Summary
Eukaryotic genomes are highly folded for packing into higher‐order chromatin structures in the nucleus. With the emergence of state‐of‐the‐art chromosome conformation capture methods and microscopic imaging techniques, the spatial organization of chromatin and its functional implications have been interrogated. Our knowledge of 3D chromatin organization in plants has improved dramatically in the past few years, building on the early advances in animal systems. Here, we review recent advances in 3D genome mapping approaches, our understanding of the sophisticated organization of spatial structures, and the application of 3D genomic principles in plants. We also discuss directions for future developments in 3D genomics in plants.
Keywords: 3D genomics, application, chromatin organization, plants, transcriptional regulation
Introduction
Eukaryotic genomes are not randomly positioned in the nucleus but are packed into higher‐order chromatin structures that have important functional implications. The spatial organization of chromatin structures allows regulatory elements that are far from the target genes in the 1D genome sequence to approach the target genes, to facilitate their transcriptional regulation (Lanctot et al., 2007). Therefore, in order to explore the functional roles of these regulatory elements, the spatial organization of chromatin in 3D has to be understood. The concept of 3D genomics encompasses chromatin interactions and 3D spatial structures of the genome in the nucleus and its effects on gene transcription, DNA replication and repair, and other biological functions in combination with linear genome sequence information (Dekker et al., 2013).
In the past decade, our understanding of the spatial organization of chromatin in the nucleus has advanced in both animal and plant systems, especially for model species such as Drosophila (Byrd & Corces, 2003; Sexton et al., 2012; Bonev & Cavalli, 2016). Studies in animals showed that the chromatin exhibits a hierarchical structure. Nucleosomes interact to form chromatin fibers, and then these chromatin fibers form chromatin loops due to physical interactions between cis‐elements (Byrd & Corces, 2003; Amano et al., 2009). Chromatin loops are stabilized to form topological associating domains (TADs) under the action of structural proteins, such as CCCTC‐binding factor (CTCF) and cohesins, and of regulatory components, such as transcription factors and heterochromatin proteins (Wang et al., 2018b). TADs in similar epigenetic landscapes interact to form chromatin compartments, and all chromatin compartments on the same chromosome merge to form chromosome territories (CTs) (Lieberman‐Aiden et al., 2009; Rao et al., 2014). Studies in plants have lagged behind those in mammals, for a number of reasons (Heger et al., 2012; Smith & Grima, 2018). Early studies on 3D genome organization used mammalian cell lines, which are relatively homogeneous, and a link between cell‐type identity and higher‐order chromatin structures could be determined. However, it is not possible to generate stable differentiated plant cell lines, so plant tissues that show high cell heterogeneity hinder the discovery of well‐defined chromatin structures, if using the same experimental approaches and data processing methods as those for animals (Smith & Grima, 2018). In addition, the major well‐known structural protein, the insulator protein CTCF that is required for TAD and loop formation and mediates the formation of higher‐order chromatin, is not found in plants (Heger et al., 2012), which makes it challenging to investigate the mechanism of 3D genome folding.
Despite this, application of 3D genomics is helpful to map the spatial chromatin conformation in plants and investigate the possible regulatory role in gene transcription (Gonzalez‐Sandoval & Gasser, 2016). This review summarizes recent advances in 3D genome mapping methods and focuses mainly on the understanding of plant 3D genome organization, in comparison with that in animals. The applications of 3D genomic principles in plants are reviewed, including in transcriptional regulation, prediction of noncoding regulatory variation, evolutionary biology and single cell 3D genomics. We also provide directions for promising future applications, such as uncovering the mechanism of 3D genomic folding, spatial haplotype chromatin interactions and dynamic 3D genome structure during plant development.
The 3D genomics techniques and application in plants
There are two main kinds of technical approaches for 3D genomic studies. The first approach is cytological evaluation and microscopy, the principle of which is to label DNA or chromatin and then use the microscope to observe the spatial organization of chromatin (Probst, 2018) (Fig. 1). The labeling techniques are divided into four categories: staining with chemical dyes (Ou et al., 2017; Poulet et al., 2017), 3D‐fluorescence in situ hybridization (3D‐FISH) (Koornneef et al., 2003; Berr & Schubert, 2007; Schubert et al., 2012), immunostaining (Fransz et al., 2002; She et al., 2013) and fluorescent protein‐tagging (FP‐tagging) (Matzke et al., 2005; Lindhout et al., 2007; Deng et al., 2015; Fujimoto et al., 2016; Ren et al., 2017; Nagaki & Yamaji, 2020). However, the main disadvantage here is that the resolution is not sufficiently high to uncover the interactions between DNA regulatory elements. In the past, widefield microscopy (WF), confocal microscopy (CLSM) and light sheet fluorescence microscopy (LSFM) combined labeling techniques could only resolve structures > 200 nm, such as CTs and nuclear bodies in the nucleus (Schwarzacher et al., 1992; Aragon‐Alcaide et al.,1997; Bass et al., 2000; Baroux & Schubert, 2018). In recent years, new advances in labeling techniques include custom oligonucleotide arrays (such as Oligopaint; Beliveau et al., 2012) and modified clustered regularly interspaced short palindromic repeats/CRISPR‐associated caspase 9 (CRISPR/Cas9) systems (Dreissig et al., 2017; Hong et al., 2018), and microscopic techniques including super resolution microscopy (SRM) (Probst, 2018) (such as stochastic optical reconstruction microscopy (STORM) (Rust et al., 2006) and photoactivated localization microscopy (PALM) (Betzig et al., 2006)) and electron microscopy imaging (ELMI) (Lobastov et al., 2005). The resolution of the SRM can reach 20 nm, and the resolution of the ELMI is even higher (Baroux & Schubert, 2018). These techniques, which reach nm‐scale resolution, have enabled researchers to visualize directly the fine structure of the genome including the structure of chromatin fiber and the dynamics of individual chromosome domains (Wang et al., 2016a, 2016b,2016a, 2016b; Ou et al., 2017; Szabo et al., 2018). For example, electron microscopy tomography (EMT) with labeling (ChromEMT) can display the ultrastructure of chromosomes in situ by using the new DNA fluorescence labeling dye DRAQ5. Using the ChromEMT technique, the researchers found that nucleosome beads of chromatin do not need to form discrete fibers of increasing diameter (e.g. 30, 120 or 320 nm) to adapt to the nucleus. Instead, it forms a semi‐flexible chain with continuous changes in length between 5 nm and 24 nm and achieves different compaction levels by bending and contracting (Ou et al., 2017).
Fig. 1.
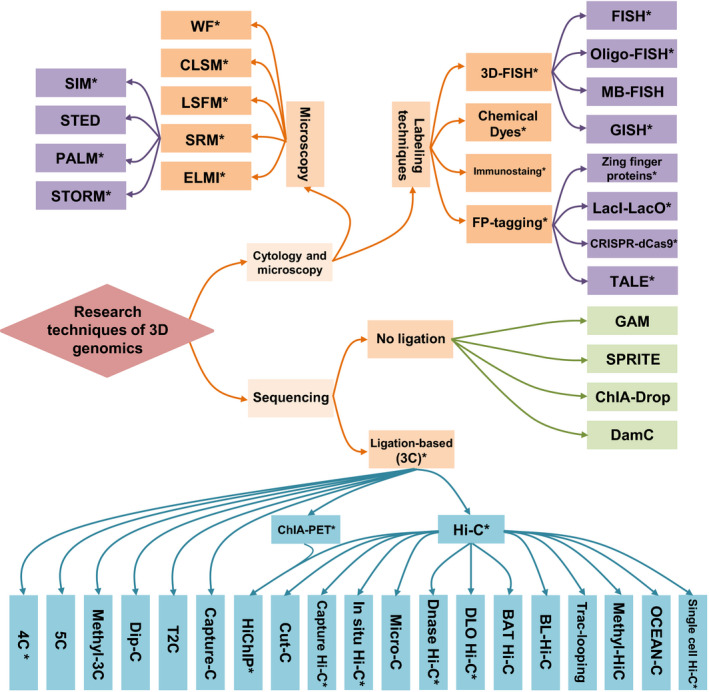
Research techniques of 3D genomics. There are mainly two kinds of technical approaches for 3D genome study: one is based on cytological evaluation and microscopy imaging; the other is based on sequencing, including techniques which require proximity ligation (chromosome conformation capture, 3C) or do not require ligation. WF, wide field microscopy (Wheeler & Tyler, 2011); CLSM, confocal microscopy (Carlsson et al., 1989); LSFM, light sheet fluorescence microscopy (Santi, 2011); SRM, super resolution microscopy (Schubert, 2017); ELMI, electron microscopy imaging (Lobastov et al., 2005); SIM, structured illumination microscopy (Fitzgibbon et al., 2010); STED, stimulated emission depletion microscopy (Dyba et al., 2003); PALM, photoactivated localization microscopy (Rust et al., 2006); STORM, stochastic optical reconstruction microscopy (Betzig et al., 2006); FISH, Fluorescence in situ hybridization (Koornneef et al., 2003); Oligo‐FISH, FISH using oligonucleotides probes (Beliveau et al., 2012); MB‐FISH, FISH using molecular beacon (Wu et al., 2010); GISH, genomic in situ hybridization (Schubert et al., 2001); FP‐tagging, fluorescent protein‐tagging (Matzke et al., 2005; Lindhout et al., 2007; Deng et al., 2015; Fujimoto et al., 2016; Ren et al., 2017; Nagaki & Yamaji, 2020); CRISPR‐dCas9, clustered regularly interspaced short palindromic repeats‐nuclease‐deficient Cas9 (Dreissig et al., 2017); TALE, transcription activator‐like effectors with a quantum dot labelling technique (Ma et al., 2017); GAM, genome architecture mapping that combines microcutting and sequencing technologies (Beagrie et al., 2017); SPRITE, split‐pool recognition of interactions by tag extension (Quinodoz et al., 2018); ChIA‐Drop, multiplex chromatin‐interaction analysis via droplet‐based and barcode‐linked sequencing (Zheng et al., 2019); DamC, methylation‐based detection of chromosomal contacts (Redolfi et al., 2019); ChIA‐PET, chromatin interaction analysis by paired‐end tag sequencing (Fullwood et al., 2009); Hi‐C, high‐throughput chromosome conformation capture (Lieberman‐Aiden et al., 2009); 4C, chromosome conformation capture‐on‐chip (Simonis et al., 2006); 5C, chromosome conformation capture carbon copy (Dostie et al., 2006); Methyl‐3C, a method that combines 3C and DNA methylation detection technology (Lee et al., 2019a); Dip‐C, a method that combines single cell 3C method with a transposon‐based whole‐genome amplification method (Tan et al., 2018); T2C, Targeted Chromatin Capture that studies chromatin organization for specific genomic regions (Kolovos et al., 2014); Capture‐C, a method that combines 3C with oligonucleotide capture technology (Hughes et al., 2014); HiChIP, a protein‐centric chromatin conformation method that combines Hi‐C with ChIA‐PET technology (Mumbach et al., 2016); Cut‐C, a method that combines antibody‐mediated cleavage by tethered nuclease with chromosome conformation capture (Shimbo et al., 2019); Capture Hi‐C, a method that combines Hi‐C with the hybridization‐based capture of targeted genomic regions (Mifsud et al., 2015); In situ Hi‐C, a method in which DNA–DNA proximity ligation is performed in intact nuclei (Rao et al., 2014); Micro‐C, a method in which chromatin is fragmented into mononucleosomes using micrococcal nuclease (Hsieh et al., 2015); Dnase Hi‐C, a method in which chromatin is fragmented by DNase I (Ma et al., 2015); DLO Hi‐C, digestion‐ligation‐only Hi‐C technology that requires two rounds of digestion and ligation, without the need for biotin labeling and pulldown (Lin et al., 2018); BAT Hi‐C, bridge linker‐Alul‐Tn5 Hi‐C method that combines Alul fragmentation with biotinylated linker‐mediated proximity ligation (Huang et al., 2020); BL‐Hi‐C, bridge linker‐Hi‐C that requires restriction enzyme targeting and two‐step proximity ligation (Liang et al., 2017); Trac‐looping, transposase‐mediated analysis of chromatin looping (Lai et al., 2018); Methyl‐HiC, a method that combines Hi‐C and DNA methylation detection technology (Li et al., 2019b); OCEAN‐C, open chromatin enrichment and network Hi‐C (Li et al., 2018); and Single cell Hi‐C, a method to perform Hi‐C in an individual nucleus (Nagano et al., 2013). Asterisks show the methods that have been applied to plants.
The other kind of technique is based on chromosome conformation capture (3C) (Dekker et al., 2002) and its derived techniques such as chromosome conformation capture‐on‐chip (4C) (Simonis et al., 2006) and chromosome conformation capture carbon copy (5C) (Dostie et al., 2006) (Fig. 1). 3C‐based methods rely on enzymatic digestion of DNA and proximal ligation. 3C can capture long‐range chromatin interaction between two specific genomic loci (Dekker et al., 2002). 4C can be used to study the interaction between a chromatin site of interest and other sites on the whole genome (Simonis et al., 2006). 5C can be used for chromatin interaction analysis between multiple genomic loci (Dostie et al., 2006). Thanks to the rapid development of high‐throughput sequencing techniques, high‐throughput chromatin conformation capture (Hi‐C) and chromatin interaction analysis based on paired‐end tag sequencing (ChIA‐PET) techniques were developed in the study of 3D genomics (Fullwood et al., 2009; Lieberman‐Aiden et al., 2009). In Hi‐C, the DNA–protein complex is crosslinked by formaldehyde, then the chromatin is fragmented by restriction endonuclease treatment and the restriction ends are filled with biotin‐labeled nucleotides. DNA fragments that are adjacent in 3D space but far away in 1D linear distance are ligated together, the sample is ultrasonically fragmented, and finally the ligated fragments containing biotin are precipitated for sequencing (Lieberman‐Aiden et al., 2009). In ChIA‐PET, the DNA–protein complex is crosslinked as in Hi‐C, then fragmented by ultrasonic treatment and captured by the target protein‐specific antibody. The captured chromatin is incorporated with a biotin‐labeled oligonucleotide linker containing an MmeI site, and adjacent linkers are connected to each other; and MmeI is used to digest the sample to obtain DNA fragments that form paired‐end tags (PETs); these PETs are sequenced (Fullwood et al., 2009). Theoretically, Hi‐C can identify the spatial proximity of any DNA fragments at the whole genome level to resolve the 3D spatial structure of the genome (Dixon et al., 2012). The ChIA‐PET approach is capable of capturing all distal interactions between DNA fragments involving a specific protein across the whole genome (Fullwood et al., 2009). Meanwhile, based on different scientific research needs, Hi‐C and ChIA‐PET have been further improved, with the emergence of methods including single‐cell Hi‐C (Nagano et al., 2013), in situ Hi‐C (a method in which DNA–DNA proximity ligation is performed in intact nuclei; Rao et al., 2014), Dnase Hi‐C (a method in which chromatin is fragmented by DNase I; Ma et al., 2015), in situ Hi‐C followed by chromatin immunoprecipitation (HiChIP) (Mumbach et al., 2016) and methyl‐3C/Hi‐C (a method that combines Hi‐C and DNA methylation detection technology; (Lee et al., 2019a; Li et al., 2019b), amongst others.
In recent years, several new 3D genomic research techniques that do not require proximal ligation have been developed, such as genome architecture mapping (GAM) (Beagrie et al., 2017), split‐pool recognition of interactions by tag extension (SPRITE) (Quinodoz et al., 2018), chromatin interaction analysis via droplet‐based and barcode‐linked sequencing (ChIA‐Drop) (Zheng et al., 2019) and DNA adenine methyltransferase identification of chromosomal interactions (DamC) (Redolfi et al., 2019) (Fig. 1). GAM is a technique that combines microsectioning and sequencing technologies to infer information about the relative location of genes and enhancers that activate them by studying the frequency of different genomic regions appearing in nuclear sections (Beagrie et al., 2017). SPRITE not only detects pairwise interactions between two loci, but also can detect multiple DNA and RNA molecules that interact simultaneously (Quinodoz et al., 2018). ChIA‐Drop uses a specific antibody to capture the target protein and its interacting DNA fragments by ChIP, and these chromatin complexes are loaded onto a microfluidic device to produce gel‐bead‐in‐emulsion droplets (Zheng et al., 2019). DamC can detect distal chromatin interaction between specific locations with a methylation status, relying on the DNA adenine methyltransferase Dam that is fused with DNA‐binding proteins and recruited to specific genomic locations (Redolfi et al., 2019).
Even though 3D genome mapping techniques have advanced over the past decade, they were developed for applications in mammalian cells and performed less in plant tissues, probably because of the presence of the plant cell wall. Currently, some methods are used successfully in plants. The traditional Hi‐C technique has been used in Arabidopsis, rice, cotton and Brassica, using the enzymes HindIII, DpnII or MboI for chromatin digestion (Grob et al., 2013; Feng et al., 2014; Grob et al., 2014; Wang et al., 2015, 2017, 2018a; Liu et al., 2016; Dong et al., 2018; Xie et al., 2019). The ChIA‐PET technique is based on the use of antibodies against chromatin‐bound proteins such as RNAPII or modified histones, and has been used to characterize chromatin in rice and maize (Li et al., 2019a; Peng et al., 2019; Zhao et al., 2019). In situ Hi‐C has been used in rice, Foxtail millet, sorghum and tomato (Dong et al., 2017; Liu et al., 2017). The improvement of in situ Hi‐C over traditional Hi‐C lays in the use of complete nuclei instead of free chromosomes for ligation, which can reduce the wrong ligation of DNA fragments from different nuclei, thus effectively reducing the background noise of the data and improving the signal‐to‐noise ratio. Capture Hi‐C is another modification of this method and has been used in Arabidopsis, and digestion‐ligation‐only Hi‐C (DLO Hi‐C) was recently used in maize (Nutzmann et al., 2020; Sun et al., 2020b). An advantage of capture Hi‐C compared with Hi‐C is that after the Hi‐C library is constructed, specific probes are used to capture the reads related to the target region, and the chromatin interaction information of the region of interest can be obtained by sequencing. In DLO Hi‐C without biotin labeling, the digested chromatin is ligated with DNA linker after enzyme digestion, and then chromatin with linkers is subjected to proximity ligation. The re‐ligated chromatin is digested by MmeI to obtain the fixed‐size DNA fragments for sequencing. HiChIP, another related technique, has been used in maize and wheat (Ricci et al., 2019; Concia et al., 2020). After using biotin to fill in the ends and ligation, the target protein‐specific antibody is used to precipitate the DNA–protein complex. Once the specific fragment containing biotin is captured, a transposase‐mediated library construction method is used to finally obtain the chromatin conformation bound by the protein of interest. HiChIP requires only a small tissue sample; compared with Hi‐C, the signal‐to‐noise ratio is significantly improved, and compared with ChIA‐PET, there are more informative reads. Of note is that a single‐cell Hi‐C technique with no biotin purification and pull‐down has been performed in rice (Zhou et al., 2019).
Hierarchical 3D chromatin structure in plants
In recent years, a large number of 3D genomic data have been generated using the powerful technologies of ultra‐high resolution microscopy and 3C‐based molecular methods at cell population‐averaged levels. In‐depth analysis of these data has provided a detailed hierarchical chromatin structure that allows us to understand how plant genomes are organized in the nucleus (Rodriguez‐Granados et al., 2016; Sequeira‐Mendes & Gutierrez, 2016; Dogan & Liu, 2018; Sotelo‐Silveira et al., 2018; Dong et al., 2020b; Ouyang et al., 2021). The hierarchical chromatin structure can be resolved and visualized at different resolutions (Fig. 2).
Fig. 2.
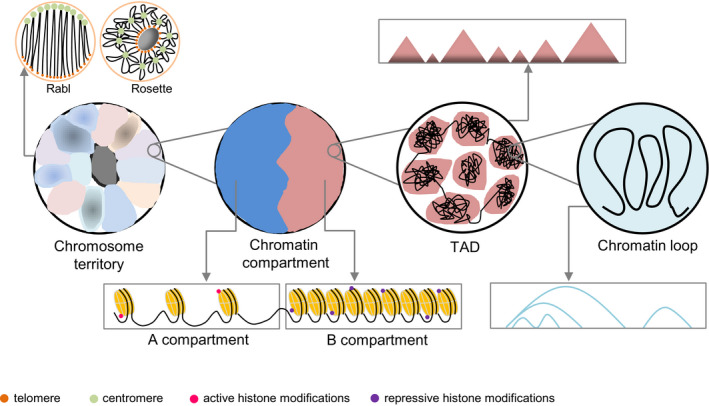
Hierarchical chromatin structure. Hierarchical chromatin structure is seen mainly at four levels: chromosome territory, chromatin compartment, topologically associating domain (TAD) and chromatin loop. In different chromosome territories, chromosomes showed different morphologies in the nucleus, such as Rabl and Rosette (Rabl, 1885; Fransz et al., 2002). In the Rabl configuration, telomeres and centromeres of chromosomes cluster at the two poles of the nucleus, respectively. In the Rosette configuration, the nucleolus is surrounded by telomeres, heterochromatin and centromeres cluster together and euchromatin emanate freely in the nucleus, forming a rosette‐like configuration. Plant chromosomes can be divided into two kinds of regions: A and B compartments. The A compartment is related to high gene density, active epigenetic modifications and active transcriptional activity; the B compartment has higher transposon density and inhibitory epigenetic modifications. TAD is a relatively independent local unit. The interaction strength within a TAD is significantly stronger than that between different TADs. The regulatory roles of cis‐regulatory elements in target genes can be established by forming chromatin loop in space.
Chromosome territories (CTs)
Chromosomes are not randomly arranged in the nucleus during mitosis interphase, but each chromosome occupies a relatively limited nuclear region, designated a chromosome territory (CT) (Fransz & de Jong, 2011). CTs can be directly visualized by cytological and microscopy techniques (Lichter et al., 1988; Pinkel et al., 1988). Meanwhile, in the Hi‐C maps, the interaction frequency of sites on the same chromosome is much higher than that of any two sites located on different chromosomes, which also can prove the existence of CTs (Lieberman‐Aiden et al., 2009). The existence of plant CTs was first confirmed by FISH observation in Arabidopsis thaliana (Lysak et al., 2001; Pecinka et al., 2004) and then found in other species (Kulikova et al., 2001; Szinay et al., 2010; Nagaki & Yamaji, 2020). The mechanisms establishing and maintaining these CTs are not fully understood in plants; however, the attachment of chromatin to the nuclear envelope via some lamin‐like proteins such as CROWDED NUCLEI 1 (CRWN1) and CRWN4 seems to have an important role (Bronshtein et al., 2016; Hu et al., 2019). In Arabidopsis interphase nuclei, centromeres are located at the nuclear periphery and telomeres are located mainly at the centrally localized nucleolus, and chromosomes form different CTs (Fransz et al., 2002; Schubert et al., 2014). The CTs between the different chromosomes only overlap at the edge, thus ensuring that each CT is relatively independent and can interact with each other (Cremer & Cremer, 2010). This model is supported by the discovery of a plant‐specific nuclear structure or ‘KNOT’ which comprises both long‐ and short‐range intra‐ and interchromosomal interactions between chromosomal arms. The genomic regions found to reside on all chromosomes, which are involved in the KNOT, are named as KNOT ENGAGED ELEMENT1 (KEE1) to KEE10 in Arabidopsis. These KEEs share a few conserved sequence motifs, exhibit an enrichment of small RNAs and H3K27me1, and represent preferred transposon landing sites (Grob et al., 2014). In rice, maize, tomato, sorghum, cotton and Brassica, both strong intra‐ and interchromosomal interactions have been observed between euchromatin arms (Dong et al., 2017, 2018; Wang et al., 2018a; Xie et al., 2019). This finding proves the existence of CTs in these plants, and raises the possibility that the KNOT structure is present in their nuclei. The identification of KNOT in rice and Brassica nuclei supports this view (Dong et al., 2018; Xie et al., 2019). Plant CTs are relatively conserved in different tissue samples, but their position in the nucleus seems to be unstable (Albert et al., 2019). Studies in Arabidopsis found that the locations of CTs are different in different organs. For example, in the root tip and rosette leaf cells, the positions of CTs are random (Pecinka et al., 2004; Berr et al., 2006); in the endosperm cells, their positions are nonrandom (Baroux et al., 2007); in the meristematic cells of roots and stems, the sister nuclei show mirror‐symmetric arrangement of the homologous CTs after mitosis (Berr & Schubert, 2007).
Plant chromosomes often show other different morphologies, such as Rabl (Rabl, 1885) and Rosette‐like structures (Fransz et al., 2002) in the nuclei (Fig. 2), which depends on the ways in which chromosome arms, centromeres and telomeres are folded and make contact with each other. In the Rabl configuration, chromosomes are arranged in a polarized manner – centromere regions are clustered at one pole of the nucleus and telomere regions are clustered at the other pore (Rabl, 1885); in the Rosette‐like configuration, heterochromatin is highly concentrated and forms chromocenters while the euchromatin emanates outward (Fransz et al., 2002). Different morphologies are related to changes in genome size. In plants with large genomes, such as barley, wheat and rye, chromosomes exhibit the Rabl configuration (Rabl, 1885; Noguchi & Fukui, 1995; Prieto et al., 2004; Concia et al., 2020); whereas in plants with smaller genomes, such as Arabidopsis, chromosomes often present the rosette‐like configuration (Fransz et al., 2002). It recently has been found that there are other factors which influence chromatin configurations, such as the content and distribution of heterochromatin, and different environmental conditions (Tessadori et al., 2007; Santos et al., 2011; Schubert & Shaw, 2011). In addition to CTs, the idea of genome territories in plants has been raised. A genomic in situ hybridization (GISH) experiment for the interphase nuclei of Hordeum chilense and Secale africanum hybrids showed that chromatin from the two parental genomes did not mix with each other (Leitch et al., 1990). Recently, wheat chromatin architecture was mapped through an analysis of intra‐ and intersubgenomic chromatin interactions (Concia et al., 2020). It is found that the three subgenomes are nonrandomly spatially distributed in the nucleus, implying that polyploid subgenomes may occupy different genome territories. In terms of the functional implications of CTs, studies in Arabidopsis interphase nuclei showed that repressed chromatin is preferentially located at the nuclear periphery, and permissive chromatin occupies more internal space in the nucleus, and this positioning may have a role in the transcriptional regulation of genes, similar to findings in animals (Fransz et al., 2002; Fransz & de Jong, 2011; Schubert et al., 2014).
Chromatin compartment
In the hierarchical structures of eukaryote genomes, the next level of CTs is the chromatin compartment (Gibcus & Dekker, 2013). At the megabase level, chromatin can be divided into the continuous regions separated from each other. Chromatin within these regions has similar chromosomal characteristics and interaction patterns, and the interaction patterns between adjacent regions vary. These chromatin regions are called chromatin compartments (Lieberman‐Aiden et al., 2009; Bonev & Cavalli, 2016). Based on principal component analysis (PCA) on Hi‐C maps, chromosomes can be divided into several A/B compartments (Lieberman‐Aiden et al., 2009). The spatial separation of A and B compartments in the nucleus was confirmed based on FISH at the single‐cell level in humans (Wang et al., 2016a, 2016b,2016a, 2016b). The characteristics of the compartments in plants are similar to those in animals. The A compartment is characterized by a high gene density and epigenetic modifications associated with active transcriptional activity, whereas the B compartment has a higher transposon density and more repressive epigenetic modifications (Dong et al., 2017; Xie et al., 2019). In Arabidopsis, individual chromosomes have regions described as loose structural domains (LSDs) and compacted structural domains (CSDs) (Grob et al., 2014). LSDs in chromosome arms (CAs) are positively correlated with active histone modifications and transcription rates; whereas CSDs in CAs are correlated with abundance of transposable elements (TEs) (Grob et al., 2014). In other plants with large genomes such as maize, tomato, sorghum and foxtail millet, chromosomes are usually divided into two A compartments found at both ends of chromosomes (global A compartment), and one B compartment in the middle of the chromosomes (global B compartment) (Dong et al., 2017). At higher resolution, these chromosomes can be further divided into local A compartments and local B compartments (Dong et al., 2017; Wang et al., 2018a; Xie et al., 2019). Notably, the local A compartments are euchromatin regions, whereas local B compartments are heterochromatin regions, suggesting that the local chromatin contact probability obtained by Hi‐C maps can be used to define the euchromatin and heterochromatin states of chromatin on cytological evaluation (Dong et al., 2017). By comparing chromatin structures of rice, maize and foxtail millet, it was found that their global A/B compartments were stable among different tissues, whereas the local A/B compartments had tissue‐specific dynamics associated with differential gene expression (Dong et al., 2020a). In addition, chromatin compartments exhibit status switching between A and B under different conditions (Wang et al., 2018a; Xie et al., 2019; Zhang et al., 2019a).
Topologically associating domain‐like structure (TAD‐like structure)
On the submegabase scale, mammalian chromatin can be further divided into topological associating domains (TADs) according to its local interaction mode. TADs are represented as continuous square domains along the diagonal in Hi‐C maps. Each TAD is a relatively independent local unit, and the interaction strength within TADs is significantly stronger than that between different TADs (Dixon et al., 2012; Bonev & Cavalli, 2016; Du et al., 2017). TADs have been shown to be the genuine structural units separated from each other in single cells of Drosophila by Oligo‐FISH and super‐resolution microscopy (Szabo et al., 2018). Generally, the boundary of a TAD in mammalian species is enriched with structural proteins, such as cohesins, CTCF and modifications associated with active transcription (Dixon et al., 2012). However, TADs in plants appear to be slightly different to those in animals. TAD is not a very obvious feature in the genome structure of Arabidopsis, which shows special structural features called ‘positive strips’ in a 2 kb resolution Hi‐C map (Wang et al., 2015). 'Positive strips' represent kbp‐sized segments exhibiting higher intrachromosome interaction rates than neighboring chromatin regions, and genes located in genomic regions of ‘positive strips’ have lower expression levels than average, accompanied by an enrichment of the repressive chromatin modification H3K27me3 (Wang et al., 2015). These 'positive strips' on the Hi‐C heatmap resemble TAD edges. At a higher resolution, a number of TAD‐boundary‐like regions were identified with a similar number of TAD‐interior‐like regions (Wang et al., 2015). In some other plants such as rice, cotton and Brassica, positive strips were not found but TAD‐like structures were identified with different sizes, accounting for 25–93% of the genomic length. These TAD‐like structures are similar to mammalian TADs, for example, cis‐interactions are enriched in the domain interior, active genes and histone modifications are enriched in the domain border (Liu et al., 2017; Wang et al., 2018a). In wheat, the term ICONS (intergenic condensed spacers) was used to represent the TAD‐like structures (Concia et al., 2020). It is interesting to investigate whether the presence of TADs depends on genome size, because the genome of Arabidopsis is much smaller than most crop plants. Overall, these observations support the view that plant genomes are packed into TAD‐like structures or other higher‐order structures, by an as yet undefined molecular mechanism.
In mammals, TADs can create insulated environments that limit the enhancer search space for target genes; and genes in the same TAD are usually co‐regulated (Dixon et al., 2012; Nora et al., 2012; Zhan et al., 2017). TAD‐like structures in plants are suggested to be separated by differential expression and epigenetic states, but genes located in the interiors of TAD‐like structures are not co‐regulated and do not tend to be associated with higher expression levels (Concia et al., 2020; Dong et al., 2020a). In plants, regulatory elements such as enhancers have been observed that can contact promoters located in different TAD‐like structures by forming long‐range chromatin loops, indicating that TAD borders in plants may not act as canonical insulators (Dong et al., 2017). The functional role of plant TAD‐like structures remains to be explored further.
Chromatin loops
It is well‐known that some cis‐regulatory elements, such as enhancers, are far from their target genes in terms of linear sequence. The regulatory roles of cis‐regulatory elements in target genes can be established by forming chromatin loops. In mammals, chromatin loops form four main types – enhancer‐promoter loops, polycomb‐mediated loops, gene loops and structural loops – of which the first three types are found in plants (Noguchi & Fukui, 1995). Gene‐centered chromatin loops in plants are formed primarily between gene islands outside the repressive domains, probably because the interactions between active transcriptional gene islands (local A compartment) are isolated from concentrated heterochromatin (local B compartment) (Dong et al., 2017). In Arabidopsis, transcription start sites (TSSs) tend to form chromatin loops with downstream regions, whereas transcription termination sites (TTSs) tend to form chromatin loops with upstream regions, known as “gene loops”, which represent a major difference with genome folding in mammals (Liu et al., 2016; Dong et al., 2018). An example is the chromatin loops for the flowering locus C (FLC). 3C analysis showed that a c. 2.7‐kb region covering the FLC promoter and transcriptional start site was in contact with the sequence directly downstream of the gene, and the 3′ flanking region of FLC had the strongest interaction with the first exon (Liu et al., 2016). Several chromatin loops that link three closely related genes encoding phosphate transporters (PHT1;1, PHT1;3 and PHT1;6), form a repressive hub with H3K27me3‐marked loci in Arabidopsis, which has similarity to the Polycomb‐mediated HOX loop cluster in mammals (Liu et al., 2016). Enhancer‐promoter loops are widespread in plants with larger genomes than Arabidopsis, such as rice and maize, and many of them can be validated using the self‐transcribing active regulatory region sequencing (STARR‐seq) method (Li et al., 2019a; Peng et al., 2019; Ricci et al., 2019; Sun et al., 2019; Zhao et al., 2019). In addition to gene‐centered loops, a recent study identified chromatin loops in centromeric regions which were mediated by circular centromeric retrotransposon 1 (CRM1) RNAs (Liu et al., 2020).
The advanced application of 3D genome principles in plants
Construction of chromatin interactome for transcriptional regulation
It is believed that chromatin conformation is closely related to gene expression and regulation, and regions with strong chromatin interaction generally show functional dependency (Spitz & Furlong, 2012; Mendes et al., 2013; Phillips‐Cremins et al., 2013) (Fig. 3). Sometimes, changes in chromatin compactness may influence the accessibility of chromatin to transcription factors and chromatin remodeling factors, which may eventually lead to changes of gene expression (Bannister & Kouzarides, 2011; Rutowicz et al., 2019). In Arabidopsis, a single‐gene resolution Hi‐C map was constructed to show that local chromatin loops between the 5′ and 3′ ends of the genes were preferentially associated with highly expressed genes (Liu et al., 2016). In maize, high‐resolution chromatin interaction maps were constructed using ChIA‐PET and DLO Hi‐C, and showed that chromatin loops were formed between the regulatory elements, and the genes with a proximal promoter interaction were co‐expressed (Li et al., 2019a; Peng et al., 2019; Sun et al., 2020b). In rice, similar to maize, researchers constructed a promoter–promoter interaction map associated with H3K4me3 and RNA polymerase II (RNAPII), and found that promoter–promoter interacting genes often were co‐transcribed (Zhao et al., 2019). In wheat, a study found that gene pairs associated with gene‐to‐gene loops tended to be co‐regulated in roots and stems (Concia et al., 2020). The chromatin interactome in each of these plants provides a valuable resource for uncovering the regulatory landscape among genes and regulatory elements.
Fig. 3.
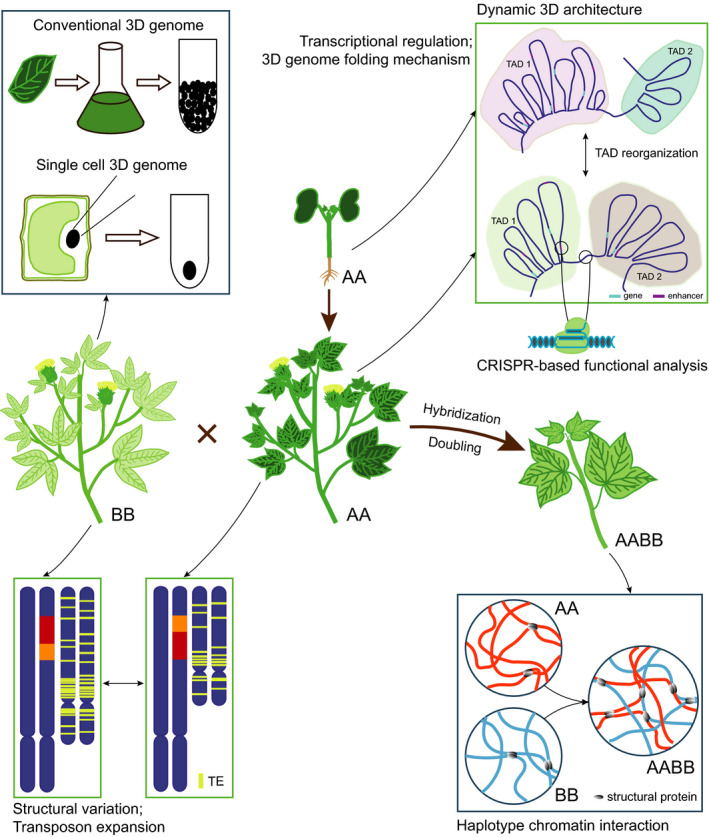
Some proposed future directions of 3D genomics research in plants. There are many possible directions of 3D genomic research using different plant samples at different scales, such as the identification of large‐scale structural variation between different plants, construction of dynamic 3D genome maps in plant cell differentiation and organ development, and 3D genome analysis from a tissue sample to a single cell. The research field of 3D genomics in plants will be expanded, for example to understand the mechanisms of 3D genome folding, the effects of transposon expansion on shaping 3D genome architecture, the spatial haplotype chromatin interaction, and the CRISPR‐based functional analysis of enhancers and boundaries of TAD‐like structures.
Enhancers are recognized as noncoding DNA elements that function independently of transcriptional direction and relative position with the target promoter, and participate in gene transcription regulation through long‐distance chromatin interactions (chromatin loops). By analysing data for histone modification, DNA methylation and chromatin accessibility, researchers were able to identify possible enhancer elements. For example, 1495 putative enhancers were identified in maize by integrating DHS, H3K9ac and LUMR datasets (Oka et al., 2017). In Arabidopsis, 10,044 DNase I hypersensitive sites (DHSs) located in the intergenic regions that are far from gene promoters, are predicted to be putative enhancers (Zhu et al., 2015). A study identified thousands of putative cis‐regulatory elements (CREs) in 13 species, and revealed the prevalence of distal CREs in plants (Lu et al., 2019). By constructing DHS maps in different tissues of three grass species, the researchers identified a large number of distal intergenic DHSs, and found that tissue‐specific DHSs may act as distal regulatory elements (Han et al., 2020). With the development of 3D genomics technologies, chromatin loops can be identified by Hi‐C or ChIA‐PET, which can help us directly identify enhancers interacting with promoters so as to accurately study the regulatory effect of enhancers on gene transcription (Liu et al., 2016; Wang et al., 2018a; Li et al., 2019a; Peng et al., 2019; Sun et al., 2020b). Some enhancers were functionally validated. In Arabidopsis, the transcriptional programming triggered by jasmonate (JA) is orchestrated by the key transcription factor MYC2, and the function of MYC2 depends on its physical interaction with MED25. By analyzing the occupancy pattern of MYC2 and MED25, a JA response‐related enhancer was identified. It was found that JA regulates the dynamic chromatin loops between enhancer and promoter in a MED25‐dependent manner, and the JA enhancer at the MYC2 site (named ME2) positively regulates the expression of the MYC2 in a short‐term JA response (Wang et al., 2019). In maize, the b1 gene encodes a transcription factor that regulates flavonoid biosynthesis, and has two epialleles (B‐I and B′) with the same DNA sequence but different expression levels. The chromatin loops of these two epialleles have been studied. In husk tissue, both the expression level and the interaction frequency of B‐I are higher than those of B′. The hepta‐repeat located upstream of the TSS acts as an enhancer to mediate the high expression of B‐I gene through a multi‐loop structure (Louwers et al., 2009). Likewise, the intergenic quantitative trait locus (QTL) KRN4 associated with kernel row number was mapped to a c. 60 kb region downstream of UNBRANCHED3 (UB3) in maize. KRN4 has been shown to act as an enhancer of the UB3 promoter to fine‐tune the expression of UB3 in the meristem of the ear inflorescence by interacting with the UB3 promoter and recruiting the UB2‐centered transcriptional complex (Du et al., 2020).
Prediction of noncoding regulatory variations for target genes
Many genome‐wide association studies (GWAS) have identified loci of interest that are located in intergenic noncoding genomic regions, which suggests that noncoding variations may have functional implications in controlling traits (Zhang & Lupski, 2015; Mullin et al., 2020). By integrating 3D genomic data and expression quantitative trait loci (eQTL) and GWAS data, the trait‐associated noncoding regulatory variation can be identified, and the spatial relationship between eQTL and genes can be explored at the 3D genomic level (Won et al., 2016; Lalonde et al., 2019). This serves as a useful methodology for linking noncoding regulatory variations to their target genes in the post‐genomics era, with some examples found in plants (Wang et al., 2017; Peng et al., 2019; Zhao et al., 2019; Sun et al., 2020b). In maize, eQTL and the regulated genes tend to approach each other in 3D space to form chromatin loops, thereby potentially defining a mechanistic basis for regulating expression. In addition, some distal elements overlapping agronomic trait QTL also were identified, and the functional role of distal elements in the regulation of agronomic traits was mediated by chromatin loops (Peng et al., 2019; Sun et al., 2020b). In rice, there is a spatial physical proximity between eQTL and the regulated genes. Combined with ChIA‐PET analysis, it is speculated that genomic variation in distal regulatory elements may contribute to phenotypic variation by influencing the expression of target genes (Zhao et al., 2019). In cotton, the effects of domestication on the divergence of distal regulatory elements were explored by using Hi‐C and population genomics data. A total of 99 709 single nucleotide polymorphisms (SNPs) were identified in the 21 409 putative enhancers, and the frequency of the sequence variation of enhancers was higher than that of the promoters and exons but was lower than introns, indicating that enhancers might evolve rapidly. In one case, a 120‐kb enhancer located upstream of the tubulin α‐3 (TUA3) gene had undergone domestication selection, probably associated with the differential expression of TUA3 between cultivated and wild cotton accessions (Wang et al., 2017).
Evolutionary view of spatial chromatin organization
Over the past decade, there have been many studies on chromosome evolution and polyploidization as well as on the evolutionary renewal and conservation of homologous gene expression (Woodhouse et al., 2014; Bottani et al., 2018). These studies were carried out primarily at the linear genome level. However, at the 3D genome level, it remains largely unknown how higher‐order genome structures may have changed during the evolution of species. By combining Hi‐C, histone modification, DNA methylation and gene expression data, one can investigate the evolutionary dynamics of chromatin structure and its effects on chromosome evolution and transcriptional regulation in different species. A combined analysis of FISH and Hi‐C data in A. thaliana and A. lyrata showed that the chromatin of A. lyrata was more compact than that of A. thaliana, occupying a smaller nuclear space and having stronger chromatin interactions. At the same time, this tight chromatin compactness was associated with an increase in frequency of the repressive chromatin modification H3K27me3 (Zhu et al., 2017). A similar observation was found in the hybrid between the two species. Because the main difference in genome architecture between A. thaliana and A. lyrata derives from the differential expansion of TEs during evolution, this study raises the question of whether similar changes have taken place in chromatin compactness in the evolution of other species (Zhu et al., 2017). Comparison of synthesized autotetraploid Arabidopsis with diploid plants revealed that autotetraploid Arabidopsis showed more interchromosomal interactions and less short‐range chromatin interactions, such as an increase in the relative amount of gene loops at the FLC locus (Zhang et al., 2019a). By comparing the 3D genome structure of each subgenome in tetraploid cotton with that of its putative diploid ancestors, it is found that genome polyploidization has contributed to the status switching of A/B compartments and the reorganization of TADs. In the process of polyploidization, the formation of TAD boundaries occurs preferentially in open chromatin, which is consistent with the deposition of active chromatin modification (Wang et al., 2018a). In an analysis of the 3D genome of Brassica rapa and B. oleracea, it was found that fully retained genes with syntenic copies among three subgenomes (LF, MF1 and MF2) had the strongest chromatin interaction in the LF subgenome, which is consistent with the dominant expression pattern of genes at the subgenome level (LF > MF1 > MF2). These observations illustrate the association between spatial chromatin organization and differential gene expression in diploidization. The KNOT structures were found to have expanded in B. rapa and contracted in B. oleracea genome evolution (Xie et al., 2019). During the evolution of species, there also is a correlation between the changes in 3D structure and in epigenetic modification. For example, in the primitive land plant Marchantia polymorpha, H3K27me3 is enriched in the repressed B compartment, whereas in flowering plants, H3K27me3 dedicated to transcriptional repression is enriched in the loose A compartment (Montgomery et al., 2020).
Single cell 3D genome
The technique of 3C and its derivatives allow us to study 3D genome structure at the cell population or tissue level, but genome structure undergoes significant changes in different cell types or stages of cell differentiation. The differences of 3D genome organization between cells can only be observed by performing single cell measurements (Nagano et al., 2013). This raises the necessity of developing single‐cell 3D genome mapping techniques (Fig. 3). In mammals, a few single cell 3D genome mapping techniques have been developed and their application shows that many TADs are reorganized in different cells, whereas by contrast, chromatin compartments and lamina‐associated domains are usually relatively stable (Stevens et al., 2017). The development of ultra‐high resolution microscopy and cytological techniques provides approaches to study 3D structure at the single cell level. Using these techniques, it is found that the high‐order chromatin structure of A/B compartments and TADs in population cells still exists in individual cells of animals (Wang et al., 2016a, 2016b,2016a, 2016b; Szabo et al., 2018). Nowadays, the study 3D structure at the single cell level is only at the CT level by microscopic technology in plants, and the visualization of fine compartments and TADs structures has not been fulfilled (Lysak et al., 2001; Pecinka et al., 2004; Szinay et al., 2010). In plants, the development of single cell 3D genome mapping techniques needs to overcome the existence of plant cell wall and obtain single cell or nucleus for further study. There are different single‐cell isolation techniques in plants: isolation of nuclei tagged in specific cell types (INTACT), and derivatives of this method using affinity to heterologous baits (Deal & Henikoff, 2011); manual selection using micropipettes (Li et al., 2015); laser capture microdissection (LCM), using a highly focused laser beam and specialized microscope (Aubry et al., 2016); and fluorescence activated cell sorting (FACS) using fluorescent tags and cytometry (Lee et al., 2019b). However, in plants, the study of the single‐cell 3D genome is still in its infancy. At present, single‐cell 3D genome mapping has been performed only in rice. The manual selection method was used to isolate rice single cells for the investigation of 3D chromatin organization and dynamics during fertilization (Zhou et al., 2019). By analyzing the eggs, sperm cells, zygotes and stem cells, it was found that the 3D chromatin structures of rice eggs and sperm cells are similar to those of mesophyll cells, and are recombined after fertilization. This study revealed characteristics of chromatin compartments and telomere/centromere structures at the single cell level in rice that are distinct from mammalian cells (Zhou et al., 2019).
Future perspectives
Uncovering mechanisms of 3D genome folding in plants
In mammals, the mechanism of the 3D genome organization has been explored preliminarily (Liu et al., 2002; Hyman et al., 2014; Fudenberg et al., 2016; Boija et al., 2018; Chong et al., 2018; Shin et al., 2019; Zhang et al., 2020). A phase separation process plays an important role in distinguishing chromatin compartments. Phase separation refers to the phenomenon that different components (proteins, nucleic acids, or other molecules) in cells collide with each other and fuse to form liquid droplets (or condensate). Owing to the difference in affinity, some components are wrapped in the droplets and some components are blocked outside the droplets (Hyman et al., 2014; Boija et al., 2018; Shin et al., 2019). The loop‐extrusion model was proposed to interpret the formation of TAD structures. In the loop‐extrusion model, an extrusion factor (e.g. a ring‐shaped cohesin complex) will extrude a chromatin loop until it encounters an extrusion barrier (CTCF and other insulating proteins) (Rao et al., 2014; Fudenberg et al., 2016). However, in plants, the mechanism of higher‐order structure formation is unknown. It has been found that there is a lack of CTCF homologs in plants, but the homologs of the subunits of cohesin are present (Liu et al., 2002; Zhang et al., 2020). At the same time, TADs in plants are not as distinct as in animals. Of note is the observation that TAD boundaries in rice and maize exhibit an enrichment of plant‐specific TCP transcriptional factor binding sites, raising the possibility that transcriptional factors may be involved in the formation of TADs, as described in mammals (Liu et al., 2017; Stadhouders et al., 2019; Sun et al., 2020b). These observations indicate that the mechanism of 3D genome folding in plants may be slightly different from that in animals, and requires further study (Fig. 3).
The effects of transposon expansion on shaping 3D genome architecture
A TE or transposon is a genetic sequence that can replicate itself or translocate to another site in the genome. TEs account for large proportions of many plant genomes, and are particularly abundant in maize, wheat and other plants with large genomes. Generally, differences in genome size in plants are closely related to either polyploidy or the expansion of transposons, or both. Even within a given plant genus, such as Oryza or Gossypium, the differential expansion of TEs also can contribute to large differences in genome size between species (Hawkins et al., 2006; Stein et al., 2018). Many studies have shown that TEs have a profound impact on the transcriptional regulation of adjacent genes and the evolution of the whole genome (Kashkush et al., 2003; Feng et al., 2013). In terms of 3D genome structure, an increasing number of studies in mammals have shown that TEs have a role in the formation of TAD boundaries, which will influence the organization of TADs (e.g. Zhang et al., 2019b). It also was found that the 3D genome conformation will affect TE insertion; for example in Arabidopsis, KEE regions, which represent heterochromatin islands, show a preference for TE insertion (Grob et al., 2014). In rice, approximately 44% (5296) of H3K9me2‐marked regions act as anchors involved in chromatin interactions, and the density of TEs in these anchors is higher than those in basal sites, which indicates that H3K9me2 binding sites with higher TE density might be involved in chromatin interactions (Zhao et al., 2019). Recently, it was found in Arabidopsis that heat stress induces TE activation, and those activated TEs were associated with changes in local chromosomal interactions (Sun et al., 2020a). The detailed relationship between TE activity and 3D genome organization remains to be explored further in plants (Fig. 3).
Spatial haplotype chromatin interaction in plants
Interchromosomal interaction represents an important regulatory level for gene transcription in eukaryotes. An analysis of interchromosomal interaction usually requires an understanding of haplotype‐specific information in the nucleus, which can help uncover the regulatory basis of many biological issues, such as heterosis in terms of superior traits in the hybrids relative to both parents (Birchler et al., 2003). In the past few years, a number of studies have found that the differential expression of genes at the haplotype level might be responsible for the appearance of heterosis (Guo et al., 2003; Stupar et al., 2007; Song et al., 2010). The underlying mechanism of haplotype‐specific expression has been proposed from epigenetic studies, including data from small RNA, DNA methylation and histone modification (Ni et al., 2009; He et al., 2010; Li et al., 2014; Ng et al., 2014). With the development of 3D genomics, these studies can be extended to the spatial haplotype or interchromosomal interaction level. 3D genome studies in humans and Drosophila provide context for related studies in plants (Sexton et al., 2012; Tang et al., 2015). More specifically, polyploid species which usually exhibit enhanced growth vitality and adaptability compared with ancestor species, are widespread in the plant kingdom. The investigation of interchromosomal interaction in polyploid plants has another meaning – studies of intersubgenomic interaction responsible for transcriptional complexity of homologous genes; these should facilitate the elaboration of molecular mechanisms of karyotype homeostasis distinct from results of traditional genetic and epigenetic studies (Ha et al., 2009; Xiong et al., 2011; Schatlowski et al., 2014). As far as we know, the study of 3D genome‐based intersubgenomic interaction at the current stage is limited in plants to cotton, and data show that spatial chromatin interaction between the two subgenomes might be related to the coordinated expression of homoeologous genes in tetraploids (Wang et al., 2018a). Collectively, the emergence of 3D genomics provides an excellent approach for exploring haplotype‐specific or interchromosomal interaction in plants (Fig. 3).
Dynamic 3D genome architecture in plant development
Based on 3D genome mapping technologies, we can explore dynamic changes of 3D chromatin structure during plant development, which is known as 4D genomics. In mammals, the 4D nucleome project has been conceived to study 3D genomics and its dynamic changes over time (Dekker et al., 2017). In humans, a large number of higher‐order structure reorganization events have been found through the analysis of chromatin interaction in the development of embryonic stem cells and the development from oocytes to fertilized eggs (Dixon et al., 2015; Flyamer et al., 2017). However, at present, there are few studies on the dynamic changes of 3D structure during plant development. In fact, in the process of plant cell differentiation, chromatin accessibility is usually changed, implying that the organization of higher‐order chromatin structures also may be a dynamic process (Wang et al., 2016a, 2016b,2016a, 2016b; Sijacic et al., 2018; Sullivan et al., 2019). In the future, the dynamics of 3D genome folding should be explored at different developmental stages. In crop plants, by integration of the transcriptome and other omics data, we can understand the effects of 3D genome dynamics on the regulation of gene transcription underpinning many important agronomic traits, and lay a foundation for further crop improvement (Fig. 3).
CRISPR‐based functional analysis of 3D genome structure
In mammals, improved CRISPR technologies have been used to investigate the fine structure of 3D genomes in combination with ultra‐high resolution microscopy. Using sgRNAs with 16 MS2 binding motifs and a catalytically inactive mutant of the Cas9 (dCas9) protein, the specific locations of transcriptionally active and inactive regions in the nucleus have been determined (Qin et al., 2017). CRISPR technology also has been used to explore the functional implications of 3D genome folding, by knock‐out or knock‐in experiments of the TAD boundary or structural proteins (such as CTCF and cohesins) responsible for chromatin loop formation (Guo et al., 2015; Lupianez et al., 2015; Fei et al., 2019). However, to our knowledge, there are few reports on the application of CRISPR technology in the study of 3D genomics in plants. By knocking out the possible 3D structural elements in plants using CRISPR technology, we can explore the mechanisms of plant‐specific 3D genomic structure formation. The TAD and loop structures could be modified through the knock‐out of large DNA fragments, to help explore the influence of the TAD or the loop change on the regulation of gene transcription. We also can knock out enhancer elements to explore the effect of enhancers on gene expression. In the future, this may become a hot topic in the field of plant 3D genomics and transcriptional regulation (Fig. 3).
Conclusions
In recent years, 3D genomics has developed rapidly, and new research technologies have emerged one after another. In particular, the development of 3C technology has revealed to us the high‐level chromatin conformation at an unprecedented scale, providing new insights into the relationship between chromatin conformation and function. In plants, these technologies are gradually being adopted, and our understanding of the 3D structure of plant genomes is getting more detailed. The technologies used in plants are mainly 3C‐based technologies, and all 3C‐based technologies use proximal ligation, and so it is difficult to completely eliminate false positives of chromatin connections through current algorithms. Therefore, in the future, the use of some nonligation technologies also is needed in plants. At present, chromatin in plants presents several classic structures. Although TAD‐like structures show similar patterns in plants and animals on Hi‐C matrices, their characteristics and functions are somewhat different. The plant TAD‐like structures lack the important insulator protein CTCF, but may involve some transcription factors such as TCP; at the same time, the boundary of plant TAD‐like structures may not have an insulating role. Plant 3D genomics has become a hot topic in recent years, and its combined analysis with multi‐omics data has revealed the interaction of regulatory elements during plant development at the spatial level, allowing us to understand the regulatory mechanism of chromatin conformation on gene expression. In the future, with the development of new technology, 3D genomics will surely be applied to plant science study on a variety of levels.
Author contributions
MW proposed the project, and LP drafted the manuscript; and MW, KL, GL and XZ revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This project was supported financially by the National Natural Science Foundation of China (31801405 and 31922069) to MW. The authors declare that they have no competing interests.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- Albert PS, Zhang T, Semrau K, Rouillard JM, Kao YH, Wang CR, Danilova TV, Jiang J, Birchler JA. 2019. Whole‐chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proceedings of the National Academy of Sciences, USA 116: 1679–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal dynamics at the Shh locus: limb bud‐specific differential regulation of competence and active transcription. Developmental Cell 16: 47–57. [DOI] [PubMed] [Google Scholar]
- Aragon‐Alcaide L, Reader S, Miller T, Moore G. 1997. Centromeric behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma 106: 327–333. [DOI] [PubMed] [Google Scholar]
- Aubry S, Aresheva O, Reyna‐Llorens I, Smith‐Unna RD, Hibberd JM, Genty B. 2016. A specific transcriptome signature for guard cells from the C4 plant Gynandropsis gynandra . Plant Physiology 170: 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Research 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Pecinka A, Fuchs J, Schubert I, Grossniklaus U. 2007. The triploid endosperm genome of Arabidopsis adopts a peculiar, parental‐dosage‐dependent chromatin organization. Plant Cell 19: 1782–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Schubert V. 2018. Technical review: microscopy and image processing tools to analyze plant chromatin: practical considerations. Methods in Molecular Biology 1675: 537–589. [DOI] [PubMed] [Google Scholar]
- Bass HW, Nagar S, Hanley‐Bowdoin L, Robertson D. 2000. Chromosome condensation induced by geminivirus infection of mature plant cells. Journal of Cell Science 113: 1149–1160. [DOI] [PubMed] [Google Scholar]
- Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR et al. 2017. Complex multi‐enhancer contacts captured by genome architecture mapping. Nature 543: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR et al. 2012. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proceedings of the National Academy of Sciences, USA 109: 21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I. 2006. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata . The Plant Journal 48: 771–783. [DOI] [PubMed] [Google Scholar]
- Berr A, Schubert I. 2007. Interphase chromosome arrangement in Arabidopsis thaliana is similar in differentiated and meristematic tissues and shows a transient mirror symmetry after nuclear division. Genetics 176: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott‐Schwartz J, Hess HF. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Auger DL, Riddle NC. 2003. In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM et al. 2018. Transcription factors activate genes through the phase‐separation capacity of their activation domains. Cell 175: 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev B, Cavalli G. 2016. Organization and function of the 3D genome. Nature Reviews Genetics 17: 661–678. [DOI] [PubMed] [Google Scholar]
- Bottani S, Zabet NR, Wendel JF, Veitia RA. 2018. Gene expression dominance in allopolyploids: hypotheses and models. Trends in Plant Science 23: 393–402. [DOI] [PubMed] [Google Scholar]
- Bronshtein I, Kanter I, Kepten E, Lindner M, Berezin S, Shav‐Tal Y, Garini Y. 2016. Exploring chromatin organization mechanisms through its dynamic properties. Nucleus 7: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd K, Corces VG. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. Journal of Cell Biology 162: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson K, Wallen P, Brodin L. 1989. Three‐dimensional imaging of neurons by confocal fluorescence microscopy. Journal of Microscopy 155: 15–26. [DOI] [PubMed] [Google Scholar]
- Chong S, Dugast‐Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X et al. 2018. Imaging dynamic and selective low‐complexity domain interactions that control gene transcription. Science 361: eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concia L, Veluchamy A, Ramirez‐Prado JS, Martin‐Ramirez A, Huang Y, Perez M, Domenichini S, Rodriguez Granados NY, Kim S, Blein T et al. 2020. Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biology 21: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. 2010. Chromosome territories. Cold Spring Harbor Perspectives in Biology 2: a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. 2011. The INTACT method for cell type‐specific gene expression and chromatin profiling in Arabidopsis thaliana . Nature Protocols 6: 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O'Shea CC, Park PJ, Ren B et al. 2017. The 4D nucleome project. Nature 549: 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti‐Renom MA, Mirny LA. 2013. Exploring the three‐dimensional organization of genomes: interpreting chromatin interaction data. Nature Reviews Genetics 14: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Deng W, Shi X, Tjian R, Lionnet T, Singer RH. 2015. CASFISH: CRISPR/Cas9‐mediated in situ labeling of genomic loci in fixed cells. Proceedings of the National Academy of Sciences, USA 112: 11870–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz‐Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan ES, Liu C. 2018. Three‐dimensional chromatin packing and positioning of plant genomes. Nature Plants 4: 521–529. [DOI] [PubMed] [Google Scholar]
- Dong P, Tu X, Chu PY, Lu P, Zhu N, Grierson D, Du B, Li P, Zhong S. 2017. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Molecular Plant 10: 1497–1509. [DOI] [PubMed] [Google Scholar]
- Dong P, Tu X, Li H, Zhang J, Grierson D, Li P, Zhong S. 2020a. Tissue‐specific Hi‐C analyses of rice, foxtail millet and maize suggest non‐canonical function of plant chromatin domains. Journal of Integrative Plant Biology 62: 201–217. [DOI] [PubMed] [Google Scholar]
- Dong P, Tu X, Liang Z, Kang BH, Zhong S. 2020b. Plant and animal chromatin 3D organization, similar structure but different function. Journal of Experimental Botany 71: 5119–5128. [DOI] [PubMed] [Google Scholar]
- Dong Q, Li N, Li X, Yuan Z, Xie D, Wang X, Li J, Yu Y, Wang J, Ding B et al. 2018. Genome‐wide Hi‐C analysis reveals extensive hierarchical chromatin interactions in rice. The Plant Journal 94: 1141–1156. [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C et al. 2006. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Research 16: 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreissig S, Schiml S, Schindele P, Weiss O, Rutten T, Schubert V, Gladilin E, Mette MF, Puchta H, Houben A. 2017. Live‐cell CRISPR imaging in plants reveals dynamic telomere movements. The Plant Journal 91: 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Liu L, Peng Y, Li M, Li Y, Liu D, Li X, Zhang Z. 2020. UNBRANCHED3 expression and inflorescence development is mediated by UNBRANCHED2 and the distal enhancer, KRN4, in maize. PLoS Genetics 16: e1008764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. [DOI] [PubMed] [Google Scholar]
- Dyba M, Jakobs S, Hell SW. 2003. Immunofluorescence stimulated emission depletion microscopy. Nature Biotechnology 21: 1303–1304. [DOI] [PubMed] [Google Scholar]
- Fei T, Li W, Peng J, Xiao T, Chen CH, Wu A, Huang J, Zang C, Liu XS, Brown M. 2019. Deciphering essential cistromes using genome‐wide CRISPR screens. Proceedings of the National Academy of Sciences, USA 116: 25186–25195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Leem YE, Levin HL. 2013. Transposon integration enhances expression of stress response genes. Nucleic Acids Research 41: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. 2014. Genome‐wide Hi‐C analyses in wild‐type and mutants reveal high‐resolution chromatin interactions in Arabidopsis. Molecular Cell 55: 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. 2010. Super‐resolution imaging of plasmodesmata using three‐dimensional structured illumination microscopy. Plant Physiology 153: 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, Razin SV, Mirny LA, Tachibana‐Konwalski K. 2017. Single‐nucleus Hi‐C reveals unique chromatin reorganization at oocyte‐to‐zygote transition. Nature 544: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. 2002. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proceedings of the National Academy of Sciences, USA 99: 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, de Jong H. 2011. From nucleosome to chromosome: a dynamic organization of genetic information. The Plant Journal 66: 4–17. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Reports 15: 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Sugano SS, Kuwata K, Osakabe K, Matsunaga S. 2016. Visualization of specific repetitive genomic sequences with fluorescent TALEs in Arabidopsis thaliana. Journal of Experimental Botany 67: 6101–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH et al. 2009. An oestrogen‐receptor‐alpha‐bound human chromatin interactome. Nature 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Dekker J. 2013. The hierarchy of the 3D genome. Molecular Cell 49: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Sandoval A, Gasser SM. 2016. On TADs and LADs: spatial control over gene expression. Trends in Genetics 32: 485–495. [DOI] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Luedtke NW, Wicker T, Grossniklaus U. 2013. Characterization of chromosomal architecture in Arabidopsis by chromosome conformation capture. Genome Biology 14: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Grossniklaus U. 2014. Hi‐C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Molecular Cell 55: 678–693. [DOI] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Danilevskaya ON, Yang X, Hu Z. 2003. Genome‐wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. The Plant Journal 36: 30–44. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington CJ, Chen X, Wang XJ, Chen ZJ. 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proceedings of the National Academy of Sciences, USA 106: 17835–17840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Wang P, Wang Q, Lin Q, Chen Z, Yu G, Miao C, Dao Y, Wu R, Schnable JC et al. 2020. Genome‐wide characterization of DNase I‐hypersensitive sites and cold response regulatory landscapes in grasses. Plant Cell 32: 2457–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. 2006. Differential lineage‐specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Research 16: 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F et al. 2010. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. 2012. The chromatin insulator CTCF and the emergence of metazoan diversity. Proceedings of the National Academy of Sciences, USA 109: 17507–17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Lu G, Duan J, Liu W, Zhang Y. 2018. Comparison and optimization of CRISPR/dCas9/gRNA genome‐labeling systems for live cell imaging. Genome Biology 19: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. 2015. Mapping nucleosome resolution chromosome folding in yeast by micro‐C. Cell 162: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang N, Bi X, Karaaslan ES, Weber AL, Zhu W, Berendzen KW, Liu C. 2019. Plant lamin‐like proteins mediate chromatin tethering at the nuclear periphery. Genome Biology 20: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Jiang Y, Zheng H, Ji X. 2020. BAT Hi‐C maps global chromatin interactions in an efficient and economical way. Methods 170: 38–47. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, De Gobbi M, Taylor S, Gibbons R, Higgs DR. 2014. Analysis of hundreds of cis‐regulatory landscapes at high resolution in a single, high‐throughput experiment. Nature Genetics 46: 205–212. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Julicher F. 2014. Liquid‐liquid phase separation in biology. Annual Review of Cell and Developmental Biology 30: 39–58. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nature Genetics 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Kolovos P, van de Werken HJ, Kepper N, Zuin J, Brouwer RW, Kockx CE, Wendt KS, van Ijcken IWF, Grosveld F, Knoch TA. 2014. Targeted Chromatin Capture (T2C): a novel high resolution high throughput method to detect genomic interactions and regulatory elements. Epigenetics & Chromatin 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Fransz P, de Jong H. 2003. Cytogenetic tools for Arabidopsis thaliana . Chromosome Research 11: 183–194. [DOI] [PubMed] [Google Scholar]
- Kulikova O, Gualtieri G, Geurts R, Kim DJ, Cook D, Huguet T, de Jong JH, Fransz PF, Bisseling T. 2001. Integration of the FISH pachytene and genetic maps of Medicago truncatula . The Plant Journal 27: 49–58. [DOI] [PubMed] [Google Scholar]
- Lai B, Tang Q, Jin W, Hu G, Wangsa D, Cui K, Stanton BZ, Ren G, Ding Y, Zhao M et al. 2018. Trac‐looping measures genome structure and chromatin accessibility. Nature Methods 15: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Codina‐Fauteux VA, de Bellefon SM, Leblanc F, Beaudoin M, Simon MM, Dali R, Kwan T, Lo KS, Pastinen T et al. 2019. Integrative analysis of vascular endothelial cell genomic features identifies AIDA as a coronary artery disease candidate gene. Genome Biology 20: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Reviews Genetics 8: 104–115. [DOI] [PubMed] [Google Scholar]
- Lee DS, Luo C, Zhou J, Chandran S, Rivkin A, Bartlett A, Nery JR, Fitzpatrick C, O'Connor C, Dixon JR et al. 2019a. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nature Methods 16: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LR, Wengier DL, Bergmann DC. 2019b. Cell‐type‐specific transcriptome and histone modification dynamics during cellular reprogramming in the Arabidopsis stomatal lineage. Proceedings of the National Academy of Sciences, USA 116: 21914–21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Mosgoller W, Schwarzacher T, Bennett MD, Heslop‐Harrison JS. 1990. Genomic in situ hybridization to sectioned nuclei shows chromosome domains in grass hybrids. Journal of Cell Science 95: 335–341. [DOI] [PubMed] [Google Scholar]
- Li A, Liu D, Wu J, Zhao X, Hao M, Geng S, Yan J, Jiang X, Zhang L, Wu J et al. 2014. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Liu H, Huang L, Zhang X, Dong X, Song W, Zhao H, Lai J. 2019a. Long‐range interactions between proximal and distal regulatory regions in maize. Nature Communications 10: 2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu Y, Zhang Y, Kubo N, Yu M, Fang R, Kellis M, Ren B. 2019b. Joint profiling of DNA methylation and chromatin architecture in single cells. Nature Methods 16: 991–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Jia L, Cao Y, Chen Q, Li C. 2018. OCEAN‐C: mapping hubs of open chromatin interactions across the genome reveals gene regulatory networks. Genome Biology 19: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Yan J. 2015. Dissecting meiotic recombination based on tetrad analysis by single‐microspore sequencing in maize. Nature Communications 6: 6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Li G, Wang Z, Djekidel MN, Li Y, Qian MP, Zhang MQ, Chen Y. 2017. BL‐Hi‐C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nature Communications 8: 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Tang CJ, Watkins PC, Manuelidis L, Ward DC. 1988. Rapid detection of human chromosome 21 aberrations by in situ hybridization. Proceedings of the National Academy of Sciences, USA 85: 9664–9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO et al. 2009. Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Hong P, Zhang S, Xu W, Jamal M, Yan K, Lei Y, Li L, Ruan Y, Fu ZF et al. 2018. Digestion‐ligation‐only Hi‐C is an efficient and cost‐effective method for chromosome conformation capture. Nature Genetics 50: 754–763. [DOI] [PubMed] [Google Scholar]
- Lindhout BI, Fransz P, Tessadori F, Meckel T, Hooykaas PJ, van der Zaal BJ. 2007. Live cell imaging of repetitive DNA sequences via GFP‐tagged polydactyl zinc finger proteins. Nucleic Acids Research 35: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cheng YJ, Wang JW, Weigel D. 2017. Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nature Plants 3: 742–748. [DOI] [PubMed] [Google Scholar]
- Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D. 2016. Genome‐wide analysis of chromatin packing in Arabidopsis thaliana at single‐gene resolution. Genome Research 26: 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D. 2002. Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. The Plant Journal 29: 405–415. [DOI] [PubMed] [Google Scholar]
- Liu Y, Su H, Zhang J, Liu Y, Feng C, Han F. 2020. Back‐spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biology 18: e3000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobastov VA, Srinivasan R, Zewail AH. 2005. Four‐dimensional ultrafast electron microscopy. Proceedings of the National Academy of Sciences, USA 102: 7069–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M. 2009. Tissue‐ and expression level‐specific chromatin looping at maize b1 epialleles. Plant Cell 21: 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Marand AP, Ricci WA, Ethridge CL, Zhang X, Schmitz RJ. 2019. The prevalence, evolution and chromatin signatures of plant regulatory elements. Nature Plants 5: 1250–1259. [DOI] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene‐enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Fransz PF, Ali HB, Schubert I. 2001. Chromosome painting in Arabidopsis thaliana . The Plant Journal 28: 689–697. [DOI] [PubMed] [Google Scholar]
- Ma W, Ay F, Lee C, Gulsoy G, Deng X, Cook S, Hesson J, Cavanaugh C, Ware CB, Krumm A et al. 2015. Fine‐scale chromatin interaction maps reveal the cis‐regulatory landscape of human lincRNA genes. Nature Methods 12: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Wang M, Li W, Zhang Z, Zhang X, Tan T, Zhang XE, Cui Z. 2017. Live cell imaging of single genomic loci with quantum dot‐labeled TALEs. Nature Communications 8: 15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJ, Huettel B, van der Winden J, Matzke M. 2005. Use of two‐color fluorescence‐tagged transgenes to study interphase chromosomes in living plants. Plant Physiology 139: 1586–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes MA, Guerra RF, Berns MC, Manzo C, Masiero S, Finzi L, Kater MM, Colombo L. 2013. MADS domain transcription factors mediate short‐range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 25: 2560–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares‐Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA et al. 2015. Mapping long‐range promoter contacts in human cells with high‐resolution capture Hi‐C. Nature Genetics 47: 598–606. [DOI] [PubMed] [Google Scholar]
- Montgomery SA, Tanizawa Y, Galik B, Wang N, Ito T, Mochizuki T, Akimcheva S, Bowman JL, Cognat V, Marechal‐Drouard L et al. 2020. Chromatin organization in early land plants reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Current Biology 30: 573–588.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin BH, Tickner J, Zhu K, Kenny J, Mullin S, Brown SJ, Dudbridge F, Pavlos NJ, Mocarski ES, Walsh JP et al. 2020. Characterisation of genetic regulatory effects for osteoporosis risk variants in human osteoclasts. Genome Biology 21: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. 2016. HiChIP: efficient and sensitive analysis of protein‐directed genome architecture. Nature Methods 13: 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Yamaji N. 2020. Decrosslinking enables visualization of RNA‐guided endonuclease‐in situ labeling signals for DNA sequences in plant tissues. Journal of Experimental Botany 71: 1792–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. 2013. Single‐cell Hi‐C reveals cell‐to‐cell variability in chromosome structure. Nature 502: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DW, Miller M, Yu HH, Huang TY, Kim ED, Lu J, Xie Q, McClung CR, Chen ZJ. 2014. A role for CHH methylation in the parent‐of‐origin effect on altered circadian rhythms and biomass heterosis in Arabidopsis intraspecific hybrids. Plant Cell 26: 2430–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, Sun Q, Chen ZJ. 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi J, Fukui K. 1995. Chromatin arrangements in intact interphase nuclei examined by laser confocal microscopy. Journal of Plant Research 108: 209–216. [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J et al. 2012. Spatial partitioning of the regulatory landscape of the X‐inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann HW, Doerr D, Ramirez‐Colmenero A, Sotelo‐Fonseca JE, Wegel E, Di Stefano M, Wingett SW, Fraser P, Hurst L, Fernandez‐Valverde SL et al. 2020. Active and repressed biosynthetic gene clusters have spatially distinct chromosome states. Proceedings of the National Academy of Sciences, USA 117: 13800–13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka R, Zicola J, Weber B, Anderson SN, Hodgman C, Gent JI, Wesselink JJ, Springer NM, Hoefsloot HCJ, Turck F et al. 2017. Genome‐wide mapping of transcriptional enhancer candidates using DNA and chromatin features in maize. Genome Biology 18: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O'Shea CC. 2017. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357: eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Xiao Q, Li G, Li X. 2021. Technologies for capturing 3D genome architecture in plants. Trends in Plant Science 26: 196–197. [DOI] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I. 2004. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR‐bearing chromosomes. Chromosoma 113: 258–269. [DOI] [PubMed] [Google Scholar]
- Peng Y, Xiong D, Zhao L, Ouyang W, Wang S, Sun J, Zhang Q, Guan P, Xie L, Li W et al. 2019. Chromatin interaction maps reveal genetic regulation for quantitative traits in maize. Nature Communications 10: 2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips‐Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. 1988. Fluorescence in situ hybridization with human chromosome‐specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proceedings of the National Academy of Sciences, USA 85: 9138–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet A, Duc C, Voisin M, Desset S, Tutois S, Vanrobays E, Benoit M, Evans DE, Probst AV, Tatout C. 2017. The LINC complex contributes to heterochromatin organisation and transcriptional gene silencing in plants. Journal of Cell Science 130: 590–601. [DOI] [PubMed] [Google Scholar]
- Prieto P, Santos AP, Moore G, Shaw P. 2004. Chromosomes associate premeiotically and in xylem vessel cells via their telomeres and centromeres in diploid rice (Oryza sativa). Chromosoma 112: 300–307. [DOI] [PubMed] [Google Scholar]
- Probst AV. 2018. A compendium of methods to analyze the spatial organization of plant chromatin. Methods in Molecular Biology 1675: 397–418. [DOI] [PubMed] [Google Scholar]
- Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M. 2017. Live cell imaging of low‐ and non‐repetitive chromosome loci using CRISPR‐Cas9. Nature Communications 8: 14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y et al. 2018. Higher‐order inter‐chromosomal Hubs Shape 3D Genome Organization in the nucleus. Cell 174: 744–757.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. 1885. Uber Zelltheilung. Morphologisches Jahrbuch 10: 214–330. [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redolfi J, Zhan Y, Valdes‐Quezada C, Kryzhanovska M, Guerreiro I, Iesmantavicius V, Pollex T, Grand RS, Mulugeta E, Kind J et al. 2019. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nature Structural & Molecular Biology 26: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Deng L, Xue Y, Suzuki K, Zhang W, Yu Y, Wu J, Sun L, Gong X, Luan H et al. 2017. Visualization of aging‐associated chromatin alterations with an engineered TALE system. Cell Research 27: 483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejia‐Guerra MK, Colome‐Tatche M et al. 2019. Widespread long‐range cis‐regulatory elements in the maize genome. Nature Plants 5: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Granados NY, Ramirez‐Prado JS, Veluchamy A, Latrasse D, Raynaud C, Crespi M, Ariel F, Benhamed M. 2016. Put your 3D glasses on: plant chromatin is on show. Journal of Experimental Botany 67: 3205–3221. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. 2006. Sub‐diffraction‐limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 3: 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutowicz K, Lirski M, Mermaz B, Teano G, Schubert J, Mestiri I, Kroten MA, Fabrice TN, Fritz S, Grob S et al. 2019. Linker histones are fine‐scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biology 20: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi PA. 2011. Light sheet fluorescence microscopy: a review. Journal of Histochemistry & Cytochemistry 59: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AP, Ferreira L, Maroco J, Oliveira MM. 2011. Abiotic stress and induced DNA hypomethylation cause interphase chromatin structural changes in rice rDNA loci. Cytogenetic and Genome Research 132: 297–303. [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Wolff P, Santos‐Gonzalez J, Schoft V, Siretskiy A, Scott R, Tamaru H, Kohler C. 2014. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell 26: 3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert I, Fransz PF, Fuchs J, de Jong JH. 2001. Chromosome painting in plants. Methods in Cell Science 23: 57–69. [PubMed] [Google Scholar]
- Schubert I, Shaw P. 2011. Organization and dynamics of plant interphase chromosomes. Trends in Plant Science 16: 273–281. [DOI] [PubMed] [Google Scholar]
- Schubert V. 2017. Super‐resolution microscopy – applications in plant cell research. Frontiers in Plant Science 8: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V, Berr A, Meister A. 2012. Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness. Chromosoma 121: 369–387. [DOI] [PubMed] [Google Scholar]
- Schubert V, Rudnik R, Schubert I. 2014. Chromatin associations in Arabidopsis interphase nuclei. Frontiers in Genetics 5: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzacher T, Anamthawat‐Jonsson K, Harrison GE, Islam AK, Jia JZ, King IP, Leitch AR, Miller TE, Reader SM, Rogers WJ et al. 1992. Genomic in situ hybridization to identify alien chromosomes and chromosome segments in wheat. Theoretical and Applied Genetics 84: 778–786. [DOI] [PubMed] [Google Scholar]
- Sequeira‐Mendes J, Gutierrez C. 2016. Genome architecture: from linear organisation of chromatin to the 3D assembly in the nucleus. Chromosoma 125: 455–469. [DOI] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three‐dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C. 2013. Chromatin reprogramming during the somatic‐to‐reproductive cell fate transition in plants. Development 140: 4008–4019. [DOI] [PubMed] [Google Scholar]
- Shimbo T, Kawamura M, Wijaya E, Takaki E, Kaneda Y, Tamai K. 2019. Cut‐C: cleavage under tethered nuclease for conformational capture. BMC Genomics 20: 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. 2019. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 176: 1518. [DOI] [PubMed] [Google Scholar]
- Sijacic P, Bajic M, McKinney EC, Meagher RB, Deal RB. 2018. Changes in chromatin accessibility between Arabidopsis stem cells and mesophyll cells illuminate cell type‐specific transcription factor networks. The Plant Journal 94: 215–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture‐on‐chip (4C). Nature Genetics 38: 1348–1354. [DOI] [PubMed] [Google Scholar]
- Smith S, Grima R. 2018. Single‐cell variability in multicellular organisms. Nature Communications 9: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GS, Zhai HL, Peng YG, Zhang L, Wei G, Chen XY, Xiao YG, Wang L, Chen YJ, Wu B et al. 2010. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super‐hybrid rice. Molecular Plant 3: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo‐Silveira M, Chávez Montes RA, Sotelo‐Silveira JR, Marsch‐Martínez N, de Folter S. 2018. Entering the next dimension: plant genomes in 3D. Trends in Plant Science 23: 598–612. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Stadhouders R, Filion GJ, Graf T. 2019. Transcription factors and 3D genome conformation in cell‐fate decisions. Nature 569: 345–354. [DOI] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, Zwickl DJ, Zhang L, Zhang C, Chougule K, Gao D, Iwata A, Goicoechea JL et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nature Genetics 50: 285–296. [DOI] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, Lee SF, Leeb M, Wohlfahrt KJ, Boucher W, O'Shaughnessy‐Kirwan A et al. 2017. 3D structures of individual mammalian genomes studied by single‐cell Hi‐C. Nature 544: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM. 2007. Nonadditive expression and parent‐of‐origin effects identified by microarray and allele‐specific expression profiling of maize endosperm. Plant Physiology 145: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Thompson A, Sandstrom R, Thurman RE, Neph S, Johnson AK, Sullivan ST, Sabo PJ, Neri FV 3rd et al. 2019. Mapping and dynamics of regulatory DNA in maturing Arabidopsis thaliana siliques. Frontiers in Plant Science 10: 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, He N, Niu L, Huang Y, Shen W, Zhang Y, Li L, Hou C. 2019. Global quantitative mapping of enhancers in rice by STARR‐seq. Genomics, Proteomics & Bioinformatics 17: 140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Jing Y, Liu X, Li Q, Xue Z, Cheng Z, Wang D, He H, Qian W. 2020a. Heat stress‐induced transposon activation correlates with 3D chromatin organization rearrangement in Arabidopsis. Nature Communications 11: 1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dong L, Zhang Y, Lin D, Xu W, Ke C, Han L, Deng L, Li G, Jackson D et al. 2020b. 3D genome architecture coordinates trans and cis regulation of differentially expressed ear and tassel genes in maize. Genome Biology 21: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M et al. 2018. TADs are 3D structural units of higher‐order chromosome organization in Drosophila. Science Advances 4: eaar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szinay D, Bai Y, Visser R, de Jong H. 2010. FISH applications for genomics and plant breeding strategies in tomato and other solanaceous crops. Cytogenetic and Genome Research 129: 199–210. [DOI] [PubMed] [Google Scholar]
- Tan L, Xing D, Chang CH, Li H, Xie XS. 2018. Three‐dimensional genome structures of single diploid human cells. Science 361: 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B et al. 2015. CTCF‐mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, Schulkes RK, van Driel R, Fransz P. 2007. Light‐regulated large‐scale reorganization of chromatin during the floral transition in Arabidopsis. The Plant Journal 50: 848–857. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. 2015. Genome‐wide analysis of local chromatin packing in Arabidopsis thaliana . Genome Research 25: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li S, Li Y, Xu Y, Wang Y, Zhang R, Sun W, Chen Q, Wang XJ, Li C et al. 2019. MED25 connects enhancer‐promoter looping and MYC2‐dependent activation of jasmonate signalling. Nature Plants 5: 616–625. [DOI] [PubMed] [Google Scholar]
- Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L et al. 2017. Asymmetric subgenome selection and cis‐regulatory divergence during cotton domestication. Nature Genetics 49: 579–587. [DOI] [PubMed] [Google Scholar]
- Wang M, Wang P, Tu L, Zhu S, Zhang L, Li Z, Zhang Q, Yuan D, Zhang X. 2016a. Multi‐omics maps of cotton fibre reveal epigenetic basis for staged single‐cell differentiation. Nucleic Acids Research 44: 4067–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang P, Lin M, Ye Z, Li G, Tu L, Shen C, Li J, Yang Q, Zhang X. 2018a. Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nature Plants 4: 90–97. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sun Q, Czajkowsky DM, Shao Z. 2018b. Sub‐kb Hi‐C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nature Communications 9: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su JH, Beliveau BJ, Bintu B, Moffitt JR, Wu CT, Zhuang X. 2016b. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G, Tyler KM. 2011. Widefield microscopy for live imaging of lipid domains and membrane dynamics. Biochimica et Biophysica Acta 1808: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre‐Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D et al. 2016. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proceedings of the National Academy of Sciences, USA 111: 5283–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Tian ZQ, Zhang ZL, Huang BH, Jiang P, Xie ZX, Pang DW. 2010. Direct fluorescence in situ hybridization (FISH) in Escherichia coli with a target‐specific quantum dot‐based molecular beacon. Biosensors & Bioelectronics 26: 491–496. [DOI] [PubMed] [Google Scholar]
- Xie T, Zhang FG, Zhang HY, Wang XT, Hu JH, Wu XM. 2019. Biased gene retention during diploidization in Brassica linked to three‐dimensional genome organization. Nature Plants 5: 822–832. [DOI] [PubMed] [Google Scholar]
- Xiong Z, Gaeta RT, Pires JC. 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus . Proceedings of the National Academy of Sciences, USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Mariani L, Barozzi I, Schulz EG, Bluthgen N, Stadler M, Tiana G, Giorgetti L. 2017. Reciprocal insulation analysis of Hi‐C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Research 27: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lupski JR. 2015. Non‐coding genetic variants in human disease. Human Molecular Genetics 24: R102–R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zheng R, Wang Y, Zhang Y, Hong P, Fang Y, Li G, Fang Y. 2019a. The effects of Arabidopsis genome duplication on the chromatin organization and transcriptional regulation. Nucleic Acids Research 47: 7857–7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng C, Su H, Liu Y, Liu Y, Han F. 2020. The Cohesin complex subunit ZmSMC3 participates in meiotic centromere pairing in maize. Plant Cell 32: 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu Y, Hu R, Lee AY et al. 2019b. Transcriptionally active HERV‐H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nature Genetics 51: 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wang S, Cao Z, Ouyang W, Zhang Q, Xie L, Zheng R, Guo M, Ma M, Hu Z et al. 2019. Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nature Communications 10: 3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Tian SZ, Capurso D, Kim M, Maurya R, Lee B, Piecuch E, Gong L, Zhu JJ, Li Z et al. 2019. Multiplex chromatin interactions with single‐molecule precision. Nature 566: 558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Jiang W, Zhao Y, Zhou DX. 2019. Single‐cell three‐dimensional genome structures of rice gametes and unicellular zygotes. Nature Plants 5: 795–800. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhang W, Zhang T, Liu B, Jiang J. 2015. Genome‐wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27: 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Hu B, Becker C, Dogan ES, Berendzen KW, Weigel D, Liu C. 2017. Altered chromatin compaction and histone methylation drive non‐additive gene expression in an interspecific Arabidopsis hybrid. Genome Biology 18: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


