Abstract
High‐grade cervical intraepithelial neoplasia (CIN2 and CIN3) represents a heterogeneous disease with varying cancer progression risks. Biomarkers indicative for a productive human papillomavirus (HPV) infection (HPV E4) and a transforming HPV infection (p16ink4a, Ki‐67 and host‐cell DNA methylation) could provide guidance for clinical management in women with high‐grade CIN. This study evaluates the cumulative score of immunohistochemical expression of p16ink4a (Scores 0‐3) and Ki‐67 (Scores 0‐3), referred to as the “immunoscore” (IS), in 262 CIN2 and 235 CIN3 lesions derived from five European cohorts in relation to immunohistochemical HPV E4 expression and FAM19A4/miR124‐2 methylation in the corresponding cervical scrape. The immunoscore classification resulted in 30 lesions within IS group 0‐2 (6.0%), 151 lesions within IS group 3‐4 (30.4%) and 316 lesions within IS group 5‐6 (63.6%). E4 expression decreased significantly from CIN2 to CIN3 (P < .001) and with increasing immunoscore group (P trend < .001). Methylation positivity increased significantly from CIN2 to CIN3 (P < .001) and with increasing immunoscore group (P trend < .001). E4 expression was present in 9.8% of CIN3 (23/235) and in 12.0% of IS group 5‐6 (38/316). Notably, in a minority (43/497, 8.7%) of high‐grade lesions, characteristics of both transforming HPV infection (DNA hypermethylation) and productive HPV infection (E4 expression) were found simultaneously. Next, we stratified all high‐grade CIN lesions, based on the presumed cancer progression risk of the biomarkers used, into biomarker profiles. These biomarker profiles, including immunoscore and methylation status, could help the clinician in the decision for immediate treatment or a “wait and see” policy to reduce overtreatment of high‐grade CIN lesions.
Keywords: cervical cancer, cervical precancer, DNA hypermethylation, HPV E4 protein, human papillomavirus, immunohistochemistry, Ki‐67, p16ink4a, productive HPV infection, transforming HPV infection
Short abstract
What's new?
Treating all high‐grade cervical intraepithelial neoplasia (CIN2/3) with excisional therapy leads to overtreatment, as these lesions have varying cancer progression risks. Here, the authors evaluated expression patterns of p16ink4a, Ki‐67 and the HPV E4 protein, and methylation of FAM19A4/miR124‐2 in high‐grade CIN. The biomarker expression patterns revealed the high degree of heterogeneity among CIN2/3 lesions. Biomarker profiles based on the presumed cancer progression risks were established and could guide clinicians in choosing whether to treat immediately or wait and see.
Abbreviations
- ACTB
ß‐actin
- CIN
cervical intraepithelial neoplasia
- Ct value
cycle threshold value
- DNA
deoxyribonucleic acid
- E4
panHPVE4 protein
- FAM19A4
family with sequence similarity 19 (chemokine [C‐C]‐motif)‐like) member A4
- FFPE
formalin‐fixed paraffin‐embedded
- H&E
haematoxylin and eosin
- HPV
human papillomavirus
- IS
immunoscore
- LLETZ
large‐loop excision of the transformation zone
- miR124‐2
microRNA 124‐2
- mAb
mouse monoclonal antibodies
1. INTRODUCTION
Cervical screening programmes aim to prevent cervical cancer by detection and treatment of the precursor lesion cervical intraepithelial neoplasia (CIN). Women with abnormal cervical scrapes are referred to the gynaecologist for colposcopy and, if indicated, a guided biopsy is taken from the most suspicious area for high‐grade CIN. Pathologists classify these CIN lesions with increasing CIN grade into grades 1 to 3 based on the degree of replacement of the normal epithelium by dysplastic epithelium and the severity of atypia of the abnormal cells in haematoxylin and eosin (H&E)‐stained sections. 1 However, classical histological grading of CIN remains subjective and is associated with a considerable interobserver and intraobserver variation and a moderate reproducibility. 2 , 3 , 4 , 5 , 6
Accurate assessment of high‐grade CIN (ie, CIN2 and CIN3) is essential since treatment depends predominantly on lesion grade. In current practice, CIN3 and the majority of CIN2 lesions are treated by large‐loop excision of the transformation zone (LLETZ) or conisation in order to prevent progression to cervical cancer. However, spontaneous regression of CIN2 (40‐50%) and CIN3 lesions (~30%) frequently occurs, 7 , 8 , 9 indicating that the current diagnostic process is associated with substantial overtreatment. Particularly women within the reproductive age would benefit from a more conservative approach to prevent obstetric complications like preterm birth due to excisional treatment. 10 There is thus a need for a classification system that can help to prevent overtreatment of high‐grade CIN.
Cervical cancer and CIN lesions are caused by a persistent infection with the human papillomavirus (HPV). High‐grade CIN lesions represent a heterogeneous group consisting of a subset of productive CIN lesions, characterised by the production of new viral particles, and a subset of transforming CIN lesions, characterised by HPV E6 and E7 deregulation. 11 Biomarkers could help to differentiate CIN lesions associated with a productive HPV infection and high regression rates from CIN lesions associated with a transforming HPV infection and a presumed high short‐term progression risk to cervical cancer. 11 An important biomarker in the detection of transforming HPV infections is p16ink4a, which is used as a surrogate marker for transforming activity of HPV E7. 12 Immunohistochemical staining of p16ink4a in clinical practice is currently only recommended by the LAST criteria in certain contexts, including cases where there is morphological doubt between low‐grade and high‐grade CIN. 13 Another potential biomarker for CIN classification is Ki‐67, a marker for cell cycle activity. 14 Coexpression of p16ink4a and Ki‐67, within the same cell, reflects cellular dysfunction and is only seen in HPV‐transformed dysplastic cells. 15 , 16 Recently, a classification system based on the cumulative score of p16ink4a (Scores 0‐3) and Ki‐67 (Scores 0‐3) expression has shown to be more accurate and reproducible for CIN grading compared with haematoxylin and eosin (H&E) assessment alone. The cumulative score also reduces the diagnosis of CIN2 lesions for which clinical management is not uniform. 17 This cumulative score of p16ink4a and Ki‐67 is further referred to as the “immunoscore” (IS). Another immunohistochemical marker that could improve the accuracy of CIN diagnosis is the panHPV E4 protein (E4). 18 E4 accumulates in cervical cells supporting HPV viral genome amplification. Therefore, it could be used as a biomarker for the identification of low‐grade lesions and productive subsets of high‐grade lesions. 19 , 20
DNA methylation of promoter regions in tumour suppressor genes is a key event during cervical carcinogenesis. 11 Several studies have shown the applicability of methylation markers for the identification of transforming CIN lesions, as well as the potential to predict regression and/or progression of CIN lesions. 21 , 22 Furthermore, methylation analysis has demonstrated that CIN2/3 lesions associated with a persistent HPV infection of at least 5 years had cancer‐like methylation patterns, suggesting a high short‐term progression risk to cervical cancer. 23 , 24 Methylation analysis in combination with immunohistochemical staining of p16ink4a, Ki‐67 and E4 might provide biomarker profiles that could improve the clinical guidance in women with high‐grade CIN to prevent overtreatment of regressive lesions and potential obstetric complications.
The current study evaluates the cumulative p16ink4a and Ki‐67 immunoscore in a large series of high‐grade CIN lesions derived from five European cervical referral or screening cohorts in relation with immunohistochemical HPV E4 expression patterns and FAM19A4/miR124‐2 methylation analysis in the corresponding cervical scrape. All high‐grade CIN lesions were stratified according to the p16ink4a and Ki‐67 immunoscore, E4 expression and FAM19A4/miR124‐2 methylation status into biomarker profiles in order to assess heterogeneity within high‐grade CIN lesions and to obtain biomarker profiles that could help to personalise treatment of high‐grade CIN lesions based on the presumed cancer progression risk.
2. METHODS
2.1. Study population and sample collection
An international, multicentre, post hoc study was designed within five European prospective referral or HPV‐based screening cohorts to assess biomarker patterns in a large series of high‐risk HPV‐positive high‐grade cervical lesions. CIN2/3 lesions were selected based on local pathology diagnosis and availability of corresponding cervical scrapes. CIN lesions were included in the study population when diagnosis of CIN2/3 was confirmed after revision by an expert pathologist based on H&E staining. Formalin‐fixed paraffin‐embedded (FFPE) cervical biopsies or LLETZ specimen were used to assess p16ink4a, Ki‐67 and E4 expression. Data on HPV status and FAM19A4/miR124‐2 methylation of corresponding cervical scrapes were obtained from the participating institutes. 25 Supplementary Table 1 shows detailed information on included samples from all participating institutes.
2.2. HPV testing and methylation analysis
HPV status and FAM19A4/miR124‐2 methylation analysis were evaluated in cervical scrapes corresponding to the selected tissue blocks. CIN lesions were only included in case the corresponding cervical scrape was high‐risk HPV positive. Participating institutes used clinically validated high‐risk HPV DNA assays to determine HPV status 26 and used the QIAsure Methylation Test (QIAGEN, Hilden, Germany) on the Rotorgene PCR‐platform (QIAGEN, Hilden, Germany) to assess FAM19A4/miR124‐2 methylation status as described previously. 25 , 27 The housekeeping gene β‐actin (ACTB) was used as a reference gene. Methylation levels were calculated as ΔΔCt values by comparing the target Ct values to the Ct values of ACTB relative to that of the calibrator. Methylation status was labelled positive if the QIAsure Methylation Test result exceeded the preset ΔΔCt value threshold for methylation positivity for FAM19A4 and/or miR124‐2 according to manufacturer's instructions.
2.3. Immunohistochemistry
Serial sections of 3 μm were cut from all selected tissue blocks. The “sandwich” technique was used to enable histopathologic review of the H&E stained tissue sections flanking the sections subjected to immunohistochemical staining. In between sections were immunostained with mouse monoclonal antibodies (mAb) against Ki‐67 antigen (Clone MIB‐1, DAKO, Agilent Technologies, Santa Clara, CA), or p16ink4a antigen (Clone E6H4, CINtec, Roche, Basel, Switzerland) by the automated IHC Ventana staining machine (Ventana Medical Systems, Roche, Oro Valley, AZ). The mAb panHPV E4 staining (developed in the laboratory of J. Doorbar, Cambridge, England, available through Labo Bio‐medical Products B.V., Rijswijk, The Netherlands) was performed as described previously. 20 , 28 In case of multiple available FFPE blocks per patient, the tissue block including the worst histology outcome was selected using H&E sections or original histology reports. Lesions with uninterpretable immunohistochemical staining were excluded (n = 3).
2.4. Scoring of Ki‐67, p16 INK4a and E4
Two expert pathologists scored Ki‐67, p16ink4a and E4, independent of HPV and methylation results, as described previously. 17 , 20 In short, nuclear Ki‐67 staining in cells of the squamous epithelium was scored positive. Score 0 was considered a normal staining pattern (ie, scattered staining of nuclei in the basal layers). Scores 1, 2 and 3 were defined as increased nuclear staining predominantly found in the lower one‐third, lower two‐third and more than two‐thirds of the epithelium, respectively. For p16ink4a scoring, diffuse or “block” staining of the cell cytoplasm and/or nucleus in squamous epithelium was considered positive. Score 0 was defined as either no p16ink4a positivity or focally scattered positive cells or small cell clusters (ie, patchy staining). Scores 1, 2 and 3 were defined as diffuse positive staining restricted to the lower one‐third of the epithelium, diffuse positive staining restricted to the lower two‐thirds of the epithelium and diffuse positive staining involving the full thickness of the epithelium, respectively. The cumulative score of Ki‐67 and p16ink4a (ranging from 0‐6) was referred to as the “immunoscore” (IS). 17 IS group 5‐6 (high immunoscore) are considered CIN3‐like lesions, IS group 3‐4 (intermediate immunoscore) are considered CIN2‐like lesions and IS group 0‐2 (low immunoscore) are considered CIN1‐like lesions. Membranous and/or cytoplasmic E4 staining was scored as either negative (Score 0), focally positive (ie, limited staining of some cells restricted to the superficial layer of the epithelium, Score 1) or extensively positive (ie, widespread positive staining in the superficial layers of the epithelium extending to half of the epithelial width, Score 2). 29 Examples of the scoring of E4 staining are shown in Supplementary Figure 2.
2.5. Statistical analysis
Methylation status and HPV E4 expression were associated with CIN grade and immunoscore groups (0‐2, 3‐4 and 5‐6). χ 2 tests and χ 2 tests for trend were conducted to assess E4 expression and methylation positivity with increasing CIN grade and immunoscore. Adjusted for CIN grade or immunoscore group, the association between age and methylation status or HPV E4 expression was examined using a Mantel‐Haenszel analysis. Extensive E4 staining was considered as E4 positive. Differences in DNA methylation levels between groups were assessed using pairwise Mann‐Whitney U‐tests. A P value of .05 was considered as statistically significant with Bonferroni adjustment for multiple testing. All P values are considered two‐sided. All statistical analyses were performed using SPSS (version 26.0, IBM Corp, Armonk, NY).
3. RESULTS
3.1. Study population
The study population consisted of 497 high‐grade CIN lesions: 262 CIN2 (52.7%) and 235 CIN3 (47.3%). Supplementary Figure 1 shows the study flowchart of five European prospective referral or HPV‐based screening cohorts from which the high‐grade CIN lesions were derived. Supplementary Table 1 shows detailed cohort information per participating centre. 30 , 31 , 32 , 33 , 34 Classification of these high‐grade CIN lesions into immunoscore (IS) groups resulted in 30 CIN lesions within IS group 0‐2 (6.0%), 151 CIN lesions within IS group 3‐4 (30.4%) and 316 CIN lesions within IS group 5‐6 (63.6%). Table 1 shows the association between H&E CIN diagnosis and immunoscore groups.
TABLE 1.
Association between H&E CIN diagnosis and immunoscore groups
| Immunoscore group | ||||||||
|---|---|---|---|---|---|---|---|---|
| CIN grade | 0‐2 | 3‐4 | 5‐6 | Total | ||||
| CIN2 | 23 | 8.8% | 105 | 40.1% | 134 | 51.1% | 262 | 52.7% |
| CIN3 | 7 | 3.0% | 46 | 19.6% | 182 | 77.4% | 235 | 47.3% |
| Total | 30 | 6.0% | 151 | 30.4% | 316 | 63.6% | 497 | 100% |
3.2. E4 expression and methylation marker status in CIN grades and immunoscore groups
Positive E4 staining was present in 17.3% (86/497) of all CIN lesions. E4 positivity was significantly higher in CIN2 lesions than CIN3 lesions (Table 2A; P < .001) and decreased significantly with increasing immunoscore group (Table 2B; P trend < .001). Women <29 years showed 26.3% (25/95) positive E4 staining compared with 15.2% (61/402) positive E4 staining in women ≥29 years (P = .010). Adjusted for CIN grade and IS group, E4 positivity was significantly higher in women <29 years than women ≥29 years (P = .020 and P = .004, respectively).
TABLE 2.
E4 expression and methylation status in CIN (A) and immunoscore groups (B)
| (A) CIN grade | E4 score | Methylation status | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Total | |||||
| CIN2 | 199 | 76.0% | 63 | 24.0% | 97 | 37.0% | 165 | 63.0% | 262 |
| CIN3 | 212 | 90.2% | 23 | 9.8% | 49 | 20.9% | 186 | 79.1% | 235 |
| Total | 411 | 82.7% | 86 | 17.3% | 146 | 29.4% | 351 | 70.6% | 497 |
| (B) Immunoscore | E4 score | Methylation status | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Total | |||||
| 0–2 | 21 | 70.0% | 9 | 30.0% | 15 | 50.0% | 15 | 50.0% | 30 |
| 3–4 | 112 | 74.2% | 39 | 25.8% | 63 | 41.7% | 88 | 58.3% | 151 |
| 5–6 | 278 | 88.0% | 38 | 12.0% | 68 | 21.5% | 248 | 78.5% | 316 |
| Total | 411 | 82.7% | 86 | 17.3% | 146 | 29.4% | 351 | 70.6% | 497 |
FAM19A4/miR124‐2 methylation in corresponding cervical scrapes was positive in 70.6% (351/497) (Table 2). Methylation positivity increased significantly with severity of CIN grade (Table 2A; P < .001) and with increasing immunoscore group (Table 2B; P trend < .001). Women <29 years were methylation positive in 57.9% (55/95) compared with 73.6% (296/402) in women ≥29 years (P = .002). Adjusted for CIN grade and IS group, methylation positivity was significantly lower in women <29 years compared with women ≥29 years (P = .005 and P = .001, respectively).
3.3. Methylation levels in E4 positive and negative CIN lesions
Overall, methylation positivity was higher in E4‐negative lesions than E4‐positive lesions (308/411, 74.9% vs 43/86, 50.0%; P < .001). Figure 1 shows methylation levels of FAM19A4 and miR124‐2 in E4‐positive and E4‐negative lesions stratified by CIN grade (A,C) and IS group (B,D). A reference population of 230 squamous cell carcinomas, 35 of which the corresponding cervical scrape was tested for FAM19A4/miR124‐2 methylation, was added to this figure to enable comparison of methylation levels between CIN2/3 lesions and cervical cancer. For every CIN grade and IS group, E4‐negative lesions showed higher methylation levels than E4‐positive lesions, although not statistically significant for some comparisons (Figure 1). E4 expression was present in 9.8% (23/235) of CIN3 and in 12.0% (38/316) of IS group 5‐6. These E4‐positive CIN3 and IS 5‐6 lesions showed methylation positivity in 60.9% (14/23) and 65.8% (25/38) of corresponding scrapes, respectively (Figure 1E). Similar findings were found in CIN2 and IS group 3‐4.
FIGURE 1.
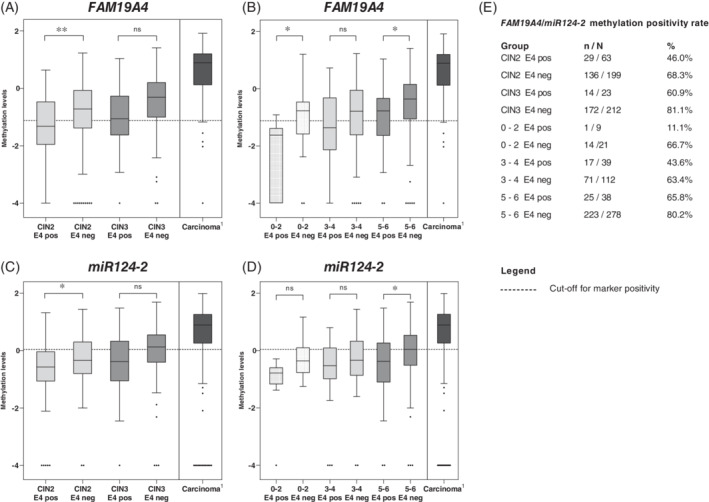
Methylation levels represented by the log10‐transformed ΔΔCt ratios of FAM19A4 stratified by E4 positivity in CIN grade (A) and immunoscore groups (B) and of miR124‐2 stratified by E4 positivity in CIN grade (C) and immunoscore groups (D). FAM19A4/miR124‐2 methylation positivity rate for each subgroup is shown in E. Superscript 1 indicates FAM19A4/miR124‐2 methylation levels of a reference population of 230 squamous cell carcinoma, in the corresponding smear originating from Reference 35. CIN, cervical intraepithelial neoplasia; pos, positive; n, number of methylation positives; N, group size; neg, negative. *P < .05, **P < .01, ***P < .001, ns: not significant
3.4. Productive and transforming characteristics in a small subset of CIN 2/3 lesions
In Table 3, we stratified all high‐grade CIN lesions according to immunoscore, methylation status and E4 positivity. The various combinations of biomarker expression patterns confirm the heterogeneity within high‐grade CIN lesions. These findings also indicate that CIN lesions can exhibit both productive (E4 positivity) and transforming characteristics (high p16ink4a and Ki‐67 scores and methylation positivity) simultaneously. Figure 2 shows examples of CIN2 and CIN3 lesions with combined productive and transforming characteristics.
TABLE 3.
Biomarker expression patterns reflect heterogeneity within high‐grade CIN lesions
| Immunoscore | IS 0–2 | IS 3–4 | IS 5‐6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM status | MM− | MM+ | MM− | MM+ | MM− | MM+ | |||||||
| E4 status | E4+ | E4− | E4+ | E4− | E4+ | E4− | E4+ | E4− | E4+ | E4− | E4+ | E4− | |
| CIN2 | n | 6 | 7 | 1 | 9 | 18 | 26 | 14 | 47 | 10 | 30 | 14 | 80 |
| CIN3 | n | 2 | 0 | 0 | 5 | 4 | 15 | 3 | 24 | 3 | 25 | 11 | 143 |
| <29 y | n | 1 | 0 | 0 | 3 | 8 | 8 | 2 | 5 | 8 | 15 | 6 | 39 |
| ≥29 y | n | 7 | 7 | 1 | 11 | 14 | 33 | 15 | 66 | 5 | 40 | 19 | 184 |
| Total | 8 | 7 | 1 | 14 | 22 | 41 | 17 | 71 | 13 | 55 | 25 | 223 | |
Note: High‐grade lesions stratified by immunoscore group, FAM19A4/miR124‐2 methylation status and E4 status.
FIGURE 2.
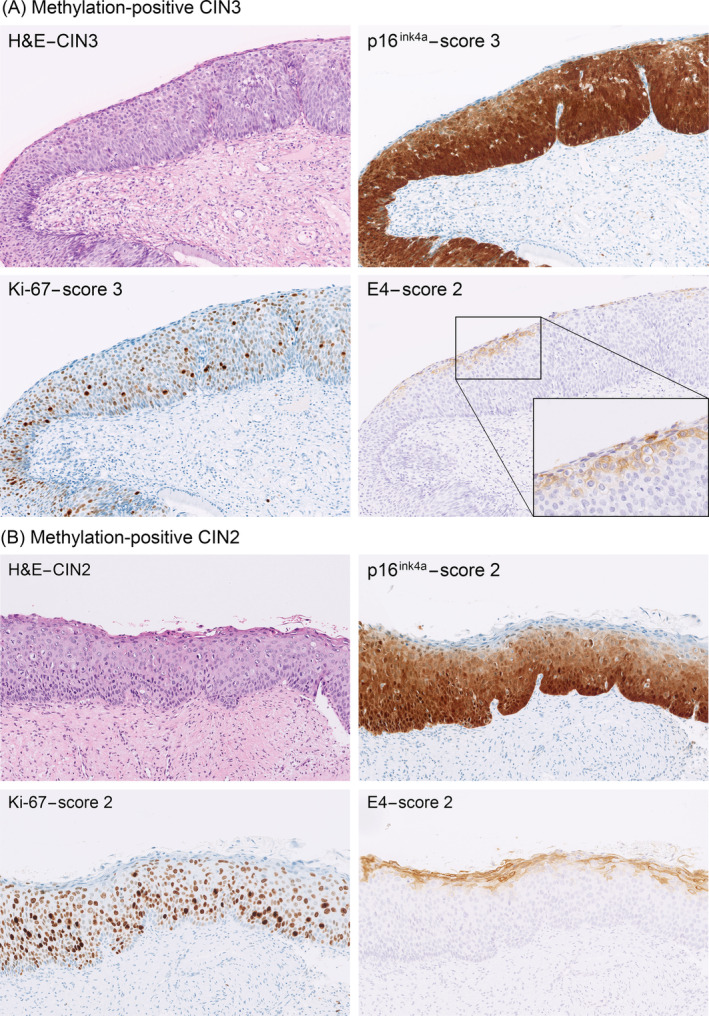
Productive and transforming characteristics can overlap within high‐grade CIN lesions. (A) A methylation‐positive CIN3 lesion with extensive E4 staining (predominantly membranous staining). Corresponding Ki‐67 and p16ink4a stainings show full‐thickness Ki‐67 staining of the epithelium (Score 3) and full‐thickness p16ink4a staining of the epithelium (Score 3). (B) A methylation‐positive CIN2 lesion with extensive E4 staining (both membranous and cytoplasmic staining). Corresponding Ki‐67 staining shows positive nuclei predominantly found in the lower two‐thirds of the epithelium (Score 2) and the p16ink4a staining shows diffuse positivity in the lower two‐thirds of the epithelium (Score 2)
4. DISCUSSION
In the current study, we assessed the cumulative p16ink4a and Ki‐67 immunoscore in a large international series of high‐grade CIN lesions in relation to immunohistochemical HPV E4 expression and FAM19A4/miR124‐2 methylation analysis in the corresponding cervical scrape. We focused on CIN2/3 and not on ≤CIN1, because especially in high‐grade lesions the differences in biomarker results could have major consequences in clinical guidance. A considerable variation in E4 expression as indicator of a productive HPV infection was found in CIN2 and CIN3 lesions, and also in p16ink4a/Ki‐67 expression and FAM19A4/miR124‐2 methylation analysis, both markers for a transforming HPV infection. The p16ink4a and Ki‐67 immunoscore grading system resulted in more CIN3‐like lesions (increase of 34%) characterised by a high immunoscore (IS group 5‐6) and in less CIN2‐like lesions (decrease of 42%) characterised by an intermediate immunoscore (IS group 3‐4). E4 expression decreased from CIN2 to CIN3 and from IS group 3‐4 lesions to IS group 5‐6 lesions while methylation positivity increased with severity of CIN grade and increasing immunoscore group. Stratifying all lesions according to immunoscore, methylation status and E4 expression confirmed the heterogeneity within high‐grade CIN. Besides, in a small subset of lesions, combined productive and transforming characteristics were found.
We used the cumulative three‐tiered immunoscore grading system with biomarkers p16ink4a and Ki‐67 as an alternative grading system for CIN lesions, because it has a higher reproducibility for CIN3‐like lesions (IS group 5‐6) and CIN1‐like lesions (IS group 0‐2) and decreases the diagnosis of CIN2‐like lesions (IS group 3‐4) for which clinical management is not uniform and less defined. 17 In line with previous findings, the immunoscore grading system classified 262 CIN2 and 235 CIN3 into 316 lesions with a high immunoscore (IS group 5‐6), 151 lesions with an intermediate immunoscore (IS group 3‐4) and 30 lesions with a low immunoscore (IS group 0‐2).
Our results showed significant increased methylation positivity with increasing CIN grade and IS group. Methylation positivity was 79.1% (186/235) in CIN3 and 78.5% (248/316) in IS group 5‐6. In addition, we found a decrease in E4 expression with increasing CIN grade and IS group. E4 expression was present in 9.8% (23/235) of CIN3 and in 12.0% (38/316) of IS group 5‐6. In earlier studies concerning E4 expression in CIN2/3 lesions, Griffin et al suggested largely mutually exclusive staining patterns between E4 and p16inka4 as neoplastic severity increases. 36 However, Leeman et al found 3% positive E4 staining in CIN3. 29 We showed that in a small number of CIN3 lesions (9.8%; 23/235) and IS 5‐6 lesions (12.0%; 38/316), E4 positivity was found, which is in agreement with our earlier findings. 20 Moreover, we now extend these findings by showing that methylation was present in 60.9% (14/23) of E4‐positive CIN3 and in 65.8% (25/38) of E4‐positive IS 5‐6 lesions. This indicates that in a minority of CIN3 and IS group 5‐6, characteristics of both a productive and transforming HPV infection can be found and that production of viral particles and induction of cellular transformation is not mutually exclusive and may overlap. Until now, this phenomenon has only been described in HIV‐positive women. 37
Previous studies have shown that CIN lesions in younger women are more likely to regress, probably because these lesions are less advanced due to a shorter duration of the associated HPV infection. 38 Indeed, we found that women <29 years showed more characteristics of productive HPV infections as expressed by a significant higher E4 positivity than women ≥29 years. Women <29 years also showed less methylation positivity than women ≥29 years. Although the numbers are small, in a subanalysis including only women ≤23 years (n = 13), 11 women (84.6%) showed extensive E4 expression and only 4 women (30.8%) were methylation positive. This lower methylation positivity and higher E4 expression in young women support the hypothesis that these lesions are less advanced CIN lesions.
Based on the results of this study, we stratified all CIN2/3 lesions according to the immunoscore and methylation status into biomarker profiles (Table 4). E4 expression was not used in these biomarker profiles. The low rate of E4 expression (17%) in high‐grade CIN lesions, the absence of E4 in cervical carcinomas and in a considerable amount of CIN1 lesions 20 , 29 , 36 and the presence of E4 in a minority of methylation‐positive CIN2/3, makes the potential clinical significance of absence or presence of E4 expression difficult to interpret in terms of regression or progression. Methylation status was included since methylation levels increase with increasing disease severity and are extremely high in cervical carcinomas (Figure 1). 35 , 39 , 40 , 41 Furthermore, when the duration of a HPV infection is taken as a proxy for CIN lesion existence, methylation positivity is higher in CIN2/3 lesions associated with a long duration of the HPV infection (>5 years) in contrast to lesions associated with a short duration of the HPV infection (<5 years). 23 In addition, a negative FAM19A4/miR124‐2 methylation test in HPV‐positive women provided a very low 14‐year cervical cancer risk. 42 In agreement with the latter finding, preliminary results of the CONCERVE study 22 show that women with CIN2/3 with a negative FAM19A4/miR124‐2 methylation test have a higher regression rate than women with a positive methylation test (Kremer, Dick, Meijer et al., unpublished data, manuscript in preparation). Therefore, it is assumed that methylation‐positive CIN2/3 lesions have a higher short‐term progression risk to cervical cancer than methylation‐negative CIN2/3 lesions.
TABLE 4.
High‐grade CIN lesions ranked according to immunoscore and FAM19A4/miR124‐2 methylation status with a potential management proposal based on the presumed progression risk to cervical cancer
| Immunoscore group | IS 0‐2 | IS 3‐4 | IS 5‐6 | ||||
|---|---|---|---|---|---|---|---|
| MM status | MM− | MM+ | MM− | MM+ | MM− | MM+ | |
| CIN2 | n | 13 | 10 | 44 | 61 | 40 | 94 |
| CIN3 | n | 2 | 5 | 19 | 27 | 28 | 154 |
| <29 y | n | 1 | 3 | 16 | 7 | 23 | 45 |
| ≥29 y | n | 14 | 12 | 47 | 81 | 45 | 203 |
| Total | 15 | 15 | 63 | 88 | 68 | 248 | |
Note: Yellow colour indicates low progression risk to cancer, follow‐up after 12 months. Orange colour indicates intermediate progression risk to cancer, follow‐up after 6 months. Red colour indicates high progression risk to cancer, LLETZ advised.
Abbreviations: CIN, cervical intraepithelial neoplasia; IS, immunoscore; MM, methylation marker; MM−, methylation marker negative; MM+, methylation marker positive; n, number of lesions.
Table 4 shows all CIN2/3 lesions stratified according to immunoscore and methylation status into three groups. The depicted colour codes within these biomarker profiles correspond to the presumed short‐term progression risk to cervical cancer based on the results of DNA methylation analysis and the immunoscore. Lesions with a presumed high short‐term progression risk to cervical cancer are depicted in red and LLETZ is advised. Lesions with a presumed low short‐term progression risk to cervical cancer are depicted in yellow and may have a “wait and see” follow‐up of 12 months, whereas the orange colour depicts the women with a presumed intermediate risk. In this proposal, no difference has been made between methylation‐positive IS group 3‐4 and methylation‐positive IS group 5‐6. Increase of p16ink4a block stain expression has been associated with severity of CIN grade, 43 but p16ink4a expression does not predict clinical behaviour or prognosis of CIN lesions. 44 , 45 , 46 For methylation‐negative women in IS group 3‐4 and IS group 5‐6, we suggest a follow‐up time of 6 months. Especially for women in child‐bearing age, the proposed management scheme provides the physician more objective arguments for a “wait and see” strategy for some high‐grade CIN lesions, thereby preventing cervical morbidity. This management proposal might be used as a clinical guidance for the physician in the treatment of high‐grade CIN. Our study shows that from the 497 high‐grade lesions, 15 lesions have an assumed low risk and 146 lesions have an intermediate risk. This proposal will result in the immediate treatment in 68% and in a “wait and see” policy in 32% of the high‐grade CIN lesions. Since clinicians do not prefer to postpone treatment of CIN3 unless in case of pregnancy or fertility treatment, the biggest advantage may be expected in the treatment of CIN2 lesions. Table 4 shows that from the 262 CIN2 lesions, 13 lesions have a supposed low risk and 94 lesions are assumed to have an intermediate risk, which supports postponement of treatment. Of course, the presumed biological behaviour of CIN2/3 lesions with the investigated biomarker profiles needs to be further evaluated in prospective studies.
The strength of the current study is the large sample size of high‐grade lesions derived from five European HPV‐based screening or referral cohorts including both women <29 and ≥29 years. A limitation is that due to the initial selection of CIN2/3 lesions only a small number of the CIN2/3 lesions were classified as IS 0‐2. Therefore, in this study, the results of these CIN2/3 lesions within IS group 0‐2 should be interpreted with care. Another limitation is that biopsy sampling error cannot be completely excluded. However, histological specimens in our study were collected by experienced gynaecologists, who routinely follow and treat women suspected for high‐grade CIN lesions. Due to the large number of samples and the initial selection of high‐grade CIN lesions, we consider this potential bias to have a very limited effect on our results.
In conclusion, we found a considerable amount of heterogeneity in biomarker expression in a large series of high‐grade CIN lesions evaluated with p16ink4a, Ki‐67 and E4 immunohistochemical staining and FAM19A/miR124‐2 methylation. We defined biomarker profiles that might help the clinician in a more personally tailored management of women with high‐grade CIN based on the presumed short‐term cancer progression risk thereby preventing overtreatment, especially in young women.
CONFLICT OF INTERESTS
CJLMM, DAMH and RDMS are minority shareholders of Self‐screen B.V., a spin‐off company of VUmc; Self‐screen B.V. develops, manufactures and licences the high‐risk HPV assay and methylation marker assays for cervical cancer screening and holds patents on these tests. Self‐screen B.V. was supported by the Valid‐screen project, funded by the SME Instrument in the Horizon 2020 Work Program of the European Commission (Valid‐screen 666 800). CJLMM is part‐time CEO of Self‐screen B.V. and has a very small number of shares of MDXHealth and previously of QIAGEN; has received speakers fees from GSK, QIAGEN and SPMSD/Merck; and served occasionally on the scientific advisory boards (expert meeting) of these companies. DAMH has been on the speakers bureau of QIAGEN, serves occasionally on the scientific advisory boards of Pfizer and Bristol‐Myers Squibb. AOV has received reimbursement of travel expenses for attending conferences and honoraria for speaking from Abbott Molecular, QIAGEN and Seegene. JHB institution has received research funding or consumables at reduced price or for free to support research from BD Diagnostics, Agena Bioscience, Genomica SAU, LifeRiver Biotech and QIAGEN. He has received honoraria for lectures from BD Diagnostics and Hologic Ltd. JHB is appointed member of the National Danish Cervical Screening Committee by the Danish Health Authority and member of the cervical screening steering committee of the Capital Region of Denmark. AF is employed by Self‐screen B.V. PH has received speaker's fees from Roche and MSD. FJV, SD, LMAS, BLW, DNTP Group, MP, NET, JB and MCGB have no conflicts of interest.
ETHICS STATEMENT
The work in this study with human‐derived material was conducted under national and international rules and legislation, as well as European standards of research ethics, as it is expressed in the applicable legislation/regulations (The Declaration of Helsinki; informed consent for participation of human subjects in medical and scientific research) and guidelines for Good Clinical Practice. The studies were approved by the local ethics committees where applicable. All subcontractors involved in the VALID‐SCREEN project have signed a written agreement in which the obligation for obtaining an informed consent form from the patient is laid down in accordance with the applicable national and European laws and regulations.
Supporting information
Supplementary Figure 1 Study flowchart. Abbreviations: CIN: cervical intraepithelial neoplasia, IHC: immunohistochemistry, FFPE: formalin‐fixed paraffin embedded, H&E; haematoxylin‐eosin, IS: immunoscore.
Supplementary Figure 2. Scoring of HPV E4 immunostaining. E4 staining is scored as negative staining (Score 0) in (A), focally positive staining (limited staining of some cells restricted to the superficial layer of the epithelium, Score 1) in (B), extensive widespread membranous and/or cytoplasmic staining in the superficial layers of the epithelium extending to half of the epithelial width, Score 2) in a methylation‐negative lesion in (C) and extensive mainly membranous staining in the superficial epithelial layers of a methylation‐positive lesion in (D).
Supplementary Table 1. Cohort information by participating centre * FFPE material from the POBASCAM and VUSA‐Screen trials was localised and retrieved from the local histopathological laboratories through the nationwide network and registry of histo‐ and cytopathology in the Netherlands (DNTP group).30 # Extraction was either done automatically with the Biorobot EZ1 (QIAGEN) or manually using the QIAamp DNA mini kit.
Abbreviations: PBS: Phosphate‐Buffered Saline, UCM: Universal Collection Medium, HC2: Hybrid Capture 2
ACKNOWLEDGEMENTS
This work is dedicated to our colleagues Prof. Dr P. J. F. Snijders, who passed away on 27 May 2018, and Prof. Dr. K. U. Petry, who passed away on 21 April 2020, and were both active investigators in this study. FFPE material from the POBASCAM and VUSA‐screen trials was localised and retrieved from the local histopathological laboratories through the nationwide network and registry of histo‐ and cytopathology in the Netherlands (DNTP group). 30 We would like to thank Dr. Snježana Frković Grazio from Slovenia for providing FFPE samples.
This project was funded by the EU Horizon 2020 program (project ID 666800), the CoheaHr research consortium (EC FP7 Health 2013 Innovation 1 CoheaHr), RISCC Network (grant number 847845) and the Dutch Cancer Society (grant number KWF VU 2014‐7238).
Vink FJ, Dick S, Heideman DAM, et al. Classification of high‐grade cervical intraepithelial neoplasia by p16ink4a, Ki‐67, HPV E4 and FAM19A4/miR124‐2 methylation status demonstrates considerable heterogeneity with potential consequences for management. Int. J. Cancer. 2021;149:707–716. 10.1002/ijc.33566
Frederique J. Vink and Stèfanie Dick contributed equally to this work.
Funding information EU Horizon 2020, Grant/Award Number: Project ID 666800; KWF Kankerbestrijding, Grant/Award Number: KWF VU 2014‐7238; RISCC Network, Grant/Award Number: Grant number 847845; the CoheaHr reserach consortium, Grant/Award Number: EC FP7 Health 2013 Innovation 1 CoheaHr
DATA AVAILABILITY STATEMENT
The data that support the findings of our study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Arbyn M, Anttila A, Jordan J, et al. European Guidelines for Quality Assurance in Cervical Cancer Screening. Second edition—summary document. Ann Oncol. 2010;21:448‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stoler MH, Schiffman M. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS‐LSIL Triage Study. JAMA. 2001;285:1500‐1505. [DOI] [PubMed] [Google Scholar]
- 3. Castle PE, Stoler MH, Solomon D, Schiffman M. The relationship of community biopsy‐diagnosed cervical intraepithelial neoplasia grade 2 to the quality control pathology‐reviewed diagnoses: an ALTS report. Am J Clin Pathol. 2007;127:805‐815. [DOI] [PubMed] [Google Scholar]
- 4. Stoler MH, Ronnett BM, Joste NE, Hunt WC, Cuzick J, Wheeler CM. The interpretive variability of cervical biopsies and its relationship to HPV status. Am J Surg Pathol. 2015;39:729‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Massad LS, Collins YC. Strength of correlations between colposcopic impression and biopsy histology. Gynecol Oncol. 2003;89:424‐428. [DOI] [PubMed] [Google Scholar]
- 6. Etherington IJ, Luesley DM, Shafi MI, Dunn J, Hiller L, Jordan JA. Observer variability among colposcopists from the West Midlands region. Br J Obstet Gynaecol. 1997;104:1380‐1384. [DOI] [PubMed] [Google Scholar]
- 7. Tainio K, Athanasiou A, Tikkinen KAO, et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: systematic review and meta‐analysis. BMJ. 2018;360:k499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ostör AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993;12:186‐192. [PubMed] [Google Scholar]
- 9. Castle PE, Schiffman M, Wheeler CM, Solomon D. Evidence for frequent regression of cervical intraepithelial neoplasia‐grade 2. Obstet Gynecol. 2009;113:18‐25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kyrgiou M, Athanasiou A, Kalliala IEJ, et al. Obstetric outcomes after conservative treatment for cervical intraepithelial lesions and early invasive disease. Cochrane Database Syst Rev. 2017;11:Cd012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV‐induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395‐405. [DOI] [PubMed] [Google Scholar]
- 12. von Knebel Doeberitz M, Reuschenbach M, Schmidt D, Bergeron C. Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev Proteomics. 2012;9:149‐163. [DOI] [PubMed] [Google Scholar]
- 13. Darragh TM, Colgan TJ, Cox JT, et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV‐associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis. 2012;16:205‐242. [DOI] [PubMed] [Google Scholar]
- 14. Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23:315‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jenkins D, Bosch F. Human Papillomavirus: Proving and Using a Viral Cause for Cancer. 1st ed. Amsterdam, The Netherlands: Elsevier; 2020. [Google Scholar]
- 16. Shiraz A, Crawford R, Egawa N, Griffin H, Doorbar J. The early detection of cervical cancer. The current and changing landscape of cervical disease detection. Cytopathology. 2020;31:258‐270. [DOI] [PubMed] [Google Scholar]
- 17. van Zummeren M, Leeman A, Kremer WW, et al. Three‐tiered score for Ki‐67 and p16(ink4a) improves accuracy and reproducibility of grading CIN lesions. J Clin Pathol. 2018;71(11):981‐988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doorbar J. The E4 protein; structure, function and patterns of expression. Virology. 2013;445:80‐98. [DOI] [PubMed] [Google Scholar]
- 19. Griffin H, Wu Z, Marnane R, et al. E4 antibodies facilitate detection and type‐assignment of active HPV infection in cervical disease. PLoS One. 2012;7:e49974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zummeren MV, Kremer WW, Leeman A, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124‐2 genes in cervical cancer and precursor lesions. Mod Pathol. 2018;31:1842‐1850. [DOI] [PubMed] [Google Scholar]
- 21. Louvanto K, Aro K, Nedjai B, et al. Methylation in predicting progression of untreated high‐grade cervical intraepithelial neoplasia. Clin Infect Dis. 2019;70(12):2582‐2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kremer WW, Berkhof J, Bleeker MC, et al. Role of FAM19A4/miR124‐2 methylation analysis in predicting regression or non‐regression of CIN2/3 lesions: a protocol of an observational longitudinal cohort study. BMJ Open. 2019;9:e029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila). 2014;7:1251‐1257. [DOI] [PubMed] [Google Scholar]
- 24. Verlaat W, Snijders PJF, Novianti PW, et al. Genome‐wide DNA methylation profiling reveals methylation markers associated with 3q gain for detection of cervical precancer and cancer. Clin Cancer Res. 2017;23:3813‐3822. [DOI] [PubMed] [Google Scholar]
- 25. Bonde J, Floore A, Ejegod D, et al. Methylation markers FAM19A4 and miR124‐2 as triage strategy for primary HPV screen positive women; a large European multi‐center study. Int J Cancer. 2020;148(2):396‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arbyn M, Snijders PJ, Meijer CJ, et al. Which high‐risk HPV assays fulfil criteria for use in primary cervical cancer screening? Clin Microbiol Infect. 2015;21:817‐826. [DOI] [PubMed] [Google Scholar]
- 27. Floore A, Hesselink A, Ostrbenk A, et al. Intra‐ and inter‐laboratory agreement of the FAM19A4/mir124‐2 methylation test: results from an international study. J Clin Lab Anal. 2019;33:e22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Baars R, Griffin H, Wu Z, et al. Investigating diagnostic problems of CIN1 and CIN2 associated with high‐risk HPV by combining the novel molecular biomarker PanHPVE4 with P16INK4a. Am J Surg Pathol. 2015;39:1518‐1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leeman A, Jenkins D, Del Pino M, et al. Expression of p16 and HPV E4 on biopsy samples and methylation of FAM19A4 and miR124‐2 on cervical cytology samples in the classification of cervical squamous intraepithelial lesions. Cancer Med. 2020;9:2454‐2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Casparie M, Tiebosch AT, Burger G, et al. Pathology databanking and biobanking in The Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell Oncol. 2007;29:19‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rijkaart DC, Berkhof J, Rozendaal L, et al. Human papillomavirus testing for the detection of high‐grade cervical intraepithelial neoplasia and cancer: final results of the POBASCAM randomised controlled trial. Lancet Oncol. 2012;13:78‐88. [DOI] [PubMed] [Google Scholar]
- 32. Rijkaart DC, Berkhof J, van Kemenade FJ, et al. Meijer CJ. HPV DNA testing in population‐based cervical screening (VUSA‐screen study): results and implications. Br J Cancer. 2012;106:975‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poljak M, Ostrbenk A, Seme K, et al. Comparison of clinical and analytical performance of the Abbott Realtime High Risk HPV test to the performance of hybrid capture 2 in population‐based cervical cancer screening. J Clin Microbiol. 2011;49:1721‐1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luyten A, Buttmann‐Schweiger N, Luyten K, et al. Early detection of CIN3 and cervical cancer during long‐term follow‐up using HPV/Pap smear co‐testing and risk‐adapted follow‐up in a locally organised screening programme. Int J Cancer. 2014;135:1408‐1416. [DOI] [PubMed] [Google Scholar]
- 35. Vink FJ, Meijer C, Clifford GM, et al. FAM19A4/miR124‐2 methylation in invasive cervical cancer: a retrospective cross‐sectional worldwide study. Int J Cancer. 2020;147:1215‐1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Griffin H, Soneji Y, Van Baars R, et al. Stratification of HPV‐induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod Pathol. 2015;28:977‐993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kremer WW, Vink FJ, van Zummeren M, et al. Characterization of cervical biopsies of women with HIV and HPV co‐infection using p16(ink4a), ki‐67 and HPV E4 immunohistochemistry and DNA methylation. Mod Pathol. 2020;33(10):1968‐1978. [DOI] [PubMed] [Google Scholar]
- 38. Bekos C, Schwameis R, Heinze G, et al. Influence of age on histologic outcome of cervical intraepithelial neoplasia during observational management: results from large cohort, systematic review, meta‐analysis. Sci Rep. 2018;8:6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Verlaat W, Van Leeuwen RW, Novianti PW, et al. Host‐cell DNA methylation patterns during high‐risk HPV‐induced carcinogenesis reveal a heterogeneous nature of cervical pre‐cancer. Epigenetics. 2018;13:769‐778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dick S, Verhoef L, De Strooper LM, et al. Evaluation of six methylation markers derived from genome‐wide screens for detection of cervical precancer and cancer. Epigenomics. 2020;12:1569‐1578. [DOI] [PubMed] [Google Scholar]
- 41. Kelly H, Benavente Y, Pavon MA, De Sanjose S, Mayaud P, Lorincz AT. Performance of DNA methylation assays for detection of high‐grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta‐analysis. Br J Cancer. 2019;121:954‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Strooper LMA, Berkhof J, Steenbergen RDM, et al. Cervical cancer risk in HPV‐positive women after a negative FAM19A4/mir124‐2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow‐up. Int J Cancer. 2018;143:1541‐1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva DC, Gonçalves AK, Cobucci RN, Mendonça RC, Lima PH, Cavalcanti GJ. Immunohistochemical expression of p16, Ki‐67 and p53 in cervical lesions—a systematic review. Pathol Res Pract. 2017;213:723‐729. [DOI] [PubMed] [Google Scholar]
- 44. Sagasta A, Castillo P, Saco A, et al. p16 staining has limited value in predicting the outcome of histological low‐grade squamous intraepithelial lesions of the cervix. Mod Pathol. 2016;29:51‐59. [DOI] [PubMed] [Google Scholar]
- 45. Miralpeix E, Genovés J, Maria Solé‐Sedeño J, et al. Usefulness of p16(INK4a) staining for managing histological high‐grade squamous intraepithelial cervical lesions. Mod Pathol. 2017;30:304‐310. [DOI] [PubMed] [Google Scholar]
- 46. Guedes AC, Brenna SM, Coelho SA, Martinez EZ, Syrjänen KJ, Zeferino LC. p16(INK4a) expression does not predict the outcome of cervical intraepithelial neoplasia grade 2. Int J Gynecol Cancer. 2007;17:1099‐1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Study flowchart. Abbreviations: CIN: cervical intraepithelial neoplasia, IHC: immunohistochemistry, FFPE: formalin‐fixed paraffin embedded, H&E; haematoxylin‐eosin, IS: immunoscore.
Supplementary Figure 2. Scoring of HPV E4 immunostaining. E4 staining is scored as negative staining (Score 0) in (A), focally positive staining (limited staining of some cells restricted to the superficial layer of the epithelium, Score 1) in (B), extensive widespread membranous and/or cytoplasmic staining in the superficial layers of the epithelium extending to half of the epithelial width, Score 2) in a methylation‐negative lesion in (C) and extensive mainly membranous staining in the superficial epithelial layers of a methylation‐positive lesion in (D).
Supplementary Table 1. Cohort information by participating centre * FFPE material from the POBASCAM and VUSA‐Screen trials was localised and retrieved from the local histopathological laboratories through the nationwide network and registry of histo‐ and cytopathology in the Netherlands (DNTP group).30 # Extraction was either done automatically with the Biorobot EZ1 (QIAGEN) or manually using the QIAamp DNA mini kit.
Abbreviations: PBS: Phosphate‐Buffered Saline, UCM: Universal Collection Medium, HC2: Hybrid Capture 2
Data Availability Statement
The data that support the findings of our study are available from the corresponding author upon reasonable request.


