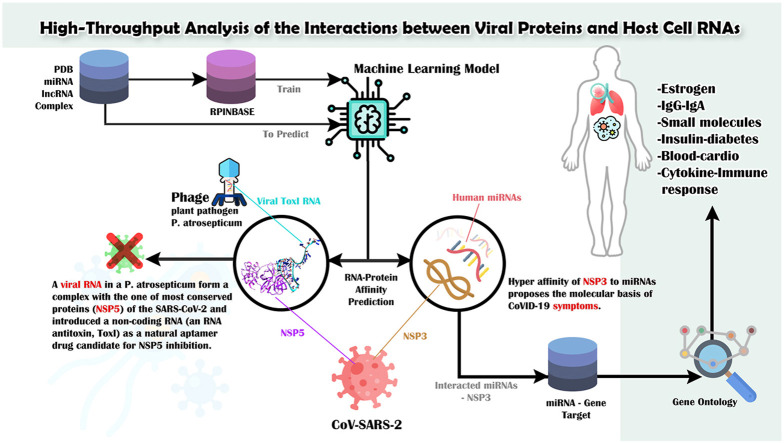
Abstract
RNA-protein interactions of a virus play a major role in the replication of RNA viruses. The replication and transcription of these viruses take place in the cytoplasm of the host cell; hence, there is a probability for the host RNA-viral protein and viral RNA-host protein interactions. The current study applies a high-throughput computational approach, including feature extraction and machine learning methods, to predict the affinity of protein sequences of ten viruses to three categories of RNA sequences. These categories include RNAs involved in the protein-RNA complexes stored in the RCSB database, the human miRNAs deposited at the mirBase database, and the lncRNA deposited in the LNCipedia database. The results show that evolution not only tries to conserve key viral proteins involved in the replication and transcription but also prunes their interaction capability. These proteins with specific interactions do not perturb the host cell through undesired interactions. On the other hand, the hypermutation rate of NSP3 is related to its affinity to host cell RNAs. The Gene Ontology (GO) analysis of the miRNA with affiliation to NSP3 suggests that these miRNAs show strongly significantly enriched GO terms related to the known symptoms of COVID-19. Docking and MD simulation study of the obtained miRNA through high-throughput analysis suggest a non-coding RNA (an RNA antitoxin, ToxI) as a natural aptamer drug candidate for NSP5 inhibition. Finally, a significant interplay of the host RNA-viral protein in the host cell can disrupt the host cell's system by influencing the RNA-dependent processes of the host cells, such as a differential expression in RNA. Furthermore, our results are useful to identify the side effects of mRNA-based vaccines, many of which are caused by the off-label interactions with the human lncRNAs.
Keywords: Host cell RNA, Viral nonstructural proteins (NSP), RNA-Protein affinity, RPINBASE, COVID-19
Graphical abstract
1. Introduction
Coronaviruses, including SARS-CoV-2 with a genome size of 30 kilobases (kb) in length, possess the largest known RNA genomes [1,2]. The structural proteins of the coronavirus are spike (S), membrane (M), envelope (E), and nucleocapsid (N) [2]. Additionally, the polyproteins of pp1a and pp1ab are encoded by two long open reading frames (ORFs), i.e., ORF1a and ORF1b. Proteins of ORF1a and ORF1ab may be involved in cellular signaling and the modification of cellular gene expression, as well as pathogenicity by mechanisms yet to be exactly determined [3]. Papain-like protease (PLpro) and 3C-like protease (3CLpro) cleave the ORF1ab polyprotein into 15–16 non-structural proteins (NSPs) [4,5]. These proteins are required for intracellular virus replication, and they play important roles in virus pathogenesis and its virulence [[6], [7], [8]]. In many viruses, the cleavage of large polyproteins by viral and/or cellular proteases is a strategy for regulating virus replication, gene expression, and maturation [9].
In the SARS-CoV life cycle, replication and transcription are mediated by a replication transcription complex (RTC). RTC drives viral genome replication and subgenomic mRNA synthesis [10]. Although the central role of NSP3 is not well recognized, it is reported that some domains of NSP3 interact with NSP5, NSP6, NSP12, NSP13, NSP14, and NSP16. Consequently, NSP3 may serve as one of the RTC scaffolding proteins there is a hypothesis of a membrane-associated scaffolding function for NSP3 in the infected cells [11]. NSP12 has little activity and its functions require accessory factors, including NSP7 and NSP8 [4,5]. According to Yin et al., the presence of NSP7 and NSP8 significantly increases NSP12 binding to the template-primer RNA. Moreover, the NSP12-NSP7-NSP8 complex shows RNA polymerization activity on a poly-U template upon the addition of adenosine triphosphate (ATP). The partial double-stranded RNA template inserts into the central channel of the RdRp in a way that it lies in the active site cleft of NSP12 [12,13]. NSP3 is the largest multi-domain protein produced by coronaviruses [[12], [13], [14]], which significantly differs between SARS-CoV and SARS-CoV-2. However, the most similar proteins are the RNA helicases (NSP 13), which are identical in all except one of their 603 amino acids; the RNA-dependent RNA polymerases (NSP 12), sharing all except 34 of 955 amino acids; and the primary protease (NSP 5), which is similar in SARS-CoV-1 and SARS-CoV-2 with only 13 different amino acids among 306 ones [15].
The replication and transcription of viruses take place in the host cell's cytoplasm; hence, it is logical to consider the probability of the host RNA-viral protein and viral RNA-host protein interactions. These types of host-virus interplay can be a pathogenic factor or a noise in the virus's vital mechanism. The interactions of viral proteins, especially the open reading frame-encoded NSPs, with host cell proteins, and the interactions of viral protein-RNA have previously been studied [[16], [17], [18], [19]]. Molecular dynamics (MD)-based methods are popular methods to investigate the protein-protein or protein-RNA interactions; however, these methods are computationally-costly simulations.
Hatton et al. have presented a model for the role of RNA and DNA-binding activities in Hepatitis B Virus (HBV) replication. As they note, the capsid protein of the virus adversely affects the reverse transcription of the viral pre-genome RNA into DNA [20]. Moreover, Johnson et al. have proposed another model for Cowpea chlorotic mottle viruses (CCMV), in which the virus RNA's interaction with the capsid protein alters the RNA structure and the pathway for in vitro assembly [21]; hence, the virus RNA-protein interactions can facilitate the virus replication process. Recently, the physical interactions of host cell lncRNAs with the SARS-CoV-2 genome (Viral RNA) have been investigated by Moazzam et al. [22].
Therefore, viral proteins’ affinity toward different classes of host RNA molecules can be one of the missing links in the molecular mechanisms associated with the pathogenesis of the viruses, especially the ongoing COVID-19 outbreak.
In this study, we focused on the interplay between the host RNAs and the virus proteins. For the first time, through high-throughput data analysis, pathway analysis, docking, and MD, we have investigated the interactions of host cell RNAs and viral proteins, as the less studied case in viral pathogenesis. We generated a predictive model using machine learning from RNA-protein complexes in the PDB (RCSB.org) database. Moreover, we predicted all interactions between the proteins of ten viruses and three categories of human RNA (i.e., complexes’ RNA, microRNA, and long non-coding RNA). Our results indicate the high potential of some NSPs to recognize human RNAs. Therefore, NSPs can play a major role during the evolution of coronaviruses and provide clues for understanding the coronavirus transcription and replication machinery.
Finally, our results describe the role of host RNA-viral protein interactions and propose that differential gene expression may not be the only process that affects the RNA system in the host cell. Thus, these findings can be beneficial in proposing the molecular basis of COVID-19 symptoms and the role of the viral proteins with a hypermutation rate in the infection. Moreover, docking and MD can help drug discovery through miRNAs as the natural candidate for the inhibition of viral proteins.
2. Methods and materials
In this study, a computational approach was applied to predict the affinity of proteins encoded by six genera of coronaviruses to three categories of RNA sequences, including RNAs involved in the protein-RNA complexes stored in the RCSB database, the human miRNAs deposited at the mirBase database, and the lncRNA deposited at the LNCipedia database. Moreover, two genera of influenza and hepatic viruses were also considered as control groups.
Here, three categories of RNAs were examined, i.e., (1) RNAs involved in the protein-RNA complexes stored in the RCSB database, (2) human miRNAs deposited at the mirBase database, and (3) the lncRNA deposited at the LNCipedia database.
The list of viruses and RNAs used in this study is presented in Table 1 , and more details about the viruses are available in Table 2 and the complete used sequences of the genomes of these viruses are presented in Supplementary file 6. However, as a limitation of the study, we did not consider the variations of the genomes; and ignored all of the mutations which cause variants among RNA virus sequences. The flowchart of our methodology is illustrated in Fig. 1 .
Table 1.
Evaluated and utilized data materials.
| Title | Type | Count | |
|---|---|---|---|
| 1 | Viruses | virus | 10 |
| 2 | Viral protein sequences | Protein | 177 |
| 3 | RNAs involved in RNA-proteins complexes | RNA | 1682 |
| 4 | Micro RNA (homo sapiens) | RNA | 2465 |
| 5 | Long non-coding RNA | RNA | 102405 |
| 6 | Positive samples | RNA-Protein Complex | 9401 |
| 7 | Negative samples | RNA-Protein Complex | 94010 |
Table 2.
The key molecular features of viruses used in this study [23]. Human Coronavirus HCoV is the abbreviation for “Human Coronavirus” and SARS-CoV-1 is the abbreviation for SARS coronavirus. H1N1 and H5N1 are influenzas A virus subtypes.
| Virus | Genome length (kbp) | Proteins count | Incubation (median) | |
|---|---|---|---|---|
| 1 | HCoV- 229E | 27.3 | 20 | 3 |
| 2 | SARS-CoV-2 | 30 | 25 | 8 |
| 3 | H1N1 | 13.6 | 12 | 1.5 |
| 4 | H5N1 | 13.6 | 12 | 4 |
| 5 | Hepatitis B Virus (HBV) | 3.2 | 7 | 115 |
| 6 | Hepatitis C Virus (HCV) | 9.4 | 10 | 87.5 |
| 7 | MERS-CoV | 30 | 24 | 8 |
| 8 | HCoV- NL63 | 28 | 20 | 4.6 |
| 9 | HCoV- OC43 | 30 | 22 | 3 |
| 10 | SARS-CoV-1 | 30 | 24 | 6 |
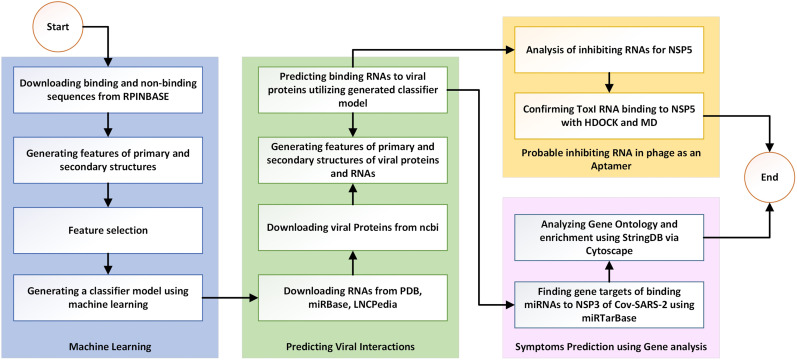
Fig. 1.
Flowchart of the methodology used in this study, including the collection of RNA and protein data, generating the dataset, training a predictive model, and the prediction steps.
2.1. Data collection
RPINBASE [24] provides positive samples using atomic distance and negative samples using the clan and family theory of proteins. Preprocessed sequences of the RNA involved in the RNA-protein complex PDB (RNA sequences involved in PDB complexes in the RCSB database) with the values of their features were downloaded from RPINBASE, which contains 1682 unique RNA sequences from 5076 RNA sequences in 2258 complexes. To train a predictor model, 9401 positive and 94010 negative samples with their feature vectors were downloaded from RPINBASE.
Moreover, 2465 unique microRNA sequences of homo sapiens were kept among 48886 sequences downloaded from miRBase (release 22) [[25], [26], [27]]. The high-confidence set (version 5.2) of lncRNA sequences was downloaded from LNCipedia with 107346 initial and 102405 unique RNA sequences with less than 5000 nucleotides [28]. In addition, 1682 RNA sequences were reused as RNAs involved in complexes. All 177 sequences of ten viral proteins were downloaded from NCBI in the FASTA format.
To evaluate the potential side effects of RNA binding to human proteins, 74823 protein sequences were downloaded from uniport [29]. By applying filters such as sequence length in the range between 20 and 4000 amino acids and omitting sequences containing unknown amino acids, 66025 appropriate sequences were kept.
2.2. Feature extraction
The primary and secondary structures of the RNA and proteins were targeted to generate a wide range of features for machine learning, as presented in Table 3 . Furthermore, hybrid features were generated using both structures. The aggregation includes count, average, minimum, maximum, and sum functions on the sets. The sequential primary structure features were generated by recoding and optimizing the “protein-encoding toolbox” [30]. Features of the secondary structure of RNAs were generated by interpreting the dot-bracket output of RNAfold of the Vienna package [31]. Following the method proposed in Torkamanian-Afshar et al. [32], the top 196 structural features of RNAs and proteins are selected. Their method uniformly picks some samples from the original dataset. It sorts features by their discriminatory power in descending order as well. Then, the method calculates the standard deviation of their ranks. Finally, the features corresponding to the highest discriminatory powers and the lowest rank deviations are retained for the training set. To evaluate the binding affinity, the RNA-protein pair dataset was generated. The secondary structures of viral proteins were generated using GOR IV [33,34], i.e., a tool for predicting the secondary structure of proteins.
Table 3.
Feature vector composition.
| Target | Length | Description |
|---|---|---|
| Protein Primary | 2010 | AAC, AAP, APAAC, CTDC, CTDD, CTriad, DC, Geary, Moran, MoreauBroto, PAAC, QSO, SOCN |
| Protein Secondary | 30 | Aggregation (AlphaHelix, BetaSheet, Coil), Parallel and Antiparallel beta-sheets |
| RNA Secondary | 44 | Aggregation (Stem, Bulge, Internal loop, Hairpin, Multiloop, Single strand) |
| RNA Hybrid | 100 | Monomer, dimer, and trimer in stems and loops |
| Total Length | 2184 |
2.3. Machine learning
In the machine learning approach, the process of training a model and the prediction of protein-interacting RNAs includes two main steps. To perform the first step and build a model, the ML.NET framework [35] was used to train and generate the model. To train a predictor model, 9401 positive and 94010 negative samples with their feature vectors were downloaded from RPINBASE [24]. Due to the lack of reported negative samples, the imbalanced training dataset was generated, and the misclassification cost parameter was set to 10 to compensate for the inequality. The RPINBASE, built from raw sequences of RNAs and proteins deposited at the NCBI database, contains preprocessed and ready-to-learn sequences of complexes with a wide range of primary and secondary structural features. In the cross-validation procedures of the training, the best accuracy of 96.9% with the F1 score of 97% was obtained using the LightGbmBinaryTrainer (Light Gradient Boosted Machine Binary Trainer) algorithm [36]. Moreover, the Light Gradient Boosted Machine classifier algorithm was used to predict the binding score for each RNA-protein pair.
2.4. Prediction
The trained model receives an RNA-protein feature vector and outputs a scalar discrimination score between 0 and 1 as a prediction result, where 0 represents the negative class and 1 is for the positive class. In these kinds of binary classifications, a score less than 0.5 for an RNA-protein pair is interpreted as a non-binding pair (negative) and scores greater than 0.5 are considered as binding pairs (positive). While a lower predicted score means a lower binding chance, a higher binding chance can be understood from the higher scores.
2.5. RNA-protein docking and molecular dynamics
We examined all the predicted RNAs that interact with the most conserved proteins of SARS-CoV-2 (i.e., NSP5, NSP12, and NSP13) to identify a putative inhibitor for these key proteins. Then, to evaluate the capacity of this RNA to be introduced as a drug or at least as a seed for aptamer design, we examined the details of the interactions of this RNA by simulating the RNA-protein docking, followed by molecular dynamic (MD) simulation.
The docking simulation was performed using default parameters on the Haddock server available at https://haddock.science.uu.nl/ [37]. The 3D structures of ToxI and COVID-19 main protease were found from the PDB files 2XDB and 7BUY, respectively, which were obtained from the RCSB database [38]. Finally, we ran the MD simulation by implementing the amber14sb_OL15 force filed, 2000 STEP EM (emtol = 0.001 and emstep = 0.01), 200000 NVT steps and 200000 NPT steps both with dt = 1 fs, and, finally, 100 ns MD with dt = 2 fs in Gromacs-2019 software [39,40]. All figures of the structures and complexes have been created using the UCSF Chimera software [41].
2.6. Gene ontology analysis
To determine the contingency pathways that may be perturbed by this RNA in the human body, the gene ontology processes were performed using String DB and the Cytoscape software. Due to the limit of 2000 proteins in String DB, all gene ontology processes were performed using the String DB [42] plugin in the Cytoscape [43] application. Moreover, miRNAs target genes were extracted from the mirTar [44] database.
3. Results
3.1. Higher affinity of Coronavirus proteins to the RNAs
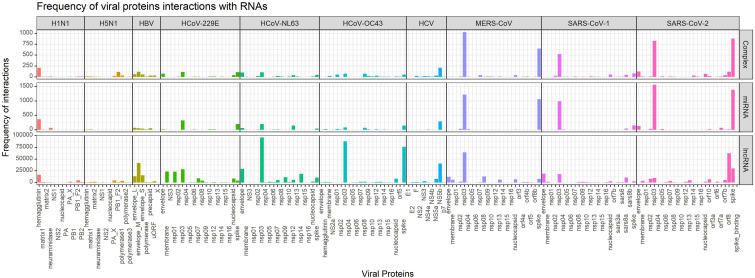
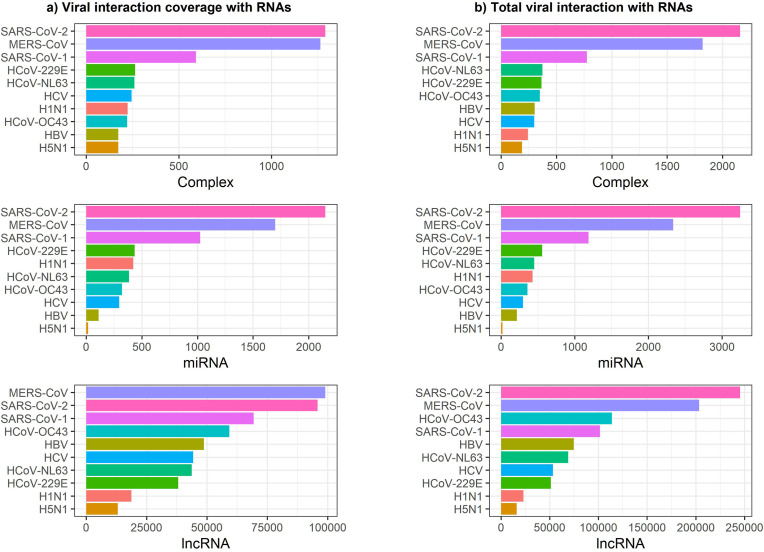
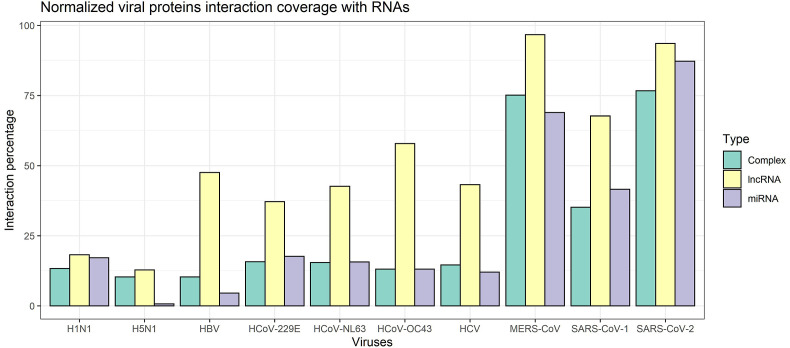
Fig. 4 shows the predicted numbers of interactions of all proteins of the ten viruses with RNAs involved in complexes (abbreviated to Complex), miRNAs, and lncRNAs. Fig. 2 -a presents the number of RNAs that interact with at least one protein of the virus, i.e., the “coverage” of a protein. The total count of interactions between any RNA with all the proteins is given in Fig. 2-b. The numerical values are presented in Supplementary File 1. Fig. 3 depicts the normalized viral proteins interaction coverage with RNAs demonstrating the percentage of total RNAs (investigated in this study) that interact with virus proteins. The predicted frequencies of interactions of all proteins of all examined viruses are presented in Fig. 4.
Fig. 4.
Viral proteins bound to RNAs involved in complexes (abbreviated to Complex), miRNAs, and lncRNAs.
Fig. 2.
Distribution of viral proteins' interactions with RNAs for each virus: a) coverage of viral proteins interacting with RNAs (at least one interaction between any viral protein and RNAs), b) total interactions between all viral proteins of any virus with RNAs.
Fig. 3.
Normalized viral proteins interaction coverage with RNAs demonstrating the percentage of total interactions of viruses with all the investigated categories of RNAs.
3.2. Hyper affinities of NSP3 of coronaviruses to the RNAs
As this study focuses on SARS-CoV-2, the exact frequency of interactions of this virus is presented in Table 4 . Interaction matrices are available in Supplementary Files 2, 3, and 4.
Table 4.
Proteins of SARS-CoV-2 and their interactions with RNAs (sorted by interactions frequency with lncRNAs).
| Protein | Len | Complex | miRNA | lncRNA | Protein | Len | Complex | miRNA | lncRNA |
|---|---|---|---|---|---|---|---|---|---|
| nsp03 | 1945 | 830 | 1565 | 88118 | nsp06 | 290 | 0 | 0 | 558 |
| orf8 | 121 | 116 | 12 | 62405 | nsp15 | 346 | 6 | 0 | 478 |
| spike | 1273 | 879 | 1388 | 29750 | orf7a | 121 | 6 | 64 | 12 |
| nsp14 | 527 | 29 | 0 | 18537 | nsp02 | 638 | 6 | 35 | 37 |
| envelope | 75 | 121 | 133 | 12120 | nsp13 | 601 | 1 | 0 | 58 |
| nucleocapid | 419 | 70 | 0 | 8495 | nsp10 | 139 | 1 | 0 | 50 |
| orf10 | 38 | 15 | 19 | 6629 | nsp04 | 500 | 1 | 0 | 18 |
| membrane | 222 | 6 | 0 | 5523 | nsp09 | 113 | 0 | 0 | 13 |
| orf7b | 43 | 40 | 4 | 5256 | orf3a | 275 | 0 | 0 | 11 |
| nsp07 | 83 | 10 | 0 | 3167 | nsp12 | 932 | 1 | 0 | 8 |
| nsp05 | 306 | 13 | 25 | 1615 | spike binding | 223 | 1 | 0 | 2 |
| orf6 | 61 | 4 | 1 | 1531 | nsp16 | 298 | 0 | 0 | 2 |
| nsp08 | 198 | 2 | 0 | 1148 | nsp01 | 180 | 0 | 0 | 0 |
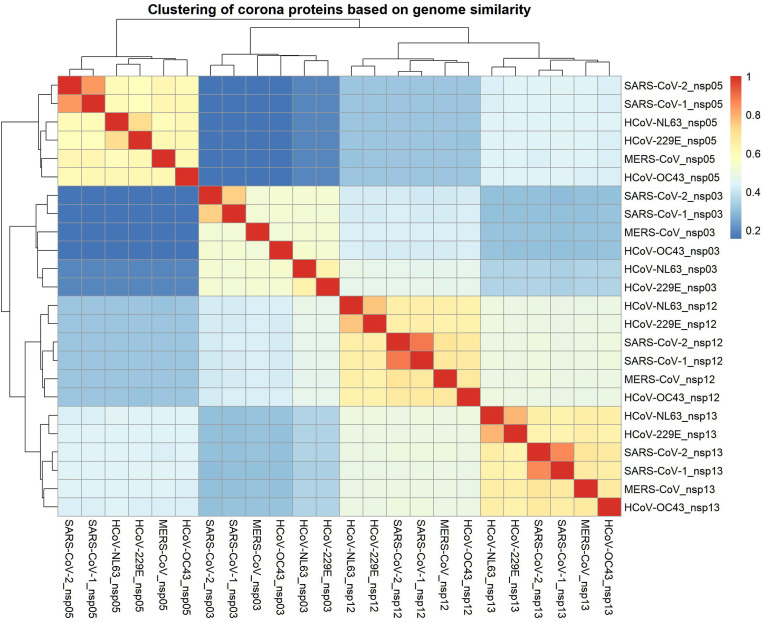
In Fig. 5 , the pairwise similarity matrix of NSP3, NSP5, NSP12, and NSP13 of coronaviruses (HCoV-229E, SARS-CoV-2, MERS-CoV, HCoV-NL63, HCoV-OC43, and SARS-CoV-1) are calculated by the Levenshtein alignment with an equal mismatch penalty of −1. The same NSPs of different genera make a block of the represented matrix; hence, the matrix is blocked. The gradual change from dark blue to red shows the increase in the similarity score. Clearly, the block related to NSP3 shows the lowest similarity. In contrast, the NSP13 block represents the highest similarity.
Fig. 5.
Clustering and correlation matrix of four proteins (NSP3, NSP5, NSP12, and NSP13) of coronaviruses (HCoV-229E, SARS-CoV-2, MERS-CoV, HCoV-NL63, HCoV-OC43, and SARS-CoV-1) based on Levenshtein similarity of primary structures of proteins.
3.3. Hypermutation rate of NSP3 of coronaviruses to the RNAs
As the similarity is the blocked matrix, we calculated the upper triangular averages of these similarity values for the corresponding blocks of each protein. The obtained values are 0.31, 0.52, 0.66, and 0.68 for the blocks related to NSP3, NSP5, NSP12, and NSP13, respectively. A lower value of the average similarity means lower mutual similarities between the same NSPs in various viruses and vice versa.
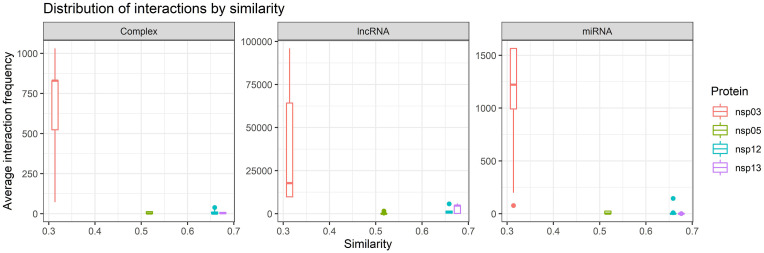
Fig. 6 illustrates the correlation of average similarities with the predicted interaction number for each NSP (percentage of the interacted number to the total number of the RNAs). NSP12 and NSP13, as the most conserved proteins (with the highest similarity), have the lowest percentage of interaction. However, the proteins with the highest percentage of predicted partners (NSP3s) have the lowest average similarity score.
Fig. 6.
Average upper triangular similarity values corresponding to NSP3, NSP5, NSP12, and NSP13 vs. the predicted interaction numbers of each NSP. The obtained similarity values are 0.31, 0.52, 0.66, and 0.68 for the blocks related to NSP3, NSP5, NSP12, and NSP13, respectively.
3.4. SARS-CoV-2 NSP5 inhibited by a viral miRNA
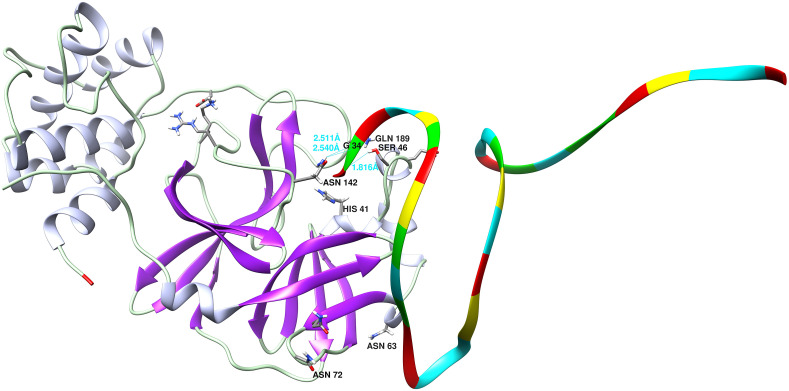
After inspecting all RNAs interacting with proteins of SARS-CoV-2, we selected the shortest non-human RNA to simulate its interaction with the NSP5; surprisingly, it was an antitoxin RNA. ToxI (chain G of PDB 2XDB) was predicted to bind to NSP5. Residues His41 & Cys145, and nucleic acids 2 & 16 of the RNA were considered as the active site of the protein and RNA respectively. For both molecules, the passive residues were selected automatically. Moreover, the total length of RNA was set to be fully flexible. All other parameters were considered as the default parameters of the Haddock2.4 server. The output of the simulation included Haddock score = −53.1+-7.3, z-score = −1.8, and cluster size of 132 from 169 total structures. The 3D structure of the complex obtained by the Haddock server after 100 ns MD simulation is presented in Fig. 7 . The 3D structure of NSP5 is shown in pink & gray and the nucleic acids of RNA are shown in cyan(U), yellow(C), red(A), and green(G). Residues of the catalytic dyad are labeled with His41/Cys145 The rmsd plot of MD simulation is shown in Supplementary File 7.
Fig. 7.
The 3D structure of NSP5 is shown in pink & gray and the nucleic acids of RNA are shown in cyan(U), yellow(C), red(A), and green(G). Residues of His41/Cys145 are catalytic dyad. The hydrogen bonds between guanine34 and Asn142 & Ser46 are clear. This figure was produced by the UCSF Chimera 1.14-linux_x86_64 (https://www.cgl.ucsf.edu/chimera/download.html).
3.5. Gene ontology analysis
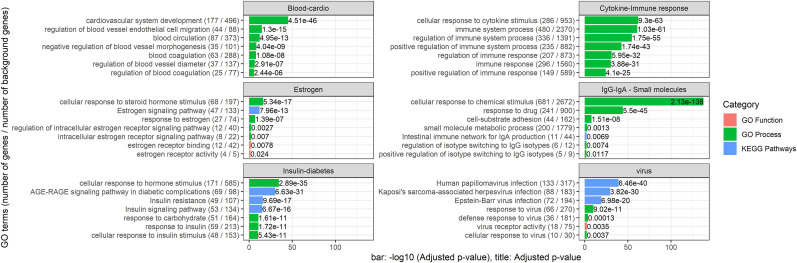
In this section, we focused on the miRNAs-NSP3 interaction and extracted the list of miRNAs that were predicted to have interaction with NSP3. Using the mirTAR database, we obtained their target in the human cells and ran gene ontology and pathway enrichment analysis. The details of the results are presented in Supplementary File 5. Fig. 8 shows the top GO terms. Some other results of the GO analysis are presented in the supplementary files.
Fig. 8.
Annotation (enriched GO terms and pathways) of the miRNA that presented affiliation to NSP3 of SARS-CoV-2.
4. Discussion
By attempting to generate a predictive machine learning model to identify the binding of the viral proteins to the host RNAs with a special emphasis on SARS-CoV-2, this study investigates the affinity of protein sequences of ten viruses to three categories of RNA sequences through computational studies. Moreover, it is addressing the important question about the role of evolutionary variation in viral proteins through a computational approach. The results can be beneficial in drug discovery and proposing a molecular basis for symptoms, especially the role of the viral proteins with a hypermutation rate in the infection.
4.1. Higher affinity of Coronavirus proteins, especially NSP3, to the RNAs
Fig. 2, Fig. 3 show that the predicted total affinities of the proteins of SARS-CoV-2 and MERS-CoV are significantly higher than those of other viruses examined in this study. However, the total predicted interaction numbers of viral proteins with different host RNA categories followed the same trends among different viruses. To examine this finding, we evaluated different proteins of the viruses one by one.
Fig. 4 shows the frequency of the predicted interaction of all proteins of the ten viruses with different categories of RNAs. It should be noted that the details of the data related to SARS-CoV-2 are presented in Table 4. Among different coronaviruses proteins, the NSP3 protein interacted with a large number of human RNA molecules, while a few RNA molecules were predicted to interact with the proteins involved in the RTC and protease. The predicted interaction numbers are independent of the lengths of the related proteins. While NSP3 has 1945 residues, NSP5 (3C-like protease, 3CLpro) and NSP12 (RNA-directed RNA polymerase) have 306 and 932 residues, respectively. It seems that it is a strategy of the virus that the key proteins in the replication, transcription, and structural formation of the virus remain intact as much as possible since these proteins facilitate the fundamental mechanisms of the viral life cycle, independent of the host cell and virus genera. Interestingly, while the receptor-binding domain of the spike protein interacts with a few human RNAs, the resting part of the spike protein has a high potential to absorb the RNAs. As shown in Table 4, the predicted number of total RNAs interacting with the spike receptor-binding domain is 3, while more than 33000 interactions were predicted for the whole spike. This again confirms our assumption that the proteins that facilitate the basic mechanism of the virus are kept away from the host cell's RNA system, and some proteins including NSP3 and spike (except for the spike receptor-binding domain) interfere with the host cell's RNAs.
4.2. Hypermutation rate of NSP3 of Coronavirus
On the other hand, NSP3 is the largest multi-domain protein produced by coronaviruses [[12], [13], [14]]. Previous studies have shown that when only 0.16% of the residues of NSP13, 3.5% of the residues of NSP12, and 4.2% of the residues of primary protease (NSP5) are different between SARS-CoV and SARS-CoV-2, this ratio is 17–26% for different domains of NSP3 [11]. In line with these findings, Fig. 5 shows that these similarities and differences can be generalized to other genera of the coronaviruses. Moreover, compared to other proteins, NSP3 of SRAS-CoV-2 has the highest distance from NSP3 of other coronavirus genera. Furthermore, the alignment of different genomes of SARS-CoV-2 shows that NSP3 has the highest rate of mutation compared to other proteins. In other words, it shows 547 missense mutations, while 3CLPro and RdRp have 67 and 194 missense mutations, respectively [45].
Fig. 6 shows the correlation of the similarity score (as an index of variation) and the predicted interaction numbers of each NSP. Interestingly, among various coronaviruses, the potential capacity of NSP for interacting with different RNA categories increased with the decrease in the sequence similarity. It seems that there is a significant relationship between the rate of interaction with RNAs and the probability of mutation in viral proteins. Additionally, not only does NSP3 play a decoy role to protect other viral proteins from the host cell's RNAs, but it also disturbs the host cell's system.
4.3. Virus against virus
Although the tendency of viral proteins for binding to the host RNA molecules can provide an evolutionary advantage for viruses, it can also be a starting point for designing virus-inhibiting aptamers that can negatively regulate the viral gene expression and its pathogenesis. Thus, we examined all NSPs-RNAs predicted interactions that can interact with the most conserved proteins of SARS-CoV-2 (i.e., NSP5, NSP12, and NSP13), and we found a miRNA (ToxI) that forms a complex with NSP5.
Small genetic elements composed of a toxin gene and its cognate antitoxin compose Toxin–antitoxin (TA) systems. Antitoxins are either proteins or non-coding RNAs that often control their cognate toxins through direct interactions, and, in conjunction with other signaling elements, through the transcriptional and translational regulation of the TA module's expression. In the cell, the antitoxin and the toxin interact with each other (ToxI–ToxN), suppressing toxicity [[46], [47], [48]]. ToxIN, encoded by a plasmid from the plant pathogen P. atrosepticum, is the first example of the type III toxin–antitoxin (TA) system, in which a protein toxin, i.e., ToxN, is inhibited by an RNA antitoxin [[46], [47], [48]]. All these results support ToxI as a potential inhibitor of NSP5, where the production of a virus (bacteriophage) can inhibit the proliferation of another virus (SARS-CoV-2).
4.4. NSP3 hyperaffinity to miRNAs and COVID-19 symptoms
According to the gene ontology and pathway analysis, it is clear that NSP3's hyperaffinity to miRNAs can influence the majority of molecular functions in the host cell that are related to COVID-19 symptoms. Here, we focused on the miRNAs-NSP3 interactions and extracted the list of miRNAs that were predicted to interact with NSP3. Using the mirTAR database, we obtained their targets in the human cells and ran gene ontology and pathway enrichment analysis. The details of the results are presented in Supplementary File 5. The meaningful GO terms and KEGG pathways were found for estrogen receptor, IgG-IgA, small molecules, cellular response to the virus, insulin-diabetes, cardio and blood, and cytokines and immune response. These perturbed functions and pathways in the host cells are related to the known outcomes of the COVID-19, indicating that patients with diabetes are at a greater risk of worse prognosis and mortality [49]. Moreover, it is shown that hypertension, obesity, and diabetes are the common comorbidities of the patients hospitalized with COVID-19 [50]. Interestingly, CXCL10, CCL7, and IL-1 receptor antagonists reported by a previous study [51] are the targets of hsa-let-7f-5p, hsa-miR-135b-3p, hsa-miR-15a-5p, hsa-miR-122-5p, hsa-miR-21-5p, hsa-miR-130a-3p, hsa-miR-142-3p, hsa-miR-877-3p, hsa-miR-346, hsa-miR-191-5p, and hsa-miR-887-3p, which were predicted by our results to interact with NSP3. We believe that the genes included in the cellular response to chemical stimulus GO process can help identify the reasons for the prevalence of smell and taste dysfunctions in COVID-19 patients [52,53].
The meaningful GO terms and KEGG pathways were identified for estrogen (receptor activity, receptor binding, and signaling pathway), IgG-IgA, small molecules (cellular response to chemical stimulus, response to drug, small molecule metabolic process), cellular response to the virus (defense response to the virus, Epstein-Barr virus infection, human papillomavirus infection, Kaposi's sarcoma-associated herpesvirus infection, modulation of host processes by the virus), insulin-diabetes (such as cellular response to glucose stimulus, insulin stimulus, low-density lipoprotein particle stimulus, insulin receptor signaling pathway, insulin receptor substrate binding, insulin resistance, insulin secretion, insulin signaling pathway, insulin-like growth factor receptor signaling pathway, negative regulation of insulin receptor signaling pathway, negative regulation of insulin secretion, regulation of insulin secretion, insulin, AGE-RAGE signaling pathway, and type II diabetes mellitus), cardio and blood (such as adrenergic signaling in cardiomyocytes, arrhythmogenic right ventricular cardiomyopathy (ARVC), blood circulation, blood coagulation, blood vessel remodeling, blood vessel morphogenesis, blood pressure, regulation of heartrate, and regulation of systemic arterial blood pressure), and cytokines and immune response.
4.5. mRNA-based vaccines
Consistent with our findings, Vandelli et al. have investigated the interactions between the SARS-CoV-2 genome and human proteins, showing that the 5′end of the viral genome is highly structured and can interact with various human proteins [54]. Here, we predicted the affinity between proteins encoded by the SARS-CoV-2 genome and human mRNA and miRNAs. Therefore, the findings can be used in mRNA-based vaccines to predict the side effects caused by the off-label interactions with these human macromolecules.
In this study, for the first time, as we know, we emphasize the role of viral protein-host RNA (especially, non-coding RNAs) physical interactions in the pathogenicities of the coronavirus infection. The detailed analysis of the RNA-Protein interaction may make a little sense and miRNA's physical interaction with the viral proteins may not seems satisfactory; however, there are pieces of evidence about this proposal. In addition to the results of molecular docking and MD simulations of one of the predicted interactions (Fig. 7), here, we address the results of some of the previous studies in support of our idea of disruption of the cell system via the viral proteins.
Many DNA and RNA viruses synthesize their ncRNAs to degrade, boost, or hijack cellular miRNAs [55,56]. According to Skalsky and Cullen; Over 200 viral miRNAs had been identified until 2010.viruses could use these miRNAs to manipulate both host and viral gene expression [57]. HSUR 1 of Herpesvirus, a viral U-rich noncoding RNA, is a well-known virus macromolecule that interacts with the host cell miRNAs and degrades host microRNA-27 [58]. According to our previous study, 160 human miRNAs have been predicted targeting the SARS-CoV-2 genome l [59]. All of these studies address the role of miRNAs in the host cell-virus interplay. Furthermore, Tomato leaf curl Palampur virus (ToLCPalV) and African cassava mosaic virus Cameroon Strain (ACMV) belong to one of the most devastating plant viruses worldwide, the Begomovirus genus. There is some direct evidence that the AC4 protein of the above-mentioned viruses is a virus-encoded protein that plays a role as a suppressor during the posttranscriptional gene-silencing process. Also, This viral protein binds to and inactivates mature host miRNAs and blocks the miRNA-mediated regulation of target mRNAs [60,61]. These results confirm the viral protein-host miRNA interaction. Furthermore, as a therapeutic strategy, Chen et al. reported a new mechanism that involves interactions between microRNA and HIV-1 Gag protein's RNA-binding (nucleocapsid) domain to inhibit Gag assembly and virus production [62].
5. Conclusion
NSPs act specifically, and they are not perturbed by unsuitable interactions with the host cell RNAs. The gene ontology and pathway analysis show that NSP3's hyperaffinity to miRNAs can influence the majority of molecular functions in the host cell that are related to COVID-19 symptoms. On the other hand, our results are in favor of a vaccine that selects the RNA part of the viral genome that is matched with the binding-site of the spike protein. Moreover, a non-coding RNA (RNA antitoxin, ToxI) was obtained as a natural candidate for NSP5 inhibition.
We believe that the significant interplay between the host cell RNA and the viral protein in the host cell can disrupt the cell's system by influencing the RNA-dependent processes of the host cells, such as a differential expression in RNA. Although this potential of interaction may be helpful in finding an RNA aptamer or drug for the inhibition of viral proteins, our findings are not limited to this application. In addition, our results can also be helpful for researchers who work on the molecular mechanisms behind different symptoms of COVID-19.
Declaration of competing interest
The authors have no competing interests.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104611.
Authors’ contributions
Hossein Lanjanian: Conceptualization, writing, editing, and revising the manuscript, formal analysis, investigation. Sajjad Nematzadeh Miandoab: Conceptualization, writing, editing, and revising the manuscript, implementation, formal analysis, investigation. Shadi Hosseini: Writing, editing, and revising the manuscript, investigation. Mahsa Torkamanian Afshar: Conceptualization, investigation, and revising the manuscript. Farzad Kiani: Conceptualization, editing, and revising the manuscript. Maryam Moazzam-Jazi: Writing, editing, and revising the manuscript, investigation. Nizamettin Aydin: Supervision, project administration, revising the manuscript. Ali Masoudi-nejad: Supervision, project administration, revising the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Result matrices (supplementary files) are available at the following link: https://github.com/sajjad-nematzadeh/Covid19RP.
Funding
No funding.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Delmas B., Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Lai B.S., Juhas M. Recent advances in aptamer discovery and applications. Molecules. 2019;24 doi: 10.3390/molecules24050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C.Y., Huang Y., Lau S.K.P., Tsoi H.W., Yuen K.Y. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol. Immunol. 2005;49:899–908. doi: 10.1111/j.1348-0421.2005.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1805–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y., Sun S.Q., Guo H.C. Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements. Virol. J. 2016;13:1–17. doi: 10.1186/s12985-016-0561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu L., Crawford S.E., Hyser J.M., Estes M.K., Prasad B.V.V. Rotavirus non-structural proteins: structure and function. Curr. Opin. Virol. 2012;2:380–388. doi: 10.1016/j.coviro.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yost S.A., Marcotrigiano J. Viral precursor polyproteins: keys of regulation from replication to maturation. Curr. Opin. Virol. 2013;3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Hemert M.J., van den Worm S.H.E., Knoops K., Mommaas A.M., Gorbalenya A.E., Snijder E.J. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lécine P., Canard B. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;80(368):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. BioRxiv. 2020:2020. doi: 10.1101/2020.04.27.063180. 04.27.063180. [DOI] [PubMed] [Google Scholar]
- 15.Frick D.N., Virdi R.S., Vuksanovic N., Dahal N., Silvaggi N.R. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59:2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar R., Cruz L., Sandhu P.K., Buchkovich N.J. UL88 mediates the incorporation of a subset of proteins into the virion tegument. J. Virol. 2020;94 doi: 10.1128/jvi.00474-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J., Liu Y., Gao P., Hu Y., Chai Y., Zhou S., Kong C., Zhou L., Ge X., Guo X., Han J., Yang H. Mapping the nonstructural protein interaction network of porcine reproductive and respiratory syndrome virus. J. Virol. 2018;92:2020. doi: 10.1128/jvi.01112-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.-T., Veit M., Su S. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94 doi: 10.1128/jvi.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanjanian H., Moazzam-Jazi M., Hedayati M., Akbarzadeh M., Guity K., Sedaghati-khayat B., Azizi F., Daneshpour M.S. SARS-CoV-2 infection susceptibility influenced by ACE2 genetic polymorphisms: insights from the Tehran Cardio-Metabolic Genetic Study. Sci. Rep. 2021;11:1529. doi: 10.1038/s41598-020-80325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatton T., Zhou S., Standring D.N. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 1992;66:5232–5241. doi: 10.1128/jvi.66.9.5232-5241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson J.M., Willits D.A., Young M.J., Zlotnick A. Interaction with capsid protein alters RNA structure and the pathway for in vitro assembly of Cowpea chlorotic mottle virus. J. Mol. Biol. 2004;335:455–464. doi: 10.1016/j.jmb.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 22.Moazzam‐Jazi M., Lanjanian H., Maleknia S., Hedayati M., Daneshpour M.S. Interplay between SARS‐CoV‐2 and human long non‐coding RNAs. J. Cell Mol. Med. 2021:16596. doi: 10.1111/jcmm.16596. jcmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks G.F., Carroll K.C., Morse S.A., Butel J.S., Mietzner T. Jawetz Melnick & Adelberg’s Medical Microbiology Twenty-Sixth. 26th. McGraw-Hill; New York, USA: 2013. [DOI] [Google Scholar]
- 24.Torkamanian-Afshar M., Lanjanian H., Nematzadeh S., Tabarzad M., Najafi A., Kiani F., Masoudi-Nejad A. RPINBASE: an online toolbox to extract features for predicting RNA-protein interactions. Genomics. 2020;112 doi: 10.1016/j.ygeno.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Kozomara A., Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A., Birgaoanu M., Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volders P.J., Anckaert J., Verheggen K., Nuytens J., Martens L., Mestdagh P., Vandesompele J. Lncipedia 5: towards a reference set of human long non-coding rnas. Nucleic Acids Res. 2019;47:D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang W., Ke M. Protein Encoding: a Matlab toolbox of representing or encoding protein sequences as numerical vectors for bioinformatics. J. Chem. Pharmaceut. Res. 2014;6:2000–2007. Www.Jocpr.Com Available Online. [Google Scholar]
- 31.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torkamanian-Afshar M., Nematzadeh S., Tabarzad M., Najafi A., Lanjanian H., Masoudi-Nejad A. In silico design of novel aptamers utilizing a hybrid method of machine learning and genetic algorithm. Mol. Divers. 2021;1:3. doi: 10.1007/s11030-021-10192-9. [DOI] [PubMed] [Google Scholar]
- 33.Kloczkowski A., Ting K.L., Jernigan R.L., Garnier J. Combining the GOR V algorithm with evolutionary information for protein secondary structure prediction from amino acid sequence. Proteins Struct. Funct. Genet. 2002;49:154–166. doi: 10.1002/prot.10181. [DOI] [PubMed] [Google Scholar]
- 34.Garnier J., Gibrat J.F., Robson B. [32] GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed Z., Amizadeh S., Bilenko M., Carr R., Chin W.-S., Dekel Y., Dupre X., Eksarevskiy V., Erhardt E., Eseanu C., Filipi S., Finley T., Goswami A., Hoover M., Inglis S., Interlandi M., Katzenberger S., Kazmi N., Krivosheev G., Luferenko P., Matantsev I., Matusevych S., Moradi S., Nazirov G., Ormont J., Oshri G., Pagnoni A., Parmar J., Roy P., Shah S., Siddiqui M.Z., Weimer M., Zahirazami S., Zhu Y. Machine learning at microsoft with ML .NET. Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 2019:2448–2458. doi: 10.1145/3292500.3330667. [DOI] [Google Scholar]
- 36.Ke G., Meng Q., Finley T., Wang T., Chen W., Ma W., Ye Q., Liu T.-Y. 2017. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. [Google Scholar]
- 37.Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., Van Dijk M., De Vries S.J., Bonvin A.M.J.J. The HADDOCK2 . 2 web Server : user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. 2000. http://www.rcsb.org/pdb/status.html [DOI] [PMC free article] [PubMed]
- 39.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 40.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Von Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu J.B., Chiu C.M., Da Hsu S., Huang W.Y., Chien C.H., Lee T.Y., Da Huang H. MiRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinf. 2011;12:1–12. doi: 10.1186/1471-2105-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koyama T., Platt D., Parida L. Variant analysis of SARS-cov-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harms A., Brodersen D.E., Mitarai N., Gerdes K. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell. 2018;70:768–784. doi: 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Blower T.R., Pei X.Y., Short F.L., Fineran P.C., Humphreys D.P., Luisi B.F., Salmond G.P.C. A processed noncoding RNA regulates an altruistic bacterial antiviral system. Nat. Struct. Mol. Biol. 2011;18:185–191. doi: 10.1038/nsmb.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unterholzner S.J., Poppenberger B., Rozhon W. Toxin–antitoxin systems. Mobile Genet. Elem. 2013;3 doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Apicella M., Campopiano M.C., Mantuano M., Mazoni L., Coppelli A., Del Prato S. Review COVID-19 in people with diabetes: understanding the reasons for worse outcomes. 2017. Www.Thelancet.Com/Diabetes-Endocrinology. 8. [DOI] [PMC free article] [PubMed]
- 50.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA, J. Am. Med. Assoc. 2020;10022:E1–E8. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 2020;20:277. doi: 10.1038/s41577-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamidreza Bagheri S., Asghari A., Farhadi M., Shamshiri A.R., Kabir A., Kamrava S.K., Jalessi M., Mohebbi A., Alizadeh R., Honarmand A.A., Ghalehbaghi B., Salimi A., Firouzabadi D. Coincidence of COVID-19 epidemic and olfactory dysfunction outbreak in Iran. Med. J. Islam. Repub. Iran. 2020;34:446–452. doi: 10.34171/mjiri.34.62. n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butowt R., von Bartheld C.S. Neuroscientist; 2020. Anosmia in COVID-19: Underlying Mechanisms and Assessment of an Olfactory Route to Brain Infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandelli A., Monti M., Milanetti E., Armaos A., Rupert J., Zacco E., Bechara E., Delli Ponti R., Tartaglia G.G. Structural analysis of SARS-CoV-2 genome and predictions of the human interactome. Nucleic Acids Res. 2020;48:11270–11283. doi: 10.1093/nar/gkaa864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tycowski K.T., Guo Y.E., Lee N., Moss W.N., Vallery T.K., Xie M., Steitz J.A. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y.E., Steitz J.A. Virus meets host MicroRNA: the destroyer, the booster, the hijacker. Mol. Cell Biol. 2014;34:3780–3787. doi: 10.1128/mcb.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo Y.E., Riley K.J., Iwasaki A., Steitz J.A. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol. Cell. 2014;54:67–79. doi: 10.1016/j.molcel.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jafarinejad-Farsangi S., Jazi M.M., Rostamzadeh F., Hadizadeh M. High affinity of host human microRNAs to SARS-CoV-2 genome: an in silico analysis. Non-Coding RNA Res. 2020;5:222–231. doi: 10.1016/j.ncrna.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chellappan P., Vanitharani R., Fauquet C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulshreshtha A., Kumar Y., Roshan P., Bhattacharjee B., Mukherjee S.K., Hallan V. AC4 protein of tomato leaf curl Palampur virus is an RNA silencing suppressor and a pathogenicity determinant. Microb. Pathog. 2019;135:103636. doi: 10.1016/j.micpath.2019.103636. [DOI] [PubMed] [Google Scholar]
- 62.Chen A.K., Sengupta P., Waki K., Van Engelenburg S.B., Ochiya T., Ablan S.D., Freed E.O., Lippincott-Schwartz J. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2676–E2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Result matrices (supplementary files) are available at the following link: https://github.com/sajjad-nematzadeh/Covid19RP.