving the colors according to the number of the relationship types.
Fig. 7.
The child-of-based layout for the weighted aggregate taxonomy of the IDO ontology (b = 2).
3.4. Measures of efficiency for a layout of a taxonomy
When considering the layout of a taxonomy figure on a screen one has to take into account its two dimensions. The vertical dimension depends on the number of levels in the taxonomy. The horizontal dimension is related to the number of partial-areas and areas in a level. For some taxonomies, typically for those with many relationship types, the vertical dimension is larger, e.g., the taxonomies for CIDO and IDO. For taxonomies with relatively low numbers of relationship types the situation is reversed, with the horizontal dimension being larger. Examples include the Biological Process hierarchy of NCIt [23] and the Specimen hierarchy of SNOMED CT [24]. For the first kind we prefer portrait layout and for the second the landscape layout is more fitting.
An important consideration for the effectiveness of a layout of a taxonomy is that the text associated with each node is legible, because the name and number associated with a node carry the knowledge represented by the taxonomy. If, for example, the number of levels is large when a taxonomy is long and narrow, then the text may not be legible when the whole taxonomy needs to fit on one screen. Of course, one can zoom in and increase the size of the text, but then only a portion of the figure is displayed and the overall view is lost. One may scroll down and study portions of the figure, one at a time, but then some child-of relationships are emanating out of the figure. Thus, we will consider layouts where the whole figure is visible on one screen.
Since it is desirable that all the levels fit onto one screen, the vertical dimension is equal to the number of the levels in a taxonomy. In the relationship-based layout, this is the number of relationship levels, since all the nodes with the same number of relationship types are at the same level. In the child-of-based layout, the levels are determined by the child-of relationships among the nodes, and if multiple child-of relationships emanate from a node, the level of the node is determined by the longest child-of path from this node to the root node of the taxonomy.
The situation regarding the horizontal dimension is more complex. One issue is that some areas of the taxonomy may have many partial-areas, which are then arranged in several layers inside an area node. For example, in Fig. 4 there is an area with 17 partial-areas and there are three areas with seven partial-areas. The 17 partial-areas in the area are arranged in four layers. This layering is performed in order not to overextend the width of the layout. In WATs (weighted aggregate taxonomies) the need for layering is minimized because the small partial-areas in an area are aggregated into larger ancestor partial-areas. Thus layering is rare in aggregate taxonomies. For example, there are only two areas with four partial-areas layered in two layers in Fig. 5.
We are interested in the measure of the horizontal dimension, namely, the maximum number of partial-areas in a level, because the layout should enable this level to fit on a screen.
Let V(T, O, L) denote the vertical dimension of a taxonomy T of an ontology O for layout L, namely the number of levels in the taxonomy.
Let Hmax(T, O, L) denote the horizontal dimension of a taxonomy T of an ontology O for layout L, namely the maximum number of partial-areas in any level.
Let R(T, O, L) denote the ratio between the horizontal and vertical dimensions for a taxonomy T of an ontology O for layout L, namely, R(T, O, L) = .
Table 1 shows those measures for the weighted aggregate partial-area taxonomy T with the relationship-based layout L1 (Fig. 5) and T with the child-of-based layout L2 (Fig. 7).
Table 1.
The horizontal and vertical dimensions and their ratio for layouts of the IDO aggregate taxonomy
| (T, L1) | (T, L2) | |
|---|---|---|
| V | 10 | 6 |
| Hmax | 6 | 10 |
| R = Hmax / V | 0.60 | 1.67 |
For an efficient layout it is required that the proportion of the vertical and horizontal dimensions corresponds to the proportion of computer screens. Reviewing several screens, we found that the ratio of the horizontal dimension to the vertical dimension is in the range of 1.0 – 1.75. For a standard 11*8.5 paper page the ratio is about 1.3 with the landscape view.
Looking at Table 1 we see a large difference between the ratios of the two layouts. The ratio 1.67 for the child-of-based layout fits the range of ratios of horizontal to vertical dimensions of screens while the 0.6 ratio for the relationship-based layout does not. However, one cannot use such a strict rule due to several reasons. When looking at Fig. 5 we see many boxes where the horizontal dimension is triple the size of the vertical dimension. This is caused by the need to list all the relationship types for each area in the relationship-based layout. In the child-of-based layout, only the newly introduced relationship types are listed in an area. As a result, we see in Fig. 7 that the average ratio of the horizontal dimension of a box to the vertical dimension is about 2. Another problem is that few areas contain several partial-areas, so the space requirement for all partial-areas are not uniform. Hence the number of levels and the maximum number of partial-areas per level are not the only factor for deciding the efficiency of a layout.
However, we can define the Ratio of the Ratios (RR) of the two layouts as a measure to compare their efficiency. Let
be the Ratio of Ratios of layouts L2 and L1 for an aggregate taxonomy T of an ontology O. If RR is larger than 1, we say that layout L2 is more efficient than layout L1. If RR is much larger than 1, then it is likely that L2 is an efficient layout while L1 is not. For the IDO RR = 1.67/0.60 = 2.78 is large and indeed viewing both figures on a landscape layout screen L2 is readable while L1 is not.
For the case of printing the taxonomy on a page the user can choose between landscape and portrait layouts. The R(T, O, L) value can guide the user which layout fits better for the taxonomy. If R(T, O, L) is larger than 1 then the landscape layout is more fitting because the horizontal dimension is larger than the vertical dimension and vice versa.
4. Results
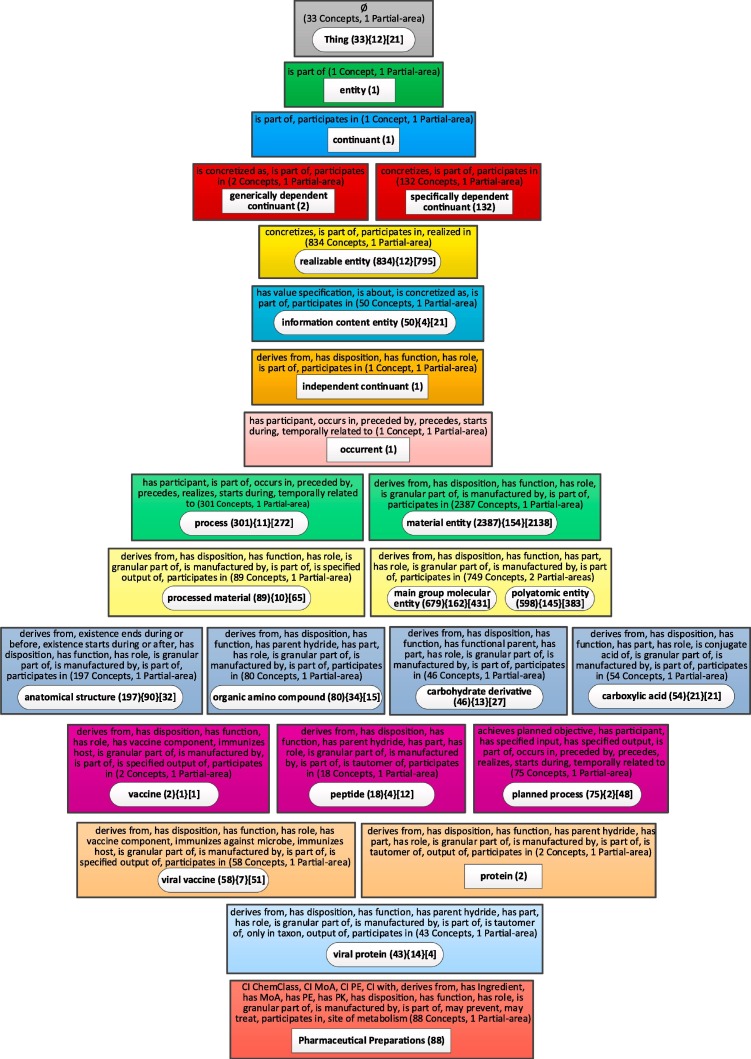
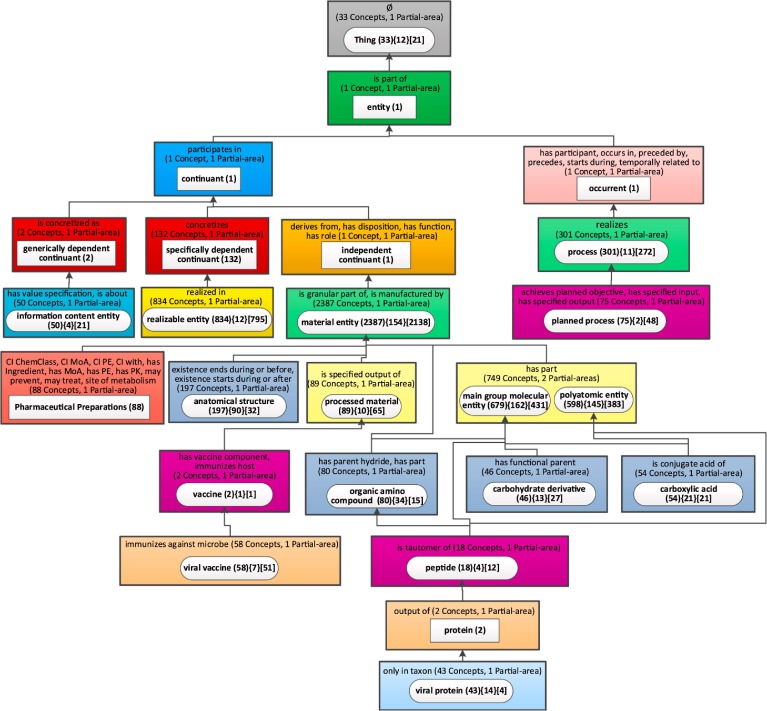
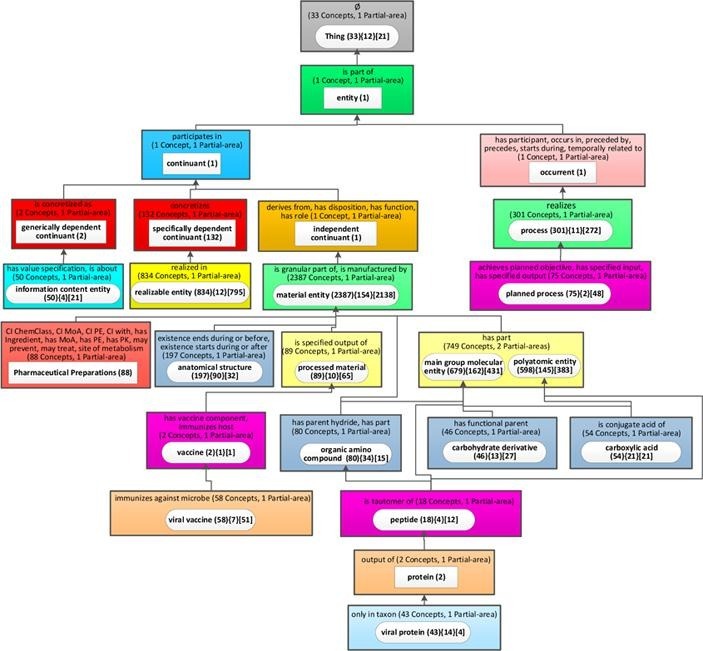
The canonical summarization network used for ontologies with relationships is the partial-area taxonomy. The partial-area taxonomy for CIDO contains 519 partial-areas in 177 areas, by far too many to fit on a screen. For ontologies such as CIDO, with a large number of partial-areas, we invented the weighted aggregate partial-area taxonomy [56], as demonstrated for IDO in the Methods section. Thus, we present the weighted aggregate taxonomy (Fig. 8 ), for CIDO with 5,138 concepts utilizing the relationship-based layout. We used a bound of b = 42, which means that all partial-areas summarizing 42 or more concepts are considered large, and the remaining partial-areas are considered small. We chose the value of 42 because it results in an aggregate taxonomy with 25 nodes.
Fig. 8.
The relationship-based layout of the weighted aggregate partial-area for CIDO (b = 42).
In Fig. 8 we see a long and narrow taxonomy, where the names of the partial-areas and the relationships of the areas are unreadable on a landscape layout screen. Most of the space is not utilized. The same figure would be barely readable if shown in portrait layout on a page, but we have chosen to present it in the way it will look on a screen to communicate the inefficiency of this layout (Please use zooming for readability.) Furthermore, in this figure, we had to omit the child-of relationships between partial-areas due to lack of space. For the child-of relationships between partial-areas in consecutive levels, this omission is acceptable. However, in many cases, the child-of relationships are directed to partial-areas several levels up, and this information is lost in the figure. For example, the partial-area ‘planned process’ in Level 12 has a child-of relationship to the partial-area ‘process’ in Level 9. ‘Pharmaceutical Preparations’ in Level 15 is child-of ‘material entity’ in Level 9 as well. The vertical dimension of this layout is 15 and the horizontal dimension is 4. The ratio R of the horizontal to the vertical dimensions is very low 4/15 = 0.27. The lack of the child-of relationships hurts orientation.
Realizing that the relationship-based layout is not efficient we now show the child-of-based layout of the same taxonomy (Fig. 9 ). This figure only has 10 levels and the partial-areas are nicely spread over the page, enabling easy reading of the names of partial-areas and of the new relationships introduced in each area. The horizontal and vertical dimensions are 5 and 10, respectively, with a ratio R of 0.5. This number is low due to the nature of the CIDO taxonomy being long and narrow. This ratio does not fit the range of corresponding ratio of screens. Nevertheless, comparing the two layouts for CIDO the ratio of ratios RR = 0.5/0.27 = 1.85 is high, implying that the child-of-based layout is much more efficient than the relationship-based layout for the aggregate taxonomy of CIDO. Comparing the actual Fig. 8, Fig. 9, one sees that only the latter is efficient.
Fig. 9.
The weighted aggregate taxonomy for CIDO in child-of-based layout (b = 42).
The child-of relationships between nodes support orientation. Furthermore the child-of hierarchy enable contextual comprehension where one proceeds from a node to its children and grandchildren which are relate to one another. Hence comprehension of a group of related nodes is easier and faster.
One issue with comprehension of an ontology is that the user “cannot see the forest for the trees.” Being occupied with the individual concepts, one misses the “big picture” of the ontology. When (YH), the curator of CIDO, saw Fig. 9 and Fig. 10 , he remarked that “it gives the 'forest' view of CIDO.”
Fig. 10.
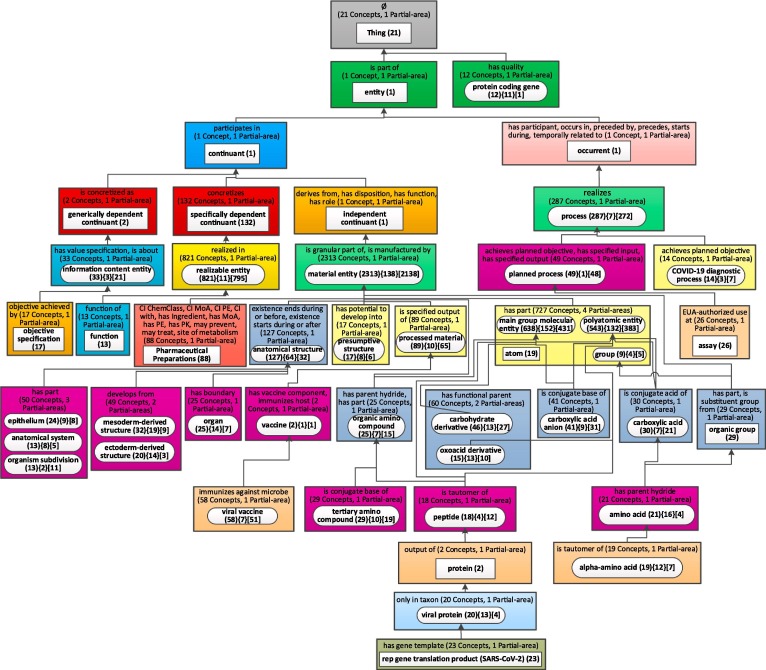
A more refined weighted aggregate taxonomy of CIDO with 46 nodes (b = 12).
What comprehension details can be garnered from Fig. 9 about the content of CIDO? The names of the top-level aggregate partial-areas are generic and not too informative. Also, the numbers of concepts listed for many of them are small. Examples are: 'entity (1)', 'continuant (1),' 'occurrent (1),' 'generally dependent continuant (2)' and 'independent continuant (1).' Those concepts were selected to represent major subjects, not due to the sizes (=numbers of concepts) of their partial-areas, but due to their weights (=numbers of all their concepts + numbers of their descendant concepts). The importance of using weight rather than size for identifying major subjects in CIDO is illustrated in the lower levels of the taxonomy, where 'Vaccine (2)' was selected due to its child 'viral vaccine (58).' Similarly, 'protein (2)' was selected due to its child 'viral protein (43).'
Some of the top-level nodes represent larger subjects: 'Thing (33),' 'specifically dependent continuant (132),' 'realizable entity (834),' and 'material entity (2387).' Those names as well as those of the smaller top major subjects mentioned above, appear in CIDO due to the CIDO design choice of extending the IDO ontology (see Methods section). Both IDO and CIDO follow the OBO Foundry principles, and many of those major subjects appear also in the IDO aggregate taxonomy (Fig. 7).
The nodes farther down represent major subjects that are obviously relevant for biomedical users. On the right of the figure appear 'process (3 0 1)' and 'planned process (75).' On the left there are 'information content entity (50),' 'Pharmaceutical Preparations (88),' 'anatomical structure (1 9 7),' 'processed material (89),' 'organic amino compound (80),' and other chemical subjects.
While one would expect these major subjects in CIDO, the number of concepts belonging to each subject is informative for users. The curators imported several subhierarchies from source ontologies such as ChEBI, NDF-RT and GO, but the user is not necessarily aware of the exact number of imported concepts from each of them. The summary display provides the curators and the users with this numeric information.
The following description illustrates how the above major subjects are relevant to a user of CIDO. 'Planned process' is defined as a process that is planned by a human and realizes a plan defined in a plan specification. For example, a specific 'planned process' is assay, which includes the 'COVID-19 RT-PCR assay'. 'Reverse Transcription Polymerase Chain Reaction (RT-PCR)' is the gold standard among the diagnosis methods for COVID-19 detection. Currently CIDO represents 24 subclasses of specific RT-PCR assays for diagnosis of COVID-19.
'Plan specification' is defined as a directive information entity that specifies the plan to achieve specific objectives. 'Infectious disease control strategy' is a specific plan specification that includes many specific strategies for controlling infectious diseases such as COVID-19. Examples of the control strategies in CIDO include 'quarantine control strategy,' 'travel-related infectious disease control strategy,' and 'place closure control strategy' (e.g., school closure).
The node 'Pharmaceutical Preparations' (from NDF-RT) represents over 100 drugs that have been experimentally found to be effective against coronavirus infections in vitro or in vivo [45]. These drugs and the evidence about their anti-coronavirus effectiveness were manually annotated from peer-reviewed articles, mapped to NDF-RT and ChEBI and then imported into CIDO. They were further interlinked through different relationships. NDF-RT provides classifications of these drugs and their related characteristics such as the mechanisms of action (MoAs). For example, there are a few controversial drugs such as Chloroquine and Hydroxychloroquine, which were initially authorized and then withdrawn for clinical usage by the FDA. However, experimental evidence did show their effectiveness against coronavirus infections in vitro [61], and therefore they are included in CIDO.
A major subhierarchy of 'processed material' in CIDO is vaccines. Under 'viral vaccine,' there is a 'coronavirus vaccine' subhierarchy. Currently CIDO includes 28 SARS vaccines and 19 MERS vaccines defined under coronavirus vaccines [46]. Details of COVID-19 vaccines are being annotated and added. Based on the OBO Foundry principles, the information about those vaccines was initially represented in the Vaccine Ontology (VO) [62] and then imported into CIDO.
CIDO represents both host and viral processes. CIDO includes over 200 human proteins as drug targets of anti-coronavirus drugs. Under 'viral protein,' and 'SARS-CoV-2 proteins,' CIDO lists more than 20 proteins produced by the SARS-CoV-2 viruses. The list of proteins includes the spike glycoprotein, commonly referred as “S protein,” which is responsible for attaching to and invading host cells. Information about these proteins was imported from the Protein Ontology (PRO) [63].
4.1. Optimizing display parameters
The question of what the optimal number of nodes in a taxonomy is, in order to support comprehension, is far from resolved in Human Computer Interaction research. There is always a hardware limitation of how many nodes (named boxes) one can display on a screen in a readable format. We hypothesize that the optimal number of nodes in a taxonomy is between 25 and 50.
There is clearly a tradeoff. When the number of displayed nodes is close to the upper limit (50), the comprehension effort required is more intense. Some users would not even attempt to comprehend a network with 50 nodes. On the other hand, a taxonomy with more nodes provides a higher granularity display, immediately exposing more major subjects of the ontology.
To illustrate this issue, Fig. 10 shows a more granular aggregate taxonomy of CIDO with 46 nodes for a bound of b = 12. The horizontal and vertical dimensions of this layout are 13 and 11 respectively, yielding a ratio of 1.18. In spite of the large number of nodes this layout is readable. As a result, some of the major subjects were refined to yield new subjects with smaller numbers of concepts, namely, partial-areas that summarize fewer than 42 but at least 12 concepts, and therefore were not aggregated into their ancestors. These partial-areas are now represented by their own nodes and may aggregate some of their smaller descendant partial-areas. Examples include 'protein coding gene' with 12 concepts, 11 of which are children aggregated into this aggregate partial-area. These 11 concepts are all the protein-coding genes of the SARS-CoV-2 virus (cause of COVID-19).
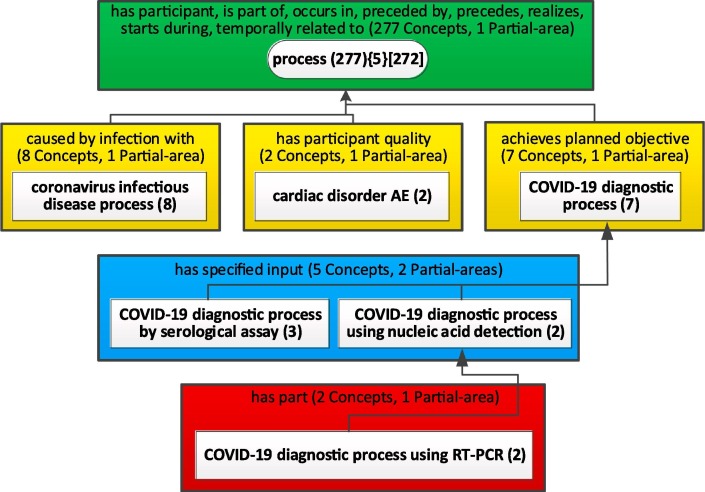
Another new node in Fig. 10 is 'COVID-19 diagnosis process (14),' which was extracted out of the 'process (3 0 1)' node in Fig. 9, leaving 'process (2 8 7)' in Fig. 10. This new node is discussed in relation to Fig. 11 below. The node 'assay (26)' was extracted from 'planned process (75)' in Fig. 9. Both were discussed above in the context of Fig. 9. The difference is that in Fig. 9 these subjects were implicit and in Fig. 10 they are explicitly displayed.
Fig. 11.
Expansion of the 'process' subject of Fig. 9.
The two nodes 'atom (19)' and 'group (9)' in Fig. 10 were extracted from 'material entity (2387)' in Fig. 9. The node 'organic group (29)' was also extracted from 'material entity' and is a child of the node 'group.' The child 'Amino acid (21)' and the grandchild 'alpha-amino acid (19)' of 'carboxylic acid' were extracted from 'carboxylic acid (54)' in Fig. 9. Another node 'oxoacid derivative (15)' was extracted from 'polyatomic entity.' Six more new nodes were extracted from the 'anatomical structure (1 9 7)' node in Fig. 9.
The node 'rep gene translation product (SARS-CoV-2) (23)' in Fig. 10 was extracted from 'viral protein (43)' in Fig. 9. This node represents a protein that is a translation product of the rep gene in SARS-CoV-2. The viral rep gene produces two translation products, ORF1ab and ORF1a, which encode replicase polyprotein 1a (PP1a) and polyprotein 1ab (PP1ab), two polyproteins that are critical for viral replication [64].
What does our approach suggest when a user wants to better understand one of the major subjects? In such cases the node of the major subject can be expanded (using our OAF software tool) into a network of secondary subjects, as was explained in the Methods section with regards to the IDO.
For example, when we expand the 'process' subject in Fig. 9, we obtain Fig. 11. This is an aggregate taxonomy with a bound b = 2. Thus, five partial-areas, each with one concept, were aggregated into the root node 'process,' but 24 concepts appear now in six partial-areas, such as 'coronavirus infectious disease process (8)' or 'COVID-19 diagnostic process (7)' with its own children such as 'COVID-19 diagnostic process using nucleic acid detection (2),' which in turn has a child 'COVID-19 diagnostic process using RT-PCR (2).' Such small secondary subjects might be highly relevant for the users of CIDO. We discussed above the importance of RT-PCR, the gold standard diagnostic method for detecting the presence of SARS-CoV-2 and its descendant concepts in CIDO. ‘Coronavirus infectious disease process’ is a CIDO-specific concept that links different entities, including the host organism, anatomical location, cause, and phenotype outcomes shown in the dynamic COVID-19 process.
This example illustrates that in a secondary subject network and even in a major subject network, some nodes may be aggregate partial-areas, while the others are “just” partial-areas. The example in Fig. 12 shows a case where all nodes are partial-areas. If a user is interested in 'process' but not in any of the other partial-areas in Fig. 11, OAF can expand the node ‘process (2 7 7){5}[2 7 2]’ into a tertiary subject network of a root partial-area of 277 concepts and 5 child partial-areas of one concept.
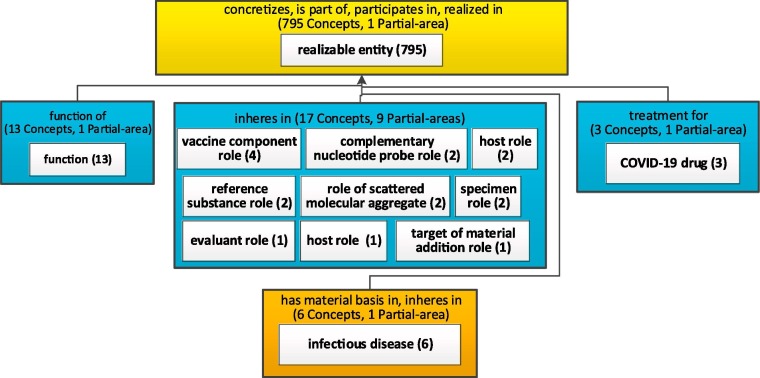
Fig. 12.
Secondary subjects of 'realizable entity' of Fig. 9.
Consider another major subject, 'realizable entity (8 3 4){12}[7 9 5]' in Fig. 9. It can be expanded into a network (Fig. 12) of secondary subjects such as 'COVID-19 drug (3),' 'infectious disease (6)' and various roles related to the coronavirus and the research on medications and vaccines for COVID-19.
In the Discussion we will raise the problem of large secondary subjects, such as 'realizable entity (7 9 5)' or 'process (2 7 7).'
5. Discussion
In previous work, we designed the Tribal Summarization Network (TAN) for ontologies without relationships, for which a WAT cannot be derived. However, one can derive a TAN for ontologies like CIDO that do have relationships by simply ignoring the relationships. The result is shown in Fig. 13 . The nodes in the first level show the children of the root ‘Thing’ enumerating the numbers of their descendants. Typically, the second level would show concepts that are descendants having multiple parents at the first level. For CIDO, this does not occur. As a result, the Tribal Summarization Network of CIDO has only three nodes and is not informative. This disappointing summarization is expected when deriving the TAN of CIDO, because it ignores the rich relationship structure of CIDO. Exactly this rich relationship structure enables the effective summarization of CIDO by the WAT.
Fig. 13.
The Tribal Summarization Network for the CIDO ontology.
The research on the aggregate taxonomy was incremental. Ochs et al. [40], [65] developed the idea of aggregation of partial-areas based on the size (=number of concepts) of a partial-area. That is, partial-areas with a size of at least a given bound b become nodes in the aggregate taxonomy and all partial-areas with sizes smaller than the bound are aggregated into the closest large ancestor partial-area. In a study testing the aggregate taxonomy for identifying major subjects [57], it was shown that using the size for selecting the nodes for the aggregate taxonomy leads to missing important major subjects. The same phenomenon is illustrated in the CIDO aggregate taxonomy, where the partial-area 'vaccine' has a size of 1 and the partial-area 'protein' has a size of 2, but they have larger weights due to their large child partial-areas 'viral vaccine' and 'viral protein,' respectively. Performing aggregation by size would miss these two small partial-areas in spite of their importance. To remedy this problem, we introduced the weighted aggregate taxonomy used in this paper. In [56] the weighted aggregate taxonomy was used for a multilevel navigation system for an ontology. We intentionally did not publish the aggregate taxonomy results in a journal paper following the early conference publications, since that research did not reach the proper maturity level at that point.
Applying the aggregate taxonomy derivation to the CIDO ontology led to the innovation of the child-of-based layout of the aggregate taxonomy, which supports both improved orientation and comprehension. For the above applications the readability of the aggregate taxonomy was not an issue, but for user comprehension and orientation, the readability is crucial. With the child-of-based layout, the research on the aggregation of partial-areas to obtain a compact comprehensible summarization of an ontology has reached maturity.
There are two factors making the child-of-based layout visualization better for comprehension and orientation of CIDO than the relationship-based layout. One factor is showing only the newly introduced relationship types within the area. The full list of relationships is obtainable by traversing the path(s) to the root partial-area while collecting the relationship types introduced along the way. This convention saves space that is instead allocated for better readability of the names of major subjects. The other factor is basing the layout on the child-of hierarchy rather than on the number of relationship types used in the relationship-based layout, as was explained in Section 3.4. The first factor helps comprehension, while the second factor supports orientation.
One advantage of the aggregate taxonomy for comprehension is the option of applying it multiple times. This was demonstrated in this paper by the expansion of a major subject node into a secondary subject network. For a large ontology such as GO or ChEBI or a large hierarchy of SNOMED CT such as the 'Procedure' or 'Clinical finding' hierarchies, two layers of subject networks may not be sufficient for comprehension. If a secondary subject network still consists of many nodes, one needs to use aggregation again. In such case a user will have to drill down further to a third or even forth aggregation layer. Because we keep the number of nodes in the subject networks at each level limited to between 25 and 50 nodes, better comprehension is supported at every level. Hence this framework is expandable for large ontologies.
It is interesting to note that the additional major subjects appearing in Fig. 10, but not in Fig. 9, are also appearing as secondary subjects when the major subjects of Fig. 9 are expanded. Examples include ‘COVID-19 diagnostic process (7)’ in Fig. 11, which combined with its descendant partial-areas has the weight 14, larger than b = 12. Another example is ‘function (13)’ in Fig. 12. Those examples illustrate the two ways of exposing the “smaller” subjects. One way is with higher granularity network of major subjects. The other is as secondary subjects of major subjects in a more restricted network of major subjects.
5.1. Generality of the Benefits of the Child-of-based layout
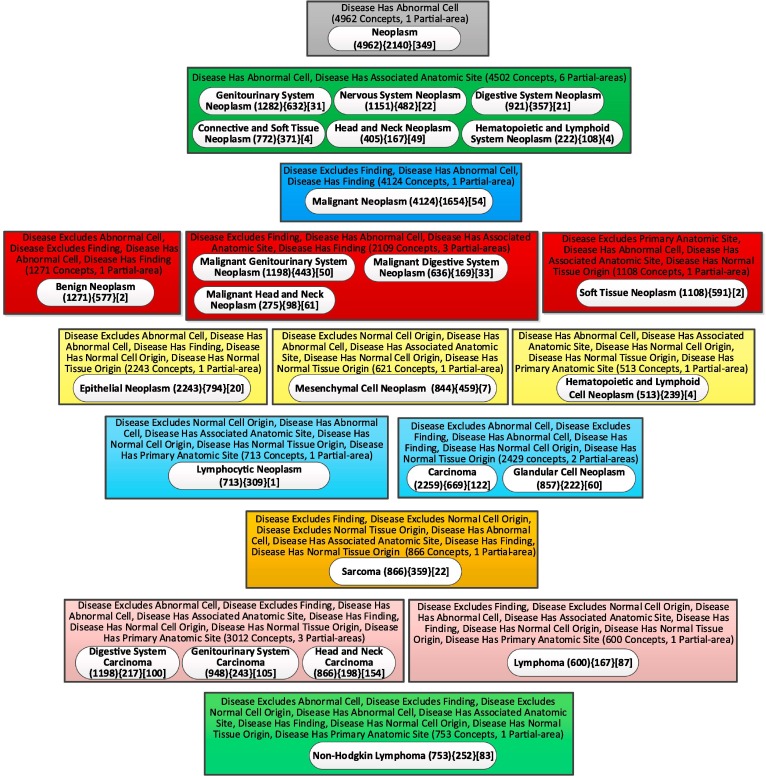
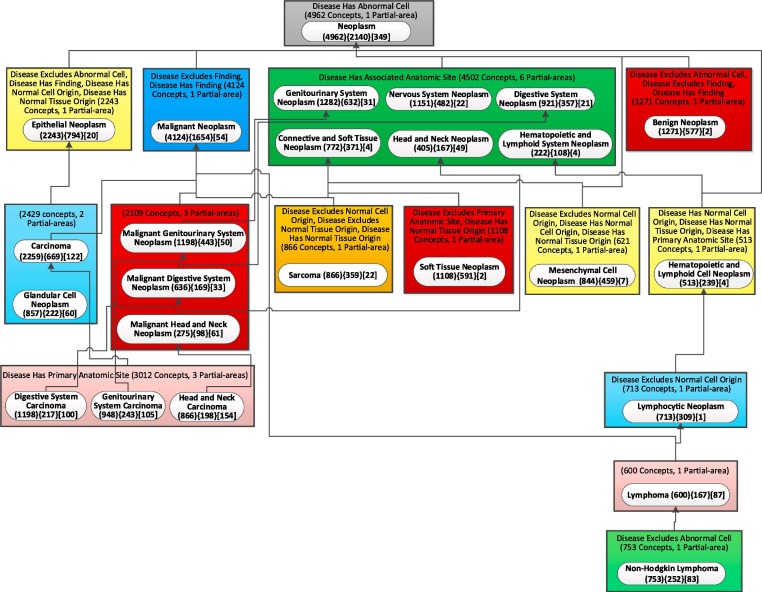
To demonstrate that the child-of-based layout is more efficient for taxonomies of other ontologies with large numbers of relationships beyond the cases of IDO and CIDO, we revisited the Neoplasm subhierarchy of NCIt, which was presented in [56] with the relationship-based layout. The 'Neoplasm' subhierarchy of the ‘Disease, Disorder or Finding’ hierarchy of NCIt, contained 8,845 concepts in the September 2016 release. In the January 2021 release the same subhierarchy grew to 12,058 concepts. Its partial-area taxonomy has 4,200 partial-areas. We will compare the two layouts for the aggregate taxonomy with 25 nodes for this subhierarchy. Fig. 14 shows the relationship-based layout and Fig. 15 shows the child-of-based layout. The ratios of the horizontal to the vertical dimension for the two layouts are 6/9 = 0.67 and 9/6 = 1.50. The Ratio of Ratios RR is 1.50/0.67 = 2.24, a high value. The child-of-based layout is more efficient than the relationship-based layout.
Fig. 14.
The relationship-based layout of the weighted aggregate partial-area for the Neoplasm subhierarchy of NCIt (b = 722).
Fig. 15.
The child-of-based layout of the weighted aggregate partial-area for the Neoplasm subhierarchy of NCIt (b = 722).
Both CIDO and the NCIt’s Neoplasm subhierarchy are characterized by many relationships, which caused the relationship-based layout to be long and narrow. Those are ontologies for which the difference between the two layouts is most obvious. For ontologies with few relationships, the number of levels of the relationship-based layout is not that large. For such ontologies the main benefit is the clear delineation of the child-of hierarchy, supporting orientation. Table 2 lists the ratio of relationship-based layout aggregate taxonomy (R1), the ratio of child-of-based layout aggregate taxonomy (R2) and the ratio of the latter to the former (RR) for the IDO, CIDO and NCIt’s Neoplasm subhierarchy.
Table 2.
The comparison of ratios for the two layouts of the aggregate taxonomy for IDO, CIDO and NCIt’s Neoplasm subhierarchy
| R1 (relationship-based layout) |
R2 (child-of-based layout) |
RR (=R2/R1) | |
|---|---|---|---|
| IDO | 0.60 | 1.67 | 2.78 |
| CIDO | 0.27 | 0.50 | 1.85 |
| NCIt Neoplasm subhierarchy | 0.67 | 1.50 | 2.24 |
Limitations and Future Work: A drawback of the visualization of CIDO shown in Fig. 9 is that some of the major subject nodes represent a large number of concepts. Examples include 'realizable entity (8 3 4),' 'material entity (2387),' and 'process (3 0 1)'. Even more problematic is that the corresponding partial-areas of the aggregate partial-areas have 795, 2138, and 272 concepts, respectively. Hence, even when considering the network of secondary subjects of each of these major subjects, a large partial-area remains. This is especially bothersome for major subjects with enigmatic names as the first two above. The phenomenon of a very large partial-area “hiding” in a major subject, which is not even exposed in the secondary aggregate taxonomy, is due to the fact that all those concepts share exactly the same set of relationship types.
It is likely that some of the concepts in such a partial-area are actually missing a relationship type. When such a relationship type is added to the concepts, this set of concepts will constitute a separate partial-area and if it has a weight large enough then a new major subject appears as a result. For example, in a later release of CIDO, the relationship type chemical has protein target as inhibitor was added to some concepts of 'material entity' and as a result a new major subject 'chemical entity (1 0 1)' emerged. This example demonstrates that one remedy for breaking monolithic large major subjects into smaller refined major subjects is by adding potentially missing relationship types to subsets of their concepts.
When no such missing relationship types are found, one needs to resort to other solutions. One observation is that the partition of an ontology into areas, in our research, was based on relationship types only. However, the properties of an ontology are of two kinds: relationships (object properties) and data properties. One can further divide large partial-areas based on their sets of data property types. Another option is to divide the partial-areas according to the targets of the relationships.
It is not known what the optimal number of nodes in a summarization network should be. In this work, we used a bound of 25 nodes, which accommodates a convenient layout on a screen and supports easy comprehension. We showed an example of an alternative, more refined, aggregate taxonomy of CIDO with 46 nodes. There is a trade-off between the advantage of exposing additional major subjects versus a denser layout making comprehension harder. In future work, we plan to investigate what the optimal range for this trade-off is.
In the current paper, we do not have a study of users that assesses the advantages of the child-of-based layout over the relationship-based layout. We are planning such a study for the future.
6. Conclusions
The intensive ongoing research on medications and vaccinations for COVID-19 requires support of a reference ontology. CIDO is the largest and fastest growing COVID ontology. Users of CIDO need support for comprehending its content. We presented in this paper the aggregate taxonomy summarization network and its child-of-based layout to support summarization and orientation of the CIDO ontology. The large number of relationship types in CIDO caused problems with readability of the previous relationship-based layout of the aggregate taxonomy for CIDO. The new child-of-based layout was shown to overcome those problems. A layout efficiency measure was introduced and shows the advantages of the child-of-based layout. Generality of the child-of-based layout was demonstrated.
CRediT authorship contribution statement
Ling Zheng: Conceptualization, Methodology, Writing - original draft. Yehoshua Perl: Conceptualization, Methodology, Writing - original draft. Yongqun He: Validation, Formal analysis. Christopher Ochs: Software, Data curation. James Geller: Software, Data curation. Hao Liu: Resources, Writing - review & editing. Vipina K. Keloth: Resources, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Research reported in this publication was supported by the National Center for Advancing Translational Sciences (NCATS), a component of the National Institute of Health (NIH) under award number UL1TR003017. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic acids research. 2011;39(Web Server issue):W541-5. [DOI] [PMC free article] [PubMed]
- 2.He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, et al. CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci Data. 2020;7(1):181. [DOI] [PMC free article] [PubMed]
- 3.He Y, Yu H, Ong E, Wang Y, Liu Y, Huffman A, et al. CIDO: The Community-Based Coronavirus Infectious Disease Ontology. 11th International Conference on Biomedical Ontologies (ICBO-2020). 2020.
- 4.Smith B., Ashburner M., Rosse C., Bard J., Bug W., Ceusters W., et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong E, Huffman A, Brunson T, Sanati N, Zheng J, Masci AM, et al. Ontology-based representation and analysis of vaccine immune response data in ImmPort. 9th International Workshop on Vaccine and Drug Ontology Studies (VDOS-2020). 2020.
- 6.Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44(D1):D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler S., Vasilevsky N.A., Engelstad M., Foster E., McMurry J., Ayme S., et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45(D1):D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown S.H., Elkin P.L., Rosenbloom S.T., Husser C., Bauer B.A., Lincoln M.J., et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Medinfo. 2004;11(477–481) [PubMed] [Google Scholar]
- 9.Hanna J., Joseph E., Brochhausen M., Hogan W.R. Building a drug ontology based on RxNorm and other sources. J Biomed Semantics. 2013;4(1):44. doi: 10.1186/2041-1480-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.COVID-19 Ontology [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/COVID-19.
- 11.COVID-19 Infectious Disease Ontology [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/IDO-COVID-19.
- 12.Cowell L.G., Smith B. Infectious disease ontology. Infectious disease informatics. 2010:373–395. [Google Scholar]
- 13.Virus Infectious Disease Ontology [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/VIDO.
- 14.WHO COVID-19 Rapid Version CRF semantic data model [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/COVIDCRFRAPID.
- 15.Ontology for Collection and Analysis of COviD-19 Data [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/CODO.
- 16.COVID-19 Surveillance Ontology [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/COVID19.
- 17.ACT COVID Ontology [2/21/2021]. Available from: https://github.com/shyamvis/ACT-COVID-Ontology.
- 18.ICD [2/21/2021]. Available from: http://www.who.int/classifications/icd/en/.
- 19.McDonald C.J., Huff S.M., Suico J.G., Hill G., Leavelle D., Aller R., et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin. Chem. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 20.CPT (Current Procedural Terminology) [2/21/2021]. Available from: https://www.ama-assn.org/amaone/cpt-current-procedural-terminology.
- 21.National Drug Code (NDC) [2/21/2021]. Available from: https://www.fda.gov/drugs/drug-approvals-and-databases/national-drug-code-directory.
- 22.Halper M., Gu H., Perl Y., Ochs C. Abstraction Networks for Terminologies: Supporting Management of “Big Knowledge”. Artif. Intell. Med. 2015;64(1):1–16. doi: 10.1016/j.artmed.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min H., Perl Y., Chen Y., Halper M., Geller J., Wang Y. Auditing as part of the terminology design life cycle. J. Am. Med. Inform. Assoc. 2006;13(6):676–690. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Halper M., Min H., Perl Y., Chen Y., Spackman K.A. Structural methodologies for auditing SNOMED. J. Biomed. Inform. 2007;40(5):561–581. doi: 10.1016/j.jbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Halper M., Wei D., Perl Y., Geller J. Abstraction of complex concepts with a refined partial-area taxonomy of SNOMED. J. Biomed. Inform. 2012;45(1):15–29. doi: 10.1016/j.jbi.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochs C., Agrawal A., Perl Y., Halper M., Tu S., Carini S., et al. Deriving an Abstraction Network to Support Quality Assurance in OCRe. AMIA Annu Symp Proc. 2012;681–9 [PMC free article] [PubMed] [Google Scholar]
- 27.Ochs C., He Z., Perl Y., Arabandi S., Geller J. Proceedings of the 4th International Conference on Biomedical Ontology. 2013. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology; pp. 84–89. [Google Scholar]
- 28.He Z., Ochs C., Soldatova L., Perl Y., Arabandi S., Geller J. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. VDOS. 2013 [Google Scholar]
- 29.Sim I., Tu S.W., Carini S., Lehmann H.P., Pollock B.H., Peleg M., et al. The Ontology of Clinical Research (OCRe): an informatics foundation for the science of clinical research. J. Biomed. Inform. 2014;52:78–91. doi: 10.1016/j.jbi.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arabandi S., Ogbuji C., Redline S., Chervin R., Boero J., Benca R., et al. Developing a Sleep Domain Ontology. AMIA Clinical Research Informatics Summit. 2010 [Google Scholar]
- 31.Zeginis D., Hasnain A., Loutas N., Deus H.F., Fox R., Tarabanis K. A collaborative methodology for developing a semantic model for interlinking Cancer Chemoprevention linked-data sources. Semantic Web. 2014;5(2):127–142. [Google Scholar]
- 32.Da Q., King R., Hopkins A., Bickerton R., Soldatova L. An ontology for description of drug discovery investigations. J Integrative Bioinformatics. 2010;7(3):126–139. doi: 10.2390/biecoll-jib-2010-126. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mungall C.J., Torniai C., Gkoutos G.V., Lewis S.E., Haendel M.A. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13(1):R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochs C., Geller J., Perl Y., Chen Y., Xu J., Min H., et al. Scalable Quality Assurance for Large SNOMED CT Hierarchies Using Subject-based Subtaxonomies. J. Am. Med. Inform. Assoc. 2014;22(3):507–518. doi: 10.1136/amiajnl-2014-003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perl Y., Geller J., Halper M., Ochs C., Zheng L., Kapusnik-Uner J. Introducing the Big Knowledge to Use (BK2U) challenge. Ann. N. Y. Acad. Sci. 2017;1387(1):12–24. doi: 10.1111/nyas.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochs C., Geller J., Perl Y., Chen Y., Agrawal A., Case J.T., et al. A Tribal Abstraction Network for SNOMED CT Hierarchies without Attribute Relationships. J. Am. Med. Inform. Assoc. 2014;22(3):628–639. doi: 10.1136/amiajnl-2014-003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu H., Halper M., Geller J., Perl Y. Benefits of an object-oriented database representation for controlled medical terminologies. J. Am. Med. Inform. Assoc. 1999;6(4):283–303. doi: 10.1136/jamia.1999.0060283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimino J.J., Clayton P.D., Hripcsak G., Johnson S.B. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J. Am. Med. Inform. Assoc. 1994;1(1):35–50. doi: 10.1136/jamia.1994.95236135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochs C., Zheng L., Perl Y., Geller J., Gu H., Kapusnik-Uner J., et al. Drug-drug Interaction Discovery Using Abstraction Networks for “National Drug File – Reference Terminology” Chemical Ingredients. AMIA Annu Symp Proc. 2015;973–82 [PMC free article] [PubMed] [Google Scholar]
- 41.Peroni S, Motta E, d’Aquin M, editors. Identifying key concepts in an ontology, through the integration of cognitive principles with statistical and topological measures. Asian Semantic Web Conference; 2008: Springer.
- 42.Pathak J., Johnson T.M., Chute C.G. Survey of modular ontology techniques and their applications in the biomedical domain. Integr Comput Aided Eng. 2009;16(3):225–242. doi: 10.3233/ICA-2009-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katifori A., Halatsis C., Lepouras G., Vassilakis C., Giannopoulou E. Ontology visualization methods—a survey. ACM Computing Surveys (CSUR) 2007 39(4):10-es. [Google Scholar]
- 44.Dudáš M., Lohmann S., Svátek V., Pavlov D. Ontology visualization methods and tools: a survey of the state of the art. The Knowledge Engineering Review. 2018;33 [Google Scholar]
- 45.Liu Y, Chan WKB, Wang Z, Hur J, Xie J, Yu H, et al. Ontological and Bioinformatic Analysis of Anti-Coronavirus Drugs and Their Implication for Drug Repurposing against COVID-19. Preprints 2020:2020030413.
- 46.Ong E., Wong M.U., Huffman A., He Y. COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. Front. Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coronavirus Infectious Disease Ontology [2/21/2021]. Available from: https://github.com/CIDO-ontology/cido.
- 48.Coronavirus Infectious Disease Ontology at BioPortal [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/CIDO.
- 49.Coronavirus Infectious Disease Ontology at Ontobee [2/21/2021]. Available from: http://www.ontobee.org/ontology/CIDO.
- 50.Gu H., Perl Y., Geller J., Halper M., Singh M. A methodology for partitioning a vocabulary hierarchy into trees. Artif. Intell. Med. 1999;15(1):77–98. doi: 10.1016/s0933-3657(98)00046-3. [DOI] [PubMed] [Google Scholar]
- 51.Cambridge Dictionary [2/21/2021]. Available from: https://dictionary.cambridge.org/us/.
- 52.BioPortal [2/21/2021]. Available from: http://bioportal.bioontology.org/.
- 53.Morrey C.P., Geller J., Halper M., Perl Y. The Neighborhood Auditing Tool: a hybrid interface for auditing the UMLS. J. Biomed. Inform. 2009;42(3):468–489. doi: 10.1016/j.jbi.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochs C., Geller J., Perl Y., Musen M.A. A Unified Software Framework for Deriving, Visualizing, and Exploring Abstraction Networks for Ontologies. J. Biomed. Inform. 2016;62:90–105. doi: 10.1016/j.jbi.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochs C., Perl Y., Halper M., Geller J., Lomax J. Gene Ontology Summarization to Support Visualization and Quality Assurance. BICoB. 2015:167–174. [Google Scholar]
- 56.Zheng L, Ochs C, Geller J, Liu H, Perl Y, Coronado SD. Multi-layer Big Knowledge Visualization Scheme for Comprehending Neoplasm Ontology Content. 2017 IEEE International Conference on Big Knowledge (ICBK). p. 127-34.
- 57.Zheng L., Perl Y., Elhanan G., Ochs C., Geller J., Halper M. Summarizing an Ontology: A “Big Knowledge” Coverage Approach. Studies in health technology and informatics. 2017;245:978–982. [PubMed] [Google Scholar]
- 58.Infectious Disease Ontology [2/21/2021]. Available from: https://bioportal.bioontology.org/ontologies/IDO.
- 59.Basic Formal Ontology [2/21/2021]. Available from: http://basic-formal-ontology.org/.
- 60.Goodrich M.T., Tamassia R. Algorithm Design: Foundations. Analysis, and Internet Examples: Wiley. 2001 [Google Scholar]
- 61.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Y, Cowell L, Diehl AD, Mobley H, Peters B, Ruttenberg A, et al. VO: Vaccine Ontology. The 1st International Conference on Biomedical Ontology (ICBO-2009). 2009.
- 63.Natale DA, Arighi CN, Barker WC, Blake JA, Bult CJ, Caudy M, et al. The Protein Ontology: a structured representation of protein forms and complexes. Nucleic acids research. 2011;39(Database issue):D539-45. [DOI] [PMC free article] [PubMed]
- 64.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis. Proofreading and Final Capping. Cells. 2020;9(5) doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ochs C., Perl Y., Geller J., Musen M.A. Using Aggregate Taxonomies to Summarize SNOMED CT Evolution. International Workshop on Biomedical and Health Informatics. 2015:1008–1015. [Google Scholar]