Abstract
The gut microbiota plays a major role in human health and an alteration in gut microbiota structure and function has been implicated in several diseases. In the colon, mucus covering the epithelium is critical to maintain a homeostatic relationship with the gut microbiota by harboring a microbial community at safe distance from the epithelium surface. The mucin glycans composing the mucus layer provide binding sites and a sustainable source of nutrients to the bacteria inhabiting the mucus niche. Access to these glycan chains requires a complement of glycoside hydrolases (GHs) produced by bacteria across the phyla constituting the human gut microbiota. Due to the increased recognition of the role of mucus-associated microbes in human health, how commensal bacteria breakdown and utilize host mucin glycans has become of increased interest and is reviewed here. This short review provides an overview of the strategies evolved by gut commensal bacteria to access this rich source of the nutrient with a focus on the GHs involved in mucin degradation.
Keywords: glycosylation, gut microbiota, mucin, mucus
Introduction
The gastrointestinal (GI) tract is home to a diverse range of microbial species collectively referred to as the gut microbiota which have a profound impact on host health. It is well established that the gut microbiota aids digestion of complex dietary polysaccharides which reach the colon undigested, enabled by the vast array of glycolytic enzymes encoded by gut symbionts (Zimmermann et al. 2019, El Kaoutari et al. 2013). In addition to dietary polysaccharides, gut microbes can utilize host glycans as a nutrient source. The ability to metabolize glycans such as human milk oligosaccharides, glycosaminoglycans and glycan moieties of glycoproteins and glycolipids found at mucosal surfaces grants bacteria a competitive advantage. This is particularly relevant to the microbial community that resides within the mucus layer of the large intestine.
The mucus layer is viewed as a defence mechanism, protecting the epithelial layer from microbes and other luminal compounds, but in the colon, mucus also plays a major biological function by harboring a distinct microbial community called the mucus-associated microbiota. This is enabled by the bilayer organization of the colonic mucus which is divided into a stratified inner layer virtually impenetrable to bacteria and a loose outer layer providing a niche to microbes adapted to this environment (Johansson et al. 2008). This microbial community is tolerated due to the mutually beneficial relationship established with the host as a result of long-term coevolution (Neish 2009). Benefit to the host includes effective mucin turnover and stimulation of mucus production through Toll-like receptor-mediated interactions with sentinel goblet cells (Birchenough et al. 2016). Continuous mucus production is essential to maintain gut barrier function and is strengthened by the production of antimicrobial compounds against pathogenic bacteria (McGuckin et al. 2011). Other benefits of the mucus-associated microbiota include colonization resistance whereby pathogenic niches are already occupied by commensal species (Sorbara and Pamer 2019), and the production of metabolites directly implicated in the communication of microbes with the host. The mucus-associated gut microbiota can also significantly affect the development of the host immune system as extensively reviewed (Pickard et al. 2017).
Mucin glycosylation and associated bacteria in the gut
Mucin glycans make up ~80% of the molecular mass of mucins, the main structural component of mucus. Mucin-type O-glycosylation is initiated by a large family of polypeptide GalNAc transferases (ppGalNAc Ts) that add α-GalNAc to the Ser and Thr residues of peptides. Mucin glycosylation is characterized by a high degree of structural diversity which is based on three elements. The first is the type of core structure. There are eight mucin core structures in humans with structures 1–4 most commonly found in intestinal mucins (Tailford et al. 2015a; Thomsson et al. 2012; Brockhausen et al. 2009). The second stage of glycan diversity is determined by the action of a range of glycosyltransferases that elongate the mucin core through the addition of galactose, N-acetylgalactosamine (GalNAc) and/or N-acetylglucosamine (GlcNAc) residues leading to linear or branched chains of up to 20 residues (Gunning et al. 2013). The third element of diversity is conferred by the peripheral epitopes that are often fucosylated, sialylated or sulphated (Tailford et al. 2015a).
At the ecological level, the diversity of mucin glycans along the GI tract contributes to shape the structure and function of the gut microbiota. While the luminal microbiota may respond primarily to diet, the mucus-associated microbiota is influenced more directly by host-related factors. Importantly, the ability to utilize host mucin glycans as a carbon source gives bacteria a sustainable and consistent nutrient supply and a competitive advantage to colonize the mucus layer (Marcobal et al. 2013). As reviewed in Tailford et al. (2015a), it is now established that mucin glycan degradation is widespread across the major phyla represented in the human gut microbiota. Akkermansia muciniphila is a mucin glycan degradation specialist and, therefore, considered as a keystone member of the mucus-associated microbiota (Shin et al. 2019) while Bacteroidetes are viewed as general glycan degraders able to switch from dietary to host glycan metabolism due to their extensive array of carbohydrate-active enzymes (Ndeh and Gilbert 2018). Actinobacteria, which are largely represented by Bifidobacteria in the human gut microbiota, are typically adapted to carbohydrates with a low degree of polymerization and mucin glycan metabolism strategy is similar to the Firmicutes (Ndeh and Gilbert 2018). Consistent with this, the presence of mucins in in vitro fermentation models leads to an increased proportion of Bacteroidetes, Akkermansia and Lachnospiraceae species that are known mucin glycan degraders, whilst levels of Lactobacillus and Bifidobacterium decrease (Tran et al. 2016). In vivo, both chronic and intermittent fiber deficiency promotes enrichment of mucin glycan degrading bacteria in mouse models, leading to a significant increase in A. muciniphila and Bacteroides caccae species accompanied by a decrease of the fiber-degrading species (Desai et al. 2016).
Mucin glycan degradation strategies by gut commensal bacteria
Microbes most adept at mucin glycan degradation often encode sulfatases, deacetylases, sialidases and fucosidases to remove terminal structures and grant greater accessibility to the extended core structures (Etienne-Mesmin et al. 2019; Ndeh and Gilbert 2018). The monosaccharides freed by the action of these enzymes may be utilized by the bacteria themselves or released in the environment for scavenging bacteria (Marcobal et al. 2013). Furthermore, in silico analysis revealed that up to 86% of the human gut microbiota encode genes for cleavage of mucin glycans, with 89% encoding genes for the metabolism of the monosaccharides released (Ravcheev and Thiele 2017). The current model for mucin glycan degradation by the gut microbiota involves the sequential action of a number of glycoside hydrolases (GHs) (www.cazy.org; Figure 1) (Lombard et al. 2014).
Fig. 1.
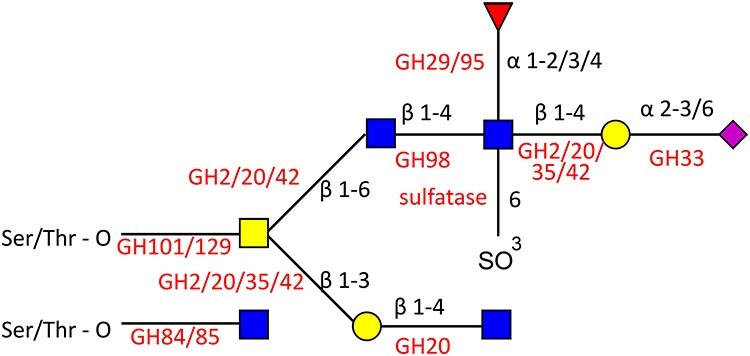
Specificity of bacterial GHs and sulfatases. A hypothetical mucin glycan is depicted with monosaccharide symbols following the symbol nomenclature for glycans (Varki et al., 2015). Linkages are shown with black text and GHs are shown in red text.
Sulfate residues terminate mucin glycans and have been proposed to prevent GHs from removing terminal sugars, thus preventing the breakdown of mucin glycans (Etienne-Mesmin et al. 2019). In addition, the release of sulfate residues has been proposed to increase the levels of sulfate-reducing bacteria in the gut, leading to the production of H2S, which can disrupt the mucus network and lead to epithelial damage (Praharaj et al. 2018; Ijssennagger et al. 2016). Mucin-desulfating enzymes have been characterized primarily from the Bacteroides genus, with examples from B. fragilis and B. thetaiotaomicron (Praharaj et al. 2018; Cartmell et al., 2017). Recent work identified a B. fragilis sulfatase that was shown to be essential for growth on mucus in vitro and robust mucosal colonization in vivo (Donaldson et al., 2020).
Exo-acting GHs are then involved in the trimming of terminal sugars from the O-glycan mucin chains, starting with the removal of fucose and sialic acid residues capping the GI mucin chains.
Fucose release involves fucosidases belonging to GH29 and GH95 families (www.cazy.org). GH95 enzymes functionally characterized so far show strict substrate specificity to the terminal Fucα1–2Gal linkage and hydrolyse the linkage via an inverting mechanism whereas GH29 enzymes show relatively relaxed substrate specificities with hydrolysis proceeding via a retaining mechanism (www.cazy.org). Fucosidases are found among numerous members of the gut microbiota, and often multiple fucosidases are found within a single genome, for example Bifidobacterium bifidum (Ashida et al., 2009), Bifidobacterium longum (Garrido et al., 2016, Bunesova et al. 2016), Ruminococcus gnavus (Crost et al. 2013) or A. muciniphila (Ottman et al. 2017). In these species, transcriptomics studies demonstrated that fucosidases were upregulated during growth on mucins, supporting their role in mucin glycan breakdown and utilization (Shin et al. 2019; Crost et al. 2016). Fucose metabolism has also been demonstrated for B. thetaiotaomicron and can trigger host fucosylation which B. thetaiotaomicron then uses as a nutrient source (Pickard and Chervonsky 2015). Fucose and mucin cross-feeding initiated by B. bifidum enables growth of Eubacterium hallii, an early occurring commensal species that produces butyrate and propionate from fermentation metabolites but that cannot degrade complex oligo- and polysaccharides (Bunesova et al. 2018; Schwab et al. 2017). However, not all fucosidases are extracellular, for example, 3 intracellular fucosidases with varying substrate specificities toward disaccharides have been characterized from lactobacilli (AlfA, AlfB and AlfC) (Rodríguez-Díaz et al. 2011), suggesting that Lactobacilli may import fucosyl-oligosaccharides.
Sialic residues are another highly sought-after source of nutrient terminating mucin glycan chains. The sialic acids comprise a family of 9-carbon sugar acids found predominantly on cell surface glycans of humans and other animals, the most common form of sialic acid in humans is N-acetylneuraminic acid (Neu5Ac). To access this carbon and nitrogen source, intestinal bacteria (both gut symbionts and pathogens) express GH33 sialidases (also known as neuraminidases), which cleave terminal sialic acid residues. Several sialidases have been functionally and structurally characterized from gut bacteria including species of Clostridia (Navarro et al. 2018) and Bacteroidetes, such as Bacteroides fragilis or Bacteroides thetaiotamicron (Juge et al. 2016), as well as specific strains of Bifidobacterium (Nishiyama et al. 2018), R. gnavus (Tailford et al. 2015b) and A. muciniphila (Huang et al. 2015a). B. fragilis sialidase preferentially cleaves the sialyl α2,8 linkage compared to sialyl α2,3 and α2,6 linkages that are more commonly targeted (Tanaka et al. 1994). These sialidases are usually extracellular so the sialic acid is released into the environment, where it can be imported by the bacteria or scavenged by other microbes including strains from the same species. The action of two sialidases from B. bifidium was shown to support the growth of Bifidobacterium breve through sialic acid crossing-feeding (Nishiyama et al. 2018). Interestingly, some bacteria, such as B. thetaiotaomicron ATCC 29148 or A. muciniphila DSM 22959 (Tailford et al. 2015b; Huang et al., 2015a) encode sialidases allowing access to the underlying sugars of the glycan chains but do not encode genes required for sialic acid utilization (Brigham et al. 2009). In certain conditions such as post-antibiotic treatment, the levels of free sialic acid can promote the expansion of pathogens such as Clostridium difficile, Salmonella and Escherichia coli that do not produce sialidases (Ng et al. 2013). Another study showed that the expansion of certain Bacteroides species in a mouse model of colitis led to increased levels of sialidases and subsequent outgrowth of E. coli, which was dependent on the ability to catabolize sialic acid (Huang et al. 2015b). It is believed that O-acetyl ester modifications of sialic acids present at high levels in the mammalian colon can help protect from the action of bacterial sialidases (Robinson et al. 2017). In turn, some gut bacteria, produce sialylate-O-acetylesterases to remove them. In vitro foraging studies demonstrated that sialidase-dependent E. coli growth on mucin is enabled by Bacteroides EstA, a sialate O-acetylesterase acting on glycosidically linked sialylate-O-acetylesterase substrates (Robinson et al. 2017). It was, therefore, proposed that EstA specifically unlocks the nutritive potential of 9-O-acetylated sialic acids in mucus for mucin glycan foraging bacteria. Interestingly, R. gnavus encodes an intramolecular-trans-sialidase which releases 2,7-anhydro-Neu5Ac instead of Neu5Ac released by hydrolytic sialidases (Tailford et al. 2015b). This is proposed to be part of a selfish mechanism employed by R. gnavus to hold on to sialic acid by releasing it in a form only it can preferentially access and utilize (Bell et al. 2020). The full metabolic pathway for the utilization of 2,7-anhydro-Neu5Ac was recently unraveled and it was shown that the pathway was intrinsically linked to mucosal colonization by R. gnavus in mouse models (Bell et al. 2020).
Following removal of terminal sugars, GHs including galactosidases (GH2, GH20, GH35, GH42, GH98), N-acetylglucosaminidases (GH84, GH85, G89) and N-acetylgalactosaminidases (GH101, GH129) can degrade the extended core structures, releasing free monosaccharides that can support growth of bacteria (Figure 1) (Tailford et al. 2015a; Marcobal et al. 2013). Recently three mucin-acting extracellular β-galactosidases (GH2, GH35) from A. muciniphila ATCC BAA-835 were characterized with varied specificity toward glycosidic linkages. Amuc_0824 (GH2) was primarily active against Galβ1-3GalNAc whereas Amuc_0771 (GH35) showed the greatest activity against lacto-N-biose and galacto-N-biose (Kosciow and Deppenmeier 2020). In contrast, Amuc_1666 (GH2) showed activity against β1–4 linkages. These linkages are highly abundant in mucin glycans showing the importance of these enzymes in the mucin degradation strategy of A. muciniphila. Such diversity in substrate specificity is also seen across the Bifidobacteria from the human infant gut. Bioinformatics analyses revealed that β-galactosidase activity is spread across Bifidobacteria with certain clusters of β-galactosidase being strain-specific while others appeared to be shared across Bifidobacteria. Characterization of representative β-galactosidases of each cluster confirmed unique patterns of substrate specificity, with broad substrate specificity enzymes found across all subspecies (Ambrogi et al. 2019). In addition to exo-acting β-galactosidases endo-acting β-galactosidases (GH98) have also been described, for example, eabC from Clostridium perfringens is shown to able to cleave off blood group antigens (see recent review, Low et al. 2020).
Glucosaminidases that act on GlcNAc residues are found in multiple GH families. Exo-β-N-acetylglucosaminidases (GH84) have been identified across members of the gut microbiota (www.cazy.org), with some species encoding multiple enzymes. The substrate specificity of GH84 enzymes can include β1–2, 1–3, 1–4 and 1–6 linkages (Pluvinage et al. 2019). Endo- β-N-acetylglucosaminidases (GH85) that cleave the chitobiose core (GlcNAc-β-1,4-GlcNac) are also widespread in bacteria, and show a strict preference for GalNAc, and both core 1 and core 3 can be cleaved by these enzymes (Koutsioulis et al. 2008). Recent crystallographic evidence showed that GlcNAc was also the natural ligand for members of the GH20 family in A. muciniphila (Chen et al. 2019), a family containing exo-acting β -N-acetylglucosaminidases, β -N-acetylgalactosamindase, β −6-SO3-N-acetylglucosaminidases, and exo-acting lacto-N-biosidases (www.cazy.org).
The GH101 family regroups enzymes responsible for cleaving the core-1 O-linked glycans (Gal-β-1,3-GalNAc-α-R) with some of the family members shown to have some degree of activity against core 2 and core 3 structures (Koutsioulis et al. 2008). The α-N-acetylgalactosaminidases belonging to family GH129 show sequence similarity to GH101 members; however, they have a distinct substrate specificity, favoring the GalNAc-α1-Ser Tn antigen structure found in mucin glycoproteins. They are abundant among Bifidobacteria species and act intracellularly suggesting transport of Tn antigen containing oligosaccharides in the bacteria (Kiyohara et al. 2012). The first crystal structure from the GH129 family showed structural similarities with GH101 but differences in substrate recognition account for the altered substrate specificity (Sato et al. 2017).
In Bacteroides, oligosaccharides are imported in the periplasm where they are further degraded, and the enzymes to do this are physically linked into loci termed polysaccharide utilization loci (PULs) (Brown and Koropatkin 2020, Lapébie et al. 2019). In addition to the exo-acting GHs reported above and consistent with the glycan degradation strategy in these species, recent studies reported endo-acting enzymes that target the polyLacNAc structures within oligosaccharide side chains of mucins. These O-glycanases are found in several Bacteroides spp. as well as A. muciniphila and are a part of the GH16 family (Crouch et al. 2019). In addition, a high throughput screening approach led to the identification of novel GH31 and GH109 enzymes with α-GalNAcase activity (Rahfeld et al. 2019). These enzymes were found to have distinct specificities toward mucin-type O-glycans and blood type A-antigens. The α-GalNAcase GH31 enzymes act solely upon the GalNAc present in core structures of mucin-type O-glycans with no activity toward blood type A-antigens. The putative PULs in which the described α-GalNAcase GH31 enzymes are located showed no similarity to known mucin-degrading PULs (Rahfeld et al. 2019). It has been proposed that GH31 family enzymes may therefore play a major role in the capacity of Bacteroides spp. to efficiently degrade mucosal glycans despite their lack of GH101 or GH129 family enzymes (described above) (Rahfeld et al. 2019).
Perspectives
With the field of gut microbiota expanding beyond association studies and the increasing acknowledgment of the role of mucus-associated bacteria in human health, it is critical to continue our effort to gain mechanistic insights into the mechanisms underpinning microbial degradation of host mucin glycans. A full integration of glycomics in the field of microbiome research is warranted to further our understanding of the function and adaptation of microbial communities within the distinct nutritional niches in the gut. Combined with relevant in vivo humanized mouse models and advanced biopsy-based in vitro organ cultures, this biochemical knowledge will help to provide tangible molecular leads for developing therapeutic strategies to modulate the gut microbiota at the mucosa surface and strengthen gut barrier function in humans. Together, these targeted and omics approaches will potentiate the translation of microbiome research for biomarker development and precision medicine.
Acknowledgements
The authors gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Gut Microbes and Health BB/R012490/1 and its constituent project BBS/E/F/000PR10353 (Theme 1, Determinants of microbe-host responses in the gut across life). A.B. was supported by the BBSRC Norwich Research Park Biosciences Doctoral Training Partnership grant number BB/M011216/1.
Contributor Information
Andrew Bell, The Gut Microbes and Health Institute Strategic Programme, Quadram Institute Bioscience, Rosalind Franklin Road Norwich Research Park, Norwich NR4 7UQ, UK.
Nathalie Juge, The Gut Microbes and Health Institute Strategic Programme, Quadram Institute Bioscience, Rosalind Franklin Road Norwich Research Park, Norwich NR4 7UQ, UK.
References
- Ambrogi V, Bottacini F, O'sullivan J, O'connell Motherway M, Linqiu C, schoemaker B, schoterman M, van Sinderen D. 2019. Characterization of Gh2 and Gh42 Beta-galactosidases derived from Bifidobacterial infant isolates. AMB Express. 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct Α-L-Fucosidases from Bifidobacterium Bifidum are essential for the utilization of Fucosylated milk oligosaccharides and Glycoconjugates. Glycobiology. 19:1010–1017. [DOI] [PubMed] [Google Scholar]
- Bell A, Brunt J, Crost E, Vaux L, Nepravishta R, Owen CD, Latousakis D, Xiao A, Li W, Chen X et al. 2020. Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus Gnavus unravels mechanisms of bacterial adaptation to the gut. Nat Microbiol. 4:2393–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchenough GM, Nyström EE, Johansson ME, Hansson GC. 2016. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science. 352:1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. 2009. Sialic acid (N-acetyl Neuraminic acid) utilization by Bacteroides Fragilis requires a novel N-acetyl Mannosamine epimerase. J Bacteriol. 191:3629–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I, Schachter H, Stanley P. 2009. O-GalNAc Glycans. In: Varki A, Cummings RD, Esko JD, et al., (eds). Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 9. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1896/?report=classic.
- Brown HA, Koropatkin NM. 2020. Host glycan utilization within the Bacteroidetes sus-like paradigm. Glycobiology. cwaa054. doi: 10.1093/glycob/cwaa054. Epub ahead of print. PMID: 32518945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova V, Lacroix C, Schwab C. 2016. Fucosyllactose and L-Fucose utilization of infant Bifidobacterium Longum and Bifidobacterium Kashiwanohense. BMC Microbiol. 16:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunesova V, Lacroix C, Schwab C. 2018. Mucin cross-feeding of infant Bifidobacteria and Eubacterium Hallii. Microb Ecol. 75:228–238. [DOI] [PubMed] [Google Scholar]
- Cartmell A, Lowe EC, Baslé A, Firbank SJ, Ndeh DA, Murray H, Terrapon N, Lombard V, Henrissat B, Turnbull JE et al. 2017. How members of the human gut microbiota overcome the Sulfation problem posed by Glycosaminoglycans. Proc Natl Acad Sci. 114:7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang J, Liu M, Yang W, Wang Y, Tang R, Zhang M. 2019. Crystallographic evidence for substrate-assisted catalysis of Β-N-Acetylhexosaminidas from Akkermansia Muciniphila. Biochem Biophys Res Commun. 511:833–839. [DOI] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. 2013. Utilisation of mucin Glycans by the human gut symbiont Ruminococcus Gnavus is strain-dependent. PLoS One. 8:e76341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, Crossman LC, Juge N. 2016. The mucin-degradation strategy of Ruminococcus Gnavus: The importance of intramolecular trans-Sialidases. Gut Microbes. 7:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch LI, Liberato MV, Urbanowicz PA, Baslé A, Lamb CA, Stewart CJ, Cooke K, Doona M, Needham S, Brady RR et al. 2019. Prominent members of the human gut microbiota express Endo-acting O-Glycanases to initiate mucin breakdown. bioRxiv. 835843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A et al. 2016. A dietary Fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 167:1339–1353.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Chou W-C, Manson AL, Rogov P, Abeel T, Bochicchio J, Ciulla D, Melnikov A, Ernst PB, Chu H et al. 2020. Spatially distinct physiology of Bacteroides Fragilis within the proximal colon of Gnotobiotic mice. Nat Microbiol. 5:746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. 2013. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 11:497–504. [DOI] [PubMed] [Google Scholar]
- Etienne-Mesmin L, Chassaing B, Desvaux M, De Paepe K, Gresse R, Sauvaitre T, Forano E, Van de Wiele T, Schüller S, Juge N et al. 2019. Experimental models to study intestinal microbes-mucus interactions in health and disease. FEMS Microbiol Rev. [DOI] [PubMed] [Google Scholar]
- Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. 2016. A novel gene cluster allows preferential utilization of Fucosylated milk oligosaccharides in Bifidobacterium Longum Subsp. Longum Sc596. Sci Rep. 6:35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning AP, Kirby AR, Fuell C, Pin C, Tailford LE, Juge N. 2013. Mining the "Glycocode"—exploring the spatial distribution of Glycans in gastrointestinal mucin using force spectroscopy. FASEB J. 27:2342–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Wang MM, Kulinich A, Yao HL, Ma HY, Martínez JER, Duan XC, Chen H, Cai ZP, Flitsch SL et al. 2015a. Biochemical characterisation of the neuraminidase pool of the human gut symbiont Akkermansia Muciniphila. Carbohydr Res. 415:60–65. [DOI] [PubMed] [Google Scholar]
- Huang YL, Chassard C, Hausmann M, Von Itzstein M, Hennet T. 2015b. Sialic acid catabolism drives intestinal inflammation and microbial Dysbiosis in mice. Nat Commun. 6:8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijssennagger N, Van Der Meer R, Van Mil SWC. 2016. Sulfide as a mucus barrier-breaker in inflammatory bowel disease? Trends Mol Med. 22:190–199. [DOI] [PubMed] [Google Scholar]
- Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 105:15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N, Tailford L, Owen CD. 2016. Sialidases from gut bacteria: A mini-review. Biochem Soc Trans. 44:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara M, Nakatomi T, Kurihara S, Fushinobu S, Suzuki H, Tanaka T, Shoda S-I, Kitaoka M, Katayama T, Yamamoto K et al. 2012. Α-N-Acetylgalactosaminidase from infant-associated Bifidobacteria belonging to novel glycoside hydrolase family 129 is implicated in alternative mucin degradation pathway. J Biol Chem. 287:693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosciow K, Deppenmeier U. 2020. Characterization of three novel Beta-galactosidases from Akkermansia Muciniphila involved in mucin degradation. Int J Biol Macromol. 149:331–340. [DOI] [PubMed] [Google Scholar]
- Koutsioulis D, Landry D, Guthrie EP. 2008. Novel Endo-alpha-N-Acetylgalactosaminidases with broader substrate specificity. Glycobiology. 18:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapébie P, Lombard V, Drula E, Terrapon N, Henrissat B. 2019. Bacteroidetes use thousands of enzyme combinations to break down Glycans. Nat Commun. 10:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (Cazy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low KE, Smith SP, Abbott DW, Boraston AB. 2020. The glycoconjugate-degrading enzymes of Clostridium perfringens: tailored catalysts for breaching the intestinal mucus barrier. Glycobiology. cwaa050. doi: 10.1093/glycob/cwaa050. Epub ahead of print. PMID: 32472136. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. 2013. A refined palate: Bacterial consumption of host Glycans in the gut. Glycobiology. 23:1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcguckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 9:265–278. [DOI] [PubMed] [Google Scholar]
- Navarro MA, Li J, McClane BA, Morrell E, Beingesser J, Uzal FA. 2018. NanI sialidase is an important contributor to clostridium perfringens type F strain F4969 intestinal colonization in mice. Infect Immun. 86:e00462-18. https://doi.org/10.1128/IAI.00462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeh D, Gilbert HJ. 2018. Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol Rev. 42:146–164. [DOI] [PubMed] [Google Scholar]
- Neish AS. 2009. Microbes in gastrointestinal health and disease. Gastroenterology. 136:65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM et al. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 502:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama K, Nagai A, Uribayashi K, Yamamoto Y, Mukai T, Okada N. 2018. Two extracellular Sialidases from Bifidobacterium Bifidum promote the degradation of Sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 52:22–28. [DOI] [PubMed] [Google Scholar]
- Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Dos Santos VAPM, Smidt H, Belzer C, de Vos WM. 2017. Genome-scale model and omics analysis of metabolic capacities of Akkermansia Muciniphila reveal a preferential mucin-degrading lifestyle. Appl Environ Microbiol. 83:e01014–e01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Chervonsky AV. 2015. Intestinal Fucose as a mediator of host-microbe Symbiosis. J Immunol. 194:5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard JM, Zeng MY, Caruso R, Nunez G. 2017. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 279:70–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage B, Massel PM, Burak K, Boraston AB. 2019. Structural and functional analysis of four family 84 glycoside hydrolases from the opportunistic pathogen clostridium perfringens. Glycobiology. 30:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praharaj AB, Dehury B, Mahapatra N, Kar SK, Behera SK. 2018. Molecular dynamics insights into the structure, function, and substrate binding mechanism of mucin desulfating sulfatase of gut microbe Bacteroides Fragilis. J Cell Biochem. 119:3618–3631. [DOI] [PubMed] [Google Scholar]
- Rahfeld P, Wardman JF, Mehr K, Huff D, Morgan-Lang C, Chen H-M, Hallam SJ, Withers SG. 2019. Prospecting for microbial alpha-N-Acetylgalactosaminidases yields a new class of Gh31 O-Glycanase. J Biol Chem. 294:16400–16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravcheev DA, Thiele I. 2017. Comparative genomic analysis of the human gut microbiome reveals a broad distribution of metabolic pathways for the degradation of host-synthetized mucin Glycans and utilization of mucin-derived monosaccharides. Front Genet. 8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LS, Lewis WG, Lewis AL. 2017. The Sialate O-Acetylesterase Esta from gut Bacteroidetes species enables Sialidase-mediated cross-species foraging of 9-O-acetylated Sialoglycans. J Biol Chem. 292:11861–11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Díaz J, Monedero V, Yebra MJ. 2011. Utilization of natural Fucosylated oligosaccharides by three novel alpha-L-Fucosidases from a probiotic lactobacillus Casei strain. Appl Environ Microbiol. 77:703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Liebschner D, Yamada Y, Matsugaki N, Arakawa T, Wills SS, Hattie M, Stubbs KA, Ito T, Senda T et al. 2017. The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors. J Biol Chem. 292:12126–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C, Ruscheweyh H-J, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. 2017. Trophic interactions of infant Bifidobacteria and Eubacterium Hallii during L-Fucose and Fucosyllactose degradation. Front Microbiol. 8:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Noh J-R, Chang D-H, Kim Y-H, Kim MH, Lee ES, Cho S, Ku BJ, Rhee M-S, Kim B-C et al. 2019. Elucidation of Akkermansia Muciniphila probiotic traits driven by mucin depletion. Front Microbiol. 10:1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara MT, Pamer EG. 2019. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol. 12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015a. Mucin glycan foraging in the human gut microbiome. Front Genet. 6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Gall GL, de Vos WM, Taylor GL, Juge N. 2015b. Discovery of intramolecular trans-Sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun. 6:7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Ito F, Iwasaki T. 1994. Two Sialidases which preferentially hydrolyze Sialyl alpha 2-8 linkage from Bacteroides Fragilis Sbt3182. J Biochem. 115:318–321. [DOI] [PubMed] [Google Scholar]
- Thomsson KA, Holmen-Larsson JM, Angstrom J, Johansson MEV, Xia LJ, Hansson GC. 2012. Detailed O-Glycomics of the Muc2 mucin from colon of wild-type, Core 1-and Core 3-transferase-deficient mice highlights differences compared with human Muc2. Glycobiology. 22:1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Boudry C, Everaert N, Thewis A, Portetelle D, Daube G, Nezer C, Taminiau B, Bindelle J. 2016. Adding mucins to an in vitro batch fermentation model of the large intestine induces changes in microbial population isolated from porcine Feces depending on the substrate. FEMS Microbiol Ecol. 92:fiv165. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T et al. 2015. Symbol nomenclature for graphical representations of Glycans. Glycobiology. 25:1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. 2019. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 570:462–467. doi: 10.1038/s41586-019-1291-3. Epub 2019 Jun 3. PMID: 31158845; PMCID: PMC6597290. [DOI] [PMC free article] [PubMed] [Google Scholar]


